Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(2):475-486. doi:10.7150/ijbs.55716 This issue Cite
Review
Application of radiomics and machine learning in head and neck cancers
Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Changsha 410078, Hunan, China.
Received 2020-11-10; Accepted 2020-12-10; Published 2021-1-1
Abstract
With the continuous development of medical image informatics technology, more and more high-throughput quantitative data could be extracted from digital medical images, which has resulted in a new kind of omics-Radiomics. In recent years, in addition to genomics, proteomics and metabolomics, radiomic has attracted the interest of more and more researchers. Compared to other omics, radiomics can be perfectly integrated with clinical data, even with the pathology and molecular biomarker, so that the study can be closer to the clinical reality and more revealing of the tumor development. Mass data will also be generated in this process. Machine learning, due to its own characteristics, has a unique advantage in processing massive radiomic data. By analyzing mass amounts of data with strong clinical relevance, people can construct models that more accurately reflect tumor development and progression, thereby providing the possibility of personalized and sequential treatment of patients. As one of the cancer types whose treatment and diagnosis rely on imaging examination, radiomics has a very broad application prospect in head and neck cancers (HNC). Until now, there have been some notable results in HNC. In this review, we will introduce the concepts and workflow of radiomics and machine learning and their current applications in head and neck cancers, as well as the directions and applications of artificial intelligence in the treatment and diagnosis of HNC.
Keywords: radiomics, machine learning, head and neck cancers, sequential treatment, big data
Introduction
Head and neck cancer (HNC) is the eighth leading cause of cancer-related deaths [1]. Now the main treatment modalities for head and neck tumors include surgery, radiotherapy, chemotherapy and immunotherapy [2]. Due to tumor heterogeneity which serve as a known prognostic factor in HNC, a uniform treatment plan is not conducive to improved patient outcomes [3]. Therefore, personalized treatment plan should be implemented for each patient to improve the survival time and minimize the side effects [4]. In addition to tissue and blood tests, the diagnosis and treatment plan of HNC are also highly dependent on imaging, including computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET) [5]. However, structural based medical images are traditionally evaluated subjectively and qualitatively, and in most cases, the experience of reader can greatly influence the results. In recent years, the emergence of radiomics has attracted attention, which can extract quantitative imaging features from conventional medical images, and these features can also be combined with pathology and molecular biomarker, so as to more accurately assess the biological state of tumors and make personalized diagnosis and treatment plans for patients [6-10].
In recent decades, radiomics has become a new and evolving field in medical imaging [11-12]. With radiomics, people have discovered new image biomarkers, by collecting high-throughput quantitative features of oncology medical images. More and more research has shown that medical images contain more information than is available to the naked eye, and that extractable image parameters will in turn have some correlation with tumor clinical characteristics [11,13-15]. Radiomics is designed to be used as a clinical decision support tool by extracting quantitative data from medical images. Mass data will also be generated in this process [7,11,16-17]. Machine learning has promising application in radiomics due to its algorithms that are best suited for analysis of high-dimensional data [18-20]. In recent years, due to the unprecedented development of machine learning algorithms, coupled with the fact that the data required already exist and are easily available, there have been many studies using radiomics in the diagnosis, treatment and prognosis of HNC. In this review, we summarized the radiomics research of HNC, and introduced its general principles and typical workflow, as well as its future prospects and limitations in the field of HNC.
Workflow of radiomics and machine learning
Over the last decade, various “-omics” concepts have emerged one by one with the progress of high-throughput computer algorithms, which referring to the collective characterization and quantification of pools of biology information (e.g. proteomics, genomics, metabolomics). In recent years, more and more attention has been paid to radiomics, which refers to the automated extraction of mathematically defined radiological features from two- or three-dimensional (2D or 3D) medical images, as well as the application of data mining and analysis techniques [21-22]. Radiomics consists of extracting hundreds of quantitative features through automated or semi-automated software. It based on a hypothesis is that mineable data can be extracted from medical images and provides additional information about tumors' phenotype, genes, and proteins for use in patients [15, 23-24]. In recent years, more and more researchers have begun to focus on predicting molecular biomarkers, predicting therapeutic responses, and predicting survival prognostics in patients with HNC by extracting radiomics information features (including shape description, intensity, and texture characteristics) from different imaging patterns (e.g. CT, MRI, PET, ultrasound images) [11,13]. Some subfields of radiomics focus on the identification and scientific exploitation of relationships between quantitative bioimaging and genomic features of tumors. Previous studies have shown that the characteristics of medical images can distinguish between the biological characteristics of some tissues, such as tumors, inflammation and necrosis. Sometimes these characteristics also could be used to study the correlation with disease diagnosis and prognosis [25-28]. At the same time, some characteristics of medical images can be reflective of molecular and genetic characteristics of tumors [29-32].
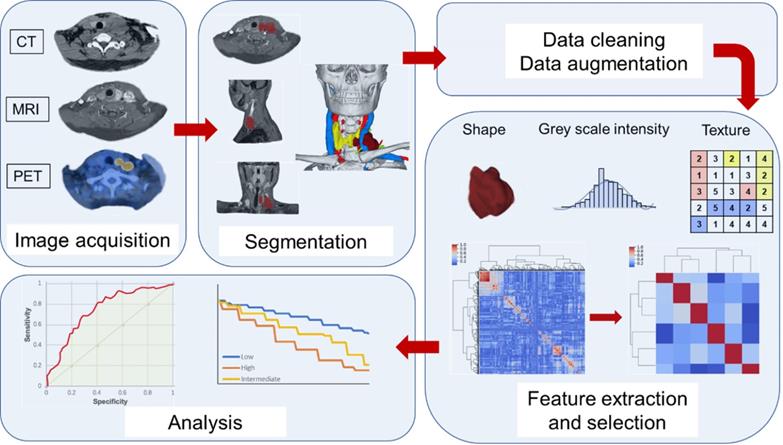
In a typical radiomics workflow, image acquisition is often the first step and a critical one [33-34]. Researchers must obtain high-quality, standardized imaging. The data source of radiomics is always obtained from retrospective medical imaging images. Different imaging techniques can lead to differences in image signals and image textures in medical imaging due to different acquisition parameters and reconstruction schemes [35-36]. If the parameters collected vary widely, this can introduce signal changes that are not caused by biological effects. For radiomics image analysis, a large number of images need to be selected. These are ideally standardized for image characteristics in resolution, reconstruction and acquisition parameters, as well as clinical characteristics such as tumor stage, tumor classification or prognosis [37-39].
The next step is to delineate (“segmentation”) the target area and volume in a medical image, generating sub-parts of the image in 2D and 3D images called areas of interest (ROI) and volume of interest (VOI) [33-34]. Segmentation must be reproducible and reliable, and it can be divided into manual, semi-automatic or automated execution. Manual separation requires two independent physicians (clinicians or imaging physicians) to complete, which can be time-consuming and labor-intensive, and the results are subject to observer variability and are not suitable for large-scale cohort studies. Semi-automatic image separation still requires human-machine coordination and an experienced physician is required to have an identification and modification of the automatically separated boundaries. Automatic image separation does not require human involvement, avoids heterogeneity between and within evaluators, and results are more repeatable, faster, and more suitable for large imaging datasets [34,40]. The raw data needs to be preprocessed to distinguish the signals from the noise, and the selection of this step is very important because it will directly affect the extracted features [41].
Then the extraction of radiomics features would be implemented, which are usually performed fully automated by professional software [42-43]. The radiomics features include shape features, which are used to represent the shape and geometry of ROI, such as head and neck tumor volume, length axis ratio, surface area/volume ratio, etc. [44]. The first-order feature is used to study the distribution of voxel values without considering spatial relationships, such as the mean, median, standard deviation, and peak of the voxels strength.
Second-order features, or texture features, are used to analyze the characteristics of the spatial distribution relationship of voxel intensity between voxels, and can be used to measure heterogeneity within tumors, such as a co-occurrence matrix (GLCM) that could calculates the correlation between two gray levels at a certain distance and a certain direction in an image, calculates the gray-level run length matrix (GLRLM) of continuous voxels with the same intensity in a fixed direction, and the neighborhood gray-level different matrix (NGLDM) between the quantized voxel intensity and the average speed-up intensity of neighboring voxel within a certain distance [45-47]. Deep learning is a sub-field of machine learning that has risen to the forefront of artificial intelligence, and one of the most popular deep learning tools available today, the convolutional neural network (CNN), can also be used to extract depth characteristics [48-49]. Convolutional analysis is performed on the image through the CNN, and the data in the fully connected layer is used as the obtained depth feature. These features can continue to be used in the CNN or in other classifiers [50-51]. In the stage of radiomics feature extraction, a large amount of data will be obtained. Before using these features, redundant features, unrelated or useless features should be excluded, leaving only a subset of features that are valid for modeling [42,44]. In Figure 1, we summarized the general workflow of radiomics.
Radiomics extracts valid, quantitative features from medical images that can be combined with other routine prognostic markers such as clinical staging, tissue molecular markers, and pathological features [10]. Various studies have shown that this type of predictive model based on medical images combined with various other data is superior in the evaluation of disease and survival prediction [13,53-55]. It has been shown that machine learning is a powerful statistical tool that is required to effectively develop and apply such large amounts of high-dimensional data. The choice of modeling method depends on the type of data and the purpose of the study. Machine learning methods include decision trees (DT), random forests (RF), logistic regression, bayesian models, support vector machines (SVM) and recently, deep learning which has gained much attention [56-59]. The technique has been widely used in the development of various predictive models for HNC.
Application of radiomics and machine learning in head and neck cancers
Multiple radiomics studies in HNC have reported in various magazines recent years. These studies generally focus on the diagnostic prediction of radiomics in HNC (pre-treatment staging, pathological subtypes, differentiation of tumors from inflammation or necrosis), and prediction of tumor status after treatments (include the status of certain pathogenic viruses, the prediction of early recurrence or lymph node metastasis), the prediction of survival and adverse reactions after treatments. Some researchers have studied radiogenomics to explore the prediction of expression of some molecules in HNC.
Typical radiomics workflow. ROI is first delineated. Then extract the features from the ROI, and finally model and analyzed.
Radiomics assesses pre-treatment grading of head and neck cancers
| Study | Number of patients | Tumor characteristics | Imaging modality | Parameter prediction | Feature selection method, model | Machine learning algorithm |
|---|---|---|---|---|---|---|
| Wang et al. 2019 [64] | Train:150pts Validation:61pts | Locally advanced LC | CE-CT | T stage | LASSO. Multivariable logistics model | LASSO, SVM, CHAID |
| Ren et al. 2018 [65] | Train: 85pts Validation: 42pts | HNSCC | T2, CE- T1 | T stage | LASSO. Rad-score | LASSO |
| Wu et al. 2019 [69] | Train: 137pts Validation: 69pts | HNSCC | CE-CT | Degree of tumor differentiation | KPCA, random forest classifier and VT. Multivariable logistics model | RF, K-PCA |
| Mukberjee et al. 2020 [67] | Train: 113pts Validation: 71pts | HNSCC | CE- CT | Histopathologic features | PCA. Regularized regression | PCA |
| Katsoulakis et al. 2020 [71] | Train: 77pts Validation: 83pts | HNSCC | CE- CT from TCIA | Molecular differences | CERR. Logistic regression | RF, deconvolution analysis, logistic regression |
| Ren et al. 2020 [70] | Train: 59pts Validation: 29pts | SCC of the oral tongue and floor of mouth | T1, T2, CE- T1, DWI | Histologic grade | LASSO. Rad-score | LASSO |
| Romeo et al. 2020 [66] | Total: 40pts | Oropharyngeal and oral cavity SCC | CE- CT | T stage and Nodal status | Heterogeneity CAD. Machine learning classifiers | Naïve bayes, KNN, RF and so on |
| Wang et al. 2019 [68] | Train: 96pts Validation: 24pts | PTC (prospective) | T2, CE-T1, DWI | Aggressiveness level | LASSO and selection operator. Machine learning classifiers | LASSO, gradient boosting classifier, logistic regression |
Train: Training dataset; Total: Only one dataset used; Validation: Validation dataset; CE-CT: Contrast-enhanced CT; CE-T1: Contrast-enhanced T1; DWI: Diffusion weighted imaging; HPV: Human Papillomavirus; PCA: Principal component analysis; CERR: Computational environment for radiological research; CAD: Computer-aided diagnosis and detection systems; CHAID: Chi-square automatic interaction detection; KNN: K-nearest neighbor.
Pre-treatment related predictive modeling
Pre-treatment staging is an important part of tumor diagnosis and a factor closely related to tumor prognosis. Studies have shown that T-stage of head and neck tumors, viral-related status, and lymph node status greatly influence the prognosis of cancer patients [60-63]. However, the current diagnostic methods focus on pre-treatment tissue biopsy, serological testing and traditional medical image diagnosis, which can determine tumor staging to a certain extent, but ultimately are local, qualitative and subjective. Reliable assessment of tumor staging by radiomics prior to treatment can help guide treatment selection and reduce recurrence and adverse event rates. Wang et al. [64] reported the use of radiomics combined with machine learning to create a T-staging model of locally advanced laryngeal cancer (LC), the performance of the model was evaluated by the area under the receiver operating characteristic curve (AUC). The predictive performance of the nomogram incorporating radiomic signature and T category reported by radiologists is the best with an AUC of 0.892 (95% CI: 0.811 to 0.974). Ren et al. [65] extracted imaging features from MRI of 85 patients in the training cohort and demonstrated that MRI radiomics signature could distinguish stage III- IV from stage I-II head and neck squamous cell carcinoma (HNSCC). Radiomics signature may serve as a complementary tool for preoperative staging. Romeo et al. [66] prediction of tumor grade and nodal status in oropharyngeal and oral cavity squamous-cell carcinoma using a radiomic approach. It has been reported that apparent diffusion coefficient based radiomics can be a useful and promising non-invasive method for predicting histologic grade of squamous cell carcinoma (SCC) of the oral tongue and tongue and floor of mouth. In HNSCC, radiological analysis was also used to design non-invasive biomarkers and to accurately distinguish well-differentiated from moderately differentiated and poorly differentiated HNSCC, with an AUC of 0.96 and an accuracy of 0.92. It has been reported that radiomics CT models have the potential to predict characteristics typically identified on pathologic assessment of HNSCC [67]. In a cohort of 96 papillary thyroid carcinoma (PTC) patients, a prospective study enrolled consecutive patients who underwent neck MR scans and subsequent thyroidectomy during the study interval. Machine learning-based MRI prediction models can distinguish between aggressive and non-aggressive PTC before surgery, and this approach facilitates the formation of personalized PTC treatment plans [68]. We identified eight studies investigated the feasibility of radiomics for the classification of HNC before treatment (Table 1). Thus far, these exploratory studies show that radiomics prediction model has the potential to become another non-invasive diagnostic tool for HNC before treatment, which can make the staging of tumors more objective and accurate, and even predict the malignancy of tumors and have a certain guiding effect on the subsequent treatment.
In the last 3 years, there have been an increasing number of studies to predict tumor response to certain treatments. It is well known that the treatment of HNC is mainly surgery, but there are also various treatment options with induction chemotherapy, concurrent chemoradiation, targeted therapy or immunotherapy [72-73]. In order to better formulate personalized treatment plans for cancer patients, a sequential treatment system can be realized as soon as possible. The establishment of a radiomics model that can predict the treatment effect or the incidence of complication after treatment plays a very important role in achieving the above goals. While Bologna et al. Wang et al. and Zhao et al. [74-76] they retrospectively extracted radiomics signatures of each type of weighted images from MRI of nasopharyngeal carcinoma (NPC) patients. Selected the useful radiomics features by least absolute shrinkage and selection operator (LASSO) to form a valid model to predict early response of NPC patients to induction chemotherapy, which helps to personalize risk stratification and treatment of NPC patients. Jin et al. [77] reported their preliminary results based on radiomics features from CT scans in 70 patients with esophageal cancer (EC), they found that the model with radiomic features combined with dosimetric parameters is promising and outperforms that with radiomic features alone in predicting the treatment response of patients with EC who underwent concurrent chemoradiation. Acute xerostomia is the most common side effect of radiation therapy for HNC. Pota et al and Liu et al. [78-79] conducted a study on HNC and NPC, respectively. They obtained CT scans of patients before, during, and after treatment to obtain imaging features and establish the best model to use during the initial treatment phase to predict the development of acute xerostomia after radiation therapy in cancer patients. Radiation induced brain injury is a relatively common brain complication in patients with NPC after radiation therapy. Although this adverse reaction is not fatal in general, but it seriously affects patients' life treatment. Researchers extracted 10320 textural features by analyzing MRI multiple-weighted images of 242 patients with NPC who had undergone radiation therapy. Three prediction models were established using the RF method, all of which could dynamically predict radiation induced brain injury in advance, enabling early detection and allowing clinicians to take preventive measures to stop or slow down the deterioration of radiation induced brain injury [80]. Eight studies explored the ability of radiomics to predict the response of HNC to certain treatments or early prediction of complication after treatment (Table 2).
Prediction models for recurrence, metastasis and survival
The most extensive research of radiomics in HNC is its prediction of prognosis, including the study of its relationship with prognostic indicators such as progression-free survival (PFS), overall survival (OS), five-year survival rate, distant metastasis (DM), and local recurrence (LR). With the development of medical diagnosis and treatment technology, HNC has made great progress in the use of therapeutic drugs [82-84]. However, due to the specificity of the growth site of HNC, many patients are already in advanced stage when they are found, which makes the prognosis of HNC still poor, with the five-year survival rate ranging from 25% for hypopharyngeal cancer to 80% for NPC [85-86]. Therefore, scientists are interested in more accurately predicting LR, lymph node metastasis (LNM) and even distant metastasis (DM) of HNC and the survival rate of patients, which can also better serve in the development of personalized treatment plans for patients.
Radiomics predicts tumor response and adverse symptom after treatment
| Study | Number of patients | Tumor characteristics | Imaging modality | Therapy/Symptom | Outcome, model | Machine learning algorithm |
|---|---|---|---|---|---|---|
| Yu et al. 2019 [81] | Train: 51pts; Validation: 19pts | NPC | T2, CE-T1 | ART | Replan status of patient. Multivariable logistics model | LASSO, logistic regression |
| Zhang et al. 2020 [80] | Total: 242pts | NPC | T2, CE-T1 | Early detection of radiation- induced brain injury | Radiation induced temporal lobe injury. Random forest | RF |
| Pota et al. 2017 [78] | Total: 37pts (74 parotid glands) | HNC | Before RT (CT1), at the middle of treatment (CT2) and after RT (CT3) | Xerostomia, shrinkage of parotid glands | Parotid shrinkage rate and 12-months xerostomia. Machine learning classifiers | Naïve bayes, LFA |
| Jin et al. 2019 [77] | Train: 70pts Validation: 24pts | EC | Before treatment (CT1), 3 months after CRT(CT2) | Concurrent chemoradiation | 3 months after CRT, Machine learning classifiers | SVM, RBF, XGBoost, PCA |
| Liu et al. 2019 [79] | Total: 35pts | NPC | Five CT sets acquired at treatment position during the RT | Acute xerostomia | Patients' saliva was collected every other 10 days during the RT. Multivariate Cox regression | Multivariate machine learning algorithms |
| Bologna et al. 2020 [74] | Total: 50pts (25 responders and 25 non-responders) | Sinonasal cancers | T1, T2, ADC | Induction chemotherapy | T1+T2+ADC model displayed the highest; radiomics score | PCA, naïve bayes, SVM, KNN and so on |
| Wang et al. 2018 [75] | Total: 120pts | NPC | T2, T2FS, CE- T1 | Induction chemotherapy | Early response to induction chemotherapy; logistic regression | Logistic regression |
| Zhao et al. 2019 [76] | Train: 100pts Validation: 23pts | NPC | T1, T2, CE- T1 | Induction chemotherapy | PFS; multivariable logistic regression | LASSO, SVM, logistic regression |
ART: Adaptive radiotherapy; LFA: Likelihood-fuzzy analysis; RBF: Radial basis function; XGBoost: Extreme gradient boosting algorithm; CRT: Concurrent chemoradiation; T2FS: T2 weighted fat-suppressed; RT: Radiation therapy; EBV: Epstein-Barr virus; ADC: Apparent diffusion coefficient.
People used MRI images from 360 patients with NPC as a training cohort for feature extraction from the maximal axis region of the tumor. Eleven features were selected to construct the radiomics score (Rad-score), which was significantly associated with local recurrence-free survival (LRFS). Rad-scores were generated using the Cox proportional hazards regression model, and can reliably predict LRFS in patients with non-metastatic T4 NPC, which might guide individual treatment decisions [87]. There are still a lot of imaging genomics combined with machine learning of various algorithms to build predictive models for LR of HNC. From the M.D. Anderson cancer center head and neck quantitative imaging working group, which analyzed CT/MRI and PET images from 465 patients with HNC. Machine learning methods were applied to yield a radiomic signature consisting of features with minimal overlap and maximum prognostic significance, and derived from pre-treatment imaging consisting of 2 radiographic signatures [88]. In thyroid and EC, predictive models of radiomics have also been used [89-94]. LNM is a significant prognostic factor in patients with HNC, and the ability to predict it accurately is essential to optimizing treatment. The accuracy of LNM identification strongly depends on the physician's experience, therefore, the establishment of an automatic prediction model for LNM can greatly help physicians in their practice [34]. Radiomics models are built based on handcrafted features, while deep learning learns the features automatically. In order to have better prediction, many researchers have proposed hybrid prediction models [95-98]. Another research group used the PyRadiomics platform, and extracted the imaging features of primary tumors in all patients who did not exhibit DM before treatment. This retrospective cohort analysis included 176 patients with NPC. Then used minimum redundancy-maximum relevance and LASSO algorithms to select the strongest features and build a logistic model for DM prediction [99]. From these existing exploratory studies, it is easy to see that most researchers have extracted radiomics signatures manually or semi-manually from various types of imaging images of training cohorts, then used machine learning algorithms to extract valid features and build predictive models, and then used independent cohorts to verify the validity of the models. Table 3 summarizes the reported studies of representative predictive models of this type in HNC.
Radiomics predicts recurrence and metastasis of head and neck cancer
| Study | Number of patients | Tumor type | Imaging modality | Outcome, feature selection method, model | Machine learning algorithm |
|---|---|---|---|---|---|
| Zhang et al. 2019 [87] | Train: 80pts Validation: 60pts | NPC | T2, CE- T1 | LR-free survival. Radiomics score, Cox regression | Logistic Regression |
| Bogowicz et al. 2017 [100] | Train: 93pts Validation: 56pts | HNSCC | CE- CT | LC and HPV status. PCA in combination with univariable logistic regression. Multivariable logistic regression | Logistic regression, PCA |
| Martens et al. 2020 [101] | Train: 103pts Validation: 71pts | HNSCC | PET, low-dose-CT | LR, DM, OS. RadCat tool Cox regression analysis. Multivariable logistic regression | Logistic regression |
| Li et al. 2018 [97] | Total: 306pts, 20 of whom developed with recurrence | NPC | CT, MR, PET | LR. PCA. Machine learning classifiers | PCA, ANN, KNN, SVM |
| Liu et al. 2019 [90] | Total: 120pts | PTC | Preoperative ultrasound images | Metastasis. Support vector machine classifier | SVM |
| Wu et al. 2020 [98] | Train: 141pts Validation: 96pts | HNC | PET, CT | LR. PCA. Multivariate Cox proportional hazards regression | PCA |
| Zhou et al. 2020 [95] | Total: 188pts | HNSCC | PET, CT | DM. Machine learning classifiers | SVM, DT and KNN |
| Bogowicz et al. 2017 [102] | Train: 128pts Validation: 50pts | HNSCC | PET, CT | LR. PCA and LASSO. Multivariable Cox regression | PCA, LASSO |
| Tan et al. 2018 [89] | Train: 154pts Validation: 76pts | ESCC | Arterial-phase CT | LMR. Rad-score, logistic regression | LASSO, logistic regression |
| Vallieres et al. 2017 [103] | Total: 300pts | HNC | Pre-treatment FDG-PET and CT | LR and DM. Machine learning classifier | Random forests |
| Kwan et al. 2018 [104] | Total: 300pts 36 DM pts | HPV-related Oropharyngeal Carcinoma | CT | DM. PyRadiomic. Radiomics score | Logistic regression |
| Park et al. 2019 [92] | Train: 400pts Validation: 368pts | PTC | Neck ultrasound | LNM. LASSO. Rad-score, LASSO regression | LASSO |
| Zhang et al. 2019 [105] | Train: 360pts Validation: 120pts | Non-metastatic T4 NPC | T1, T2, CE- T1 | LR, Rad-score, cox proportional hazards regression | Logistic regression |
| Zhang et al. 2019 [99] | Total: 176pts | NPC | PET, CT | DM. LASSO. Multivariate logistic regression | Logistic regression |
| M.D. Anderson Cancer [88] | Train: 255pts Tune: 165pts Validation: 45pts | HNC | CT, MRI, PET | 5-year LCR. Multivariable Cox regression | DT |
| Bahig et al. 2019 [106] | Total: 176pts, 20 supraglottic , 5 pyriform sinus tumors | LHSCC | DECT | LRR. Univariate Cox regression | DT |
| Lu et al. 2019 [93] | Train: 154pts Validation: 67pts | PTC | non-contrast and venous CE- CT | LNM. SVM. Multivariable logistic regression | SVM, logistic regression |
| Qu et al. 2018 [94] | Train: 90pts Validation: 91pts | EC | MRI | LNM. LASSO. Multivariable logistic regression | LASSO, elastic net regression, logistic regression |
| Martens et al. 2020 [107] | Train: 103pts Validation: 71pts | HNSCC | 18F-FDG-PET, CT | LR, DM, OS. Rad- score. Multivariable survival regression | Logistic regression |
ANN: Artificial neural network; LCR: Local control rate; DECT: Dual-energy computed tomography; LHSCC: Larynx and hypopharynx squamous cell carcinoma.
In research reports on the use of radiomics in HNC, radiomics models related to predicting survival are the most numerous. Shen et al. [108] aimed to explore the predictive value of MRI-based radiomic model for PFS in nonmetastatic NPC. They collected the clinical and MRI data from 327 patients with NPC, and five models were established. The prognostic performances of these models were evaluated by Harrell's concordance index (C-index). They find that the model incorporating radiomics, overall stage, and EBV DNA showed better performance for predicting PFS in nonmetastatic NPC patients. In HNSCC, Yuan et al. [109] consisted of a training cohort (n = 85), and LASSO Cox regression model was used to select the most useful prognostic features with their coefficients, upon which a radiomic signature was generated. They find that MRI-based radiomic signature is an independent prognostic factor for HNSCC patients. Another study identified prognostic and reliable machine-learning methods for the prediction of overall survival of head and neck cancer patients [110]. Others have used pre- and post-operative PET/CT radiomics features for HNSCC and found that combining clinicopathological characteristics with radiomics features of pre-treatment PET/CT or post-treatment PET/CT assessment of primary tumor sites as positive or negative may substantially improve prediction of OS and DFS of HNSCC patients [111-112]. The predictions of radiomics signature models based on various types of imaging sequences in various types of HNC are represented in Table 4. The main types of survival values predicted by each type of model and which machine learning algorithms were employed are specified in the table.
Other predictive models
Tumor heterogeneity is a well-known prognostic factor in HNC. A major limitation of tissue- and blood-derived tumor markers is the lack of spatial resolution to image tumor heterogeneity. Due to the hidden growth sites of HNC, it is difficult to obtain biopsies before and after treatment. At the same time, issue markers derived from tumor biopsies usually represent only a small tumor subregion at a single timepoint and are therefore often not representative of the tumors' biology or the biological alterations during and after treatment. This has also been noted by researchers, Gu et al. [138] showed that a radiomics model with excellent performance prediction of the presence of cytokeratin 19, galectin 3, and thyro-peroxidase based on CT images. This model may be used to identify benign and malignant thyroid nodules. Chen et al. [31] investigated the correlation between programmed cell death protein 1 ligand (PD-L1) immunohistochemical expression and PET/CT radiomics and found that p16 and Ki-67 staining percentages and several PET/CT-derived textural features could provide additional information to identify tumor PD-L1 expression in HNC. There are also several researchers have done studies correlating radiomics features with molecular features of HNC [10,32,139]. In recent years, some researchers have begun to focus on the comparison of the predictive performance of radiomics models of different image modalities in the same disease [36, 53,140].
Discussion
In recent years, numerous literatures revealed that radiomics has been studied in the pre-treatment diagnosis of head and neck cancer, including the prediction of efficacy and the prediction of survival. These studies have yielded promising results and have drawn good lessons for subsequent researchers. However, there is still a lack of large-scale multicenter validation in existing exploratory radiomics studies, and the vast majority of validation cohorts are still derived from retrospective data from a single independent unit. A data platform such as the cancer imaging archive (TCIA) has been created, but the quality of the data profile is mixed [67]. Although relatively reliable conclusions can be drawn from some of the mixed data by relying on big data techniques, however, differences in parameters during image acquisition or noise on the images can cause serious interference with the radiomics features extracted from them. This interference will inevitably affect the model's ability to generalize to other databases as well.
Radiomics predicts the survival of head and neck cancer
| Study | Tumor characteristics | Imaging modality | Outcome | Machine learning methods |
|---|---|---|---|---|
| Shen et al. 2020 [108] | NPC | MRI | PFS | LASSO |
| Xu et al. 2020 [113] | NPC | PET | PFS | / |
| Ouyang et al. 2017 [114] | NPC | MRI | PFS | / |
| Zhang et al. 2017 [115] | Advanced NPC | MRI | PFS | LASSO |
| Lv et al. 2019 [53] | NPC | PET | PFS | / |
| Peng et al. 2019 [52] | NPC | PET | DFS | Deep learning |
| Yuan et al. 2019 [109] | HNSCC | MRI | OS | LASSO |
| Ming et al. 2019 [116] | NPC | MRI | DFS, OS, LRFS, DMFS | LASSO |
| Mao et al. 2019 [117] | NPC | MRI | PFS | / |
| Chen et al. 2020 [118] | LC | CT | OS | LASSO |
| Folkert et al.2017 [119] | OC | PET | ACM, DM | Multiparameter logistic regression |
| Foley et al.2018 [120] | OC | PET | OS | / |
| Chen et al.2019 [121] | EC | PET | DFS, OS | Multivariate logistic regression |
| Xiong et al. 2017 [122] | EC | PET | PFS | SVM, RF |
| Feliciani et al. 2018 [123] | HNC | PET | PFS | LASSO |
| Liao et al. 2019 [124] | Oropharyngeal and hypopharyngeal cancer | PET | OS, PRFS, DFS | / |
| Lv et al. 2019 [125] | HNC | PET/CT | RFS, MFS, OS | / |
| Yang et al. 2019 [126] | Advanced NPC | MRI | PFS | LASSO |
| Leijenaar et al. 2015 [127] | OSCC | CT | OS | / |
| Agarwal et al. 2020 [111] | LC | CT | LFS | / |
| Zhong et al. 2020 [128] | T3N0M0 NPC | MRI | DFS | Deep learning |
| Parmar et al. 2015 [110] | HNC | CT | OS | Different machine-learning classifiers |
| Liu et al. 2020 [112] | HNSCC | PET | OS, DFS | / |
| Pan et al. 2019 [130] | Oral tongue cancer | CT | Survival time | PCA |
| Xie et al. 2020 [129] | HNC | PET | OS, DFS | LR, SVM, RF, XG boost classifier |
| Cozzi et al. 2019 [131] | HNC | CT | OS, PFS | / |
| Legar et al. 2018 [132] | HNC | CT | OS, LRC | Six machine learning algorithms |
| Sörensen et al. 2019 [133] | HNC | PET | OS | / |
| Haider et al. 2020 [134] | OSCC | PET | OS, PFS | / |
| Ou et al. 2017 [135] | HNC | CT | PFS, OS | PCA |
| Miller et al. 2019 [136] | OPSCC | CT | PFS | / |
| Mes et al. 2020 [137] | HNSCC | MRI | RFS, OS | / |
OPSCC: Oropharyngeal Squamous Cell Carcinoma; RFS: Relapse-free survival; ACM: All-cause mortality; LFS: Laryngectomy free survival; PRFS: Primary relapse-free survival; RFS: Recurrence-free survival; MFS: Metastasis-free survival; LRC: Locoregional tumor control; DMFS: Disease distant metastasis-free survival; OC: Oropharyngeal carcinoma.
The relationship between radiomics and clinical symptoms has been widely documented, but other data types, such as genomics, transcriptomics, proteomics, and metabolomics, have been less studied in relation to radiomics. In HNC, correlation studies between imaging and genomics are now available, as important molecular markers such as PD-L1/TP53/FAT1/KMT2D/NOTCH1/Ki-67 can be predicted by predictive models of imaging features [10,32,141]. At present, the relationship between imaging and transcriptomics has been studied in other tumors, but its combination with proteomics and metabolomics is still less studied. This may be related to the fact that currently the histological data are independent of each other, and samples with these histological data do not have radiomics data.
The next milestone in radiomics is undoubtedly the creation of decision support and predictive tool models. In order to achieve this goal, having big data of all types of data is a sine qua non, and a strong and comprehensive common database is an effective solution. To achieve this goal, in addition to the involvement of different medical centers from all over the world to provide data, a worldwide accepted standard should be developed first. This standard should establish more uniform regulations in radiomics from the acquisition of source data, segregation of regions of interest, extraction of features to the development of predictive models. Although the difficulty and cost of creating and managing high-quality public data is enormous, the benefits to human medicine are also enormous.
People have investigated the association between PD-L1 expression in HNC patients and PET/CT, but did not delve into the efficacy of immunotherapy [31]. In other tumors, such as glioma and non-small-cell lung cancer, the models found in these studies have potential important translational implications to identify highly vulnerable patients treated with immunotherapy that experience rapid disease progression and survival poor outcomes [142-143]. These studies demonstrated that clinical data combined with radiomics performed better than traditional clinical data in predicting the efficacy of immunotherapy. Immunotherapy, as a new therapy in modern cancer treatment, has been shown to be less effective in many solid tumors, such as HNC, and therefore the development of appropriate models to predict the efficacy of immunotherapy prior to treatment would be of great help in avoiding the waste of medical resources and developing more accurate and personalized treatment plans.
In the current study of radiomics in HNC, it can be found that the vast majority of studies are still based on a single imaging modality, with few studies combining multiple imaging modality characterization. The predictive power of multiple imaging modalities in the same disease is still unknown, and our current research direction is trying to fill this vacancy. At the same time, as mentioned above, the development of predictive models by combining imaging modalities with multi-modality studies is still in its infancy, and there is still a lot of room for improvement, which is the direction we are working on at our medical center. The easier access to the data required for radiomics, unlike routine biopsies or other histology, provides new directions for otolaryngologists and craniofacial surgeons to study underlying tumors of the skull (in addition to routine HNC). As we all know, compared with HNC, skull base tumors are more difficult to biopsy and diagnose, and because of their insidious development, patients often do not show symptoms until later stages. In addition, the special and complex anatomical structure of the skull base often makes it more difficult for skull base surgeons to estimate the nature of the tumor and determine the scope of resection before surgery. Although relevant studies have been done by researchers, such as Li et al. 11 selected features were finally selected from skull base MRI of 210 patients to establish a radiomics model to differentiate between skull base chordoma and chondrosarcoma [144]. Other researchers have used MRI radiomics to predict the likelihood of early progression or recurrence in a subset of patients with skull base meningiomas due to incomplete resection [145-146]. However, the application of radiomics in skull base tumors is still rare, which may be due to the special location of skull base tumors, and the image range including various neurovascular, brain tissue, bone and even nasal and orbital conditions, this results in a complex image texture. Because of these complexities, it is necessary to develop radiomics, which can be used to obtain objective information through non-invasive testing, combined with machine learning to build pathological classification prediction models or conventional prognostic models, to guide the selection of treatment, design the scope of surgery, and even guide the postoperative comprehensive treatment. This is also very much in line with the concept of sequential cancer treatment.
With the enhancement of radiomics technology, the expansion of public databases, and the advancement of deep learning algorithms, radiomics will certainly play an important role in the future clinical diagnosis, treatment and prognosis. Radiomics is expected to lay the foundation for the future personalized treatment of otolaryngology patients and the sequential treatment of tumors.
Abbreviations
HNC: head and neck cancers; CT: computed tomography; MRI: magnetic resonance imaging; PET: positron emission tomography; 2D or 3D: two- or three-dimensional; ROI: areas of interest; VOI: volume of interest; GLCM: co-occurrence matrix; GLRLM: gray-level run length matrix; NGLDM: neighborhood gray-level different matrix; CNN: convolutional neural network; DT: decision tree; RF: random forests; SVM: support vector machines; LC: laryngeal cancer; AUC: area under the receiver operating characteristic curve; HNSCC: head and neck squamous cell carcinoma; SCC: squamous cell carcinoma; PTC: papillary thyroid carcinoma; NPC: nasopharyngeal carcinoma; PD-L1: programmed cell death protein 1 ligand; LASSO: least absolute shrinkage and selection operator; EC: esophageal cancer; PFS: progression-free survival; OS: overall survival; DM: distant metastasis; LR: local recurrence; LNM: lymph node metastasis; Rad-score: radiomics score; LRFS: local recurrence-free survival; DFS: disease-free survival; TCIA: The Cancer Imaging Archive; CE-CT: contrast- enhanced CT; CE-T1: contrast-enhanced T1; DWI: diffusion weighted imaging; HPV: human Papillomavirus; PCA: principal component analysis; VT: variance-threshold; CERR: computational environment for radiological research; CHAID: chi-square automatic interaction detection; KNN: k-nearest neighbor; CAD: computer-aided diagnosis; ART: adaptive radiotherapy; LFA: likelihood-fuzzy analysis; RBF: radial basis function; CRT: concurrent chemoradiation; T2FS: T2 weighted fat-suppressed; RT: radiation therapy; EBV: Epstein-Barr virus; ADC: apparent diffusion coefficient; ANN: artificial neural network; LCR: local control rate; DECT: dual-energy computed tomography; LHSCC: larynx and hypopharynx squamous cell carcinoma; OPSCC: Oropharyngeal Squamous Cell Carcinoma; RFS: relapse-free survival; ACM: all-cause mortality; PRFS: primary relapse-free survival; RFS: recurrence-free survival; MFS: metastasis-free survival; DMFS: disease distant metastasis-free survival; OC: oropharyngeal carcinoma.
Acknowledgements
This Research was funded by the National Natural Science Foundation of China (81770985); the Hunan Province Graduate Education Innovation Project (CX20200386); and the Independent Exploration and Innovation Project for Graduate Students of Central South University (506021703). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7-30
2. Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer [published correction appears in Nat Rev Cancer. 2018 Oct;18(10):662]. Nat Rev Cancer. 2018;18(5):269-282
3. Puram SV, Tirosh I, Parikh AS. et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. 2017;171(7):1611-1624
4. Driehuis E, Kolders S, Spelier S. et al. Oral Mucosal Organoids as a Potential Platform for Personalized Cancer Therapy [published correction appears in Cancer Discov. 2020 Mar;10(3):476]. Cancer Discov. 2019;9(7):852-871
5. Hsu D, Chokshi FH, Hudgins PA. et al. Predictive Value of First Posttreatment Imaging Using Standardized Reporting in Head and Neck Cancer. Otolaryngol Head Neck Surg. 2019;161(6):978-985
6. Huang Y, Liu Z, He L. et al. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology. 2016;281(3):947-957
7. Dalal V, Carmicheal J, Dhaliwal A, Jain M, Kaur S, Batra SK. Radiomics in stratification of pancreatic cystic lesions: Machine learning in action. Cancer Lett. 2020;469:228-237
8. Trebeschi S, Drago SG, Birkbak NJ. et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann Oncol. 2019;30(6):998-1004
9. Kickingereder P, Neuberger U, Bonekamp D. et al. Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro Oncol. 2018;20(6):848-857
10. Huang C, Cintra M, Brennan K. et al. Development and validation of radiomic signatures of head and neck squamous cell carcinoma molecular features and subtypes. EBioMedicine. 2019;45:70-80
11. Lambin P, Leijenaar RTH, Deist TM. et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749-762
12. Limkin EJ, Sun R, Dercle L. et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol. 2017;28(6):1191-1206
13. Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology. 2016;278(2):563-577
14. Lambin P, Rios-Velazquez E, Leijenaar R. et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441-446
15. Zwanenburg A, Vallières M, Abdalah MA. et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology. 2020;295(2):328-338
16. Kunapuli G, Varghese BA, Ganapathy P. et al. A Decision-Support Tool for Renal Mass Classification. J Digit Imaging. 2018;31(6):929-939
17. Feng QX, Liu C, Qi L. et al. An Intelligent Clinical Decision Support System for Preoperative Prediction of Lymph Node Metastasis in Gastric Cancer. J Am Coll Radiol. 2019;16(7):952-960
18. Holzinger A, Haibe-Kains B, Jurisica I. Why imaging data alone is not enough: AI-based integration of imaging, omics, and clinical data. Eur J Nucl Med Mol Imaging. 2019;46(13):2722-2730
19. Wei J, Jiang H, Gu D. et al. Radiomics in liver diseases: Current progress and future opportunities. Liver Int. 2020;40(9):2050-2063
20. Parekh VS, Jacobs MA. Multiparametric radiomics methods for breast cancer tissue characterization using radiological imaging. Breast Cancer Res Treat. 2020;180(2):407-421
21. Yang L, Yang J, Zhou X. et al. Development of a radiomics nomogram based on the 2D and 3D CT features to predict the survival of non-small cell lung cancer patients. Eur Radiol. 2019;29(5):2196-2206
22. Fan L, Fang M, Li Z. et al. Radiomics signature: a biomarker for the preoperative discrimination of lung invasive adenocarcinoma manifesting as a ground-glass nodule. Eur Radiol. 2019;29(2):889-897
23. Sun R, Limkin EJ, Vakalopoulou M. et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19(9):1180-1191
24. Pinker K, Chin J, Melsaether AN, Morris EA, Moy L. Precision Medicine and Radiogenomics in Breast Cancer: New Approaches toward Diagnosis and Treatment. Radiology. 2018;287(3):732-747
25. Huang YQ, Liang CH, He L. et al. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer [published correction appears in J Clin Oncol. 2016 Jul 10;34(20):2436]. J Clin Oncol. 2016;34(18):2157-2164
26. Wilson R, Devaraj A. Radiomics of pulmonary nodules and lung cancer. Transl Lung Cancer Res. 2017;6(1):86-91
27. Jiang Y, Chen C, Xie J. et al. Radiomics signature of computed tomography imaging for prediction of survival and chemotherapeutic benefits in gastric cancer. EBioMedicine. 2018;36:171-182
28. Wu S, Zheng J, Li Y. et al. Development and Validation of an MRI-Based Radiomics Signature for the Preoperative Prediction of Lymph Node Metastasis in Bladder Cancer. EBioMedicine. 2018;34:76-84
29. Grossmann P, Stringfield O, El-Hachem N. et al. Defining the biological basis of radiomic phenotypes in lung cancer. Elife. 2017;6:e23421
30. Di Stefano AL, Picca A, Saragoussi E, et al. Clinical, molecular and radiomic profile of gliomas with FGFR3-TACC3 fusions [published online ahead of print, 2020 May 15]
31. Chen RY, Lin YC, Shen WC. et al. Associations of Tumor PD-1 Ligands, Immunohistochemical Studies, and Textural Features in 18F-FDG PET in Squamous Cell Carcinoma of the Head and Neck. Sci Rep. 2018;8(1):105
32. Zwirner K, Hilke FJ, Demidov G. et al. Radiogenomics in head and neck cancer: correlation of radiomic heterogeneity and somatic mutations in TP53, FAT1 and KMT2D. Radiogenomics bei Kopf-Hals-Tumoren: Korrelation von bildgebender Heterogenität und somatischen Mutationen in TP53, FAT1 und KMT2D. Strahlenther Onkol. 2019;195(9):771-779
33. Benelli M, Barucci A, Zoppetti N. et al. Comprehensive Analysis of Radiomic Datasets by RadAR. Cancer Res. 2020;80(15):3170-3174
34. Mayerhoefer ME, Materka A, Langs G. et al. Introduction to Radiomics. J Nucl Med. 2020;61(4):488-495
35. Liang ZG, Tan HQ, Zhang F. et al. Comparison of radiomics tools for image analyses and clinical prediction in nasopharyngeal carcinoma. Br J Radiol. 2019;92(1102):20190271
36. Bogowicz M, Riesterer O, Stark LS. et al. Comparison of PET and CT radiomics for prediction of local tumor control in head and neck squamous cell carcinoma. Acta Oncol. 2017;56(11):1531-1536
37. Papp L, Rausch I, Grahovac M, Hacker M, Beyer T. Optimized Feature Extraction for Radiomics Analysis of 18F-FDG PET Imaging. J Nucl Med. 2019;60(6):864-872
38. Liu Z, Zhang XY, Shi YJ. et al. Radiomics Analysis for Evaluation of Pathological Complete Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Clin Cancer Res. 2017;23(23):7253-7262
39. Dalal V, Carmicheal J, Dhaliwal A, Jain M, Kaur S, Batra SK. Radiomics in stratification of pancreatic cystic lesions: Machine learning in action. Cancer Lett. 2020;469:228-237
40. Aerts HJ, Velazquez ER, Leijenaar RT. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006
41. Larue RT, Defraene G, De Ruysscher D, Lambin P, van Elmpt W. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol. 2017;90(1070):20160665
42. Lambin P, Rios-Velazquez E, Leijenaar R. et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441-446
43. Chaddad A, Kucharczyk MJ, Daniel P. et al. Radiomics in Glioblastoma: Current Status and Challenges Facing Clinical Implementation. Front Oncol. 2019;9:374
44. Traverso A, Wee L, Dekker A, Gillies R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int J Radiat Oncol Biol Phys. 2018;102(4):1143-1158
45. Jiang Y, Yuan Q, Lv W. et al. Radiomic signature of 18F fluorodeoxyglucose PET/CT for prediction of gastric cancer survival and chemotherapeutic benefits. Theranostics. 2018;8(21):5915-5928
46. Zhang Y, Oikonomou A, Wong A, Haider MA, Khalvati F. Radiomics-based Prognosis Analysis for Non-Small Cell Lung Cancer. Sci Rep. 2017;7:46349
47. Ibrahim A, Vallières M, Woodruff H. et al. Radiomics Analysis for Clinical Decision Support in Nuclear Medicine. Semin Nucl Med. 2019;49(5):438-449
48. Xu Y, Hosny A, Zeleznik R. et al. Deep Learning Predicts Lung Cancer Treatment Response from Serial Medical Imaging. Clin Cancer Res. 2019;25(11):3266-3275
49. Podgorsak AR, Rava RA, Shiraz Bhurwani MM. et al. Automatic radiomic feature extraction using deep learning for angiographic parametric imaging of intracranial aneurysms. J Neurointerv Surg. 2020;12(4):417-421
50. Spuhler KD, Ding J, Liu C. et al. Task-based assessment of a convolutional neural network for segmenting breast lesions for radiomic analysis. Magn Reson Med. 2019;82(2):786-795
51. Choi YS, Bae S, Chang JH. et al. Fully Automated Hybrid Approach to Predict the IDH Mutation Status of Gliomas via Deep Learning and Radiomics. Neuro Oncol. 2020 noaa177
52. Peng H, Dong D, Fang MJ. et al. Prognostic Value of Deep Learning PET/CT-Based Radiomics: Potential Role for Future Individual Induction Chemotherapy in Advanced Nasopharyngeal Carcinoma. Clin Cancer Res. 2019;25(14):4271-4279
53. Lv W, Yuan Q, Wang Q. et al. Radiomics Analysis of PET and CT Components of PET/CT Imaging Integrated with Clinical Parameters: Application to Prognosis for Nasopharyngeal Carcinoma. Mol Imaging Biol. 2019;21(5):954-964
54. Trebeschi S, Drago SG, Birkbak NJ. et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann Oncol. 2019;30(6):998-1004
55. Ger RB, Zhou S, Elgohari B. et al. Radiomics features of the primary tumor fail to improve prediction of overall survival in large cohorts of CT- and PET-imaged head and neck cancer patients. PLoS One. 2019;14(9):e0222509
56. Lu CF, Hsu FT, Hsieh KL. et al. Machine Learning-Based Radiomics for Molecular Subtyping of Gliomas. Clin Cancer Res. 2018;24(18):4429-4436
57. Park YW, Oh J, You SC. et al. Radiomics and machine learning may accurately predict the grade and histological subtype in meningiomas using conventional and diffusion tensor imaging. Eur Radiol. 2019;29(8):4068-4076
58. Gu Q, Feng Z, Liang Q. et al. Machine learning-based radiomics strategy for prediction of cell proliferation in non-small cell lung cancer. Eur J Radiol. 2019;118:32-37
59. Ji GW, Zhu FP, Xu Q. et al. Machine-learning analysis of contrast-enhanced CT radiomics predicts recurrence of hepatocellular carcinoma after resection: A multi-institutional study. EBioMedicine. 2019;50:156-165
60. Azad AK, Bairati I, Samson E. et al. Genetic sequence variants and the development of secondary primary cancers in patients with head and neck cancers. Cancer. 2012;118(6):1554-1565
61. Seeburg DP, Baer AH, Aygun N. Imaging of Patients with Head and Neck Cancer: From Staging to Surveillance. Oral Maxillofac Surg Clin North Am. 2018;30(4):421-433
62. Ho AS, Kim S, Tighiouart M. et al. Metastatic Lymph Node Burden and Survival in Oral Cavity Cancer. J Clin Oncol. 2017;35(31):3601-3609
63. Chen CC, Lin JC, Chen KW. Lymph node ratio as a prognostic factor in head and neck cancer patients. Radiat Oncol. 2015;10:181
64. Wang F, Zhang B, Wu X. et al. Radiomic Nomogram Improves Preoperative T Category Accuracy in Locally Advanced Laryngeal Carcinoma. Front Oncol. 2019;9:1064
65. Ren J, Tian J, Yuan Y. et al. Magnetic resonance imaging based radiomics signature for the preoperative discrimination of stage I-II and III-IV head and neck squamous cell carcinoma. Eur J Radiol. 2018;106:1-6
66. Romeo V, Cuocolo R, Ricciardi C. et al. Prediction of Tumor Grade and Nodal Status in Oropharyngeal and Oral Cavity Squamous-cell Carcinoma Using a Radiomic Approach. Anticancer Res. 2020;40(1):271-280
67. Mukherjee P, Cintra M, Huang C. et al. CT-based Radiomic Signatures for Predicting Histopathologic Features in Head and Neck Squamous Cell Carcinoma. Radiol Imaging Cancer. 2020;2(3):e190039
68. Wang H, Song B, Ye N. et al. Machine learning-based multiparametric MRI radiomics for predicting the aggressiveness of papillary thyroid carcinoma. Eur J Radiol. 2020;122:108755
69. Wu W, Ye J, Wang Q, Luo J, Xu S. CT-Based Radiomics Signature for the Preoperative Discrimination Between Head and Neck Squamous Cell Carcinoma Grades. Front Oncol. 2019;9:821
70. Ren J, Qi M, Yuan Y, Tao X. Radiomics of apparent diffusion coefficient maps to predict histologic grade in squamous cell carcinoma of the oral tongue and floor of mouth: a preliminary study [published online ahead of print, 2020 Jun 14]. Acta Radiol. 2020 284185120931683
71. Katsoulakis E, Yu Y, Apte AP. et al. Radiomic analysis identifies tumor subtypes associated with distinct molecular and microenvironmental factors in head and neck squamous cell carcinoma. Oral Oncol. 2020;110:104877
72. Yokota T, Hamauchi S, Shirasu H. et al. How Should We Approach Locally Advanced Squamous Cell Carcinoma of Head and Neck Cancer Patients Ineligible for Standard Non-surgical Treatment? Curr Oncol Rep. 2020;22(12):118
73. Dionisi F, Fiorica F, D'Angelo E. et al. Organs at risk's tolerance and dose limits for head and neck cancer re-irradiation: A literature review. Oral Oncol. 2019;98:35-47
74. Bologna M, Calareso G, Resteghini C. et al. Relevance of apparent diffusion coefficient features for a radiomics-based prediction of response to induction chemotherapy in sinonasal cancer. NMR Biomed. 2020 e4265
75. Wang G, He L, Yuan C, Huang Y, Liu Z, Liang C. Pretreatment MR imaging radiomics signatures for response prediction to induction chemotherapy in patients with nasopharyngeal carcinoma. Eur J Radiol. 2018;98:100-106
76. Zhao L, Gong J, Xi Y. et al. MRI-based radiomics nomogram may predict the response to induction chemotherapy and survival in locally advanced nasopharyngeal carcinoma. Eur Radiol. 2020;30(1):537-546
77. Jin X, Zheng X, Chen D. et al. Prediction of response after chemoradiation for esophageal cancer using a combination of dosimetry and CT radiomics. Eur Radiol. 2019;29(11):6080-6088
78. Pota M, Scalco E, Sanguineti G. et al. Early prediction of radiotherapy-induced parotid shrinkage and toxicity based on CT radiomics and fuzzy classification. Artif Intell Med. 2017;81:41-53
79. Liu Y, Shi H, Huang S. et al. Early prediction of acute xerostomia during radiation therapy for nasopharyngeal cancer based on delta radiomics from CT images. Quant Imaging Med Surg. 2019;9(7):1288-1302
80. Zhang B, Lian Z, Zhong L. et al. Machine-learning based MRI radiomics models for early detection of radiation-induced brain injury in nasopharyngeal carcinoma. BMC Cancer. 2020;20(1):502
81. Yu TT, Lam SK, To LH. et al. Pretreatment Prediction of Adaptive Radiation Therapy Eligibility Using MRI-Based Radiomics for Advanced Nasopharyngeal Carcinoma Patients. Front Oncol. 2019;9:1050
82. Concha-Benavente F, Kansy B, Moskovitz J, Moy J, Chandran U, Ferris RL. PD-L1 Mediates Dysfunction in Activated PD-1+ NK Cells in Head and Neck Cancer Patients. Cancer Immunol Res. 2018;6(12):1548-1560
83. Ang KK, Zhang Q, Rosenthal DI. et al. Randomized phase III trial of concurrent accelerated radiation plus cisplatin with or without cetuximab for stage III to IV head and neck carcinoma: RTOG 0522. J Clin Oncol. 2014;32(27):2940-2950
84. Colevas AD, Bahleda R, Braiteh F. et al. Safety and clinical activity of atezolizumab in head and neck cancer: results from a phase I trial. Ann Oncol. 2018;29(11):2247-2253
85. Jakobsen KK, Wingstrand VL, Jensen JS. et al. Incidence and survival of hypopharyngeal cancer: a Danish Nation-Wide Study from 1980 to 2014. Acta Oncol. 2019;58(11):1570-1576
86. Dwijayanti F, Prabawa A. et al. The Five-Year Survival Rate of Patients with Nasopharyngeal Carcinoma Based on Tumor Response after Receiving Neoadjuvant Chemotherapy, Followed by Chemoradiation, in Indonesia: A Retrospective Study. Oncology. 2020;98(3):154-160
87. Zhang L, Zhou H, Gu D. et al. Radiomic Nomogram: Pretreatment Evaluation of Local Recurrence in Nasopharyngeal Carcinoma based on MR Imaging. J Cancer. 2019;10(18):4217-4225
88. M. D. Anderson Cancer Center Head and Neck Quantitative Imaging Working Group. Investigation of radiomic signatures for local recurrence using primary tumor texture analysis in oropharyngeal head and neck cancer patients. Sci Rep. 2018;8(1):1524
89. Tan X, Ma Z, Yan L, Ye W, Liu Z, Liang C. Radiomics nomogram outperforms size criteria in discriminating lymph node metastasis in resectable esophageal squamous cell carcinoma. Eur Radiol. 2019;29(1):392-400
90. Liu T, Ge X, Yu J. et al. Comparison of the application of B-mode and strain elastography ultrasound in the estimation of lymph node metastasis of papillary thyroid carcinoma based on a radiomics approach. Int J Comput Assist Radiol Surg. 2018;13(10):1617-1627
91. Liu T, Zhou S, Yu J. et al. Prediction of Lymph Node Metastasis in Patients with Papillary Thyroid Carcinoma: A Radiomics Method based on Preoperative Ultrasound Images. Technol Cancer Res Treat. 2019;18:1533033819831713
92. Park VY, Han K, Kim HJ. et al. Radiomics signature for prediction of lateral lymph node metastasis in conventional papillary thyroid carcinoma. PLoS One. 2020;15(1):e0227315
93. Lu W, Zhong L, Dong D. et al. Radiomic analysis for preoperative prediction of cervical lymph node metastasis in patients with papillary thyroid carcinoma. Eur J Radiol. 2019;118:231-238
94. Qu J, Shen C, Qin J. et al. The MR radiomic signature can predict preoperative lymph node metastasis in patients with esophageal cancer. Eur Radiol. 2019;29(2):906-914
95. Zhou Z, Wang K, Folkert M. et al. Multifaceted radiomics for distant metastasis prediction in head & neck cancer. Phys Med Biol. 2020;65(15):155009
96. Beichel RR, Ulrich EJ, Smith BJ. et al. FDG PET based prediction of response in head and neck cancer treatment: Assessment of new quantitative imaging features. PLoS One. 2019;14(4):e0215465
97. Li S, Wang K, Hou Z. et al. Use of Radiomics Combined With Machine Learning Method in the Recurrence Patterns After Intensity-Modulated Radiotherapy for Nasopharyngeal Carcinoma: A Preliminary Study. Front Oncol. 2018;8:648
98. Wu A, Li Y, Qi M. et al. Dosiomics improves prediction of locoregional recurrence for intensity modulated radiotherapy treated head and neck cancer cases. Oral Oncol. 2020;104:104625
99. Zhang L, Dong D, Li H. et al. Development and validation of a magnetic resonance imaging-based model for the prediction of distant metastasis before initial treatment of nasopharyngeal carcinoma: A retrospective cohort study. EBioMedicine. 2019;40:327-335
100. Bogowicz M, Riesterer O, Ikenberg K. et al. Computed Tomography Radiomics Predicts HPV Status and Local Tumor Control After Definitive Radiochemotherapy in Head and Neck Squamous Cell Carcinoma. Int J Radiat Oncol Biol Phys. 2017;99(4):921-928
101. Martens RM, Koopman T, Noij DP. et al. Predictive value of quantitative 18F-FDG-PET radiomics analysis in patients with head and neck squamous cell carcinoma. EJNMMI Res. 2020;10(1):102
102. Bogowicz M, Leijenaar RTH, Tanadini-Lang S. et al. Post-radiochemotherapy PET radiomics in head and neck cancer - The influence of radiomics implementation on the reproducibility of local control tumor models. Radiother Oncol. 2017;125(3):385-391
103. Vallières M, Kay-Rivest E, Perrin LJ. et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci Rep. 2017;7(1):10117. Published 2017 Aug 31
104. Kwan JYY, Su J, Huang SH. et al. Radiomic Biomarkers to Refine Risk Models for Distant Metastasis in HPV-related Oropharyngeal Carcinoma. Int J Radiat Oncol Biol Phys. 2018;102(4):1107-1116
105. Zhang LL, Huang MY, Li Y. et al. Pretreatment MRI radiomics analysis allows for reliable prediction of local recurrence in non-metastatic T4 nasopharyngeal carcinoma. EBioMedicine. 2019;42:270-280
106. Bahig H, Lapointe A, Bedwani S. et al. Dual-energy computed tomography for prediction of loco-regional recurrence after radiotherapy in larynx and hypopharynx squamous cell carcinoma. Eur J Radiol. 2019;110:1-6
107. Roland M, Thomas K, Daniel P. et al. Predictive value of quantitative 18F-FDG-PET radiomics analysis in patients with head and neck squamous cell carcinoma. EJNMMI Research. 2020;10(102):2-15
108. Shen H, Wang Y, Liu D. et al. Predicting Progression-Free Survival Using MRI-Based Radiomics for Patients With Nonmetastatic Nasopharyngeal Carcinoma. Front Oncol. 2020;10:618
109. Yuan Y, Ren J, Shi Y, Tao X. MRI-based radiomic signature as predictive marker for patients with head and neck squamous cell carcinoma. Eur J Radiol. 2019;117:193-198
110. Parmar C, Grossmann P, Rietveld D, Rietbergen MM, Lambin P, Aerts HJ. Radiomic Machine-Learning Classifiers for Prognostic Biomarkers of Head and Neck Cancer. Front Oncol. 2015;5:272
111. Agarwal JP, Sinha S, Goda JS. et al. Tumor radiomic features complement clinico-radiological factors in predicting long-term local control and laryngectomy free survival in locally advanced laryngo-pharyngeal cancers. Br J Radiol. 2020;93(1109):20190857
112. Liu Z, Cao Y, Diao W, Cheng Y, Jia Z, Peng X. Radiomics-based prediction of survival in patients with head and neck squamous cell carcinoma based on pre- and post-treatment 18F-PET/CT. Aging (Albany NY). 2020;12(14):14593-14619
113. Xu H, Lv W, Feng H. et al. Subregional Radiomics Analysis of PET/CT Imaging with Intratumor Partitioning: Application to Prognosis for Nasopharyngeal Carcinoma. Mol Imaging Biol. 2020;22(5):1414-1426
114. Ouyang FS, Guo BL, Zhang B. et al. Exploration and validation of radiomics signature as an independent prognostic biomarker in stage III-IVb nasopharyngeal carcinoma. Oncotarget. 2017;8(43):74869-74879
115. Zhang B, He X, Ouyang F. et al. Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett. 2017;403:21-27
116. Ming X, Oei RW, Zhai R. et al. MRI-based radiomics signature is a quantitative prognostic biomarker for nasopharyngeal carcinoma. Sci Rep. 2019;9(1):10412
117. Mao J, Fang J, Duan X. et al. Predictive value of pretreatment MRI texture analysis in patients with primary nasopharyngeal carcinoma. Eur Radiol. 2019;29(8):4105-4113
118. Chen L, Wang H, Zeng H, Zhang Y, Ma X. Evaluation of CT-based radiomics signature and nomogram as prognostic markers in patients with laryngeal squamous cell carcinoma. Cancer Imaging. 2020;20(1):28
119. Folkert MR, Setton J, Apte AP. et al. Predictive modeling of outcomes following definitive chemoradiotherapy for oropharyngeal cancer based on FDG-PET image characteristics. Phys Med Biol. 2017;62(13):5327-5343
120. Foley KG, Shi Z, Whybra P. et al. External validation of a prognostic model incorporating quantitative PET image features in oesophageal cancer. Radiother Oncol. 2019;133:205-212
121. Chen YH, Lue KH, Chu SC. et al. Combining the radiomic features and traditional parameters of 18F-FDG PET with clinical profiles to improve prognostic stratification in patients with esophageal squamous cell carcinoma treated with neoadjuvant chemoradiotherapy and surgery. Ann Nucl Med. 2019;33(9):657-670
122. Xiong J, Yu W, Ma J, Ren Y, Fu X, Zhao J. The Role of PET-Based Radiomic Features in Predicting Local Control of Esophageal Cancer Treated with Concurrent Chemoradiotherapy. Sci Rep. 2018;8(1):9902
123. Feliciani G, Fioroni F, Grassi E. et al. Radiomic Profiling of Head and Neck Cancer: 18F-FDG PET Texture Analysis as Predictor of Patient Survival. Contrast Media Mol Imaging. 2018;2018:3574310
124. Liao KY, Chiu CC, Chiang WC. et al. Radiomics features analysis of PET images in oropharyngeal and hypopharyngeal cancer. Medicine (Baltimore). 2019;98(18):e15446
125. Lv W, Ashrafinia S, Ma J, Lu L, Rahmim A. Multi-Level Multi-Modality Fusion Radiomics: Application to PET and CT Imaging for Prognostication of Head and Neck Cancer. IEEE J Biomed Health Inform. 2020;24(8):2268-2277
126. Yang K, Tian J, Zhang B. et al. A multidimensional nomogram combining overall stage, dose volume histogram parameters and radiomics to predict progression-free survival in patients with locoregionally advanced nasopharyngeal carcinoma. Oral Oncol. 2019;98:85-91
127. Leijenaar RT, Carvalho S, Hoebers FJ. et al. External validation of a prognostic CT-based radiomic signature in oropharyngeal squamous cell carcinoma. Acta Oncol. 2015;54(9):1423-1429
128. Zhong LZ, Fang XL, Dong D. et al. A deep learning MR-based radiomic nomogram may predict survival for nasopharyngeal carcinoma patients with stage T3N1M0 [published online ahead of print, 2020 Jul 4]. Radiother Oncol. 2020;151:1-9
129. Xie C, Du R, Ho JW. et al. Effect of machine learning re-sampling techniques for imbalanced datasets in 18F-FDG PET-based radiomics model on prognostication performance in cohorts of head and neck cancer patients. Eur J Nucl Med Mol Imaging. 2020;47(12):2826-2835
130. Pan X, Zhang T, Yang Q, Yang D, Rwigema JC, Qi XS. Survival prediction for oral tongue cancer patients via probabilistic genetic algorithm optimized neural network models. Br J Radiol. 2020;93(1112):20190825
131. Cozzi L, Franzese C, Fogliata A. et al. Predicting survival and local control after radiochemotherapy in locally advanced head and neck cancer by means of computed tomography based radiomics. Vorhersage von Überleben und lokaler Kontrolle nach Radiochemotherapie bei lokal fortgeschrittenen Kopf-Hals-Tumoren mittels computertomographiebasierter Radiomics-Technik. Strahlenther Onkol. 2019;195(9):805-818
132. Leger S, Zwanenburg A, Pilz K. et al. CT imaging during treatment improves radiomic models for patients with locally advanced head and neck cancer. Radiother Oncol. 2019;130:10-17
133. Sörensen A, Carles M, Bunea H. et al. Textural features of hypoxia PET predict survival in head and neck cancer during chemoradiotherapy. Eur J Nucl Med Mol Imaging. 2020;47(5):1056-1064
134. Haider SP, Zeevi T, Baumeister P. et al. Potential Added Value of PET/CT Radiomics for Survival Prognostication beyond AJCC 8th Edition Staging in Oropharyngeal Squamous Cell Carcinoma. Cancers (Basel). 2020;12(7):1778
135. Ou D, Blanchard P, Rosellini S. et al. Predictive and prognostic value of CT based radiomics signature in locally advanced head and neck cancers patients treated with concurrent chemoradiotherapy or bioradiotherapy and its added value to Human Papillomavirus status. Oral Oncol. 2017;71:150-155
136. Miller TA, Robinson KR, Li H. et al. Prognostic value of pre-treatment CT texture analysis in combination with change in size of the primary tumor in response to induction chemotherapy for HPV-positive oropharyngeal squamous cell carcinoma. Quant Imaging Med Surg. 2019;9(3):399-408
137. Mes SW, van Velden FHP, Peltenburg B. et al. Outcome prediction of head and neck squamous cell carcinoma by MRI radiomic signatures. Eur Radiol. 2020;30(11):6311-6321
138. Gu J, Zhu J, Qiu Q, Wang Y, Bai T, Yin Y. Prediction of Immunohistochemistry of Suspected Thyroid Nodules by Use of Machine Learning-Based Radiomics. AJR Am J Roentgenol. 2019;213(6):1348-1357
139. Yoon JH, Han K, Lee E. et al. Radiomics in predicting mutation status for thyroid cancer: A preliminary study using radiomics features for predicting BRAFV600E mutations in papillary thyroid carcinoma. PLoS One. 2020;15(2):e0228968
140. Liang ZG, Tan HQ, Zhang F. et al. Comparison of radiomics tools for image analyses and clinical prediction in nasopharyngeal carcinoma. Br J Radiol. 2019;92(1102):20190271
141. Shi Z, Jiang W, Chen X, Xu M, Wang X, Zha D. Prognostic and clinicopathological value of Ki-67 expression in patients with nasopharyngeal carcinoma: a meta-analysis. Ther Adv Med Oncol. 2020;12:1758835920951346
142. Han W, Qin L, Bay C. et al. Deep Transfer Learning and Radiomics Feature Prediction of Survival of Patients with High-Grade Gliomas. AJNR Am J Neuroradiol. 2020;41(1):40-48
143. Tunali I, Gray JE, Qi J. et al. Novel clinical and radiomic predictors of rapid disease progression phenotypes among lung cancer patients treated with immunotherapy: An early report. Lung Cancer. 2019;129:75-79
144. Li L, Wang K, Ma X. et al. Radiomic analysis of multiparametric magnetic resonance imaging for differentiating skull base chordoma and chondrosarcoma. Eur J Radiol. 2019;118:81-87
145. Wei W, Wang K, Liu Z. et al. Radiomic signature: A novel magnetic resonance imaging-based prognostic biomarker in patients with skull base chordoma. Radiother Oncol. 2019;141:239-246
146. Zhang Y, Chen JH, Chen TY. et al. Radiomics approach for prediction of recurrence in skull base meningiomas. Neuroradiology. 2019;61(12):1355-1364
Author contact
![]() Corresponding author: Weihong Jiang, E-mail: 401429edu.cn.
Corresponding author: Weihong Jiang, E-mail: 401429edu.cn.

 Global reach, higher impact
Global reach, higher impact