10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(2):651-669. doi:10.7150/ijbs.52266 This issue Cite
Review
Advancement and properties of circular RNAs in prostate cancer: An emerging and compelling frontier for discovering
1. Department of Pharmacy, Anhui Provincial Cancer Hospital, The First Affiliated Hospital of USTC West District, University of Science and Technology of China, Hefei 230031, China.
2. Inflammation and Immune Mediated Diseases Laboratory of Anhui Province, Anhui Institute of Innovative Drugs, School of Pharmacy, Anhui Medical University, Hefei, 230032, China.
3. Institute for Liver Diseases of Anhui Medical University, Hefei 230032, China.
4. Department of Urology, the Fourth Affiliated Hospital of Anhui Medical University, Hefei, 230011, China.
5. Department of Urology, The Second Hospital of Anhui Medical University, Hefei 230601, China.
6. Department of Clinical Trial Research Center, The Second Hospital of Anhui Medical University, Hefei, 230601, China.
#These authors contributed equally to this work.
Received 2020-8-21; Accepted 2020-12-18; Published 2021-1-19
Abstract
Prostate cancer (PC) is the most common carcinoma among men worldwide which results in 26% of leading causes of cancer-related death. However, the ideal and effective molecular marker remains elusive. CircRNA, initially observed in plant-infected viruses and Sendai virus in 1979, is generated from pre-mRNA back-splicing and comes in to play by adequate expression. The differential expression in prostate tissues compared with the control reveals the promising capacity in modulating processes including carcinogenesis and metastasis. However, the biological mechanisms of regulatory network in PC needs to systemically concluded. In this review, we enlightened the comprehensive studies on the definite mechanisms of circRNAs affecting tumor progression and metastasis. What's more, we validated the potential clinical application of circRNAs serving as diagnostic and prognostic biomarker. The discussion and analysis in circRNAs will broaden our knowledge of the pathogenesis of PC and further optimize the current therapies against different condition.
Keywords: circular RNA, prostate cancer (PC), biomarker, microRNA, tumorigenesis
Introduction
Prostate cancer (PC) is the most common carcinoma among men worldwide which results in 26% of leading causes of cancer-related death [1]. It is reported that the incidence and mortality of PC is noteworthy higher [2]. Accounts for 1.3 million new cases of PC is diagnosed and almost 359,000 associated deaths are responsible for PC development all over the world in 2018 [3]. Although diagnostic techniques and treatment regimens become increasingly complete and systematic, and a number of patients have achieved a better prognosis and survival rate after androgen stripping therapy, the recurrence, metastasis and castration resistance of PC still bring considerable pressure to patients [4]. Therefore, it is urgency to find a novel molecular biomarker for helping to understand the pathological bases of PC and improving the prognosis after comprehensive treatment.
Accumulating data have proved that an intricate interplay among diverse RNA species, including protein-coding messenger RNAs and non-coding RNAs such as long non-coding RNAs (IncRNAs), pseudogenes and circular RNAs, furnishes novel insight in gene regulatory network and significant implication of human disease by acting as competing endogenous RNAs (ceRNAs) or natural microRNA (miRNA) sponge [5]. Of note, circRNAs, structurally construct covalent closed loop from 5' to 3'polarity, were originally seen as represent errors in splicing and considered to below abundance. However, informed researches verified that the progression of tumor was in alignment with the amount of circRNAs, which highlighted the regulatory role in carcinogenesis.
Timeline of circRNAs and its researches on prostate cancer.
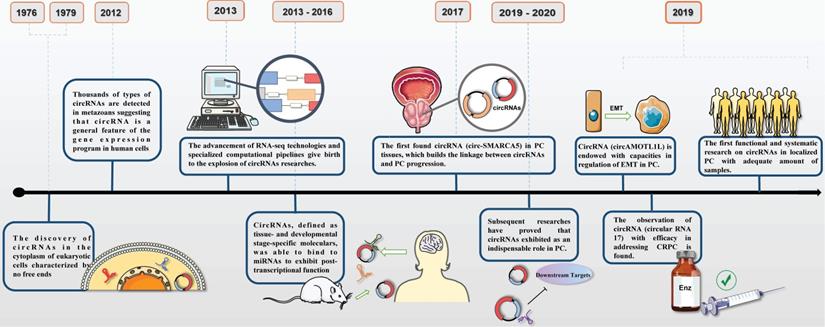
The discoveries of circRNAs in PC underwent a tough and twisted period of time. The first observation of circRNA was in 1979 experimented by Hsu and Coca-Prados under electron microscopy [6]. Then, early researches on lncRNAs, a particular type of non-coding RNA, elucidated the underlying therapeutic function in progression of PC, which hinted of the same usage in circRNAs. With sufficient efforts on discovering, dysregulation of circRNAs were reported as the leading force in affecting tumor growth, cell proliferation, invasion, migration and apoptosis in PC via modifying the expression of oncogenes andsuppressor genes [7]. Most recently, circRNAs have become promising candidates in a vast number of cellular processes and been implicated in epigenetic regulation in PC (Figure 1). FOXP4, a member of family of forkhead box transcription factors, was initially reported to be a critical regulator in NSCLC and FOXP4 silencing implied a practical role in carcinogenesis. It had been demonstrated that circABCC4 overexpression was especially common in PC tissues and exerted detrimental function via binding miR-1182, which indirectly inhibited the expression of FOXP4 [8]. Similar function was also observed in circ0005276 in PC. X-linked inhibitor of apoptosis protein (XIAP), exerted as the critical regulator in promoting tumorigenesis, was the host gene for circ0005276. And further research detected the positive interaction between circ0005276 and XIAP. This intense relevance implied us that circRNAs could be able to directly or indirectly modulate and affect the process of PC [9]. Exactly, all these parallel studies were required to testify the hypothesis and set up the regulatory network of circRNAs in PC.
Therefore, in this review, we outline that circRNAs are capable of eliciting a sequence of cellular processes consist of tumorigenesis, metastasis, epithelial-mesenchymal transition (EMT) and castration resistance in PC. Subsequently, we will discuss the potential application of circRNAs as promising targets and biomarkers in clinical strategy. Persistent observations in ascertaining regulatory role of circRNAs help us to figure out the pathogenesis of cancer generation in prostate and provide eventually treatment strategy in clinical application.
Overview of prostate cancer
PC is the most common non-skin carcinoma among men worldwide, which exerts significant burdens for patients and health-care systems in many countries [10]. Globally, PC is the sixth most common cause of cancer-related death [3]. Age, race, and family history are currently recognized risk factors for PC. However, a great deal of epidemiologic studies had revealed that the pathogenesis and progress of PC are closely related to alcohol drinking, obesity, prostatitis, benign prostatic hyperplasia (BPH). Digital rectal examination of the prostate (DRE), PSA, prostate biopsy, transrectal ultrasonography (TRUS) and multiparametric magnetic resonance imaging (mpMRI) were the frequently-used diagnostic methods of PC by clinician. However, due to the elevated level in non-neoplastic conditions, including BPH, prostatitis and prostate infarction, the sensitivity and specificity of PSA remained controversial. Besides, beyond the strategies for conservative deferred treatment and radical surgery, the androgen deprivation therapy establishes a golden standard in treating advanced PC. Consequently, it is critical to identify biomarkers with enhanced sensitivity and specificity to diagnose prostate cancer at earlier stages of the disease, and to develop therapeutic tactics that are both safer and more effective than those currently available.
The occurrence of PC follows a multistep process, moving from prostatic intraepithelial neoplasia (PIN) to advance prostate adenocarcinoma with local invasion, and enduringly to culminate in metastatic PC. The Gleason score is the strongest prognostic factor currently, which has shown a positively correlation in disease-free survival [11]. To be extent, special consideration has been given to the latest insights regarding the crucial role of inflammation for driving PC. The inflammatory mediators including lymphocytes and macrophages play impeccable role on giving rise to the initiation, induction of aggressive PC phenotype, promotion of tumor metastasis and resistance to chemotherapy of PC. The carcinogenesis of PC may involve in numerous genetic and epigenetic alterations. Many Noncoding RNAs (ncRNAs) are aberrantly expressed, and show evidence of function in oncogenesis or tumor progression. Recent studies have highlighted the prevalence and highly tissue/cell type-specific expression of circRNAs in PC. Specifically, the downregulation or upregulation of circRNAs expression attributes a tumor-suppressor or an oncogenic role to influence the clinicopathological appearance, prognosis and outcome of PC. For instance, circABCC4 could regulate PC progression by modulating FOXP4 signalling pathway [8]. CircAMOTL1L significantly decreased the migration and invasion in PC-3 and DU145 cells [12]. Therefore, it's worth mentioning that the role of these circRNAs in PC is reviewed in greater detail.
Overview of Circular RNAs
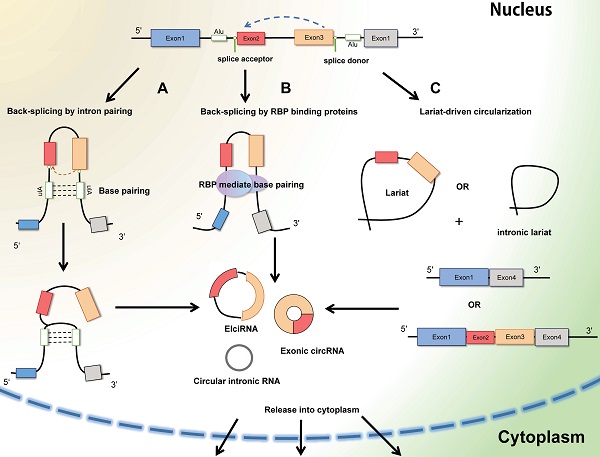
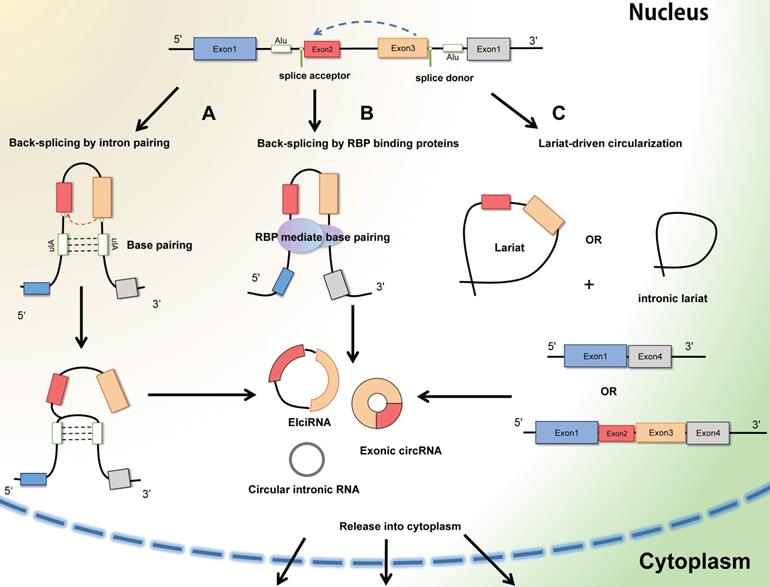
Circular RNAs (circRNAs) are generally defined as non-coding RNAs which are endogenous single-stranded circular molecules without either poly-adenylated tails in 3' ends or the cap structure at 5' ends. And it was initially observed in plant-infected viruses and Sendai virus in 1979 by Hsu et al. [6]. The circRNAs in host genes accumulate in various types of extracellular body (urine, saliva, and blood) [13], and the amount of circRNA is far more than the linear mRNAs of the corresponding host genes. Considering their multiple localization, circRNA can be segmented into exoniccircRNA (ecircRNA), circular intronic RNA (ciRNA), exon-intron RNA (EIciRNA) and intregenic RNA (icircRNA) (Figure 2). In most cases, the lariat-driven circularization model contains direct circularization of intronic lariat and circularization through exon skipping, facilitates the formation of circRNA. The hetero-lariat is formed including both introns and exons (ElcircRNA) in exon skipping. In other condition, direct back-splicing generates the formation of ciRNAs, EcircRNAs and EIcircRNAs [14]. Aforementioned features and abundant exist to make circRNAs become promising candidates in diagnosis and prognosis in cancer disease.
CircRNAs have been implicated in multiple cancer processes, including cellular activity activation, EMT regulation and nuclear and cytoplasmic trafficking. Recent studies have indicated that circRNAs can mediate a “sponge” regulatory network (sequestering microRNAs), which can differentially affect the expression of many protein-coding cancer-driving genes and key components of cancer-driving pathways during carcinogenesis. Recent independent reports have identified the circSMARCA5 was a circRNA derived from exons 15 and 16 of the SMARCA5 gene. The downregulation of circSMARCA5 in hepatocellular carcinoma (HCC) is significantly correlated with aggressive characteristics and serve as an independent risk factor for HCC patients after hepatectomy [15]. In bladder cancer, circSLC8A1 may act as sponge to regulate the expression level of miR-130b/miR-494 [16]. Moreover, some circRNAs are linked to reactivation of the androgen receptor signaling axis in PC [17]. Considering their dynamic role in cancer, circRNAs may be regarded as therapy target, helping to maintain stable disease and prohibit metastatic spread in tumorigenesis.
The correlation between Circular RNAS and prostate cancer
With the detection of new technology, the amount of identification of circRNAs in PC is increasing. Because of their abundance, stability and evolutionary conservation, people suggest that they may also have various efficacies in regulating the progression of PC. Besides, it is universally acknowledged that the androgen deprivation therapy ranked to the most prevalent treatment on PC therapeutic measures strongly subjecting to the effect of transformation from androgen dependent to androgen independent. However, with the concomitant increase of statistical correlation between PC and circRNAs, genetic and epigenetic researches are constructed to analyze the potential targets of circRNAs in alleviating the procession of tumorigenesis. Nevertheless, the generally accepted perspective of the correlation with circRNAs and PC is certified, the integrated signaling pathways and molecular mechanism of circRNA-modulated gene regulation are still remaining indistinct (Table 1).
The oncogenic or tumor suppressive roles of Circular RNAs in prostate cancer
Novel study had investigated that circ0005276, located at chromosome X, resided in both cytoplasm and nucleus. According to microarray analysis applied to detect the differential expression of mRNA in PC tissues, mRNA XIAP acquired sufficient attention with the highest fold change. Besides, the uplifted level of XIAP was found in 90 PC tissues and University of California Santa Cruz (UCSC, http://genome.ucsc.edu/) confirmed that XIAP was the host gene of circ0005276. In addition, the relative high expression of circ0005276 was also determined the production of XIAP suggesting that circ0005276 might exhibit oncogenic function in PC progression via enhancing the expression of XIAP. What's more, CCK-8 and EdU assays were carried out to validated that tumor growth was stagnated after silencing circ0005276 or XIAP [9]. Furthermore, Feng et al uncovered the tight conjunction between circ0005276 and Fused in sarcoma (FUS) by RIP assay. FUS was firstly known as an RBP contributing to the development of gastric cancer [9]. Both of Circ0005276 and FUS protein were identified as up-regulated component in PC tissues. Notably, they detected the effect of transfecting with sh-circ#1 and sh-FUS#1, finding that the levels of XIAP were consistently decreased in either ways. Precisely, circ0005276 and FUS interacted with XIAP mRNA by binding the promoter sequences of it and subsequent researches validated that the luciferase activity of XIAP was attenuated after silencing circ0005276 or FUS. Finally, rescue experiments revealed that over-expressing XIAP reversed the negative efficacy of cell proliferation induced by inhibiting circ0005276 or FUS. Thus, it was indicated that circ0005276 cooperated with FUS exerted oncogenic regulation to modulate the tumorigenesis via XIAP.
The formation of CircRNAs-Back-splicing and Lariat-driven circularization. A: Conventional and traditional formation of circRNAs model manifests that circRNAs (Exon-intron circRNAs and ExoniccircRNAs) are mainly produced by direct back splicing with the connection of complementary base pairing between inverted repeat elements (Alu elements). B: Besides, trans-acting RNA binding proteins (RBPs) can also generate circRNAs through dimerization between introns during back splicing. C: In some other cases, the mechanism of Lariat-driven circularization model prompts the formation of ExoniccircRNAs and circular intronic RNAs during exon skipping and canonical linear splicing.
The expression of circRNAs in PC
| circRNA | Genome Location | PCcells or tissues | Expression change | Function in PC | References (PMID) |
|---|---|---|---|---|---|
| circABCC4 | chr13:95813442-95840796 | PC-3, DU145, VCaP and LNCaP cells | + | Oncogene | 31270953 |
| circHIPK3 | chr11:33307958-33309057 | 22RV1, PC-3, DU145 and LNCaP cells | + | Oncogene | 30863152, 31118680 |
| circSMARCA5 | chr4:144464662-144465125 | 22RV1, PC-3, DU145 and LNCaP cells | + | Oncogene | 28765045 |
| circZNF609 | chr15:64791491-64792365 | PC-3 and LNCaP cells | + | Oncogene | 31387394 |
| hsa_circ_0001206 | chr22:21288066-21288532 | PC-3, DU145, and LNCaPcells | - | Suppressor gene and biomarker | 31198063, 32919302 |
| hsa_circ_0001633 | chr6:107824860-107827631 | PC tissues | - | Suppressor gene | 31198063 |
| hsa_circ_0009061 | chr1:23356961-23377013 | PC tissues | - | Suppressor gene | 31198063 |
| circAMOTL1L | chr22:35948707-35948901 | PC-3 and DU145 cells | - | Suppressor gene | 30531834 |
| circ102004 | chr17 | PC-3 and 22RV1 cells | + | Oncogene | 30219508 |
| circMYLK | chr3:123332641-123332832 | PC-3, DU145, LNCaP and PC-3MIE8 cells | - | Suppressor gene | 29798970 |
| circRNA17 | chr4:95561426-95578698 | EnzR-C4-2 cells | + | Oncogene | 30674872 |
| circFoxo3 | chr6:108984657-108986092 | PC-3, DU145, VCaP and LNCaP cells | - | Suppressor gene | 31593800 |
| circ0005276 | chrX:123022468-123026623 | PC-3, DU145, VCaP and LNCaP cells | + | Oncogene | 31624242 |
| hsa_circ_0044516 | chr17:48263677-48271402 | PC-3, 2B4 and 22RV1 cells | + | Oncogene and biomarker | 31625175 |
| circ-UCK2 | chr1:165859440-165877108 | EnzR-C4-2 cells | - | Suppressor gene | 31844675 |
| circ_KATNAL1 | chr13:30801548-30857928 | 22Rv1, DU145, LNCaP and PC-3 cells | - | Suppressor gene | 31800303 |
| hsa_circ_0001165 | chr20:46252654-46262380 | PC-3M IE8 cells | + | Oncogene | 31882179 |
| hsa_circ_0001085 | chr2:191765289-191789319 | PC-3M IE8 cells | + | Oncogene | 31882179 |
| hsa_circ_0004916 | chr2:44428324-44436466 | PC-3M IE8 cells | + | Oncogene | 31882179 |
| circFMN2 | chr1:240458121-240497529 | PC-3, LNCaP, VCaP and DU145 cells | + | Oncogene | 32526477 |
| circ0016068 | chr1:203274663-203278729 | PC-3, DU145, VCaP and 22RV1 cells | + | Oncogene | 32984325 |
| circMBOAT2 | chr2:9083315-9098771 | PC-3, LNCaP, VCaP, DU145 and C4-2B cells | + | Oncogene | 32645691 |
| circLMTK2 | chr7:98190727-98194572 | PC-3 and LNCaP cells | - | Suppressor gene | 31760099 |
CircABCC4 (circBase ID: hsa_circ_0030586; chr13:95813442-95840796), located on chromosome 13q32.1, took crucial roles in PC carcinogenesis. According to Huang's report, the expression of circABCC4 was differently high in both PC and cell lines compared to the control [8]. Amusingly, the rate of 5-year in patients showed strong correlation with the high expression of circABCC4 by Kaplan-Meier survival, which was often applied to estimate the probability of survival [19]. Besides, Huang et al also revealed that the number of circABCC4-deficient PC3 and DU145 cell lines derived from PC was stagnating at the G0/G1 stage of the cell cycle while few cells are found in the S and G2/M stages, which made the conclusion that the circABCC4 promoted the proliferation of PC cells. After that, they conducted vivo experiment and the result made the same [8]. All aforementioned experiments showed that circABCC4 regulated the process of the carcinoma. However, the molecular mechanism of circABCC4 in regulating carcinogenesis still remained undefined.
CircHIPK3 (circRNA ID: hsa_circ_0000284), located at chromosome 11, was firstly found to be of high abundance and have significantly regulatory potency in non-small cell lung cancer (NSCLC) [20, 21]. According to previous reports [22-27], the expression level of circHIPK3 was relatively higher in various tumor progression than the control and only circHIPK3 functioned as oncogenic factor by targeting (such as miR-558, miR-124, miR-379, miR-654). A novel research revealed that circHIPK3 could exhibit oncogenic properties by promoting the proliferation, migration, and invasion of PC. Cai et al indicated that remarkable over-expression of circHIPK3 was detected by RT-qPCR in PC tissues and cell lines (PC-3 and DU145) [28]. Besides, Chen et al found that the migration and invasion of PC was decreased in circHIPK3 knockdown group by Wound-healing assays and Transwell invasion assays. The result of CCK-8 and colony formation assays indicated the circHIPK3 silencing inhibits the PC proliferation [29]. In conclusion, circHIPK3 exerted oncogenic properties in PC cell proliferation and invasion.
Numerous studies had proved that Androgen receptor (AR) signaling pathway played an important role in progression of PC by regulating miRNAs and lncRNAs [30-33]. SMARCA5 had the potential capacity of regulating PSA post-transcriptionally in PC progression [34]. Due to the GSE18684 dataset, SMARCA5 gene and Quaking (QKI) were androgen-responsive genes. QKI generated the formation of SMARCA5. Circ-SMARCA5, encoded by the gene SMARCA5, was located at chr4:144464662-144465125. In PC tissues, circ-SMARCA5 was overexpressed detected by RT-qPCR and acted as an oncogenic circRNA. Amusingly, with the stimulation of DHT, the expression of circ-SMARCA5 was increasing, which revealed that the circ-SMARCA5 was androgen regulated gene. The conclusion certified the foregoing hypothesis. Additionally, Kong et al. confirmed that circ-SMARCA5 enhanced the proliferation and cell apoptosis in PC. Helping by flow cytometry, they found that knock-down circSMARCA5 increased the percentage of cell cycle in G1 phase and decreased it in S phase. It made the conclusion that circSMARCA5 functioned as oncogenic circRNA in promoting cell cycle progression in PC [35]. In short, androgen treatment stimulated the expression of circ-SMARCA5 and the elevated level of circ-SMARCA5 enhanced the cell proliferation and progression in PC.
CircZNF609, located at chromosome15, was initially proved to play a promoting tumor efficacy in breast cancer [36], colorectal cancer [37], renal carcinoma [38] by binding miRNAs. Soon later Jin et al. found that circZNF609 had the same function in PC. CircZNF609 was highly expressed in PC tissues and si-circZNF609 suppressed cell viability and colony formation and enhanced apoptosis in PC cell lines (PC-3 and LNCaP) by Western blot. They also found that the expression level of MMP-9 and Vimentin decreased after silencing circ-ZNF609 [39]. MMP-9 degraded the collagen of basement membrane, which destruction was usually an essential step on supporting tumor invasion and metastases [40]. Emerging evidences have shown that MMP-9 played a role in tumor invasion, metastasis and angiogenesis and mediated tumor microenvironment in basal-like triple negative breast cancer, Lewis lung carcinoma etc. [41-46]. It indicated that si-circZNF609 restrained cell migration and invasion in PC [39]. Taken together, up-regulated circZNF609 enhanced proliferation, migration, invasion and apoptosis of PC cells.
Hsa_circ_0001206, located at chromosome 22, expressed a low level of PC cell lines compared with the control. According to Song's report, they utilized hsa_circ_0001206-overexpression lentivirus to validate its dramatical down-regulated level in PC tissues and cell lines (PC-3, DU145 and LNCaP) by qRT-PCR. Moreover, Song et al revealed that the migration ability and cell proliferation ability was weaker than NC cells in over-expression hsa_circ_0001206 cells [47]. To discover its mechanism of down-regulated level of hsa_circ_0001206, they focused on DHX9. DHX9, a member of DExD/H-box helicase family, appeared to play a significant role in many biological processes including regulation of DNA replication, transcription, translation, microRNA biogenesis, RNA processing and transport, and maintenance of genomic stability [48]. Previous studies have shown that DHX9 is positively correlated with the development of colorectal cancer, hepatocellular carcinoma, Ewing sarcoma and PC [47, 49-51]. Song et al used DHX9-siRNAs to knock down DHX9 and detected its efficiency by qRT-PCT in DU145 cells. A marked reduction in hsa_circ_0001206 was observed when transfected cells with DHX9-siRNA2 compared with siRNA-negative control [47]. In short, hsa_circ_0001206 functioned as suppressive ability in PC cell proliferation, migration and invasion modulated by DHX9.
CircAMOTL1L, located at chromosome 22:35948707-35948901, was produced by exon 3 of angiomotin-like 1 gene (Amotl1) and mainly existed in cytoplasm. As initial studies had revealed, circAMOTL1 was seen as protective factor in human cardiac tissues by reducing apoptosis induced by doxorubicin (Dox) and enhancing cardiac repair through binding to and activating AKT phosphorylation and nuclear localization [52]. Inspired by aforementioned experiments, Yang et al. discovered circAMOTL1L circularized from exon-2 and exon-3 of gene Amotl1 [53]. They speculated that circAMOTL1L might be involved in migration of PC based on previous study shown that AMOTL1 was important in cadherin-11-meidated cell migration [54]. Therefore, they confirmed it through constructing recombinant plasmid and fortunately they found that circAMOTL1L downregulated in prostate tissues and functioned as suppressor in carcinogenesis and progression. Additionally, they revealed that p53 modulated the expression of RBM25 to activate circAMOTL1L biogenesis found by transfecting siRNA and using lentiviral vector system in PC-3 cells [12]. P53 played momentous role in development and progression in various cancers due to its abilities of losing and mutating [55, 56]. And novel researches demonstrated that RBM25 participated in large fraction of alternatively spliced exons throughout the human genome by interacting with the exonic splicing enhancer, CGGGCA sequence, which located within exon [57, 58]. In sum, circAMOTL1L mediated by p53-RBM25 postponed the PC progression.
Circ-102044, located at chromosome17, was highly up-regulated in PC. A novel study had demonstrated that circ-102044 functioned as oncogene in promoting migration and invasion in progression of PC and joins in a head-to-tail manner at the slice junction [59]. Besides, western blotting also revealed that the up-regulation of BCL2 and MMP2 were observed in both cell lines. The high expression level of MMP2 was positively related to metastasis of PC and BCL-2 was a protective factor that prevented PC from apoptosis through the combination of molecules and signaling pathways, such as in PTEN loss and p53 inactivation, PI3K/AKT phosphorylation, activation of RTK/STAT3/NF-kB, Ras/Raf1/MEK/ERK, microRNAs (miR-24, miR-31, miR-34, miR-195, miR-204, miR-205 and lncRNA MEG3), autophagy proteins (Beclin1 and AMBRA1) and other potent molecules (FKBP38/NR4A1/GAL3) [60, 61]. But the molecular details of pathways by which circ-102044 modulates PC still remain unknown.
CircRNA-MYLK (circRNA ID: hsa_circ_0141940), located at chromosome: 123332641-123332832, was significantly up-regulated in PC tissues and cell lines (DU145, LNCaP, PC-3 and PC-3MIE8) compared with the control (WPMY-1 cells) by RT-qPCR. To deeply clarify its potential functionsand regulatory mechanisms, Dai et al conducted functional experiments and revealed that circRNA-MYLK overexpression improved cells proliferation and colony formation, whereas circRNA-MYLK acted as suppressor in cell apoptosis [62]. Furthermore, the invasion and migration abilities of overexpressing-circRNA-MYLK were significantly stronger than the control by detecting the invasive and migratory number of PC cells (PC-3MIE8), which were reversed after silencing circRNA-MYLK. In sum, circRNA-MYLK exhibited as oncogene in prompting PC progression, migration and invasion.
Researchers identified a novel circRNA, named circMBOAT2, which was increased significantly in PC tissues and PC cell lines (VCaP, LNCaP, C4-2B, DU145 and PC-3) [63]. When circMBOAT2 was silenced, the ability of proliferation in PC-3 and DU145 cells was inhibited. Meanwhile, migratory and invasive capability of PC-3 and DU145 cells were significantly attenuated. To explore the effect of circMBOAT2 in vivo, luciferase-labeled PC-3 cells transfected with short hairpin circMBOAT2 (sh-circMBOAT2) were subcutaneously injected into BALB/c nude mice. Unsurprisingly, the volume and weight of tumors were notably decreased in the sh-circMBOAT2 group. And thus, circMBOAT2 may impair proliferation of PC cells and act as an oncogenic gene. In addition, Jinet aldiscovered that circLMTK2 was significantly downregulated in PC patients' tissues. Further results confirmed that circLMTK2 overexpression could suppress proliferation in LNCaP and PC-3 cells, while markedly elevate the apoptosis-related protein levels, including Bax, Cleaved-caspase-9 and Cleaved-caspase-3. Furthermore, Western blotting assay results showed that the cell migration and invasion in LNCaP and PC-3 cells were remarkably inhibited when the expression level of circLMTK2 was increased. Increasing evidences showed that miRNA could bind to the sites of circRNAs. MiR-183 is a well-conserved microRNA across many species from invertebrates to humans. Many studies have unveiled the functions of miR-183 in different types of tumors, such as colorectal cancer (CRC) [65], non-small cell lung cancer (NSCLC) [66], osteosarcoma [67], and so on. MiR-183 involved in the progression of LNCaP and PC-3 cells, and might be reduced by increasing the expression level of circLMTK2. Taken together, circRNAs could be regarded as oncogene or suppressor gene in tumorigenesis.
As stated, circRNAs are appropriate to be used as oncogenic stimuli or suppressive factor in modulating PC progress, which may be hinted as exhibiting possible biomarker and therapeutic targets in the future.
The epigenetical roles of Circular RNAs in prostate cancer
CircRNAs extensively participate in gene expression process among numerous animal species with gene-specific and cell-type specific manner [68]. With the increasingly functional researches launched, the epigenetic function of circRNAs is under investigation. It is ubiquitous that circRNAs have numerous miRNA binding sites which can be attached to miRNAs like a sponge due to the observation of structure [69] (Table 2). Helped by the construction of absorbing miRNAs, downstream targets are activated or suppressed to regulate the progression of PC (Table 3). In addition to that, the epigenetic features of circRNAs contains regulation of selective splicing or transcription; acting as Sponges for RBPs; protein translation modification; circRNA-derived pseudogenes; involving in the process of exosome function [70].
CircRNAs function as microRNA sponges in PC
| circRNA | microRNA sponged | PC cells | References (PMID) |
|---|---|---|---|
| circABCC4 | miR-1182 | PC-3, DU145 cells | 31270953 |
| circHIPK3 | miR-338-3p | PC-3, DU145 cells | 31118688 |
| circHIPK3 | miR-193a-3p | PC-3, DU145 cells | 30863152 |
| circZNF609 | miR-186-5p | PC-3, LNCAP cells | 31387394 |
| hsa_circ_0001206 | miR-1285-5p | DU145 cells | 31198063 |
| circAMOTL1L | miR-193a-5p | PC-3 cells | 30531834 |
| circMYLK | miR-29a | PC-3, PC-3M IE8 cells | 29798970 |
| hsa_circ_0044516 | miR-29a-3p | PC-3,2B4, RV221cells | 31625175 |
| circ_KATNAL1 | miR-145-3p | 22Rv1, DU145, LNCaP and PC-3 cells | 31800303 |
| circFMN2 | miR-1238 | PC-3, LNCaP, VCaP and DU145 cells | 32526477 |
| circ0016068 | miR-330-3p | PC-3, DU145, VCaP and 22RV1 cells | 32984325 |
| circMBOAT2 | miR-1271-5p | PC-3, LNCaP, VCaP, DU145 and C4-2B cells | 32645691 |
| circLMTK2 | miR-183 | PC-3 and LNCaP cells | 31760099 |
It had been identified that miRNAs functioned as oncogenes or suppressors in human cancers [71]. MiR-1182 was initially been regarded as tumor suppressor in gastric cancer proliferation by negatively modulating hTERT [72]. CircABCC4, also known as hsa_circ_0030586, was highly up-regulated in PC tissues according to a public PC database (GSE77661). Firstly, miR-1182 was significantly down-regulated in PC tissues, whereas circABCC4 was obviously up-regulated. Furthermore, Luciferase assays demonstrated that mutation of the AGACCUC motify impeded the connection between circABCC4 and miR-1182. The continuous knockdown results favored the view that the deficiency of circABCC4 caused a increase in miR-1182 expression leading to inhibit the cell proliferation, migration and invasion of PC. Then, Luciferase experiments provided the evidence that the FOXP4 expression exhibited the negative correlation with miR-1182 [8]. FOXP4, a forkhead transcription factor, has been demonstrated to play significant roles in embryonic development, cell cycle regulation, and oncogenesis and it is fully expressed in heart, brain, lung, liver, kidney, and testis [73]. Finally, Huang et al reported that silencing circABCC4 could dramatically reduce the expression of FOXP4 and overexpression of FOXP4 reversed the inhibitory effect of miR-1182 on FOXP4 expression in miR-1182-overexpressing PC cells (PC-3 and DU145). Functional assays were also conducted to elucidate that overexpression of FOXP4 partly rescued the proliferation, migration and invasion defect in circABCC4-deficient cells. They also found that circABCC4 played an important part in tumor growth by regulating miR-1182/FOXP4 signaling pathway [8]. Conclusion, circABCC4 accelerated the pace of progression of PC by down-regulating the expression of miR-1182 and up-regulating the expression of FOXP4.
The targets of circRNAs in PC cell lines
| circRNA | Target | PC cells | References (PMID) |
|---|---|---|---|
| circABCC4 | FOXP4 | PC-3, DU145 cells | 31270953 |
| circHIPK3 | MCL1, ADAM17 | PC-3, DU145 cells | 31118688, 30863152 |
| circZNF609 | MMP-9, vimentin, YAP1, AMPK | PC-3, LNCAP cells | 31387394 |
| hsa_circ_0001206 | Smad4 | DU145 cells | 31198063 |
| circAMOTL1L | Pcdha8, E-cadherin, vimentin, β-catenin | PC-3 cells | 30531834 |
| circRNA17 | ARv7 | C4-2, CWR22Rv1 cells | 30674872 |
| circ_KATNAL1 | WISP1, MMP-2, MMP-9, Caspase-3, Caspase-8, Caspase-9, PARP | 22Rv1, DU145, LNCaP and PC-3 cells | 31800303 |
| circFMN2 | LHX2 | PC-3 cells | 32526477 |
Previous studies had demonstrated that extracellular vehicles (EVs) embraced several biological molecular such as non-coding RNAs mediated the interaction between cancer cells and their microenvironment [74, 75]. To validate the internal mechanism of EVs in PC, numerous exosomes were extracted from patients and its healthy control, finding that 42 downregulated and 69 upregulated circRNAs in the heat map and circ-0044516 exhibited the most promising candidate for lucubrating the post-transcriptional activity with the significantly increased expression in prostate tissues. In addition, the result of colony formation assay showed that silencing circ-0044516 caused a severe reduction in colony numbers in PC cells (PC-3, 2B4 and 22RV1). The similar trend was observed in Transwell chamber with or without Matriger treatment suggesting that inhibiting circ-0044516 had negative effect on PC cell metastasis. What's more, Luciferase reporter assay revealed that circ-0044516 harbored miR-29a-3p to act as a sponge in PC. Interestingly, further study detected an inversely result that serum samples from PC patients also verified the mutually-exclusive relationship between circ-0044516 and miR-29a-3p [76]. According to this observation, circ-0044516 was proposed to process predictable function as biomarker in clinical management.
Through the bioinformatics prediction and function analysis, Cai et al suggested that circHIPK3 had the binding site with miR-338-3p. To identify the molecular mechanism of the interaction, they detected the luciferase activity and RT-qPCR data to confirm that circRNA had a negative correlation with the miR-338-3p in PC cells (PC-3 and DU145). Additionally, they indicated that ADAM17, an essential role of the ADAMs family [77], was the targeting gene of miR-338-3p. ADAM17 was becoming a ubiquitous component with tumor cell specificity, which extensively existed in malignant carcinoma [78]. Interestingly, the suppressed expression of ADAM17 by knock-down circHIPK3 is reversed by transfection of miRNA-338-inhibitor. What's more, the result of CCK-8 assay demonstrated that the proliferative and invasive capacities of circHIPK3-deficient cell lines (PC-3 and DU-145) were reversed by co-transfecting si-circHIPK3 and miRNA-338-3p inhibitor, which led to the conclusion that circHIPK3 regulated the proliferative and invasive potentials of PC through miRNA-338-3p/ADAM17 axis [28]. After that, Chen et al. [29] confirmed the hypothesis that circHIK3 also exerted its role in tumorigenesis by sponging miR-193a-3p. Ensuing Luciferase reports validated that silencing circHIPK3 suppressed the expression of MCL1 mRNA and protein while inhibiting miR-193a-3p elevated it. Furthermore, the results of CCK-8, Wound-healing and Transwell assays indicated that MCL1 overexpression increased the proliferation, migration and invasion of PC cell lines (PC3 and DU145), which was targeted by miR-193a-3p. Myeloid cell leukemia 1 (MCL1), an anti-apoptotic member of the BCL-2 protein family, suppressed intrinsic apoptosis or programmed cell death in previous study. Therefore, they identified that circHIPK3 functioned as miR-193a-3p sponge which regulated the expression of MCL1 oncogene.
Recent studies had shown that miR-186-5p participated in various occurrence of PC [79, 80]. Jin et al also found that miR-186-5p presented the protective efficacy in PC and it was modulated by circ-ZNF609. In addition, transfecting miR-186-5p inhibitor reversed the reduction in cell colony and viability and rise in cell apoptosis induced by circZNF609 while western blot favored the consistent result, which meant si-circZNF609 restrained the cell migration and invasion by up-regulating miR-186-5p. What's more, silencing circZNF609 caused relevant reductions in the level of YAP1, whereas transfecting miR-186-5p enhanced it. Then, the similar result was obtained in AMPK signaling pathway. The YAP1 and AMPK signaling pathway activated in PC and had strong association with pathogenesis [81, 82], which furnished fresh thought to Jinet al. Fortunately, they had explored that the ratio of p/t-AMPK and level of YAP1 was declined when sh-circZNF609 involved and the result was reversed with the effect of transfection of miR-186-5p [46]. Taken together, YAP1 and AMPK-catenin signaling pathways improved carcinogenesis through miR-186-5p regulated by circZNF609.
Hsa_circ_0001206 had been proved its function in carcinogenesis. To explore its potential interaction with miRNAs, miR-1285-5p was predicted by home-made microRNA target prediction software from Array star and sooner it was confirmed by dual-luciferase. MiR-1285-5p was systematically defined as regulatory factor in tumor progression [83]. Previous reports had indicated that miR-1285-5p could influence the overall survival in breast cancer and infiltrative growth of follicular varients of papillary thyroid carcinomas [84, 85]. In addition, Smad4 was identified as a potential target of miR-1285-5p by TargetScan database. To sustain the assumption, Song et al. constructed co-transfection model with miR-1285-5p mimic to observe whether the alteration of Smad4 mRNA occurred compared to the initial state in hsa_circ_0001206 overexpressing cells. Fortunately, the level of Smad4 mRNA expression elevated in overexpression of hsa_circ_0001206 cells while transfecting miR-1285-5p declined it [47]. Emerging evidences had elucidated that Smad4 exerted a suppressive effect on the progression and metastasis of PC [86]. Therefore, these results suggested that hsa_circ_0001206 regulated the tumor growth by competitively binding miR-1285-5p through modulating Smad4 expression. Noteworthily, according to the result of GEPIA web ted, lower Smad4 levels predicted better disease-free survival, which gave focus on the prognostic role of Smad4 or hsa_circ_0001206 as biomarker [47]. In summary, combined with aforementioned study, hsa_circ_0001206 intervened with the PC cell proliferation, migration and invasion by interacting with miR-1285-5p, which was negatively regulated by DHX9 protein.
In Yang's report, they searched for the correlation between miRNAs and circAMOTL1L via bioinformatics analysis and they discovered that circAMOTL1L has miR-193a-5p binding sites [12]. MiR-193a-5p was upregulated in PC tissues and PC cell lines, with significant suppression of PC3 cell apoptosis induced by oxidative stress. Then, Luciferase assays and RNA in situ hybridization presented the presence of combination between circAMOTL1L and miR-193a-5p. Additionally, RBPs (RNA-binding proteins) played significant role in forming circRNAs because of the non-canonical form of alternative splicing [57, 58]. Specially, Yang et al discovered that RBM25, one type of RBPs, directly bound to poly-G sequences or the exon splicing enhancer 5'-CGGGCA-3' motif of circAMOTL1L. Moreover, they had validated that miR-193-5p might target Pcdha 3'-UTR, which contained highly conversed miR-193-5p binding sites, to regulate the progression of PC [12]. Previous reports had manifested that Pcdha gene cluster functioned as tumor suppressor in tumor growth and metastasis [87-90]. In short, it was confirmed that the circAMOTL1L-miR-193a-5p axis suppressed the PC progression by up-regulation Pcdha.
Additionally, based on the miRBase prediction, a potential linkage between miR-29a and circRNA-MYLK on a complementary sequence was discovered and sooner confirmed by transfecting miR-29a mimics [62]. High level of miR-29a was initially reported as suppressor in PC progression [91]. Besides, the expression of miR-29a was markedly decreased in PC cells transfected with circ-MYLK overexpression vector while si-circRNA-MYLK presented opposite function in PC cells [62]. In conclusion, circRNA-MYLK promoted PC proliferation, colony formation, invasion, and migration by downregulating the expression level of miR-29a.
To obtain underlying conjunction between circRNA and miRNA, scientists attempted to find a snap path focused on miR-145. MiR-145, initially seen as participator in improving proliferation and invasion in both PC LNCaP cells and IncRNAPCGEMI, were proposed as potential cooperator with circRNAs [92]. He et al conducted microarray analysis to demonstrate five puissant candidates including hsa-circRNA-101981, hsa-circRNA-101996, hsa-circRNA-091420, hsa-circRNA-008068 and hsa-circRNA-406557. Moreover, the results of RT-PCR had validated the downregulation of hsa-circRNA-101981, hsa-circRNA-008068, hsa-circRNA-406557 and upregulation of hsa-circRNA-091420, hsa-circRNA-101996. Besides, hsa-circRNA-101981, hsa-circRNA-101996, hsa-circRNA-091420 were the three circRNAs bound to hsa-miR-145-5p and the others concentrated to hsa-miR-145-3p were all confirmed by dual-luciferase reporter assays [93].
Amusingly, both miR-145-5p and miR-145-3p were defined as anti-cancer molecules to regulate downstream proteins (MELK, NCAPG, BUB1 and CDK1) in PC [94]. Inspired by the thoughts of He et al., scientists paid much attention on the regulatory effect of miR-145-3p that was proved to be capable of suppressive role in PC. Due to the result of sequencing, a compact and complementary connection between circ_KATNAL1 and miR-145-3p was found but either of them was significantly depleted in PC cells (PC-3 and LNCaP cells). So, it was deduced that there was synergistic anti-cancer property in molecular combination of circ_KATNAL1 and miR-145-3p in PC which was further confirmed by the results of Pull-down experiments. In addition, the function of transfecting with miR-145-3p and overexpressing circ_KATNAL1 impeded the expression of WISP1 that was a key factor regulating in several types of cancers, such as hepatocellular carcinoma, breast cancer, lung cancer and colon cancer [95]. So, it was manifested that circ_KATNAL1 exerted suppressive role in PC via miR-145-3p/WISP1 pathway. What's more, declined activities of MMP-2, MMP-9, Caspase-3, Caspase-8 and Caspase-9 were observed when transfecting circ_KATNAL1 or miR-145-3p, which provided the evidence that these apoptosis-related pathways and invasive enzymes could be activated by circ_KATNAL1/miR-145-3p/WISP1 pathway [96]. Taken together, circ_KATNAL1 functioned as suppressor in PC progression by absorbing miR-145-3p to target downstream proteins.
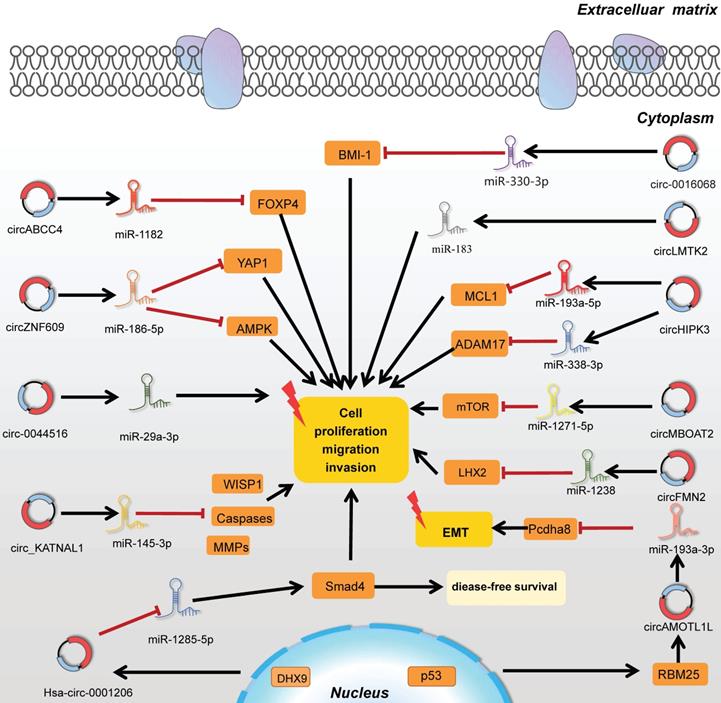
Taken together, the studies on circRNA-miRNA-mRNA pattern were indeed extraordinarily displayed in epigenetic regulations, revealing the abundant intramural network of tumorigenesis and providing a wide range of novel clues for the progression of PC. CircRNAs exerted as miRNA sponges in PC were vividly listed in Figure 3.
Roles of Circular RNAs in epithelial-mesenchymal transition
Epithelial-mesenchymal initially transition is defined as a complex molecular process required during the embryonic development for morphogenetic changes [97]. To achieve this transition, epithelial, stromal settings and microenvironment molecules that able to accommodate cellular and tissue growth in normal or altered conditions are required [98]. Several molecular factors participate in this process such as Twist, Snail, Slug and Zeb1/2. Additionally, numerous studies have demonstrated the presence of EMT-like states in PC, suggesting its involvement in PC development and metastasis [98, 99]. Besides, EMT present obvious efficacy in changing from primary PC to invasive advanced cancer. Moreover, Due to the modification of abnormal miRNAs levels can make resistant PC cells more sensitive to small molecule drugs, the combination of these innovative agents (such as small molecules and miRNAs) may represent a favorable approach for the therapeutic treatment of resistant PC.
AMOTL1L gene was initially reported as a partner of N-cadherin complex which contained tight junctions and it involved in controlling paracellular permeability and cell polarity [53, 54]. Previous studies had revealed that AMOTL1L gene might be closely related to PC progression [12]. And according to Yang's report, circAMOTL1L, expressed by gene AMOTL1L, could act as miR-193a- 5p sponge and modified the biogenesis of Pcdha expression. Furthermore, knock-down of miR-193a-5p could partly rescue the increased expression levels of mesnchymal cells (vimentin) and the decreased expression levels of epithelial cells (E-cadherin). Additionally, the transformation effect would be further enhanced with the addition of circAMOTL1L overexpression. E-cadherin was responsible for adherens junction and functioned as extracellular phenotype maintainer. Vimentin and β-catenin contributed to cellular migration and were seen as mesenchymal markers [100]. Down-regulation of E-cadherin and up-regulation of vimentin and β-catenin played momentous role in EMT, which led to reduction of intercellular adhesion and increased in cell migration. So, it was proposed that miR-193a-5p promotes the PC progression, migration and invasion through the induction of EMT [101]. Combined with aforementioned researches in yang's report, p53-RBM25 down-regulation attenuated the expression of clustered Pcdha gene to accelerate the PC progression. To figure out whether p53 exerted indispensable role in driving EMT, a rescued experiment was performed via over-expressing circAMOTL1L in p53-depleted PC3 cells and fortunately subsequent researches revealed that the decreased level of E-cadherin and increased level of vimentin and β-catenin were reversed after amplifying the expression of circAMTOL1L. Amusingly, the function of over-expression of circAMOTL1L was opposed to RBM25 knock-down and overexpression of circAMOTL1L further enhanced the expression level of RBM25 protein, which suggested that RBM25 and circAMOTL1L formed a feedback loop to regulate the EMT-related gene expression in PC cells [101].
CircRNAs function as miRNA sponges by targeting related genes in tumorigenesis.
circFoxo3, located at chromosome16, was initially seen as a suppressive regulator in breast cancer by promoting its parent gene Foxo3's expression to inhibit the level of MDM2 induced by Foxo3 ubiquitination. Recently, Shen et al elucidated the consistent result by establishing circFoxo3 siRNA transfected PC cells. Besides, immunoblotting indicated that an increased level of Foxo3 and E-cadherin and a reduction of N-cadherin and vimentin levels were observed in DU-145 cells transfected with circFoxo3. On the contrary, the appearance opposite to result mentioned above was gained when they silenced circFoxo3 and Foxo3 with siRNAs [102]. Taken together, all these data firmly confirmed that circFoxo3 exerted as androgen responsive circular RNA repressed PC viability by enhancing Foxo3 and inhibiting EMT. Surprisingly, regardless of its function in EMT, scientists also revealed that the delivery of circFoxo3 prompted the apoptosis of tumor tissues of docetaxel (DOX) injection group mice and this opposite effect was prevented by injecting circFoxo3 siRNAs, which suggesting that circFoxo3 might be correlated to PC androgen independence and provided an applicable strategy for using Dox associated medicine.
CircFMN2, located at chromosome 1, was primordially discovered in CRC and PC [103, 104]. The high expression level of circFMN2 in CRC was relevant to advanced tumor stage and tumor node metastasis (TNM) stage. Subsequently, Shan and his colleagues found that circFMN2 was upregulated in PC tissues. The expression level of circFMN2 in PC cell lines (PC-3, LNCaP, VCaP and DU145) was higher than that in the human normal prostate epithelial cell line (RWPE-1). The results showed that circFMN2 knockdown was remarkably increased the expression level of E-cadherin and greatly decreased the expression level of N-cadherin and Vimentin in PC-3 and DU145 cells. Correspondingly, the expression level of E-cadherin was decreased, and the expression level of N-cadherin and Vimentin were increased in VCaP cell when the expression level of circFMN2 was upregulated. CircFMN2 could play an oncogenic role in the pathogenesis and progression of PC. In addition, the expression level of circ-0016068 was significantly elevated in PC cell lines (DU 145, 22RV1, PC-3, and VCaP) in a separate study [103, 104]. The results discovered that the growth of 22RV1 and DU 145 cells were simultaneously inhibited when circ-0016068 was downregulated. Furthermore, circ-0016068 silence stimulated the expression level of E-cadherin and diminished the expression level of Vimentin and Snail, while circ-0016068 overexpression generated the opposite effect in 22RV1 and DU 145 cells. Moreover, nude mouse tumorigenicity assay were used to verify roles of circ-0016068 in vivo. The results showed that circ-0016068 could accelerate the expression level of Vimentin and Snail whereas suppress the expression level of E-cadherin. Therefore, circ-0016068 could serve as an oncogene regulator in the course of PC.
It was commonly considered that several signaling pathways were involved in the process of EMT including PI3K-Akt signaling pathway, TGF-β signaling pathway and MAPK signaling pathway. These cascade activations became the backbone of subsequent event of alteration in cytoskeleton, contributing to the changes in polarity, increases in the mobility, and invasiveness of tumor cells, which was called EMT. Recently, Yan et al. [106] revealed the high quantities of hsa_circ_0001085, hsa_circ_0004916 and hsa_circ_0001165 by utilizing high-throughput sequencing techniques. Amusingly, circRNAs mentioned above were capable of the change in the level of essential proteins associated with EMT. Hsa_circ_0001165 was able to absorb miR-187-3p to regulate the expression of TNF. What's more, hsa_circ_0001085 was shown to be an effective molecular in the process of PI3K-Akt signaling pathway by indirectly regulating the AKT1 and PIK3CG expression level. And the similar appearance was observed in MAPK signaling pathway and TGF-β signaling pathway via binding miR-451a and miR-196b-5p to modifying the expression of MAPK1 and TGFBR2 proteins. What was noticeable that IFN-γ firstly referred in U.G.Lo's report was performed to induce EMT in PC during this research, which provided novel avenues for the mechanism researches in the future [107].
Roles of Circular RNAs in development of castration-resistant prostate cancer
To completely cure or delay the disease, people have developed the treatments of PC including surveillance, radical local treatment, and androgen-deprivation therapy (ADT). Notably, medical (LHRH agonist) therapy and surgical castration both had efficient efficacy in advanced PC. Despite ADT therapy brought survival light to patients with advanced PC, ADT failed to complete their wishes result from its limited function only for 2-3 years and consequently led to castration-resistant PC (CRPC) [108]. Therefore, it furnished fresh thoughts to public on how to treat advanced PC avoiding castration-resistant.
Emerging studies had revealed that alternative androgen receptor (AR) and its splicing variants presented critical role in development of CRPC because of their general lack of the androgen-binding domain [109-111]. ARv7 had a conspicuous function among other 15 AR-Vs in progression of PC [112-114]. To be extent, shorter survival ability and biochemical recurrence were found in PC patients with high expression of ARv7, which indicated that elevated level of ARv7 was remarkably correlated with the stage of CRPC [115, 116]. So Wu et al. discovered the importance of predicting the expression level of ARv7 after radical prostatectomy due to its efficacy on developing Enzalutamide resistant prostate cancer (EnzRPCa). Furthermore, in Wu's report, circRNA 17, located at chromosome4, was negatively related to the expression of ARv7. They confirmed that knock-down circRNA 17 enhanced the C4-2 parental cells' resistance to Enz treatment and cell invasion through increasing the expression of ARv7 by Matrigel-chamber assay30674872. Besides, the result of Pull-down assay validated that circRNA 17 directly functioned as a reservoir or stabilized the statue of miR-181c-5p prevented from the degradation from nucleases such as Tudor-SN endonuclease to regulate the combination of miR-181C-5p with 3'UTR of ARv7, which indirectly alters the Enz-resistant and cell invasion30674872. MiR-181c-5p played momentous role in cell proliferation, cell cycle and cancer progression in cancer stem cell formation. Moreover, Transwell and 3D invasion assay indicated that Enz altered the expression of PDLIM5 (host gene of circRNA 17) through circRNA17/miR-181c-5p/ARv7 pathway to modulate the level of ARV7, which eventually resulted in CRPC cell invasion and Enz resistance30674872. And the similar result was obtained in vivo experiment by using vivo imaging system, used to monitor the PC progression23792449, for proceeding noninvasive monitoring. Taken together, circRNA17/miR-181c-5p/ARV7 axis presented a novel target that might prevent PC from being castration-resistant cancer.
CircUCK2, located at chr1:165859440 -165877108, was identified as candidate in uncovering secrets and addressing issues on castration resistant effect after long-term anti-androgen therapy in PC patients. According to Xiang's report [118], circUCK2 was shown to be elevated in EnzR-C4-2 cells and was conformed to be related to the invasion and metastasis of PC. Besides, the result of Pull-down assay concentrated the tight connection between circUCK2 and miR-767-5p and revealed that miR-767-5p presented the opposite effect to circUCK2 on cell proliferation and cell invasion by using biotinylated oligo to display results in PC. Initially reports had validated that decreased TET activity was correlated with the progression cancer, which could be induced by activation of miR-767 to repress TET1/3 mRNA, protein expression and to regulate genomic 5hmC levels [119]. Moreover, due to the literature and bioinformatics tools (miRbase, Target-scan and RNA22) provided online, TET1 was selected to be further investigated and subsequently scientists detected that the expression level of TET1 was elevated after overexpressing circUCK2 but reversed overexpressing miR-767-5p. Worthily, the decreased level of TET1 accelerated the progression of PC, which brought brand new insight for future clinical treatment by utilizing TET1 agonists or other specific biological products to delay the process of CRPC.
Roles of Circular RNAs as diagnostic markers in prostate cancer
PC is the most common non-skin carcinoma among men worldwide affecting patients and health-care systems in many countries with low relevant ratio and poor tolerance. Managements consists of digital rectal examination (DRE), PSA measurement, imaging in the form of transrectal ultrasound-guided scan (TRUS) and multiparametric magnetic resonance imaging scan (mpMRI) were deputed to screen and diagnose the potential cases with systematic conjunction [120]. Although the quantification of PSA retained the indispensable function in clinical application, it was unable to differentiate between aggressive PC and indolent disease and further associated with a high risk of over-diagnosis and overtreatment [121, 122]. What's more, sustaining debates on prostate biopsy rose from the considerations among public leading to concentrate on its controversial patient tolerance and morbidity rather than its golden standard for detection. Besides, combined with other technologies, the Gleason grade group, proposed by International Society of Urological Pathology (ISUP) in 2014, remained the backbone of clinical outcome and treatment response and the clinical features including the Gleason score became the imperative characteristics as idea and competent biomarker [123]. Owing to their role of assessing the risk of biochemical progression post curative therapy, the Gleason grade group method prevented unnecessary treatment and informed patients of their actual risk level to evaluate prognostic risk. Hitherto, there was still no reliable molecular for predicting the outcome of PC patients to distinguish patients who need definite treatment from patients who have latent disease [124]. And it was indubitable that determining clinical stages contributed to better overall survival and longer disease-free survival then subsequently led a reduction in morbidity among PC patients. Moreover, the current prognostic system still needed to complete on account of immature prognostic precision [125]. Recently, circRNAs correlated with cancer pathogenesis had become a compelling field for seeking ideal diagnostic and prognostic markers [126]. The characteristics of tissue specificity, stability and evolutionary conservation rose from the extensive concerns in scientific researches resulting in a new dimension to clinical strategies.
Current reports investigated that circ-ITCH, mapped to chromosome20, were differently expressed in PC tissues compared with adjacent noncancerous prostate tissues [127]. The level of circ-ITCH showed great efficacy on distinguishing cancer from paired adjacent tissues through ROC curve with the area under curve (AUC) being 0.812. The sensitivity and specificity reach their peak values of 88.3% and 61.7% respectively. Thus, all of these facts drew a conclusion that circ-ITCH could be used as remarkable biomarker to conform to contemporary technical improvement and result in effective screening. To explore the potential mechanism of its relation with clinicopathological development, the expression of circ-ITCH was divided into low expression (N=circ-ITCH<0.764, N=162) and high expression (N=circ-ITCH>0.764, N=162) groups. In addition, Kaplan-Meier curve analysis and log-rank test were conducted to elucidate that high expression of circ-ITCH was correlated with longer DFS (disease-free survival) (p<0.01) and better OS (overall survival) (p<0.01). Besides, the difference between high expression and low expression of circ-ITCH also included several clinical parameters containing age>60 years, PSA, Gleason score<7, pT3, non-lymph node metastasis, lymph node metastasis and surgical margin-positive. Moreover, Univariate cox's regression analysis was performed to validate the factors that affecting DFS and OS. The results presented the significance of circ-ITCH (high vs low) (HR=0.462 in DFS group and 0.399 in OS group, p<0.01) and other clinicopathological parameters including Gleason score(>7 vs <7) (HR=2.615 in shorter DFS and 3.222 in worse OS, p<0.001), pathologic T stage (pT3 vs. pT2) (HR=2.293 in shorter DFS, p<0.001) and surgical margin status (positive vs. negative) (HR=1.907 in shorter DFS and 2.283 in worse OS, p=0.001), and multivariate cox's regression revealed that circ-ITCH played momentous role in predicting longer DFS and better OS. Taken together, Huang et al. manifested that circ-ITCH exerts as suppressor genes in PC progression through regulating cells activities and signaling pathways in correlation with lower pathological stage and worse lymph node metastasis, which enabled to function as prognostic marker.
Roc curves was also conducted to assess the usage of hsa_circ_0001633, hsa_circ_0001206 and hsa_circ_0009061 as diagnostic marker and the result demonstrated that the area under the curve (AUC) for hsa_circ_0001633, hsa_circ_0001206 and hsa_circ_0009061 was respectively 0.809, 0.774, and 0.711. Furthermore, the clinical features tests for circRNAs indicated that the hsa_circ_0001206 and hsa_circ_0009061 were strongly correlated with Gleason score (G<7 or G>7) (p=0.0018 in hsa_circ_0001206) and pathological stage of patients (PT2, PT3-4) (p=0.045 in hsa_circ_0001206 and p=0.01 in hsa_circ_0009061), suggesting that hsa_circ_0001633, hsa_circ_0001206 and hsa_circ_0009061 had diagnostic and clinical value in PC [47].
Regulations roles of Circular RNAs in prostate cancer
Attempting to clarify the molecular mechanism of tumorigenesis, multitude of experiments have been established on signaling pathways of process involved in initiation and progression of PC. There are seven imperative signaling cascades prompting the development and progression of PC: Androgen receptor (AR) mediated signaling pathway; NF-kB signaling pathway; Growth factor signaling pathway; Phosphoinositide-3-kinase/AKT signaling pathway; Janus Kinase/signaling transducers and activators of transcription (JAK/STAT) signaling; MAPK pathway; Wnt/β-catenin signaling pathway.
Androgen receptor (AR), a member of the steroid hormone receptor family of ligand-activated nuclear transcription factors, was constitutive of four functional domains responsible for specific ligand binding. The AR signaling pathway played momentous role in normal function of prostate, initiation and maintenance of spermatogenesis [128, 129]. Nevertheless, galvanized by stimuli such as androgen production, transforming growth factor, splice variants of full length AR, transcription factors and AR mutants, the aberrant activation of AR signaling pathway contributed to promote the progression of PC and castration-resistant state due to the overexpression and amplification of AR target gene [130]. Specially, the clinical effect of inhibiting AR signaling pathway represented a novel aspect of treatment and enzalutamide had been approved by food and drug administration for interrupting the mechanism of AR signaling pathway despite the possibility for developing towards CRPC. Moreover, reactivating the AR signaling pathway also presented irreplaceable role in progression of CRPC and researches focused on the efficacy of therapy in CRPC by targeting AR signaling pathway. To date, accumulating potential targets were disclosed by abundant tests such as YAP signaling and AMPK signaling, which added on the understanding of brand-new mechanism of action in process of PC. To be exactly, YAP, the transcriptional co-activator YES-associated protein, might function as a tumor suppressor in PC and losing of YAP Protein in PC is strongly correlated with Gleason Score increase [130]. 5'AMP-activated kinase (AMPK) constituted a hub for cellular metabolic and growth control the essential role of lipogenesis in PC maintenance and progression and pointed out that AMPK-mediated repression of lipogenesis acted compelling function in tumor growth inhibition [132]. The discovery of new signal targets enabled the development of much needed new treatment of PC.
The interaction between signaling pathways collectively and invariably regulated the progress of PC. The cross-talk among aforementioned signaling pathways activated or inhibited the downstream signaling molecular and target genes to interfere the tumorigenesis [133]. Host of cross-talk capable of mutually influencing the progression of cancer had been demonstrated recent years and AR signaling pathway remained the central and imperative role among all pathways. Several studies had observed the vital role of circRNAs acting as miRNA sponge or indirect modulator for prompting the PC cell proliferation in AR signaling pathway. CircSMARCA5 had been reported to impact the cell cycle and hamper the apoptosis of carcinoma cells after DHT stimulation, which was strongly correlated with AR signaling pathway. Inhibiting circ-SMARCA5 elicited the increased level of cells in G1 phase and decreased level of cells in S phase compared with the control, which indicated that circSMARCA5, an androgen-induced circRNA, exhibited as oncogene in promoting progression of PC. Aforementioned results highlighted the function of circRNA in modulating tumorigenicity via AR signaling pathway, otherwise the potential molecular mechanism between circSMARCA5and AR pathway was required to be further explored [35]. CircRNA17 acting as miR-181c-5p reservoir was currently been dissected to be impeded by Enz treatment leading to increase the expression of ARv7. Specially, there were three androgen-response elements (AREs) in the 2kb promoter region in host gene PDLIM5 and ChIP assay found that AR could correlated with the AREs located at 870-884bp upstream of the transcription start site of PDLIM5. Together, Enz suppressed host gene PDLIM5 to decrease the expression of circRNA17 via transcriptional regulation consistent with the traditional AR signaling pathway [134]. In addition, Jie et al discovered the upregulation of P-ERK, P-AKT, P-JNK, JNK, β-catenin and GL11 that played crucial role in multiple signaling pathways in overexpression of circ-102004 in PC3 and 22RV1cell lines. Meanwhile, the increased expression of BCL2 and MMP2 was also been found in transfected circ-102004 cell lines resulting in the invasiveness of PC [134]. Although a sequence of signaling pathways was potentially associated with the level of circ-102004, the contribution of that in modifying complex signaling networks needed to be further elucidated. Moreover, down-regulating the level of circZNF609 restrained the expression of YAP1, p-AMPK, t-AMPK in PC3 and LNCAP cell lines, which revealed that circZNF609 participated in the YAP and AMPK signaling pathways [39].
Conclusions and expectations
From the statistics of PC in China and other parts of the world [3, 135], PC remained the most prevalent and vicious cancer threatening human health. The primary diagnostic rate was lower compared with other malignant tumors due to the obsolete specificity and accuracy of blood PSA. However, rapid advances in RNA sequencing and technologies prompted the detection of circRNAs representing a novel aspect of molecular mechanism in tumorigenesis and held some interesting clues about biomarker role in management. N. Vo et al. [136] developed an openly available circRNA compendium, named MiOncoCirc. It could use capture RNA sequencing and include data from clinical cancer samples. For instance, they generated three libraries with exome capture RNA sequencing and detected 1,092 circRNAs from urine samples of PC patients. The results completely overlapped with circRNAs identified in primary prostate adenocarcinoma tissue samples from the MiOncoCirc compendium. It's a reminder that exome capture RNA sequencing may be a promising assay for profiling circRNAs of PC patients in a noninvasive manner. The discoveries of circRNAs went through all the vicissitudes of lives with abundant efforts on attempts and trails and we hoped that not only the existing functions that circRNAs presented but the promising capacities would dedicate to the robust blooming of managements in figuring out conundrums from PC.
CircRNAs were characterized by transcriptional regulation for binding miRNA and modulating gene expression. And with its stability and specificity, circRNAs added on a new dimension to primary diagnose and provided new direction for therapeutic measure. To date, the identification and functional research of circRNAs in PC remained limited and elusive. In this review, we summarized the promoting and suppression role in carcinogenic process as well as its additional mechanism in EMT and signaling pathways. We also described the potential efficacy in prognosis, diagnosing and treating. Nevertheless, numerous outstanding issues needed to be confirmed in PC at the area of circRNAs (Figure 4).
Recent studies suggested that specific circRNA exonic sequences impact immunity and specifically suggest endogenous m6A modification dampens innate immunity [137]. Mice were vaccinated with circFOREIGN, ovalbumin and ovalbumin-expressing B16 melanoma cells successively. The results discovered that mice receiving circFOREIGN have lower tumor growth and have nearly doubled overall survival. Chen et al. [138] discovered that circMALAT1 functioned as a brake in ribosomes to retard Paired box 5 (PAX5) mRNA translation and promote self-renewal of hepatocellular cancer (HCC) stem cells by forming a specific ternary (ribosomecirc-MALAT1-PAX5 mRNA) complex. Recently, Yang et al conducted EpCAM-/- mice via CRISPR/Cas9 technology, which leading to the result that the expression of certain development and glycogen-associated genes was altered in the livers. They elucidated the more integrated mechanism of liver development and carcinogenesis as well as predict the target correlation of the circRNA-miRNA-mRNA network [139]. What's more, CRISPR/Cas9 technology was used to be utilized for genome engineering [140]. Taken together, could potential targets regulated by circRNAs be processed by CRISPR/Cas9 technology for genome treatment in PC? Could we take advantage of this technology for completing the underling circRNA-miRNA-mRNA network, which might broaden our knowledge of the pathogenesis of PC and eventually improved its treatment strategy.
The expectation of circRNAs in prostate cancer. Firstly, abundant and adequate quantities of circRNAs are measured in prostate cancer (PC) from fluid and tissues biopsies with noticeable stability and specificity, which can be conferred the capacities of prognosis and diagnosis. In addition, accumulating researches proved that circRNAs in exosomes exhibit indispensable properties in cancer progression via the function of concentration and targeting. So detecting the level of distinct circRNA in exosomes creates and extends the region of cancer screening and diagnosis in PC. Moreover, it is of importance and well established that CRISPR/Cas9 system directly modify the mutations and mistakes leading to cancer. This novel and practical technology may become a promising candidate with clinical potential in addressing therapeutic issues in PC by suppressing and activating specific molecules.
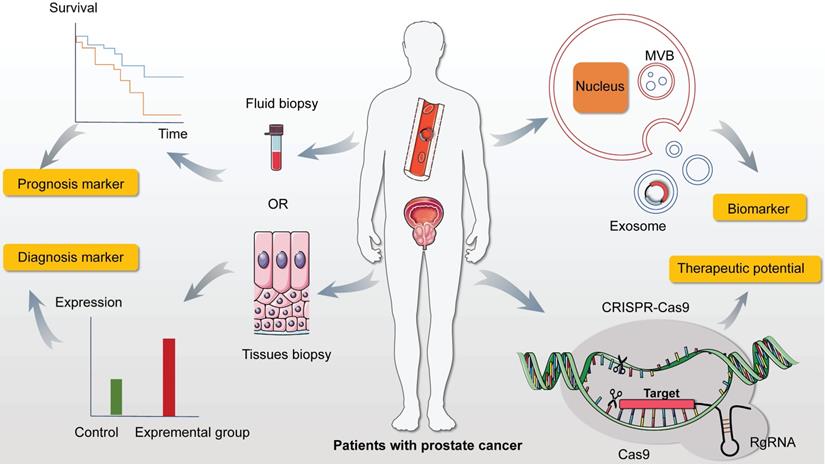
Exosomes, originated from late endosomes of the endocytic system, are initially discovered by Pan et al. as they research on extracellular cytoplasmic fusion of reticulocyte poly-vesicles [74]. The cells communication transferred by mRNA and non-coding RNA including miRNA was mediated by exosomes and that transferred nucleic acids were involved in various life processes. Specially, previous data validated the abundant quantity of circRNAs in exosomes compared to parental cells and more than 1000 exosome circRNAs in serum were represented in discriminating between patients and control [75]. Though, Shen et al. [102] elucidated the driving power of circFoxo3 in EMT from exosomes extracted from PC patients, the precise regulatory network of circRNAs in exosomes still needed to be tunneled. Thus, could we discover more typical and stable circRNAs in exosomes exhibiting as cancer-related biomarker? And it is wondering that whether circRNA presents underling oncogenic or suppressive role through targeting exclusive molecules in PC progression?
Other than diverse non-coding RNA, circRNAs covalently were bonded at 3'and 5'ends for forming a successive loop, which was more stable than liner RNAs. The mighty possibility of being novel diagnostic and prognostic marker added on a new dimension to clinical trials in PC. However, the existing discoveries of systematic regulatory network of circRNAs in PC still needed to be further explored and detailed targeting molecules elicited more expansive challenge to scientists. It would be significant to generate promising candidates for targeting therapeutic in PC to release the extensive burden in public health.
Abbreviations
AR: androgen receptor; ADT: androgen deprivation therapy; Amotl1: angiomotin-like 1 gene; AUC: area under the curve; AREs: androgen-response elements; AMPK: 5'AMP-activated kinase; BPH: benign prostatic hyperplasia; PSA: blood prostate specific antigen; CircRNAs: circular RNAs; CRPC: castration resistant prostate cancer; ciRNA: circular intronic RNA; DOX: Docetaxel; DFS: disease-free survival; ecircRNA: exoniccircRNA; EIciRNA: exon-intron RNA; EMT: epithelial-mesenchymal transition; EVs: extracellular vehicles; EnzRPCa: enzalutamide resistant prostate cancer; FUS: fused in sarcoma; FDA: food and drug administration; icircRNA: intregenic RNA; JAK/STAT: Janus Kinase/signal transducers and activators of transcription; IncRNAs: long non-coding RNAs; miRNA: microRNA; MREs: microRNA response elements; MCL1: myeloid cell leukemia 1; OS: overall survival; PC: prostate cancer; PIN: prostatic intraepithelial neoplasia; QKI: Quaking; RBPs: RNA binding-proteins; XIAP: X-linked inhibitor of apoptosis protein.
Acknowledgements
Funding
This project was supported by the National Natural Science Foundation of China (Nos. 81700522, 81602344), Natural Science Foundation of Anhui Province (1808085MH235).
Author's Contributions
Each author made substantial contributions to the manuscript in writing and editing. All authors read and approved the final manuscript.
Competing Interests
Hong Zhou, Xu-dong Zheng, Chang-ming Lin, Jie Mine, Shuang Hu, Liang-yun Li, Jia-si Chen, Yu-min Liu, Hao-dong Li, Xiao-ming Meng, Jun Li, Ya-ru Yang and Tao Xu declare that they have no conflict of interest.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34
2. Noguchi M, Koga N, Igawa T, Itoh K. Clinical development of immunotherapy for prostate cancer. Int J Urol. 2017;24:675-680
3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
4. Schymura MJ, Sun L, Percy-Laurry A. Prostate cancer collaborative stage data items-their definitions, quality, usage, and clinical implications: a review of SEER data for 2004-2010. Cancer-Am Cancer Soc. 2014;120(Suppl 23):3758-3770
5. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344-352
6. Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339-340
7. Greene J, Baird AM, Brady L, Lim M, Gray SG, McDermott R, Finn SP. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front Mol Biosci. 2017;4:38
8. Huang C, Deng H, Wang Y, Jiang H, Xu R, Zhu X, Huang Z, Zhao X. Circular RNA circABCC4 as the ceRNA of miR-1182 facilitates prostate cancer progression by promoting FOXP4 expression. J Cell Mol Med. 2019;23:6112-6119
9. Feng Y, Yang Y, Zhao X, Fan Y, Zhou L, Rong J, Yu Y. Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell Death Dis. 2019;10:792
10. Pishgar F, Ebrahimi H, Saeedi MS, Fitzmaurice C, Amini E. Global, Regional and National Burden of Prostate Cancer, 1990 to 2015: Results from the Global Burden of Disease Study 2015. J Urol. 2018;199:1224-1232
11. Epstein JI, Allsbrook WJ, Amin MB, Egevad LL. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am J Surg Pathol. 2005;29:1228-1242
12. Yang Z, Qu CB, Zhang Y, Zhang WF, Wang DD, Gao CC, Ma L, Chen JS, Liu KL, Zheng B. et al. Dysregulation of p53-RBM25-mediated circAMOTL1L biogenesis contributes to prostate cancer progression through the circAMOTL1L-miR-193a-5p-Pcdha pathway. Oncogene. 2019;38:2516-2532
13. Yao T, Chen Q, Fu L, Guo J. Circular RNAs: Biogenesis, properties, roles, and their relationships with liver diseases. Hepatol Res. 2017;47:497-504
14. Braicu C, Zimta AA, Gulei D, Olariu A, Berindan-Neagoe I. Comprehensive analysis of circular RNAs in pathological states: biogenesis, cellular regulation, and therapeutic relevance. Cell Mol Life Sci. 2019;76:1559-1577
15. Li Z, Zhou Y, Yang G, He S, Qiu X, Zhang L, Deng Q, Zheng F. Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma. Clin Chim Acta. 2019;492:37-44
16. Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen W, Jiang B, Qin H, Guo X, Liu M. et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18:111
17. Li S, Yu C, Zhang Y, Liu J, Jia Y, Sun F, Zhang P, Li J, Guo L, Xiao H. et al. Circular RNA cir-ITCH Is a Potential Therapeutic Target for the Treatment of Castration-Resistant Prostate Cancer. Biomed Res Int. 2020;2020:7586521
18. Sun HD, Xu ZP, Sun ZQ, Zhu B, Wang Q, Zhou J, Jin H, Zhao A, Tang WW, Cao XF. Down-regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells. Sci Rep. 2018;8:10111
19. Koletsi D, Pandis N. Survival analysis, part 2: Kaplan-Meier method and the log-rank test. Am J Orthod Dentofacial Orthop. 2017;152:569-571
20. Tian F, Wang Y, Xiao Z, Zhu X. [Circular RNA CircHIPK3 Promotes NCI-H1299 and NCI-H2170 Cell Proliferation through miR-379 and its Target IGF1]. Zhongguo Fei Ai Za Zhi. 2017;20:459-467
21. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-338
22. Zhang Y, Li C, Liu X, Wang Y, Zhao R, Yang Y, Zheng X, Zhang Y, Zhang X. circHIPK3 promotes oxaliplatin-resistance in colorectal cancer through autophagy by sponging miR-637. Ebiomedicine. 2019;48:277-288
23. Ke Z, Xie F, Zheng C, Chen D. CircHIPK3 promotes proliferation and invasion in nasopharyngeal carcinoma by abrogating miR-4288-induced ELF3 inhibition. J Cell Physiol. 2019;234:1699-1706
24. Liu N, Zhang J, Zhang LY, Wang L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22:3713-3718
25. Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9:175
26. Wang J, Zhao SY, Ouyang SS, Huang ZK, Luo Q, Liao L. [Circular RNA circHIPK3 acts as the sponge of microRNA-124 to promote human oral squamous cell carcinoma cells proliferation]. Zhonghua Kou Qiang Yi Xue Za Zhi. 2018;53:546-551
27. Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, Liu D, Wang M, Wang L, Zeng F, Jiang G. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. Embo Rep. 2017;18:1646-1659
28. Cai C, Zhi Y, Wang K, Zhang P, Ji Z, Xie C, Sun F. CircHIPK3 overexpression accelerates the proliferation and invasion of prostate cancer cells through regulating miRNA-338-3p. Onco Targets Ther. 2019;12:3363-3372
29. Chen D, Lu X, Yang F, Xing N. Circular RNA circHIPK3 promotes cell proliferation and invasion of prostate cancer by sponging miR-193a-3p and regulating MCL1 expression. Cancer Manag Res. 2019;11:1415-1423
30. Wan X, Pu H, Huang W, Yang S, Zhang Y, Kong Z, Yang Z, Zhao P, Li A, Li T, Li Y. Androgen-induced miR-135a acts as a tumor suppressor through downregulating RBAK and MMP11, and mediates resistance to androgen deprivation therapy. Oncotarget. 2016;7:51284-51300
31. Wan X, Huang W, Yang S, Zhang Y, Zhang P, Kong Z, Li T, Wu H, Jing F, Li Y. Androgen-induced miR-27A acted as a tumor suppressor by targeting MAP2K4 and mediated prostate cancer progression. Int J Biochem Cell Biol. 2016;79:249-260
32. Zhang Y, Zhang P, Wan X, Su X, Kong Z, Zhai Q, Xiang X, Li L, Li Y. Downregulation of long non-coding RNA HCG11 predicts a poor prognosis in prostate cancer. Biomed Pharmacother. 2016;83:936-941
33. Wan X, Huang W, Yang S, Zhang Y, Pu H, Fu F, Huang Y, Wu H, Li T, Li Y. Identification of androgen-responsive lncRNAs as diagnostic and prognostic markers for prostate cancer. Oncotarget. 2016;7:60503-60518
34. Sun D, Lee YS, Malhotra A, Kim HK, Matecic M, Evans C, Jensen RV, Moskaluk CA, Dutta A. miR-99 family of MicroRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res. 2011;71:1313-1324
35. Kong Z, Wan X, Zhang Y, Zhang P, Zhang Y, Zhang X, Qi X, Wu H, Huang J, Li Y. Androgen-responsive circular RNA circSMARCA5 is up-regulated and promotes cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;493:1217-1223
36. Wang S, Xue X, Wang R, Li X, Li Q, Wang Y, Xie P, Kang Y, Meng R, Feng X. CircZNF609 promotes breast cancer cell growth, migration, and invasion by elevating p70S6K1 via sponging miR-145-5p. Cancer Manag Res. 2018;10:3881-3890
37. Wu L, Xia J, Yang J, Shi Y, Xia H, Xiang X, Yu X. Circ-ZNF609 promotes migration of colorectal cancer by inhibiting Gli1 expression via microRNA-150. J Buon. 2018;23:1343-1349
38. Xiong Y, Zhang J, Song C. CircRNA ZNF609 functions as a competitive endogenous RNA to regulate FOXP4 expression by sponging miR-138-5p in renal carcinoma. J Cell Physiol. 2019;234:10646-10654
39. Jin C, Zhao W, Zhang Z, Liu W. Silencing circular RNA circZNF609 restrains growth, migration and invasion by up-regulating microRNA-186-5p in prostate cancer. Artif Cells Nanomed Biotechnol. 2019;47:3350-3358
40. Huang H: Matrix Metalloproteinase-9 (MMP-9) as a Cancer Biomarker and MMP-9 Biosensors: Recent Advances.Sensors (Basel). 2018; 18
41. Maso G, Piccoli M, Parolin S, Restaino S, Alberico S: Diabetes in pregnancy: timing and mode of delivery.Curr Diab Rep 2014, 14:506
42. Xu D, McKee CM, Cao Y, Ding Y, Kessler BM, Muschel RJ. Matrix metalloproteinase-9 regulates tumor cell invasion through cleavage of protease nexin-1. Cancer Res. 2010;70:6988-6998
43. Pego ER, Fernandez I, Nunez MJ. Molecular basis of the effect of MMP-9 on the prostate bone metastasis: A review. Urol Oncol. 2018;36:272-282
44. Itoh T, Tanioka M, Matsuda H, Nishimoto H, Yoshioka T, Suzuki R, Uehira M. Experimental metastasis is suppressed in MMP-9-deficient mice. Clin Exp Metastasis. 1999;17:177-181
45. Wang X, Nagase H, Watanabe T, Nobusue H, Suzuki T, Asami Y, Shinojima Y, Kawashima H, Takagi K, Mishra R. et al. Inhibition of MMP-9 transcription and suppression of tumor metastasis by pyrrole-imidazole polyamide. Cancer Sci. 2010;101:759-766
46. Chou CH, Teng CM, Tzen KY, Chang YC, Chen JH, Cheng JC. MMP-9 from sublethally irradiated tumor promotes Lewis lung carcinoma cell invasiveness and pulmonary metastasis. Oncogene. 2012;31:458-468
47. Song Z, Zhuo Z, Ma Z, Hou C, Chen G, Xu G. Hsa_Circ_0001206 is downregulated and inhibits cell proliferation, migration and invasion in prostate cancer. Artif Cells Nanomed Biotechnol. 2019;47:2449-2464
48. Lee T, Pelletier J. The biology of DHX9 and its potential as a therapeutic target. Oncotarget. 2016;7:42716-42739
49. Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, Sun SH, Yang F, Zhou WP. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68:1214-1227
50. Mi J, Ray P, Liu J, Kuan CT, Xu J, Hsu D, Sullenger BA, White RR, Clary BM. In vivo Selection Against Human Colorectal Cancer Xenografts Identifies an Aptamer That Targets RNA Helicase Protein DHX9. Mol Ther Nucleic Acids. 2016;5:e315
51. Fidaleo M, Svetoni F, Volpe E, Minana B, Caporossi D, Paronetto MP. Genotoxic stress inhibits Ewing sarcoma cell growth by modulating alternative pre-mRNA processing of the RNA helicase DHX9. Oncotarget. 2015;6:31740-31757
52. Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, Yang W, Zhang C, Yang Q, Yee A. et al. A Circular RNA Binds To and Activates AKT Phosphorylation and Nuclear Localization Reducing Apoptosis and Enhancing Cardiac Repair. Theranostics. 2017;7:3842-3855
53. Zheng Y, Vertuani S, Nystrom S, Audebert S, Meijer I, Tegnebratt T, Borg JP, Uhlen P, Majumdar A, Holmgren L. Angiomotin-like protein 1 controls endothelial polarity and junction stability during sprouting angiogenesis. Circ Res. 2009;105:260-270
54. Ortiz A, Lee YC, Yu G, Liu HC, Lin SC, Bilen MA, Cho H, Yu-Lee LY, Lin SH. Angiomotin is a novel component of cadherin-11/beta-catenin/p120 complex and is critical for cadherin-11-mediated cell migration. Faseb J. 2015;29:1080-1091
55. Ren D, Wang M, Guo W, Zhao X, Tu X, Huang S, Zou X, Peng X. Wild-type p53 suppresses the epithelial-mesenchymal transition and stemness in PC-3 prostate cancer cells by modulating miR145. Int J Oncol. 2013;42:1473-1481
56. Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, Yuan A, Lin CW, Yang SC, Chan WK. et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol. 2009;11:694-704
57. Carlson SM, Soulette CM, Yang Z, Elias JE, Brooks AN, Gozani O. RBM25 is a global splicing factor promoting inclusion of alternatively spliced exons and is itself regulated by lysine mono-methylation. J Biol Chem. 2017;292:13381-13390
58. Zhou A, Ou AC, Cho A, Benz EJ, Huang SC. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5' splice site selection. Mol Cell Biol. 2008;28:5924-5936
59. Si-Tu J, Cai Y, Feng T, Yang D, Yuan S, Yang X, He S, Li Z, Wang Y, Tang Y. et al. Upregulated circular RNA circ-102004 that promotes cell proliferation in prostate cancer. Int J Biol Macromol. 2019;122:1235-1243
60. Kim JH, Lee H, Shin EA, Kim DH, Choi JB, Kim SH. Implications of Bcl-2 and its interplay with other molecules and signaling pathways in prostate cancer progression. Expert Opin Ther Targets. 2017;21:911-920
61. Dayal S, Zhou J, Manivannan P, Siddiqui MA, Ahmad OF, Clark M, Awadia S, Garcia-Mata R, Shemshedini L, Malathi K. RNase L Suppresses Androgen Receptor Signaling, Cell Migration and Matrix Metalloproteinase Activity in Prostate Cancer Cells. Int J Mol Sci. 2017 18
62. Dai Y, Li D, Chen X, Tan X, Gu J, Chen M, Zhang X. Circular RNA Myosin Light Chain Kinase (MYLK) Promotes Prostate Cancer Progression through Modulating Mir-29a Expression. Med Sci Monit. 2018;24:3462-3471
63. Shi J, Liu C, Chen C, Guo K, Tang Z, Luo Y, Chen L, Su Y, Xu K. Circular RNA circMBOAT2 promotes prostate cancer progression via a miR-1271-5p/mTOR axis. Aging (Albany NY). 2020;12:13255-13280
64. Jin C, Zhao W, Zhang Z, Liu W. CircLMTK2 acts as a tumor suppressor in prostate cancer via regulating the expression of microRNA-183. Life Sci. 2020;241:117097
65. Zheng S, Zhong YF, Tan DM, Xu Y, Chen HX, Wang D. miR-183-5p enhances the radioresistance of colorectal cancer by directly targeting ATG5. J Biosci. 2019 44
66. Wang H, Ma Z, Liu X, Zhang C, Hu Y, Ding L, Qi P, Wang J, Lu S, Li Y. MiR-183-5p is required for non-small cell lung cancer progression by repressing PTEN. Biomed Pharmacother. 2019;111:1103-1111
67. Yang X, Wang L, Wang Q, Li L, Fu Y, Sun J. MiR-183 inhibits osteosarcoma cell growth and invasion by regulating LRP6-Wnt/beta-catenin signaling pathway. Biochem Biophys Res Commun. 2018;496:1197-1203
68. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. Plos Genet. 2013;9:e1003777
69. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388
70. Wu J, Qi X, Liu L, Hu X, Liu J, Yang J, Yang J, Lu L, Zhang Z, Ma S. et al. Emerging Epigenetic Regulation of Circular RNAs in Human Cancer. Mol Ther Nucleic Acids. 2019;16:589-596
71. Montani F, Bianchi F. Circulating Cancer Biomarkers: The Macro-revolution of the Micro-RNA. Ebiomedicine. 2016;5:4-6
72. Zhang D, Xiao YF, Zhang JW, Xie R, Hu CJ, Tang B, Wang SM, Wu YY, Hao NB, Yang SM. miR-1182 attenuates gastric cancer proliferation and metastasis by targeting the open reading frame of hTERT. Cancer Lett. 2015;360:151-159
73. Teufel A, Wong EA, Mukhopadhyay M, Malik N, Westphal H. FoxP4, a novel forkhead transcription factor. Biochim Biophys Acta. 2003;1627:147-152
74. Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101:942-948
75. Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG, Zhang B. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982
76. Li T, Sun X, Chen L. Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J Cell Biochem. 2020;121:2118-2126
77. Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6:32-43
78. Yamamoto K, Trad A, Baumgart A, Huske L, Lorenzen I, Chalaris A, Grotzinger J, Dechow T, Scheller J, Rose-John S. A novel bispecific single-chain antibody for ADAM17 and CD3 induces T-cell-mediated lysis of prostate cancer cells. Biochem J. 2012;445:135-144
79. Jones DZ, Schmidt ML, Suman S, Hobbing KR, Barve SS, Gobejishvili L, Brock G, Klinge CM, Rai SN, Park J. et al. Micro-RNA-186-5p inhibition attenuates proliferation, anchorage independent growth and invasion in metastatic prostate cancer cells. Bmc Cancer. 2018;18:421
80. Hua X, Xiao Y, Pan W, Li M, Huang X, Liao Z, Xian Q, Yu L. miR-186 inhibits cell proliferation of prostate cancer by targeting GOLPH3. Am J Cancer Res. 2016;6:1650-1660
81. Sheng X, Li WB, Wang DL, Chen KH, Cao JJ, Luo Z, He J, Li MC, Liu WJ, Yu C. YAP is closely correlated with castration-resistant prostate cancer, and downregulation of YAP reduces proliferation and induces apoptosis of PC-3 cells. Mol Med Rep. 2015;12:4867-4876
82. Chen RJ, Hung CM, Chen YL, Wu MD, Yuan GF, Wang YJ. Monascuspiloin induces apoptosis and autophagic cell death in human prostate cancer cells via the Akt and AMPK signaling pathways. J Agric Food Chem. 2012;60:7185-7193
83. Zhou S, Zhang Z, Zheng P, Zhao W, Han N. MicroRNA-1285-5p influences the proliferation and metastasis of non-small-cell lung carcinoma cells via downregulating CDH1 and Smad4. Tumour Biol. 2017;39:1393384151
84. Hironaka-Mitsuhashi A, Matsuzaki J, Takahashi RU, Yoshida M, Nezu Y, Yamamoto Y, Shiino S, Kinoshita T, Ushijima T, Hiraoka N. et al. A tissue microRNA signature that predicts the prognosis of breast cancer in young women. Plos One. 2017;12:e187638
85. Borrelli N, Denaro M, Ugolini C, Poma AM, Miccoli M, Vitti P, Miccoli P, Basolo F. miRNA expression profiling of 'noninvasive follicular thyroid neoplasms with papillary-like nuclear features' compared with adenomas and infiltrative follicular variants of papillary thyroid carcinomas. Mod Pathol. 2017;30:39-51
86. Horvath LG, Henshall SM, Kench JG, Turner JJ, Golovsky D, Brenner PC, O'Neill GF, Kooner R, Stricker PD, Grygiel JJ, Sutherland RL. Loss of BMP2, Smad8, and Smad4 expression in prostate cancer progression. Prostate. 2004;59:234-242
87. Dallosso AR, Oster B, Greenhough A, Thorsen K, Curry TJ, Owen C, Hancock AL, Szemes M, Paraskeva C, Frank M. et al. Long-range epigenetic silencing of chromosome 5q31 protocadherins is involved in early and late stages of colorectal tumorigenesis through modulation of oncogenic pathways. Oncogene. 2012;31:4409-4419
88. Yu J, Cheng YY, Tao Q, Cheung KF, Lam CN, Geng H, Tian LW, Wong YP, Tong JH, Ying JM. et al. Methylation of protocadherin 10, a novel tumor suppressor, is associated with poor prognosis in patients with gastric cancer. Gastroenterology. 2009;136:640-651
89. Dallosso AR, Hancock AL, Szemes M, Moorwood K, Chilukamarri L, Tsai HH, Sarkar A, Barasch J, Vuononvirta R, Jones C. et al. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms' tumor. Plos Genet. 2009;5:e1000745
90. Waha A, Guntner S, Huang TH, Yan PS, Arslan B, Pietsch T, Wiestler OD, Waha A. Epigenetic silencing of the protocadherin family member PCDH-gamma-A11 in astrocytomas. Neoplasia. 2005;7:193-199
91. Sengupta S, den Boon JA, Chen IH, Newton MA, Stanhope SA, Cheng YJ, Chen CJ, Hildesheim A, Sugden B, Ahlquist P. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc Natl Acad Sci USA. 2008;105:5874-5878
92. He JH, Zhang JZ, Han ZP, Wang L, Lv YB, Li YG. Reciprocal regulation of PCGEM1 and miR-145 promote proliferation of LNCaP prostate cancer cells. J Exp Clin Cancer Res. 2014;33:72
93. He JH, Han ZP, Zhou JB, Chen WM, Lv YB, He ML, Li YG. MiR-145 affected the circular RNA expression in prostate cancer LNCaP cells. J Cell Biochem. 2018;119:9168-9177
94. Goto Y, Kurozumi A, Arai T, Nohata N, Kojima S, Okato A, Kato M, Yamazaki K, Ishida Y, Naya Y. et al. Impact of novel miR-145-3p regulatory networks on survival in patients with castration-resistant prostate cancer. Br J Cancer. 2017;117:409-420
95. Ono M, Inkson CA, Sonn R, Kilts TM, de Castro LF, Maeda A, Fisher LW, Robey PG, Berendsen AD, Li L. et al. WISP1/CCN4: a potential target for inhibiting prostate cancer growth and spread to bone. Plos One. 2013;8:e71709
96. Zheng Y, Chen CJ, Lin ZY, Li JX, Liu J, Lin FJ, Zhou X. Circ_KATNAL1 regulates prostate cancer cell growth and invasiveness through the miR-145-3p/WISP1 pathway. Biochem Cell Biol. 2020;98:396-404
97. Zhang Y, Weinberg RA. Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front Med. 2018;12:361-373
98. Montanari M, Rossetti S, Cavaliere C, D'Aniello C, Malzone MG, Vanacore D, Di Franco R, La Mantia E, Iovane G, Piscitelli R. et al. Epithelial-mesenchymal transition in prostate cancer: an overview. Oncotarget. 2017;8:35376-35389
99. Liu Q, Tong D, Liu G, Xu J, Do K, Geary K, Zhang D, Zhang J, Zhang Y, Li Y. et al. Metformin reverses prostate cancer resistance to enzalutamide by targeting TGF-beta1/STAT3 axis-regulated EMT. Cell Death Dis. 2017;8:e3007
100. Lee HM, Hwang KA, Choi KC: Diverse pathways of epithelial mesenchymal transition related with cancer progression and metastasis and potential effects of endocrine disrupting chemicals on epithelial mesenchymal transition process.Mol Cell Endocrinol. 2017; 457:103-113
101. Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15-33
102. Shen Z, Zhou L, Zhang C, Xu J. Reduction of circular RNA Foxo3 promotes prostate cancer progression and chemoresistance to docetaxel. Cancer Lett. 2020;468:88-101
103. Shan G, Shao B, Liu Q, Zeng Y, Fu C, Chen A, Chen Q. circFMN2 Sponges miR-1238 to Promote the Expression of LIM-Homeobox Gene 2 in Prostate Cancer Cells. Mol Ther Nucleic Acids. 2020;21:133-146
104. Li Y, Li C, Xu R, Wang Y, Li D, Zhang B. A novel circFMN2 promotes tumor proliferation in CRC by regulating the miR-1182/hTERT signaling pathways. Clin Sci (Lond). 2019;133:2463-2479
105. Li Q, Wang W, Zhang M, Sun W, Shi W, Li F. Circular RNA circ-0016068 Promotes the Growth, Migration, and Invasion of Prostate Cancer Cells by Regulating the miR-330-3p/BMI-1 Axis as a Competing Endogenous RNA. Front Cell Dev Biol. 2020;8:827
106. Yan Z, Xiao Y, Chen Y, Luo G. Screening and identification of epithelial-to-mesenchymal transition-related circRNA and miRNA in prostate cancer. Pathol Res Pract. 2020;216:152784
107. Lo UG, Pong RC, Yang D, Gandee L, Hernandez E, Dang A, Lin CJ, Santoyo J, Ma S, Sonavane R. et al. IFNgamma-Induced IFIT5 Promotes Epithelial-to-Mesenchymal Transition in Prostate Cancer via miRNA Processing. Cancer Res. 2019;79:1098-1112
108. Egan A, Dong Y, Zhang H, Qi Y, Balk SP, Sartor O. Castration-resistant prostate cancer: adaptive responses in the androgen axis. Cancer Treat Rev. 2014;40:426-433
109. Shah K, Gagliano T, Garland L, O'Hanlon T, Bortolotti D, Gentili V, Rizzo R, Giamas G, Dean M. Androgen receptor signaling regulates the transcriptome of prostate cancer cells by modulating global alternative splicing. Oncogene. 2020;39:6172-6189
110. Liu LL, Xie N, Sun S, Plymate S, Mostaghel E, Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014;33:3140-3150
111. Cao B, Qi Y, Zhang G, Xu D, Zhan Y, Alvarez X, Guo Z, Fu X, Plymate SR, Sartor O. et al. Androgen receptor splice variants activating the full-length receptor in mediating resistance to androgen-directed therapy. Oncotarget. 2014;5:1646-1656
112. Zhang H, Zhan Y, Liu X, Qi Y, Zhang G, Sartor O, Dong Y. Splicing variants of androgen receptor in prostate cancer. Am J Clin Exp Urol. 2013;1:18-24
113. Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, Chen H, Kong X, Melamed J, Tepper CG. et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305-2313
114. Sun M, Abdollah F. Re: AR-V7 and Resistance to Enzalutamide and Abiraterone in Prostate Cancer. Eur Urol. 2015;68:162-163
115. Qu Y, Dai B, Ye D, Kong Y, Chang K, Jia Z, Yang X, Zhang H, Zhu Y, Shi G. Constitutively active AR-V7 plays an essential role in the development and progression of castration-resistant prostate cancer. Sci Rep. 2015;5:7654
116. Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, Han M, Partin AW, Vessella RL, Isaacs WB. et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16-22
117. Wang R, Sun Y, Li L, Niu Y, Lin W, Lin C, Antonarakis ES, Luo J, Yeh S, Chang C. Preclinical Study using Malat1 Small Interfering RNA or Androgen Receptor Splicing Variant 7 Degradation Enhancer ASC-J9((R)) to Suppress Enzalutamide-resistant Prostate Cancer Progression. Eur Urol. 2017;72:835-844
118. Xiang Z, Xu C, Wu G, Liu B, Wu D. CircRNA-UCK2 Increased TET1 Inhibits Proliferation and Invasion of Prostate Cancer Cells Via Sponge MiRNA-767-5p. Open Med (Wars). 2019;14:833-842
119. Loriot A, Van Tongelen A, Blanco J, Klaessens S, Cannuyer J, van Baren N, Decottignies A, De Smet C. A novel cancer-germline transcript carrying pro-metastatic miR-105 and TET-targeting miR-767 induced by DNA hypomethylation in tumors. Epigenetics-Us. 2014;9:1163-1171
120. Gosselaar C, Roobol MJ, Roemeling S, Schroder FH. The role of the digital rectal examination in subsequent screening visits in the European randomized study of screening for prostate cancer (ERSPC), Rotterdam. Eur Urol. 2008;54:581-588
121. Hugosson J, Carlsson S. Overdetection in screening for prostate cancer. Curr Opin Urol. 2014;24:256-263
122. Sanda MG, Dunn RL, Michalski J, Sandler HM, Northouse L, Hembroff L, Lin X, Greenfield TK, Litwin MS, Saigal CS. et al. Quality of life and satisfaction with outcome among prostate-cancer survivors. N Engl J Med. 2008;358:1250-1261
123. Varma M, Berney D, Oxley J, Trpkov K. Gleason score assignment is the sole responsibility of the pathologist. Histopathology. 2018;73:5-7
124. Klotz L. Prostate cancer overdiagnosis and overtreatment. Curr Opin Endocrinol Diabetes Obes. 2013;20:204-209
125. Ondracek RP, Kattan MW, Murekeyisoni C, Yu C, Kauffman EC, Marshall JR, Mohler JL. Validation of the Kattan Nomogram for Prostate Cancer Recurrence After Radical Prostatectomy. J Natl Compr Canc Netw. 2016;14:1395-1401
126. Lim M, Baird AM, Aird J, Greene J, Kapoor D, Gray SG, McDermott R, Finn SP: RNAs as Candidate Diagnostic and Prognostic Markers of Prostate Cancer-from Cell Line Models to Liquid Biopsies.Diagnostics (Basel). 2018; 8
127. Huang E, Chen X, Yuan Y. Downregulated circular RNA itchy E3 ubiquitin protein ligase correlates with advanced pathologic T stage, high lymph node metastasis risk and poor survivals in prostate cancer patients. Cancer Biomark. 2019;26:41-50
128. Lonergan PE, Tindall DJ. Androgen receptor signaling in prostate cancer development and progression. J Carcinog. 2011;10:20
129. Ramalingam S, Ramamurthy VP, Njar V. Dissecting major signaling pathways in prostate cancer development and progression: Mechanisms and novel therapeutic targets. J Steroid Biochem Mol Biol. 2017;166:16-27
130. Schalken J, Fitzpatrick JM. Enzalutamide: targeting the androgen signalling pathway in metastatic castration-resistant prostate cancer. Bju Int. 2016;117:215-225
131. Hu X, Jia Y, Yu J, Chen J, Fu Q. Loss of YAP protein in prostate cancer is associated with Gleason score increase. Tumori. 2015;101:189-193
132. Zadra G, Photopoulos C, Tyekucheva S, Heidari P, Weng QP, Fedele G, Liu H, Scaglia N, Priolo C, Sicinska E. et al. A novel direct activator of AMPK inhibits prostate cancer growth by blocking lipogenesis. Embo Mol Med. 2014;6:519-538
133. Hu R, Denmeade SR, Luo J. Molecular processes leading to aberrant androgen receptor signaling and castration resistance in prostate cancer. Expert Rev Endocrinol Metab. 2010;5:753-764
134. Wu G, Sun Y, Xiang Z, Wang K, Liu B, Xiao G, Niu Y, Wu D, Chang C. Preclinical study using circular RNA 17 and micro RNA 181c-5p to suppress the enzalutamide-resistant prostate cancer progression. Cell Death Dis. 2019;10:37
135. Liu X, Yu C, Bi Y, Zhang ZJ. Trends and age-period-cohort effect on incidence and mortality of prostate cancer from 1990 to 2017 in China. Public Health. 2019;172:70-80
136. Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X. et al. The Landscape of Circular RNA in Cancer. Cell. 2019;176:869-881
137. Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, Broughton JP, Kim J, Cadena C, Pulendran B. et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol Cell. 2019;76:96-109
138. Chen L, Kong R, Wu C, Wang S, Liu Z, Liu S, Li S, Chen T, Mao C, Liu S: Circ-MALAT1 Functions as Both an mRNA Translation Brake and a microRNA Sponge toPromote Self-Renewal of Hepatocellular Cancer Stem Cells.Adv Sci (Weinh). 2020; 7:1900949
139. Yang Y, Liu S, Lei Z, Chen G, Huang L, Yang F, Lei Y, Liu Y, Yang L, Liu W. et al. Circular RNA profile in liver tissue of EpCAM knockout mice. Int J Mol Med. 2019;44:1063-1077
140. Zhen S, Hua L, Liu YH, Sun XM, Jiang MM, Chen W, Zhao L, Li X. Inhibition of long non-coding RNA UCA1 by CRISPR/Cas9 attenuated malignant phenotypes of bladder cancer. Oncotarget. 2017;8:9634-9646
Author contact
![]() Corresponding authors: Ya-ru Yang, PhD, Department of Clinical Trial Research Center, The Second Hospital of Anhui Medical University, Hefei. Tel./Fax: +13866729433. E-mail: yangyaruedu.cn. Tao Xu, PhD, School of Pharmacy, Anhui Medical University, 81 Meishan Road, Hefei, Anhui Province, 230032, China. Tel./Fax: +86 551 65172131. E-mail: xutaoedu.cn.
Corresponding authors: Ya-ru Yang, PhD, Department of Clinical Trial Research Center, The Second Hospital of Anhui Medical University, Hefei. Tel./Fax: +13866729433. E-mail: yangyaruedu.cn. Tao Xu, PhD, School of Pharmacy, Anhui Medical University, 81 Meishan Road, Hefei, Anhui Province, 230032, China. Tel./Fax: +86 551 65172131. E-mail: xutaoedu.cn.

 Global reach, higher impact
Global reach, higher impact