10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(6):1521-1529. doi:10.7150/ijbs.59151 This issue Cite
Research Paper
Developing effective siRNAs to reduce the expression of key viral genes of COVID-19
Faculty of Health Sciences, University of Macau, Macao SAR, China
Abstract
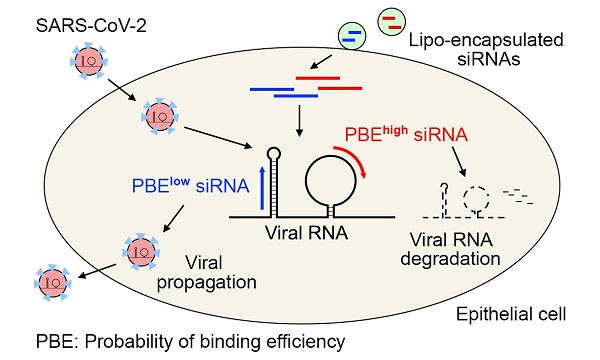
The COVID-19 pandemic has been raging worldwide for more than a year. Many efforts have been made to create vaccines and develop new antiviral drugs to cope with the disease. Here, we propose the application of short interfering RNAs (siRNAs) to degrade the viral genome, thus reducing viral infection. By introducing the concept of the probability of binding efficiency (PBE) and combining the secondary structures of RNA molecules, we designed 11 siRNAs that target the consensus regions of three key viral genes: the spike (S), nucleocapsid (N) and membrane (M) genes of SARS-CoV-2. The silencing efficiencies of the siRNAs were determined in human lung and endothelial cells overexpressing these viral genes. The results suggested that most of the siRNAs could significantly reduce the expression of the viral genes with inhibition rates above 50% in 24 hours. This work not only provides a strategy for designing potentially effective siRNAs against target genes but also validates several potent siRNAs that can be used in the clinical development of preventative medication for COVID-19 in the future.
Keywords: COVID-19, SARS-CoV-2, RNA secondary structure, siRNA, gene silencing

 Global reach, higher impact
Global reach, higher impact