10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(7):1682-1692. doi:10.7150/ijbs.56251 This issue Cite
Review
Regulation of RNA N6-methyladenosine modification and its emerging roles in skeletal muscle development
1. Guangdong Provincial Key Laboratory of Animal Molecular Design and Precise Breeding, Key Laboratory of Animal Molecular Design and Precise Breeding of Guangdong Higher Education Institutes, School of Life Science and Engineering, Foshan University, Foshan 528231, Guangdong, China.
2. Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, 518124, China.
3. State Key Laboratory of Animal Nutrition; Key Laboratory of Animal Genetics Breeding and Reproduction, Ministry of Agriculture and Rural Affairs, Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing 100193, China.
Received 2020-12-8; Accepted 2021-3-27; Published 2021-4-12
Abstract
N6-methyladenosine (m6A) is one of the most widespread and highly conserved chemical modifications in cellular RNAs of eukaryotic genomes. Owing to the development of high-throughput m6A sequencing, the functions and mechanisms of m6A modification in development and diseases have been revealed. Recent studies have shown that RNA m6A methylation plays a critical role in skeletal muscle development, which regulates myoblast proliferation and differentiation, and muscle regeneration. Exploration of the functions of m6A modification and its regulators provides a deeper understanding of the regulatory mechanisms underlying skeletal muscle development. In the present review, we aim to summarize recent breakthroughs concerning the global landscape of m6A modification in mammals and examine the biological functions and mechanisms of enzymes regulating m6A RNA methylation. We describe the interplay between m6A and other epigenetic modifications and highlight the regulatory roles of m6A in development, especially that of skeletal muscle. m6A and its regulators are expected to be targets for the treatment of human muscle-related diseases and novel epigenetic markers for animal breeding in meat production.
Keywords: RNA N6-methyladenosine, development, skeletal muscle, myogenesis
Introduction
RNA post-transcriptional modifications exist widely in eukaryotic genomes and play important roles in many biological processes such as development, reproduction, and disease [1, 2]. To date, more than 100 types of chemical modifications have been identified in cellular RNAs of eukaryotic genomes, including N7-methylguanine (m7G), N6-methyladenosine (m6A), 5-methylcytosine (m5C), N1-methyl-methylguanine (m1A), pseudouridine (ψ), and 5'-O-methylation (Nm). Among these, the m6A modification is the most common and abundant form of RNA methylation and is mainly located at the nitrogen atom in the sixth position of adenosine [3]. The m6A modification is enriched around the start and stop codons/3′-untranslated regions (UTRs) containing an RRACH consensus motif (where R is G or A, and H is A, C, or U), which is conserved across species [4]. The m6A modification was originally discovered in bacterial DNA [5] but was found for the first time in the 1970s in mRNA of Novikoff hepatoma cells, with an average of three to five m6A sites per mRNA unit [6]. In 2011, the first m6A demethylase fat mass and obesity-associated (FTO) gene was discovered, indicating for the first time that the m6A modification was dynamic and reversible [7]. This study became a milestone in RNA epigenetics research and initiated a surge in the investigation of m6A methylation. Owing to the development of high-throughput sequencing technology in combination with immunoprecipitation methods, the genome-wide presence of m6A modification in RNAs can be profiled and its dynamic regulation is explored [3, 8-12], greatly accelerating the discovery the m6A modification sites in genome of various species, and making it possible to further study the function and mechanism of m6A in development and other biological processes.
Skeletal muscle development is a complex process involving multiple stages of proliferation and differentiation. The current research on skeletal muscle development is mainly focused on molecular regulation at the levels of protein-coding genes and non-coding RNAs (such as miRNAs, lncRNAs, and circRNAs) [13]. The post-transcriptional mechanisms of skeletal muscle development remain largely unknown. Recent studies have suggested that m6A methylation regulates myoblast proliferation and differentiation during myogenesis and plays an important role in key biological processes and regulatory pathways during skeletal muscle development [14]. In the present paper, we summarize the current studies on the global landscape of m6A modifications in mammals and review the biological functions and mechanisms of enzymes regulating m6A RNA methylation. We emphasize the regulatory roles of m6A modifications in development, especially that of skeletal muscle, which may help to further understand the mechanism underlying the epigenetic regulation of skeletal muscle development and muscle-related diseases at the post-transcriptional level.
Genome-wide m6A methylation map in mammals
Genome-wide m6A methylation studies have shown that m6A methylation exists widely in the genomes of prokaryotes, eukaryotes, and viruses. In addition to those in mRNAs, a large number of m6A methylation modifications have been found in non-coding RNAs (ncRNAs), such as long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), and splice RNAs [6, 15]. It has been reported that m6A-modified adenosine accounts for 0.1-0.4% of the total adenosine content of RNA, with approximately 12,000 m6A methylation sites existing in the genome [8]. In the human genome, m6A modifications exist on more than 7,000 mRNAs and 300 ncRNAs, and are located in the 3'UTR of mRNA and the stop codon region of coding sequences (CDS), distributed mainly within G(M6A)C (70%) or A(M6A)C (30%) conservative sequences [3]. Recent studies have also suggested that the m6A modification is widespread in circRNAs and is expressed in a highly cell-specific pattern. Despite sharing m6A readers (binding proteins) and writers (methyltransferases), m6A circRNAs are frequently derived from exons that are not methylated in mRNAs [16]. Moreover, m6A modifications in circRNAs have been reported to modulate circRNA biogenesis and function [17]. Studies have shown that the genome-wide distribution of the m6A modification in embryonic stem cells and somatic cells is highly conserved across species. Approximately 70% of the transcripts modified by m6A in humans are also modified by m6A in mice, and roughly 46% of these sites are identical, indicating that m6A displays a certain degree of species conservation [8]. The whole-transcriptome landscape of m6A modifications in human fetal tissues uncovered high tissue variation. It was found that the m6A modification is positively correlated with the dynamic balance of gene expression and preferentially occupies genes rich in CpG promoters that regulate RNA transcript m6A, indicating that m6A is widely regulated by human genetic variation and promoter activity [18]. A study on the m6A modification map of the cerebellum in postnatal mice showed that m6A methylation and demethylation occur widely in both proliferating and fully differentiated neurons during differentiation in vivo. Functional analysis of genes containing on/off switches for m6A has demonstrated that m6A is a prerequisite for these genes to perform their functions at each developmental stage. RNA m6A methylation is controlled in a precise spatiotemporal manner and is involved in the regulation of postnatal cerebellar development in mice [19]. Taken together, the results shed light on the prevalence and characterizations of RNA m6A modification in different tissues, development stages and species, and provide a rich resource for in-depth exploring the mechanistic role of m6A in various biological processes.
Enzymes regulating RNA m6A methylation
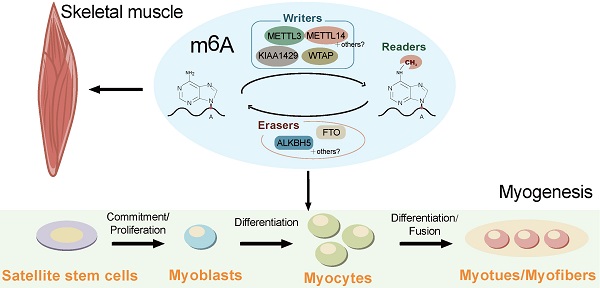
Similar to DNA methylation, the RNA m6A modification is dynamic and reversible. The m6A modification is generated by the METTL3/14 methyltransferase complex using the co-factor S-adenosyl methionine (SAM) as the major biological methyl donor [20]. The m6A modification is finely balanced through an interplay among m6A methyltransferases (writers) [21], demethylases (erasers), and binding proteins (readers) (Figure 1) [7, 22].
RNA methyltransferases include methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), Wilms tumor 1-associated protein (WTAP), Vir-like m6A methyltransferase-associated (VIRMA) protein (also named as KIAA1420), and RNA binding motif protein 15 (RBM15) [9, 20, 23-26]. METTL3 and METTL14 act as a stable heterodimer core of the RNA m6A methyltransferase complex to recognize methylated adenosine bases on RNA and perform catalytic function [20, 27]. WTAP, a mammalian splicing factor, is the third indispensable component of the m6A methyltransferase complex in vivo [28]. WTAP itself has no methylation activity; however, its interaction with the METTL3/14 heterodimer can significantly affect cellular m6A deposition [20, 28]. WTAP and METTL3/14 colocalize in nuclear speckles and participate in RNA m6A methylation [29]. In addition, recent studies have found that the zinc finger structure (ZC3H13) is a critical regulator of RNA m6A methylation and helps to anchor the Zc3h13-WTAP-Virilizer-Hakai complex, an important regulatory component of RNA m6A, in the nucleus [30].
RNA demethylases (erasers) can reverse the methylation at N6A sites [7, 22], among which FTO was the first to be discovered [7]. It has since been shown that inhibition of FTO can increase the level of m6A modification across all mRNAs [31]. In most cell lines, FTO is mainly localized in the nucleus and mediates approximately 5-10% of the total mRNA m6A demethylation, while in some leukemia cells, FTO mediates up to 40% of the total mRNA m6A demethylation [32]. AlkB homolog 5 (ALKHB5) was the second demethylase to be discovered, the gene for which belongs to the same Alkb gene family as that for FTO; however, its demethylase function was not determined until 2013, when it was shown to remove the m6A modification from single-stranded DNA and RNA [22]. The actions of FTO and ALKHB5 indicate that m6A modification is a dynamic and reversible process.
The downstream functions of m6A are mediated by reader proteins that recognize m6A and regulate mRNA processing [33], which mainly include YT521-B homology (YTH) domain-containing family 1-3 (YTHDF1-3), YTH domain-containing 1-2 (YTHDC1-2), heterogeneous ribonucleoproteins (HNRNPA2B1, HNRNPC), insulin-like growth factor 2 mRNA binding proteins (IGF2BP1/2/3), and eIF3 [34]. Remarkably, the m6A modification-related enzymes are also reported to be regulated by other genes and small-molecule compounds [35-39]. These findings greatly expand our understanding of the regulation of m6A, which might help to develop new targets for diseases and breeding.
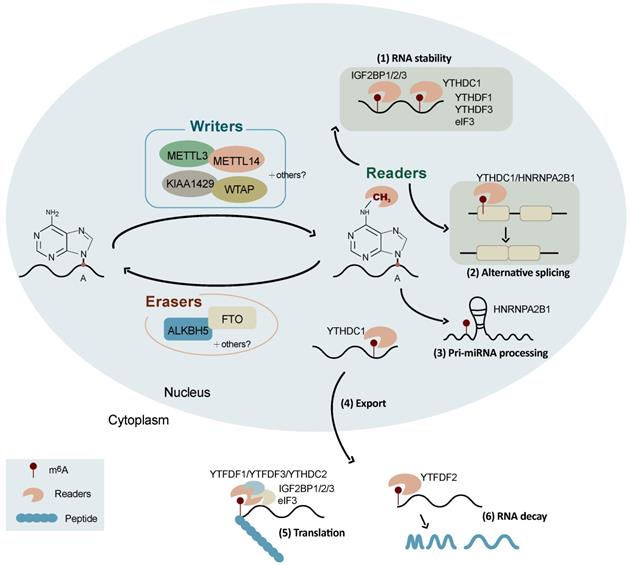
Schematic representation of the regulation and functions of m6A RNA modification. The reversible m6A RNA modification is dynamically regulated by m6A methyltransferases (writers) and demethylases (erasers), and recognized binding proteins (readers). Depending on the cellular context and their position in RNAs, m6A is involved in a number of RNA processes including alternative splicing, RNA stability, pri-miRNA processing, RNA nuclear export, RNA decay, and translation.
Interplay between m6A methylation and other epigenetic modifications
Genome-wide m6A methylation analysis has suggested that there is an interaction between m6A modification and other post-transcriptional regulatory modifications [40-43]. m6A methylation and adenosine-inosine (A-to-I) editing are two distinct and abundant RNA modifications at adenosine, between which there exists a negative correlation. A global A-to-I difference has been observed between m6A-positive and m6A-negative RNA populations. Knockdown of m6A writers or erasers leads to a global change in A-to-I editing [40]. The landscapes and distribution patterns of the m6A and m6Am methylomes across human and mouse tissues suggest that these two RNA modifications are highly specific in the brain. The overall m6A and m6Am levels are partially correlated with those of their writers and erasers [41]. Loss of YTHDF2 stabilizes lysine demethylase 6B (KDM6B), promoting histone H3 lysine-27 trimethylation (H3K27me3) of multiple proinflammatory cytokines and subsequently enhancing their transcription. Moreover, H3K27me3 is also a barrier to m6A modification during transcription. These results suggest an interplay between m6A and H3K27me3 during bacterial infection [42]. In addition, a recent study suggested that the gut microbiota affects the m6A epitranscriptome of mouse cecum and liver, mediating pathways related to metabolism, inflammation, and antimicrobial response [43]. Overall, these findings suggest a previously underappreciated interplay between m6A and other epigenetic modifications. Further studies are required to explore the interaction mechanisms between various epigenetic modifications in more detail with improved approaches and analysis methods.
Functions of m6A methylation in RNA metabolism
Substantial progress in the knowledge of the functions of m6A modification has been witnessed in recent years. It is well known that m6A is involved in a number of RNA processes including RNA nuclear export, splicing, stability, degradation, and translation (Figure 1) [32]. The initiation of mRNA export from the nucleus to the cytoplasm by m6A modification is an important way of regulating gene expression, which is mediated by METTL3 [44], ALKBH5 [22], and YTHDC1 [45]. The combination of YTHDC1, SRF3, and NXF1 (nuclear RNA output factor) can promote the nucleation of m6A-modified mRNA [46]. The main role of ALKBH5 is to demethylate m6A-modified mRNA and exerts its function by participating in the regulation of mRNA nucleation and other metabolic processes [22].
The m6A modification is also a splicing regulator and significantly affects alternative splicing patterns (Figure 1). It has been reported that mRNAs undergoing alternative splicing have greater numbers of METTL3 binding and m6A modification sites [8]. FTO targets pre-mRNAs in intronic regions and regulates alternative splicing and 3'-end processing [47]. The demethylation activity of ALKBH5 affects the nuclear speckle localization of several splicing factors and alters more than 3,000 mRNA isoforms, suggesting a role in splicing [22]. m6A modifications alter the structure of mRNAs and lncRNAs, affecting their binding to HNRNPC and therefore influencing the abundance and alternative splicing of target mRNAs in the nucleus [48]. YTHDC1 affects the nucleation and splicing of m6A-modified mRNA mainly by binding to specific m6A sites or splicing regulatory factors [49]. HNRNPA2B1 can directly bind to a set of nuclear transcripts within the transcriptome and regulate their alternative splicing in a manner similar to that of METTL3 [50]. HNRNPA2B1 also acts as a nuclear reader of the m6A marker and partially mediates its effect on primary microRNA processing and alternative splicing [50]. Moreover, m6A has been reported to be associated with alternative polyadenylation (APA) usage, which is coupled to the splicing of the last intron [51].
Recent studies have suggested that the m6A modification promotes mRNA translation through several distinct mechanisms (Figure 1) [52] including the YTHDF1-eIF3 pathway [52], cap-independent translation [53, 54], and IGF2BP-mediated translation [34]. YTHDF1 can bind to m6A-modified mRNA and recruit translation initiation factor complex eIF3 to promote mRNA translation, which is mediated by METTLE3 methylase activity and the YTHDF1-eIF3 pathway [55, 56]. YTHDF1 also recognizes m6A-modified lysosomal protease mRNA, the binding of which promotes the translation of lysosomal protease, thus inhibiting its antigenic cross-presentation ability [57]. YTHDF2 can affect RNA stability and accelerate the decay of m6A-modified RNAs [58, 59]. It has been found that the YTH and R3H domains of YTHDC2 facilitate its binding to cellular RNA and interact with small ribosomes, promoting mRNA translation [60]. YTHDF3 facilitates translation and promotes protein synthesis in synergy with YTHDF1, and affects methylated mRNA decay mediated by YTHDF2 [61]. The m6A-driven translation of circRNAs requires initiation factor eIF4G2 (a non-canonical eIF4G protein) and YTHDF3, which is enhanced by METTL3/14 and inhibited by FTO [62]. IGF2BPs enhance mRNA stability and translation in an m6A-dependent manner. In contrast to the mRNA decay-promoting function of YTHDF2, IGF2BPs promote the stability and storage of their target mRNAs, globally affecting gene expression [34]. The m6A modification also promotes the initiation of miRNA biogenesis and regulates nuclear mRNA processing events [63]. In addition, the m6A modification controls the RNA structure-dependent accessibility of RNA binding motifs to affect RNA-protein interactions for biological regulation, which is referred to as the m6A switch and widely exists throughout the transcriptome [48].
RNA m6A modification in developmental processes
The m6A modification is involved in the control of delicate gene expression patterns and regulates many developmental processes. During stem cell development, m6A can accurately regulate stem cell differentiation and reprogramming by modulating the expression of genes involved in the corresponding processes [64]. There exist several METTL3 binding sites on the mRNA of pluripotent factor NANOG in mouse embryonic stem cells. Knockout of METTL3 decreases the m6A methylation level of NANOG and suppresses the self-renewal and differentiation ability of embryonic stem cells. Moreover, METTL3 is also a regulator of murine naïve pluripotency termination. The m6A modification predominantly and directly reduces the mRNA stability of key naïve pluripotency-promoting transcripts [65]. Conditional knockout of METTL3 in bone marrow mesenchymal stem cells (MSCs) results in pathological features such as osteoporosis in mice [66]. Overexpression of METTL3 in bone marrow MSCs has a protective effect against estrogen-deficiency osteoporosis in mice. Knockout of METTL3 also reduces the translation efficiency of the MSC distributor Pth1r, inhibiting osteogenesis and adipogenesis induced by PTH in vivo [66]. LincRNA1281 can regulate the differentiation of mouse embryonic stem cells through a competitive endogenous RNA (ceRNA) model mediated by m6A methylation [67]. METTL3 is also reported as a negative regulator of autophagy of cardiomyocytes. METTL3 and ALKBH5 can oppositely regulate m6A modification of transcription factor EB (TFEB) mRNA. In turn, TFEB regulates the expression levels of METTL3 and ALKBH5 in opposite directions, which uncovers a critical link between METTL3-ALKBH5 and autophagy [68].
The m6A modification is highly enriched in the mammalian brain [3] and is essential for brain development and function [69]. Methylated RNA immunoprecipitation sequencing (MeRIP-seq) analysis has suggested that thousands of coding genes and hundreds of non-coding genes are modified by m6A in brain tissue [3]. The transcriptomic profiles of m6A are spatially regulated in different brain regions. The m6A modification is highly enriched in genes related to neuronal functions, such as synaptic release and uptake [70]. Moreover, m6A signaling is required for robust axon regeneration in the adult mammalian nervous system. METTL3 knockdown results in a prolonged cell cycle in cortical neural progenitor cells and decreased differentiation of radial glial cells. Knockout of METTL14 in mouse embryos leads to prolongation of cortical neurogenesis to the postnatal stage [71]. These results indicate a pivotal regulatory role of m6A during brain development.
The m6A modification has also been reported to be a critical regulator of adipogenesis. In rat adipocytes, the m6A modification strongly stimulates glucose oxidation [72]. FTO-mediated m6A demethylation is closely related to upregulated adipogenesis, fat mass, and body weight. FTO is highly abundant in adipose tissue and targets multiple genes to regulate adipogenesis. For instance, FTO directly targets Atg5 and Atg7 and mediates their expression in an m6A-dependent manner during adipogenesis. These two genes are also targets of YTHDF2 [73]. In porcine intramuscular pre-adipocytes, FTO can promote adipogenesis by inhibiting the Wnt/β-catenin signaling pathway [74]. The effect of FTO on adipogenesis also appears to be regulated by enhanced expression of the pro-adipogenic short isoform of runt-related transcription factor 1 (RUNX1), which can promote adipocyte proliferation [75]. FTO can affect the binding of alternative splicing regulator SRSF2 to m6A by regulating the m6A methylation level, thus mediating the alternative splicing of key genes involved in adipogenesis and regulating the differentiation of preadipocytes [76]. In addition, FTO regulates adipogenesis by controlling cell cycle progression in an m6A-YTHDF2-dependent manner [77]. The transcription factor zinc finger protein (Zfp217) modulates m6A mRNA methylation by activating FTO and CCND1 expression to promote adipocyte differentiation in an m6A-dependent manner [78, 79]. In contrast, WTAP, METTL3, and METTL14 are negatively related to adipogenesis by promoting cell cycle transition in mitotic clonal expansion [80]. METTL3 expression increases significantly in interscapular brown adipose tissue (iBAT) after birth and is an essential regulator of iBAT postnatal development and energy homeostasis [81]. In porcine bone marrow stem cells, METTL3 represses adipogenesis via an m6A-dependent pattern [82]. These results provide insight into the critical roles of m6A methylation during adipogenesis and suggest that m6A may be a novel potential biomarker of obesity.
Taken together, these findings demonstrate pivotal roles of the m6A modification in development. In the remainder of this review, we focus on the current understanding of the roles of m6A in skeletal muscle development and discuss potential future research directions.
The regulation of skeletal muscle development
Skeletal muscle is the most abundant tissue, accounting for 30-40% of body mass in adult humans [83] and 35-60% of that in domesticated animals [84]. In animal husbandry, the development of skeletal muscle is directly associated with meat production, which is one of the most important economical traits [85]. It is well known that skeletal muscle development is an extremely complex regulatory process orchestrated by myogenic genes and transcription factors (Figure 2). These transcription factors precisely control the proliferation, differentiation, and fusion of muscle cells according to strict time-sequence expression [86-88]. The myogenic regulatory factor (MRF) family plays a decisive regulatory role in the fate of myoblasts and muscle development, constituting a family of basic helix-loop-helix transcription factors. The four members of this family are myogenic differentiation 1 (MyoD), myogenic factor 5 (MyF5), myogenic protein (Myogenin), and MRF4 [89]. MyF5 and MyoD are muscle-derived determinants of myoblast regulation and differentiation, while Myogenin and MRF4 are muscle-derived differentiation factors that induce terminal differentiation and play an important role in the development of skeletal muscle in the early embryo [90]. Myogenin is the downstream gene of MyF5 and MyoD and is necessary for the fusion of mononuclear myoblasts to form multinucleated myotubes [91]. MRF4 plays an important role in the terminal differentiation of muscle cells and in the maintenance of muscle fibers [92].
In addition, non-coding RNAs (ncRNAs) also play important regulatory roles in skeletal muscle development (Figure 2). The miRNAs, miR-1, -133, and -206, are specifically expressed in skeletal muscle and myocardium (myomiR). miR-1 and miR-206 promote skeletal muscle cell differentiation by inhibiting the expression of Pax7 [93]. miRNA-133 inhibits the differentiation of myoblasts and promotes myoblast proliferation by targeting the serum response factor (SRF) gene [94]. Other miRNA, such as miR-148a and miR-21 are also reported to regulate skeletal muscle development [95, 96].
The epigenetics regulation of skeletal muscle development. The development of skeletal muscle is tightly controlled by multiple epigenetic modifications at both the DNA (e.g., histone modification and DNA methylation) and the RNA (e.g., m6A methylation, non-coding RNAs, and RNA editing) levels. The interplay between m6A and other epigenetic modifications orchestrate the expression of myogenic genes during commitment, proliferation, differentiation, and fusion of skeletal muscle.
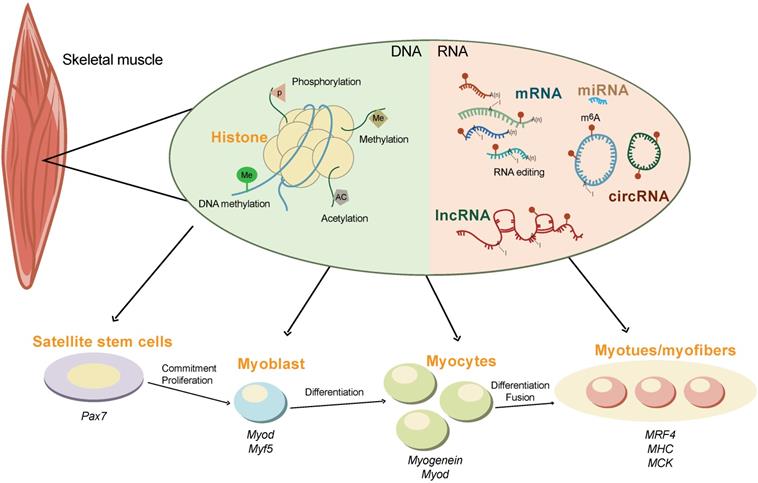
LncRNAs account for the vast majority of transcripts in the mammalian genome. LncRNA-H19 was the first lncRNA to be identified in mammals. The first exon of lncRNA-H19 can transcribe miR-675-3p and miR-675-5. These two miRNAs can downregulate the expression of Smad family members 1 and 5 (SMAD1 and SMAD5) and cell division cycle 6 (CDC6), inhibiting the proliferation and promoting the differentiation of C2C12 myoblasts [97, 98] Linc-MD1, the first identified muscle-specific lncRNA, is required for myoblast differentiation by regulating the levels of myocyte enhancer factor 2C (MEF2C) and mastermind-like protein 1 (MAML1) via the sponging of endogenous miR-133 and miR-135 in the cytoplasm [99].
A diagram showing the functions and regulation of m6A and its regulators in myogenesis and skeletal muscle development.
CircRNAs exist widely in eukaryotes and lacks a 5'-terminal cap and a 3'-terminal poly (A) tail, forming a circular structure with covalent bonds [100]. Studies have found that circRNAs are mainly involved in the regulation of muscle development by acting as molecular sponges of miRNA and encoding small peptides [101]. For example, myocyte-specific circRNA circZNF609 encodes small peptides that participate in muscle differentiation and myoblast proliferation. Downregulation of circZNF609 expression during myoblast differentiation can promote myoblast proliferation [102].
These information provide a comprehensive profile for understanding the mechanisms underlying skeletal muscle development, and highlight the important functions of ncRNAs in skeletal muscle development. ncRNAs have a great potential to become therapeutic targets for muscular diseases and molecular markers for animal breeding. However, in comparison with the above regulators, the precise roles of m6A modification in the skeletal muscle and its temporal control during skeletal muscle development are still limited.
The function and mechanism of m6A during myogenesis in vitro
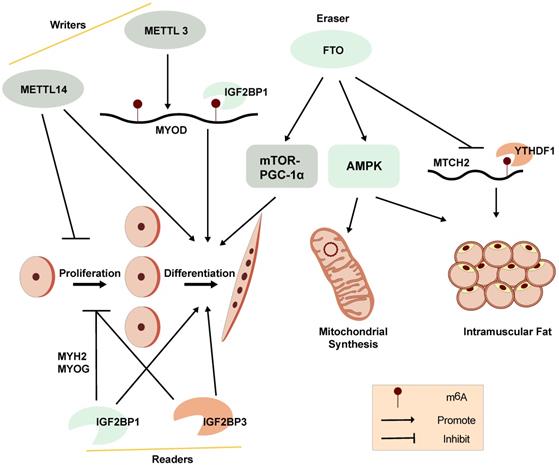
Using mouse C2C12 myoblasts and muscle-specific adult stem cells (MuSCs) as models, recent studies have suggested that RNA m6A methylation plays an important role in myoblast proliferation and differentiation in vitro (Figure 3). The m6A modifications are closely linked to genes associated with transcriptional regulation during proliferation, and positively regulate myogenic differentiation in C12C12 skeletal myoblasts [103, 104].
A distinct m6A modification profile between proliferating and differentiating C2C12 myoblasts is observed. The global m6A levels decrease when C2C12 myoblasts transition from proliferation to differentiation. METTL3 regulates m6A levels in MuSCs and myoblasts, controlling their transition to different cell states. Knockdown of METTL3 can slow the proliferation of primary MuSCs in mouse and enhance their engraftment [103]. Meanwhile, m6A modifications have been found to be enriched within the 5'-UTR of MyoD mRNA. METTL3 can stabilize MyoD mRNA by promoting mRNA processing in proliferative myoblasts for skeletal muscle differentiation. Knockdown of METTL3 can specifically downregulate the expression of processed MyoD mRNA in skeletal myoblast (Figure 3) [105]. In addition, knockdown of METTL14 inhibits the differentiation and promotes the proliferation of C2C12 myoblasts [14].
Demethylase FTO is not only associated with obesity but can also regulate mTOR-PGC-1α-mediated mitochondrial synthesis and promote muscle cell differentiation through its m6A demethylase activity (Figure 3). The expression of FTO increases during myoblast differentiation, while silencing of FTO inhibits differentiation. Depletion of FTO leads to impaired differentiation and fusion in vitro [106]. Meanwhile, overexpression of FTO could suppress the expression of mitochondrial carrier homology 2 (MTCH2), which is a target of YTHDF1. m6A enhances MTCH2 protein expression via an YTHDF1-dependent pathway and promotes adipogenesis in porcine muscles [107]. The energy sensor AMP-activated protein kinase (AMPK) gene is a key regulator of skeletal muscle lipid metabolism. The level of mRNA m6A methylation in skeletal muscle is negatively correlated with skeletal muscle lipid content. AMPK regulates skeletal muscle lipid accumulation through fat quality and obesity-related protein by FTO-dependent m6A demethylation [108]. Our recent studies suggest that the m6A methylation readers, IGF2BP1 and IGF2BP3, both promote myoblast differentiation and repress proliferation in C2C12 myoblast cells [14, 109]. These findings provide further understanding of the potential functions of RNA m6A methylation on myogenesis.
However, it's confused that the m6A regulators, including m6A methyltransferase (METTL3 and METTL14), demethylase FTO and readers (IGF2BP1 and IGF2BP3), are reported to promote myoblast differentiation in C2C12 myoblasts (Figure 3) [14, 106, 109], indicating that the epigenetic regulation of m6A modifications in myogenesis is a complex regulatory networks and might be affected by other genes and regulatory elements at multi-levels. Due to the contradictory regulation of m6A modification on myogenesis, the correlation between m6A modification and myogenesis is still needed to be further explored. The function of other m6A regulators in myogenesis should also be studied.
The dynamic change and potential roles of m6A methylation in skeletal muscle development
To date, a few studies have evaluated the genome-wide m6A modification on skeletal muscle tissue in vivo, the dynamic change of m6A methylation in skeletal muscle development and the correlation between m6A and skeletal muscle are explored [14, 103, 110]. Many muscle-specific m6A sites have been identified in mammals [41, 110, 111]. In mouse, it's reported that the maternal high-fat diet alters the m6A pattern in skeletal muscle and adipose in a development-dependent way [112]. Using liquid chromatography/mass spectrometry (LC-MS) and MeRIP-seq analysis, they find that global m6A levels increase during the early stages of skeletal muscle regeneration. In the longissimus dorsi muscle of Landrace pigs (lean-type breed), the global m6A level and the expression of METTL3 at both mRNA and protein levels are much higher than those of Jinhua pigs (obese-type breed), while the level of FTO shows the opposite trend, suggesting that the m6A level is negatively associated with fat deposition in skeletal muscle [107]. Depletion of FTO impairs postnatal skeletal muscle development in vivo [106]. The MeRIP-seq analysis of skeletal muscle in wild boar, Landrace and Rongchang (obese-type breed) pigs suggests that m6A modification occurs in a breed-differential pattern [110].
Moreover, using a refined MeRIP-seq method that is optimal for use with samples containing small amounts of RNA, our group recently performed transcriptome-wide m6A profiling at six prenatal developmental stages in porcine skeletal muscle, ranging from 33 to 90 days post coitus (dpc) [14]. A highly dynamic change in genome-wide m6A modification is observed across different developmental stages. RNA immunoprecipitation assay reveals that IGF2BP1 targets many myogenesis genes in skeletal muscle, such as MYH2 and MyoG, suggesting that IGF2BP1 regulates skeletal muscle development via binding to m6A-modified target genes [14]. The results also show that most of the m6A modified genes are commonly methylated throughout prenatal skeletal muscle development, and the differentially methylated genes are enriched in pathways related to skeletal muscle development [14]. Interestingly, we also find that DNA methylation regulates the expression and function of IGF2BP3 by modulating transcription factor SP1 binding in skeletal muscle [109].
Overall, these studies generally profiled the dynamic change of m6A modification in skeletal muscle and highlighted their correlation between m6A modification and skeletal muscle development. However, skeletal muscle is highly heterogeneous and consists of a variety of cells that have a network of crosstalk [113, 114], the function and regulation of m6A modification in vivo and in vitro might be biased. Single-cell analysis will be helpful to understand the roles of m6A modification in different skeletal muscle cell types in the future, thought this technology is currently not available in m6A. In addition, further in vivo studies in individuals using genetically modified mouses of m6A regulators as models are necessary to determine the function and mechanism of m6A in skeletal muscle development.
Closing remarks and outlook
Here, we summary the function and mechanism of m6A modification, and highlight the current status of knowledge regarding the roles of m6A in myogenesis skeletal muscle development, which helps to further understand the mechanism underlying the epigenetic regulation of skeletal muscle development and muscle-related diseases. We suggest that the m6A modification can be used as a target for the therapy of muscle diseases and a biomarker in animal breeding for the improvement of meat production traits in the future.
However, in comparison with other tissues, such as the brain and adipose, knowledge regarding the biological function and regulatory mechanism of m6A in skeletal muscle remain limited. Several avenues of research should be explored in the future. Firstly, although several studies have revealed the dynamic RNA m6A profiles during skeletal muscle development, only a few developmental stages or samples were used. For example, the largest study assayed 18 samples at six prenatal developmental stages spanning 33-90 dpc in pigs [14]. Future studies should elucidate the m6A methylome profiles of skeletal muscle spanning a wider range of developmental stages, including both prenatal and postnatal stages in mammals. Secondly, considering the difficulties in sampling skeletal muscle at the embryonic stages, future studies should develop novel methods that use small amounts of RNA to profile the genome-wide m6A maps at a higher resolution. Moreover, only bulk skeletal muscle can currently be used for genome-wide m6A methylation analysis. The fiber composition and non-muscle cells such as vasculature and immune cells may partly influence the measurement of m6A modifications. It is the hope that single-cell sequencing technology may be used to profile the m6A transcriptome map at the genome-wide level in the near future. Thirdly, recent studies performed in skeletal muscle have mainly focused on the function of the m6A modification in mRNA; however, the distribution and biological function of m6A in ncRNAs (such as lncRNAs and circRNAs) and regulatory elements (such as enhancers and promoters) remain unclear. Fourthly, adenine methylation is well known as a common mechanism for controlling gene expression. Knockout or overexpression of FTO and ALKBH5 only slightly changes the overall level of m6A, suggesting that there exist other unknown m6A demethylases that await discovery. The upstream regulators and small molecules of m6A enzymes are needed to be identified. Moreover, the regulatory mechanisms underlying the actions of m6A methyltransferases, demethylases, and binding proteins on m6A to spatiotemporally regulate downstream pathways and mediate skeletal muscle development remain largely unclear. Further investigation into targeted genes and m6A modifications using gene editing and epigenetic tools, such as CRISPR/cas9 technology, will be required to elucidate the mechanisms of dynamic m6A modification during skeletal muscle development. Finally, in addition to m6A, there exist many other epigenetic modifications in the genome and transcriptome; therefore, the interplay between m6A and other epigenetic modifications during skeletal muscle development should be explored. Integrative multi-omics will be a useful approach to comprehensively understand the regulatory mechanism of skeletal muscle development.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31902133), Guangdong Provincial key Laboratory of Animal Molecular Design and Precise Breeding Research Grant (2019B030301010), and the Key Laboratory of Animal Molecular Design and Precise Breeding of Guangdong Higher Education Institutes (2019KSYS011).
Authors Contribution
YY conceived and managed the project. JL, YP, RZ, YY wrote the manuscript. ZT and YY revised the paper. All authors approved the final version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Tong J, Flavell RA, Li HB. RNA m(6)A modification and its function in diseases. Front Med. 2018;12:481-9
2. Wu X, Sang L, Gong Y. N6-methyladenine RNA modification and cancers. Am J Cancer Res. 2018;8:1957-66
3. Meyer Kate D, Saletore Y, Zumbo P, Elemento O, Mason Christopher E, Jaffrey Samie R. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3' UTRs and near Stop Codons. Cell. 2012;149:1635-46
4. Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nature Reviews Molecular Cell Biology. 2016;18:31-42
5. Dunn D, Smith J. The occurrence of 6-methylaminopurine in microbial deoxyribonucleic acids. Biochemical journal. 1955;68:627-36
6. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71:3971-5
7. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y. et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nature Chemical Biology. 2011;7:885-7
8. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201-6
9. Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A. et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155:1409-21
10. Chen T, Hao Y-J, Zhang Y, Li M-M, Wang M, Han W. et al. m6A RNA Methylation Is Regulated by MicroRNAs and Promotes Reprogramming to Pluripotency. Cell Stem Cell. 2015;16:289-301
11. Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767-72
12. He C, Zeng Y, Wang S, Gao S, Soares F, Ahmed M. et al. Refined RIP-seq protocol for epitranscriptome analysis with low input materials. PLoS Biology. 2018;16:e2006092
13. Escriva H, Zhao X, Mo D, Li A, Gong W, Xiao S. et al. Comparative Analyses by Sequencing of Transcriptomes during Skeletal Muscle Development between Pig Breeds Differing in Muscle Growth Rate and Fatness. PLoS One. 2011;6:e19774
14. Zhang X, Yao Y, Han J, Yang Y, Chen Y, Tang Z. et al. Longitudinal epitranscriptome profiling reveals the crucial role of N6-methyladenosine methylation in porcine prenatal skeletal muscle development. Journal of Genetics and Genomics. 2020 p: 1673-8527
15. Dubin DT, Taylor RH. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975;2:1653-68
16. Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N. et al. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Reports. 2017;20:2262-76
17. Di Timoteo G, Dattilo D, Centrón-Broco A, Colantoni A, Guarnacci M, Rossi F. et al. Modulation of circRNA Metabolism by m6A Modification. Cell Reports. 2020;31:107641
18. Xiao S, Cao S, Huang Q, Xia L, Deng M, Yang M. et al. The RNA N6-methyladenosine modification landscape of human fetal tissues. Nature Cell Biology. 2019;21:651-61
19. Ma C, Chang M, Lv H, Zhang ZW, Zhang W, He X. et al. RNA m(6)A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018;19:68
20. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93-5
21. Fu Y, Dominissini D, Rechavi G, He CJNRG. Gene expression regulation mediated through reversible m 6 A RNA methylation. Nature. 2014;15:293-306
22. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18-29
23. Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T. et al. Identification of Wilms' Tumor 1-associating Protein Complex and Its Role in Alternative Splicing and the Cell Cycle. Journal of Biological Chemistry. 2013;288:33292-302
24. Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG. et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301-4
25. Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH. et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature. 2016;540:242-7
26. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z. et al. Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575-8
27. Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman FJJoBC. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. The Journal of biological chemistry. 1994;269:17697-704
28. Ping X-L, Sun B-F, Wang L, Xiao W, Yang X, Wang W-J. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Research. 2014;24:177-89
29. Schller E, Weichmann F, Treiber T, Ringle S, Meister GJR-aPotRS. Interactions, localization and phosphorylation of the m6A generating METTL3-METTL14-WTAP complex. RNA. 2018;24:499-512
30. Wen J, Lv R, Ma H, Shen H, He C, Wang J. et al. Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Molecular Cell. 2018;69:1028-38.e6
31. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y. et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell. 2018;172:90-105
32. Shi H, Wei J, He C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Molecular Cell. 2019;74:640-50
33. Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29:1343-55
34. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H. et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285-95
35. Zhou Z, Lv J, Yu H, Han J, Yang X, Feng D. et al. Mechanism of RNA modification N6-methyladenosine in human cancer. Molecular Cancer. 2020;19:1-20
36. Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L. et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell stem cell. 2018;22:191-205 e9
37. Zhang J, Tsoi H, Li X, Wang H, Gao J, Wang K. et al. Carbonic anhydrase IV inhibits colon cancer development by inhibiting the Wnt signalling pathway through targeting the WTAP-WT1-TBL1 axis. Gut. 2016;65:1482-93
38. Selberg S, Blokhina D, Aatonen M, Koivisto P, Siltanen A, Mervaala E. et al. Discovery of small molecules that activate RNA methylation through cooperative binding to the METTL3-14-WTAP complex active site. Cell reports. 2019;26:3762-71 e5
39. Chen B, Ye F, Yu L, Jia G, Huang X, Zhang X. et al. Development of cell-active N 6-methyladenosine RNA demethylase FTO inhibitor. Journal of the American Chemical Society. 2012;134:17963-71
40. Xiang JF, Yang Q, Liu CX, Wu M, Chen LL, Yang L. N(6)-Methyladenosines Modulate A-to-I RNA Editing. Mol Cell. 2018;69:126-35
41. Liu J, Li K, Cai J, Zhang M, Zhang X, Xiong X. et al. Landscape and Regulation of m(6)A and m(6)Am Methylome across Human and Mouse Tissues. Mol Cell. 2020;77:426-40
42. Wu C, Chen W, He J, Jin S, Liu Y, Yi Y. et al. Interplay of m6A and H3K27 trimethylation restrains inflammation during bacterial infection. Science advances. 2020 19;6: eaba0647
43. Jabs S, Biton A, Bécavin C, Nahori M-A, Cossart P. Impact of the gut microbiota on the m6A epitranscriptome of mouse cecum and liver. Nature Communications. 2020;11:1344
44. Fustin J-M, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M. et al. RNA-Methylation-Dependent RNA Processing Controls the Speed of the Circadian Clock. Cell. 2013;155:793-806
45. Roundtree IA, Luo G-Z, Zhang Z, Wang X, Zhou T, Cui Y. et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife. 2017;6:e31311
46. Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y. et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014;10:927-9
47. Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, Vanacova S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing. Nucleic Acids Research. 2017;45:11356-70
48. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560-4
49. Xiao W, Adhikari S, Dahal U, Chen Y-S, Hao Y-J, Sun B-F. et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Molecular cell. 2016;61:507-19
50. Claudio R A, Goodarzi H, Lee H. HNRNPA2B1 Is a Mediator of m6A-Dependent Nuclear RNA Processing Events. Cell. 2015;6:1299-308
51. Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A. et al. A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes & Development. 2015;29:2037-53
52. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H. et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388-99
53. Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian S-B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591-4
54. Meyer Kate D, Patil Deepak P, Zhou J, Zinoviev A, Skabkin Maxim A, Elemento O. et al. 5' UTR m6A Promotes Cap-Independent Translation. Cell. 2015;163:999-1010
55. Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T. et al. The epitranscriptome m6A writer METTL3 promotes chemo-and radioresistance in pancreatic cancer cells. International journal of oncology. 2018;52:621-9
56. Shi H, Zhang X, Weng Y-L, Lu Z, Liu Y, Lu Z. et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018;563:249-53
57. Han D, Liu J, Chen C, Dong L, Liu Y, Chang R. et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature. 2019;566:270-4
58. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117-20
59. Li Z, Qian P, Shao W, Shi H, He XC, Gogol M. et al. Suppression of m 6 A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell research. 2018;28:904-17
60. Kretschmer J, Rao H, Hackert P, Sloan KE, Höbartner C, Bohnsack MT. The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5'-3' exoribonuclease XRN1. Rna. 2018;24:1339-50
61. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ. et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Research. 2017;27:315-28
62. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y. et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Research. 2017;27:626-41
63. Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482-5
64. Wang Y, Zeng L, Liang C, Zan R, Ji W, Zhang Z. et al. Integrated analysis of transcriptome-wide m(6)A methylome of osteosarcoma stem cells enriched by chemotherapy. Epigenomics. 2019;11:1693-715
65. Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M. et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002-6
66. Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang Y. et al. Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat Communications. 2018;9:4772
67. Yang D, Qiao J, Wang G, Lan Y, Li G, Guo X. et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018;46:3906-20
68. Song H, Feng X, Zhang H, Luo Y, Huang J, Lin M. et al. METTL3 and ALKBH5 oppositely regulate m6A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. 2019;15:1419-37
69. Livneh I, Moshitch-Moshkovitz S, Amariglio N, Rechavi G, Dominissini D. The m6A epitranscriptome: transcriptome plasticity in brain development and function. Nature Reviews Neuroscience. 2019;21:36-51
70. Chang M, Lv H, Zhang W, Ma C, He X, Zhao S. et al. Region-specific RNA m6A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biology. 2017;7:170166
71. Weng Y-L, Wang X, An R, Cassin J, Vissers C, Liu Y. et al. Epitranscriptomic m6A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron. 2018;97:313-25.e6
72. Souness JE, Stouffer JE, Sanchez VCD. Effect of N6-methyladenosine on fat-cell glucose metabolism. Evidence for two modes of action. Biochemical Pharmacology. 1982;31:3961-71
73. Wang X, Wu R, Liu Y, Zhao Y, Bi Z, Yao Y. et al. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy. 2019;16:1221-35
74. Chen X, Luo Y, Jia G, Liu G, Zhao H, Huang Z. FTO Promotes Adipogenesis through Inhibition of the Wnt/β-catenin Signaling Pathway in Porcine Intramuscular Preadipocytes. Animal Biotechnology. 2017;28:268-74
75. Merkestein M, Laber S, McMurray F, Andrew D, Sachse G, Sanderson J. et al. FTO influences adipogenesis by regulating mitotic clonal expansion. Nature Communications. 2015;6:6792
76. Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W. et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403-19
77. Wu R, Liu Y, Yao Y, Zhao Y, Bi Z, Jiang Q. et al. FTO regulates adipogenesis by controlling cell cycle progression via m6A-YTHDF2 dependent mechanism. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2018;1863:1323-30
78. Song T, Yang Y, Wei H, Xie X, Lu J, Zeng Q. et al. Zfp217 mediates m6A mRNA methylation to orchestrate transcriptional and post-transcriptional regulation to promote adipogenic differentiation. Nucleic Acids Research. 2019;47:6130-44
79. Liu Q, Zhao Y, Wu R, Jiang Q, Cai M, Bi Z. et al. ZFP217 regulates adipogenesis by controlling mitotic clonal expansion in a METTL3-m6A dependent manner. RNA Biology. 2019;16:1785-93
80. Masatoshi K, Mitsuru O, Takayoshi S, Motoharu A, Toshihiro U, Aya I. et al. The RNA methyltransferase complex of WTAP, METTL3, and METTL14 regulates mitotic clonal expansion in adipogenesis. Molecular Cellular Biology. 2018;38:00116-18
81. Wang Y, Gao M, Zhu F, Li X, Yang Y, Yan Q. et al. METTL3 is essential for postnatal development of brown adipose tissue and energy expenditure in mice. Nature Communications. 2020;11:1648
82. Yao Y, Bi Z, Wu R, Zhao Y, Liu Y, Liu Q. et al. METTL3 inhibits BMSC adipogenic differentiation by targeting the JAK1/STAT5/C/EBPb pathway via an m6A-YTHDF2-dependent manner. The FASEB Journal. 2019;33:7529-44
83. Janssen I, Heymsfield SB, Wang ZM, Ross R. Skeletal muscle mass and distribution in 468 men and women aged 18-88 yr. Journal of Applied Physiology. 2000;89:81-8
84. Listrat A, Lebret B, Louveau I, Astruc T, Bonnet M, Lefaucheur L. et al. How Muscle Structure and Composition Influence Meat and Flesh Quality. The Scientific World Journal. 2016;2016:3182746
85. Berendsen AD, Olsen BR. Bone development. Bone. 2015;80:14-8
86. Tajbakhsh S. Skeletal muscle stem cells in developmental versus regenerative myogenesis. Journal of Internal Medicine. 2009;266:372-89
87. Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the Genetic Hierarchies Controlling Skeletal Myogenesis: Pax-3 and Myf-5 Act Upstream of MyoD. Cell. 1997;89:127-38
88. Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S. et al. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. The Journal of Cell Biology. 2006;172:91-102
89. Moncaut N, Rigby PWJ, Carvajal JJ. Dial M(RF) for myogenesis. Febs Journal. 2013;280:3980-90
90. Endo T. Molecular mechanisms of skeletal muscle development, regeneration, and osteogenic conversion. Bone. 2015;80:2-13
91. Yan X, Zhu MJ, Dodson MV, Du M. Developmental programming of fetal skeletal muscle and adipose tissue development. J Genomics. 2013;1:29-38
92. Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V. et al. Mrf4 determines skeletal muscle identity in Myf5: Myod double-mutant mice. Nature. 2004;431:466-71
93. Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X. et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. The Journal of cell Biology. 2010;190:867-79
94. Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM. et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228-33
95. Zhang J, Ying Z, Tang Z, Long L, Li K. MicroRNA-148a Promotes Myogenic Differentiation by Targeting the ROCK1 Gene. Journal of Biological Chemistry. 2012;287:21093-101
96. Bai L, Liang R, Yang Y, Hou X, Wang Z, Zhu S. et al. MicroRNA-21 Regulates PI3K/Akt/mTOR Signaling by Targeting TGFβI during Skeletal Muscle Development in Pigs. PLoS One. 2015;10:e0119396
97. Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L. et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101-12
98. Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491-501
99. Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M. et al. A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell. 2011;147:358-69
100. Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P. et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Molecular Cancer. 2017;16:94
101. Wei X, Li H, Yang J, Hao D, Dong D, Huang Y. et al. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 2017;8:e3153
102. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O. et al. Circ-ZNF609 is a Circular RNA that can be Translated and Functions in Myogenesis. Mol Cell. 2017;66:22-37
103. Gheller BJ, Blum JE, Fong EHH, Malysheva OV, Cosgrove BD, Thalacker-Mercer AE. A defined N6-methyladenosine (m6A) profile conferred by METTL3 regulates muscle stem cell/myoblast state transitions. Cell Death Discovery. 2020;6:95
104. Chen JN, Chen Y, Wei YY, Raza MA, Zou Q, Xi XY. et al. Regulation of m6A RNA Methylation and Its Effect on Myogenic Differentiation in Murine Myoblasts. Molecular Biology. 2019;53:384-92
105. Kudou K, Komatsu T, Nogami J, Maehara K, Harada A, Saeki H. et al. The requirement of Mettl3-promoted MyoD mRNA maintenance in proliferative myoblasts for skeletal muscle differentiation. Open Biology. 2017;7:170119
106. Wang X, Huang N, Yang M, Wei D, Tai H, Han X. et al. FTO is required for myogenesis by positively regulating mTOR-PGC-1alpha pathway-mediated mitochondria biogenesis. Cell Death Dis. 2017;8:e2702
107. Jiang Q, Sun B, Liu Q, Cai M, Wu R, Wang F. et al. MTCH2 promotes adipogenesis in intramuscular preadipocytes via an m(6)A-YTHDF1-dependent mechanism. FASEB J. 2019;33:2971-81
108. Wu W, Feng J, Jiang D, Zhou X, Jiang Q, Cai M. et al. AMPK regulates lipid accumulation in skeletal muscle cells through FTO-dependent demethylation of N(6)-methyladenosine. Sci Rep. 2017;7:41606
109. Yang Y, Fan X, Yan J, Chen M, Zhu M, Tang Y. et al. A comprehensive epigenome atlas reveals DNA methylation regulating skeletal muscle development. Nucleic Acids Research. 2021;49:1313-29
110. Tao X, Chen J, Jiang Y, Wei Y, Chen Y, Xu H. et al. Transcriptome-wide N (6) -methyladenosine methylome profiling of porcine muscle and adipose tissues reveals a potential mechanism for transcriptional regulation and differential methylation pattern. BMC Genomics. 2017;18:336
111. Zhang H, Shi X, Huang T, Zhao X, Chen W, Gu N. et al. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 2020;48:6251-64
112. Li X, Yang J, Zhu Y, Liu Y, Shi X, Yang G. Mouse Maternal High-Fat Intake Dynamically Programmed mRNA m(6)A Modifications in Adipose and Skeletal Muscle Tissues in Offspring. Int J Mol Sci. 2016;17:1336
113. Xi H, Langerman J, Sabri S, Chien P, Young CS, Younesi S. et al. A Human Skeletal Muscle Atlas Identifies the Trajectories of Stem and Progenitor Cells across Development and from Human Pluripotent Stem Cells. Cell Stem Cell. 2020;27:158-76
114. De Micheli AJ, Spector JA, Elemento O, Cosgrove BD. A reference single-cell transcriptomic atlas of human skeletal muscle tissue reveals bifurcated muscle stem cell populations. Skelet Muscle. 2020;10:19
Author contact
![]() Corresponding author: E-mail: yangyalancn.
Corresponding author: E-mail: yangyalancn.

 Global reach, higher impact
Global reach, higher impact