10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(12):3188-3208. doi:10.7150/ijbs.62573 This issue Cite
Review
LncRNA H19: A novel oncogene in multiple cancers
1. Department of Gastroenterology, Shengjing Hospital of China Medical University, Shenyang 110004, China
2. Department of Clinical Genetics, Shengjing Hospital of China Medical University, Shenyang 110004, China
3. Department of Urology, Shengjing Hospital of China Medical University, Shenyang 110004, China
Received 2021-5-10; Accepted 2021-7-14; Published 2021-7-25
Abstract
Long non-coding RNAs (lncRNAs) are a series of non-coding RNAs that lack open reading frameworks. Accumulating evidence suggests important roles for lncRNAs in various diseases, including cancers. Recently, lncRNA H19 (H19) became a research focus due to its ectopic expression in human malignant tumors, where it functioned as an oncogene. Subsequently, H19 was confirmed to be involved in tumorigenesis and malignant progression in many tumors and had been implicated in promoting cell growth, invasion, migration, epithelial-mesenchymal transition, metastasis, and apoptosis. H19 also sequesters some microRNAs, facilitating a multilayer molecular regulatory mechanism. In this review, we summarize the abnormal overexpression of H19 in human cancers, which suggests wide prospects for further research into the diagnosis and treatment of cancers.
Keywords: lncRNA, H19, oncogene, cancers, metastasis
Introduction
More than 90% of human genomic DNA is transcribed into RNAs, but less than 2% of these nucleotide sequences encode proteins [1]. Most of the transcribed RNAs are non-coding RNAs (ncRNAs), which lack of the capability to be translated into a protein. These ncRNAs are grouped according to length as long ncRNAs (lncRNAs; >200 nucleotides) and small ncRNAs (<200 nucleotides). LncRNAs are transcribed by RNA polymerase II and classified into enhancer lncRNAs, antisense lncRNAs, bidirectional lncRNAs, large intergenic ncRNAs, and intronic transcript lncRNAs [2-3]. LncRNAs modulate gene expression at three levels: post-transcriptional, transcriptional, and epigenetic. Accumulating evidence has demonstrated that lncRNAs participate in many physiological and pathological processes, including apoptosis, cell proliferation, invasion, and carcinogenesis [4-5]. Moreover, some lncRNAs have been identified to encode proteins.
LncRNA H19 (H19) was one of the first discovered lncRNAs and is encoded by the H19 gene [6]. The H19 gene is located in the region of chromosome 11p15.5 and transcribed by RNA polymerase II, spliced, and polyadenylated [7]. The 2.3 kb lncRNA is then exported from the nucleus to the cytoplasm. H19 is usually expressed in fetal tissues, and its expression is greatly reduced after birth. Recently, H19 was found to take part in a variety of pathological processes, such as inflammatory reactions, angiogenesis, neurogenesis, and fibrosis progression. Additionally, abnormal H19 overexpression is thought to be involved in the development and progression of cancer in many systems of human body, such as the digestive system, the respiratory system, the breast, the genitourinary system, the nervous system, and others.
The aim of this manuscript was to summarize recent findings on H19 expression in cancer and to clarify the impact of imprinting on cancer.
H19 in various human cancers
H19 has been found to be ectopically expressed in many tumors, where it facilitates several oncogenic behaviors, such as increased cell viability, motility, growth, migration, invasion, metastasis, epithelial-mesenchymal transition (EMT), autophagy, cell cycle progression, colony formation, and glucose metabolism [8-11]. H19 also exhibits anti-oncogenic properties in a small percentage of tumors, such as pituitary adenomas [12-14]. Additionally, H19 performs different roles in different clinical stages of the same disease, such as thyroid carcinoma [15, 16], and retinoblastoma [17, 18]. These findings imply that H19 expression might be different depending on the histological and cellular context of individual tumors.
Recently, several studies have demonstrated that H19 is involved in the clinicopathological progression of many different tumor types and is associated with clinical parameters such as tumor size, clinical stage, lymph node metastasis, distant metastasis, and overall survival (OS) [19, 20]. H19 has also been found to take part in the microRNA (miRNA)-mediated network of gene regulation by influencing the activity of the downstream mRNAs that facilitate the aggressive phenotypes of these tumors. Furthermore, H19 plays a vital role in the chemotherapeutic resistance of some tumors and can be used as a potential therapeutic target [21, 22]. The specific mechanisms and functional characterizations of H19 in tumors of each human system are shown in Tables 1-8. The mechanisms of H19 in various tumors will be further clarified below.
Functional characterization of H19 in digestive system tumors.
| Tumor types | Expression | Role | Function role | miRNAs | Related genes | References |
|---|---|---|---|---|---|---|
| Esophageal cancer | upregulation | oncogene | proliferation and metastasis | let-7c | STAT3/EZH2/Catenin | [28] |
| Esophageal cancer | upregulation | oncogene | proliferation, migration, and stemness | miR-22-3p | WNT1 | [29] |
| Gastric cancer | upregulation | oncogene | proliferation, migration, invasion, and metastasis | miR-675 | CALN1 | [20] |
| Gastric cancer | upregulation | oncogene | proliferation | miR-675 | RUNX1 | [37] |
| Gastric cancer | upregulation | oncogene | proliferation and invasion | miR-675 | RUNX1 | [38] |
| Gastric cancer | upregulation | oncogene | proliferation and apoptosis | miR-675 | FADD | [39] |
| Gastric cancer | upregulation | oncogene | proliferation and invasion | miR-141 | ZEB1 | [40] |
| Gastric cancer | upregulation | oncogene | / | let-7c | HER2 | [41] |
| Gastric cancer | upregulation | oncogene | proliferation, invasion, migration, and EMT | miR-22-3p | Snail1 | [42] |
| Colorectal cancer | upregulation | oncogene | proliferation | miR-675 | RB | [44] |
| Colorectal cancer | upregulation | oncogene | EMT | miR-138/200a | ZEB1/ZEB2 | [45] |
| Colorectal cancer | upregulation | oncogene | migration and invasion | / | RAS-MAPK | [48] |
| Colorectal cancer | upregulation | oncogene | migration, invasion, and EMT | / | hnRNPA2B1/Raf-1/ERK | [50] |
| Colorectal cancer | upregulation | oncogene | proliferation | miR-200a | β-Catenin | [51] |
| Colorectal cancer | upregulation | oncogene | motility, EMT, invasion, and migration | miR-29b-3p | PGRN | [52] |
| Colorectal cancer | upregulation | oncogene | invasion, migration, and EMT | miR-194-5p | FoxM1 | [53] |
| Colorectal cancer | upregulation | oncogene | EMT and metastasis | miR-22-3p | MMP14 | [54] |
| Colorectal cancer | upregulation | oncogene | migration and invasion | miR-138 | HMGA1 | [55] |
| Colorectal cancer | upregulation | oncogene | apoptosis | miR-141 | β-Catenin | [56] |
| Colorectal cancer | upregulation | oncogene | proliferation and migration | miR-675-5p | VDR | [57] |
| Colorectal cancer | upregulation | oncogene | autophagy | miR-194-5p | SIRT1 | [11] |
| Hepatocellular cancer | upregulation | oncogene | proliferation, migration, invasion, and EMT | miR-22 | / | [60] |
| Hepatocellular cancer | upregulation | oncogene | proliferation, migration, and invasion | miR-326 | TWIST1 | [61] |
| Hepatocellular cancer | upregulation | oncogene | proliferation, migration, invasion, and apoptosis | miR-15b | CDC42/PAK1 | [62] |
| Hepatocellular cancer | upregulation | oncogene | migration and invasion | miR-193b | MAPK1 | [63] |
| Hepatocellular cancer | upregulation | oncogene | proliferation, metastasis, and apoptosis | miR-520a-3p | LIMK1 | [64] |
| Gallbladder cancer | upregulation | oncogene | proliferation and invasion | miR-194-5p | AKT2 | [69] |
| Gallbladder cancer | upregulation | oncogene | invasion and proliferation | miR-342-3p | FOXM1 | [71] |
| Cholangiocarcinoma | upregulation | oncogene | migration and invasion | let-7a/b | IL-6 | [72] |
| Cholangiocarcinoma | upregulation | oncogene | migration and invasion | miR-372/373 | CXCR4 | [73] |
| Cholangiocarcinoma | upregulation | oncogene | proliferation, migration, and invasion | miR-612 | BCL-2 | [74] |
| Pancreatic cancer | upregulation | oncogene | migration, invasion, and EMT | let-7 | HMGA2 | [76] |
| Pancreatic cancer | upregulation | oncogene | proliferation and apoptosis | / | E2F1 | [77] |
| Pancreatic cancer | upregulation | oncogene | proliferation | miR-675 | E2F1 | [78] |
| Pancreatic cancer | upregulation | oncogene | proliferation and invasion | miR-194 | PFTK1 | [81] |
| Pancreatic cancer | upregulation | oncogene | migration, invasion, and EMT | miR-675-3p | SOCS5/STAT3 | [82] |
| Pancreatic cancer | upregulation | oncogene | proliferation, migration, and invasion | / | VGF/PI3K/AKT/CREB | [83] |
Main characteristics of the studies included in the review of digestive system tumors
| Study | Tumor types | Sample size (Normal: Tumor) | Detection Method | P value (p value) | TNM (p value) | LNM (p value) | DM (p value) | OS (p value) | DFS (p value) | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Huang | Esophageal cancer | (133 : 133) | qRT-PCR | p<0.05 | p=0.001 | / | p=0.000 | / | / | [26] |
| Tan | Esophageal cancer | (64 : 64) | qRT-PCR | p<0.01 | p=0.01 | p=0.007 | / | / | / | [27] |
| Chen | Esophageal cancer | (30 : 30) | qRT-PCR | p<0.05 | / | / | / | / | / | [28] |
| Zhang | Gastric cancer | (80 : 80) | qRT-PCR | p<0.001 | p=0.016 | p=0.002 | / | p=0.007 | / | [31] |
| Zhou | Gastric cancer | (70 : 70) | qRT-PCR | p<0.0001 | / | / | / | / | / | [32] |
| Chen | Gastric cancer | (128 : 128) | qRT-PCR | p<0.01 | p=0.005 | / | p=0.041 | p<0.001 | / | [33] |
| Hashad | Gastric cancer | (32 : 30) | qRT-PCR | p<0.001 | p=0.014 | / | / | / | / | [34] |
| Jia | Gastric cancer | (284 : 284) | qRT-PCR | p<0.05 | p=0.026 | p<0.001 | / | p=0.001 | / | [21] |
| Li | Gastric cancer | (74 : 74) | qRT-PCR | p=0.017 | / | p=0.027 | p=0.001 | p=0.036 | / | [20] |
| Zhou | Gastric cancer | (15 : 15) | qRT-PCR | p<0.05 | / | / | / | / | / | [40] |
| Wei | Gastric cancer | (24 : 24) | qRT-PCR | p<0.001 | / | p=0.015 | / | / | / | [41] |
| Gan | Gastric cancer | (40 : 40) | qRT-PCR | p<0.05 | p<0.05 | p<0.05 | / | / | / | [42] |
| Tsang | Colorectal cancer | (40 : 40) | qRT-PCR | p=0.001 | / | / | / | / | / | [44] |
| Liang | Colorectal cancer | (30 : 30) | qRT-PCR | p<0.05 | / | / | / | / | / | [45] |
| Han | Colorectal cancer | (83 : 83) | qRT-PCR | p<0.01 | p=0.008 | / | / | p=0.002 | p=0.029 | [46] |
| Chen | Colorectal cancer | (96 : 96) | qRT-PCR | p<0.01 | p=0.046 | / | / | / | p<0.01 | [47] |
| Zhang | Colorectal cancer | (60 : 60) | qRT-PCR | p<0.001 | / | p=0.010 | p=0.042 | p=0.013 | / | [50] |
| Yang | Colorectal cancer | (30 : 30) | qRT-PCR | p<0.01 | / | / | / | / | / | [51] |
| Ding | Colorectal cancer | (185 : 185) | qRT-PCR | p<0.05 | p=0.033 | p=0.002 | / | p<0.001 | / | [53] |
| Li | Colorectal cancer | (214 : 214) | qRT-PCR | p<0.05 | p=0.011 | p<0.001 | / | p<0.001 | / | [54] |
| Ren | Colorectal cancer | (10 : 10) | qRT-PCR | p=0.0169 | / | / | / | / | / | [56] |
| Li | Hepatocellular cancer | (36 : 36) | qRT-PCR | p<0.01 | p=0.044 | p=0.018 | p=0.007 | / | / | [60] |
| Zhou | Hepatocellular cancer | (46 : 46) | qRT-PCR | p<0.01 | / | / | / | / | / | [62] |
| Ding | Hepatocellular cancer | (42 : 42) | qRT-PCR | p<0.01 | / | / | / | / | / | [65] |
| Wang | Gallbladder cancer | (20 : 20) | qRT-PCR | p<0.05 | / | / | / | / | / | [69] |
| Wang | Gallbladder cancer | (24 : 24) | qRT-PCR | p<0.05 | / | p=0.017 | / | / | / | [70] |
| Wang | Gallbladder cancer | (36 : 36) | qRT-PCR | p<0.05 | / | / | / | / | / | [71] |
| Xu | Cholangiocarcinoma | (56 : 56) | qRT-PCR | p<0.001 | p=0.0145 | / | / | p=0.0007 | / | [73] |
| Ma | Pancreatic cancer | (20 : 20) | qRT-PCR | p<0.05 | / | / | / | / | / | [76] |
| Ma | Pancreatic cancer | (30 : 30) | qRT-PCR | p=0.007 | / | / | / | / | / | [77] |
| Sun | Pancreatic cancer | (45 : 45) | qRT-PCR | p<0.01 | p<0.001 | / | p<0.001 | p=0.024 | / | [81] |
| Ji | Pancreatic cancer | (39 : 39) | qRT-PCR | p<0.01 | p<0.001 | p=0.044 | p=0.001 | / | p<0.001 | [83] |
Functional characterization of H19 in respiratory, genitourinary, and nervous system tumors.
| Tumor types | Expression | Role | Function role | miRNAs | Related genes | References |
|---|---|---|---|---|---|---|
| Nasopharyngeal cancer | upregulation | oncogene | invasion | miR-630 | EZH2 | [87] |
| Nasopharyngeal cancer | upregulation | oncogene | proliferation, migration, and invasion | let-7 family | HRAS | [88 ] |
| Laryngeal cancer | upregulation | oncogene | proliferation, migration, and invasion | miR-148a-3p | DNMT1 | [90] |
| Lung cancer | upregulation | oncogene | cell cycle | miR-107 | / | [91] |
| Lung cancer | upregulation | oncogene | migration, invasion, and EMT | / | CDH1 | [96] |
| Lung cancer | upregulation | oncogene | proliferation and migration | miR-107 | NF1 | [97] |
| Lung cancer | upregulation | oncogene | proliferation | miR-138 | PDK1 | [98] |
| Lung cancer | upregulation | oncogene | viability, proliferation, and apoptosis | miR-29b-3p | STAT3 | [99] |
| Lung cancer | upregulation | oncogene | proliferation, migration, and invasion | miR-200a | ZEB1/ZEB2 | [100] |
| Lung cancer | upregulation | oncogene | proliferation, migration, and invasion | miR-615-3p | ATG7 | [101] |
| Lung cancer | upregulation | oncogene | proliferation, migration, and invasion | miR-148b-3p | DDAH1 | [102] |
| Endometrial cancer | upregulation | oncogene | viability | miR-612 | HOXA10 | [126] |
| Endometrial cancer | upregulation | oncogene | proliferation, migration, EMT, and apoptosis | miR-20b-5p | AXL/HIF-1α | [127] |
| Ovarian cancer | upregulation | oncogene | EMT | miR-370-3p | TGF-β | [129] |
| Ovarian cancer | upregulation | oncogene | Warburg effect | miR-324-5p | PKM2 | [130] |
| Renal cancer | upregulation | oncogene | migration and invasion | miR-29a-3p | E2F1 | [133] |
| Bladder cancer | upregulation | oncogene | migration | / | EZH2 | [134] |
| Bladder cancer | upregulation | oncogene | proliferation | / | ID2 | [136] |
| Bladder cancer | upregulation | oncogene | proliferation, migration, invasion, mobility, and EMT | miR-29b-3p | DNMT3B | [138] |
| Seminoma | upregulation | oncogene | chemotherapeutic sensitivity | miR-106b-5p | TDRG1 | [140] |
| Glioma | upregulation | oncogene | viability, migration, and invasion | / | NKD1 | [144] |
| Glioma | upregulation | oncogene | proliferation, migration, invasion, cell cycle, and apoptosis | / | Wnt/β-Catenin | [145] |
| Glioma | upregulation | oncogene | invasion | miR-675 | Cadherin | [146] |
| Glioma | upregulation | oncogene | proliferation | miR-675 | / | [147] |
| Glioma | upregulation | oncogene | proliferation and invasion | miR-152 | / | [148] |
| Glioma | upregulation | oncogene | proliferation, migration, and tube formation | miR-29a | VASH2 | [149] |
| Glioma | upregulation | oncogene | proliferation | miR-140 | iASPP | [150] |
| Glioma | upregulation | oncogene | migration and invasion | miR-181d | β-catenin | [151] |
| Glioma | upregulation | oncogene | migration, invasion, and EMT | miR-130a-3p | SOX4 | [152] |
| Glioma | upregulation | oncogene | proliferation, migration, invasion, and angiogenesis | miR-138 | HIF-1α | [153] |
| Glioma | upregulation | oncogene | proliferation, migration, and angiogenesis | miR-342 | Wnt5a/β-catenin | [154] |
Main characteristics of the studies included in the review of respiratory, genitourinary, and nervous system tumors.
| Study | Tumor types | Sample size (Normal: Tumor) | Detection Method | P value (p value) | TNM (p value) | LNM (p value) | DM (p value) | OS (p value) | DFS (p value) | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Li | Nasopharyngeal cancer | (30 : 31) | qRT-PCR | p<0.05 | / | / | / | / | / | [87] |
| Zhang | Nasopharyngeal cancer | (17 : 48) | qRT-PCR | p<0.01 | / | / | / | p=0.0195 | / | [88] |
| Wu | Laryngeal cancer | (82 : 82) | qRT-PCR | p<0.01 | p<0.01 | p<0.01 | / | p=0.003 | / | [90] |
| Cui | Lung cancer | (30 : 30) | qRT-PCR | p<0.01 | / | / | / | / | / | [91] |
| Zhang | Lung cancer | (70 : 70) | qRT-PCR | p<0.001 | p<0.001 | / | / | / | / | [92] |
| Xu | Lung cancer | (48 : 48) | qRT-PCR | p<0.001 | / | / | / | p=0.0125 | / | [95] |
| Gao | Lung cancer | (60 : 60) | qRT-PCR | p<0.05 | / | / | / | / | / | [96] |
| Qian | Lung cancer | (36 : 36) | qRT-PC | p<0.05 | / | / | / | / | / | [97] |
| Huang | Lung cancer | (20 : 20) | qRT-PCR | p<0.001 | / | / | / | / | / | [98] |
| Liu | Lung cancer | (305 : 305) | qRT-PCR | p<0.05 | p=0.026 | / | / | p=0.001 | / | [99] |
| Zhao | Lung cancer | (22 : 22) | qRT-PCR | p<0.001 | / | / | / | / | / | [100] |
| Zhang | Endometrial cancer | (43 : 43) | qRT-PCR | p<0.001 | / | / | / | p=0.0489 | / | [126] |
| Zhu | Endometrial cancer | (36 : 36) | qRT-PCR | p<0.001 | / | / | / | / | / | [127] |
| Zhu | Ovarian cancer | (70 : 70) | qRT-PCR | p<0.01 | / | / | / | / | / | [128] |
| Wang | Renal cancer | (92 : 92) | qRT-PCR | p<0.05 | p=0.023 | p=0.013 | p=0.002 | p<0.05 | / | [132] |
| He | Renal cancer | (30 : 30) | qRT-PCR | p<0.01 | / | / | / | / | / | [133] |
| Luo | Bladder cancer | (41 : 41) | qRT-PCR | p=0.0034 | / | / | / | / | / | [134] |
| Zhu | Bladder cancer | (48 : 48) | qRT-PCR | p<0.01 | / | p<0.01 | p<0.01 | / | / | [135] |
| Luo | Bladder cancer | (24 : 24) | qRT-PCR | p=0.0015 | / | / | / | / | / | [136] |
| Li | Bladder cancer | (19 : 40) | qRT-PCR | p<0.05 | / | / | / | / | / | [137] |
| Lv | Bladder cancer | (35 : 35) | qRT-PCR | p<0.001 | p=0.0154 | p=0.0456 | / | / | / | [138] |
| Wang | Bladder cancer | (52 : 52) | qRT-PCR | p<0.001 | p=0.009 | / | / | p=0.002 | / | [139] |
| Jiang | Glioma | (30 : 30) | qRT-PCR | p<0.0001 | / | / | / | / | / | [143] |
| Guan | Glioma | (60 : 60) | qRT-PCR | p<0.05 | / | / | / | p<0.05 | / | [145] |
| Zhang | Glioma | (35 : 35) | qRT-PCR | p<0.001 | / | / | / | p<0.005 | / | [147] |
| Zhao | Glioma | (28 : 28) | qRT-PCR | p<0.01 | / | / | / | / | / | [150] |
| Wu | Glioma | (15 : 22) | qRT-PCR | p=0.0003 | / | / | / | / | / | [151] |
| Zhou | Glioma | (30 : 30) | qRT-PCR | p<0.001 | / | / | / | / | / | [154] |
Functional characterization of H19 in breast cancer.
| Tumor types | Expression | Role | Function role | miRNAs | Related genes | References |
|---|---|---|---|---|---|---|
| Breast cancer | upregulation | oncogene | proliferation, migration, invasion, and cell cycle | / | TNFAIP8 | [114] |
| Breast cancer | upregulation | oncogene | clonogenicity, migration, and mammosphere-forming ability | let-7 | LIN28 | [116] |
| Breast cancer | upregulation | oncogene | autophagy and EMT | let-7 | LIN28 | [117] |
| Breast cancer | upregulation | oncogene | EMT | miR-200b/c and let 7b | GIT2 and CYTH3 | [118] |
| Breast cancer | upregulation | oncogene | proliferation and invasion | miR-152 | DNMT1 | [119] |
| Breast cancer | upregulation | oncogene | viability | miR-675-5p | CBL | [120] |
| Breast cancer | upregulation | oncogene | migration and invasion | miR-93-5p | STAT3 | [121] |
| Breast cancer | upregulation | oncogene | proliferation, migration, invasion, cell cycle, EMT, and apoptosis | miR-138 | SOX4 | [122] |
| Breast cancer | upregulation | oncogene | proliferation, metastasis, invasion, EMT, and apoptosis | miR-340-3p | YWHAZ | [123] |
| Breast cancer | upregulation | oncogene | proliferation, migration, invasion, and apoptosis | miR-491-5p | ZNF703 | [124] |
| Breast cancer | upregulation | oncogene | proliferation, migration, invasion, and apoptosis | miR-130a-3p | SATB1 | [125] |
Main characteristics of the studies included in the review of breast cancer.
| Study | Tumor types | Sample size (Normal: Tumor) | Detection Method | P value (p value) | TNM (p value) | LNM (p value) | DM (p value) | OS (p value) | DFS (p value) | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Zhang | Breast cancer | (24 : 24) | qRT-PCR | p=0.018 | / | / | / | / | / | [105] |
| Li | Breast cancer | (60 : 60) | qRT-PCR | p<0.001 | / | / | / | / | / | [114] |
| Wang | Breast cancer | (69 : 69) | qRT-PCR | p<0.05 | / | / | / | / | / | [115] |
| Peng | Breast cancer | (20 : 20) | qRT-PCR | p<0.01 | / | / | / | p=0.004 | / | [116] |
| Zhou | Breast cancer | (48 : 48) | qRT-PCR | p<0.05 | / | / | / | / | / | [118] |
| Li | Breast cancer | (45 : 45) | qRT-PCR | p<0.05 | / | / | / | / | / | [119] |
| Si | Breast cancer | (40 : 40) | qRT-PCR | p<0.01 | / | / | / | p<0.05 | / | [122] |
| Wang | Breast cancer | (20 : 20) | qRT-PC | p<0.05 | / | / | / | / | / | [124] |
| Zhong | Breast cancer | (50 : 50) | qRT-PCR | p<0.01 | / | / | / | / | / | [125] |
Functional characterization of H19 in other system tumors.
| Tumor types | Expression | Role | Function role | miRNAs | Related genes | References |
|---|---|---|---|---|---|---|
| Lymphoma | upregulation | oncogene | proliferation | / | AKT | [158] |
| Myeloma | upregulation | oncogene | proliferation and apoptosis | miR-152-3p | BRD4 | [161] |
| Myeloma | upregulation | oncogene | proliferation, migration, viability, cell cycle, and apoptosis | miR-29b-3p | MCL-1 | [8] |
| Myeloma | upregulation | oncogene | chemotherapeutic sensitivity | / | AKT | [162] |
| Melanoma | upregulation | oncogene | glucose metabolism and growth | miR-106a-5p | E2F3 | [9] |
| Melanoma | upregulation | oncogene | proliferation, migration, and invasion | / | NF‑κB | [165] |
| Melanoma | upregulation | oncogene | chemotherapeutic sensitivity | miR-18b | IGF | [22] |
| Leukemia | upregulation | oncogene | proliferation | miR-19a/b | ID2 | [167] |
| Leukemia | upregulation | oncogene | proliferation and apoptosis | / | ID2 | [168] |
| Leukemia | upregulation | oncogene | proliferation and apoptosis | miR-29a-3p | Wnt/β-Catenin | [169] |
| Osteosarcoma | / | oncogene | migration and invasion | miR-200s | ZEB1/ZEB2 | [171] |
| Osteosarcoma | upregulation | oncogene | migration and invasion | / | NF‑κB | [172] |
Main characteristics of the studies included in the review of other system tumors.
| Study | Tumor types | Sample size (Normal: Tumor) | Detection Method | P value (p value) | TNM (p value) | LNM (p value) | DM (p value) | OS (p value) | DFS (p value) | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Wang | Lymphoma | (60 : 40) | qRT-PCR | p<0.001 | / | / | / | p=0.0269 | / | [158] |
| Pan | Myeloma | (80 : 67) | qRT-PCR | p<0.0001 | / | / | / | / | / | [159] |
| Zheng | Myeloma | (30 : 30) | qRT-PCR | p<0.01 | / | / | / | / | / | [161] |
| Wang | Myeloma | (50 : 60) | qRT-PCR | p<0.0001 | / | / | / | / | / | [162] |
| Luan | Melanoma | (30 : 30) | qRT-PCR | p<0.0001 | p<0.001 | / | / | p<0.05 | / | [9] |
| Liao | Melanoma | (49 : 49) | qRT-PCR | p<0.05 | / | / | / | / | / | [165] |
| Shi | Melanoma | (82 : 82) | qRT-PCR | p<0.01 | p=0.0076 | p=0.0481 | p=0.0153 | p=0.021 | / | [166] |
| An | Melanoma | (30 : 30) | qRT-PC | p<0.01 | / | / | / | p=0.012 | / | [20] |
| Zhao | Leukemia | (53 : 46) | qRT-PCR | p<0.001 | / | / | / | / | / | [167] |
| Zhang | Leukemia | (36 : 161) | qRT-PCR | p=0.003 | / | / | / | p=0.02 | / | [168] |
| Zhao | Leukemia | (40 : 40) | qRT-PCR | p<0.01 | / | / | / | / | / | [169] |
| Liao | Osteosarcoma | (40 : 40) | qRT-PCR | p<0.05 | / | / | p=0.01 | p=0.00322 | / | [172] |
The role of H19 in digestive system tumors
H19 in esophageal cancer (EC)
EC is a relatively rare cancer of the digestive system. Esophageal squamous cell carcinoma (ESCC) is the main pathological subtype, accounting for 90% of the global incidence of EC [23]. The 5-year OS rate of early-stage ESCC is >90%, but <10% for patients with lymph node metastasis [24]. However, many patients miss the opportunity for early detection because of atypical symptoms. Therefore, it is necessary to explore early diagnostic biomarkers in ESCC.
Emerging evidence suggests that H19 is highly expressed in EC and plays an important role in EC development [25]. The study by Huang et al. [26] reported that H19 expression was associated with tumor metastasis and depth in EC samples. Increased H19 expression promoted EMT, growth, and invasion of EC cells. Conversely, decreasing H19 expression inhibited the growth, migration, and invasion of ESCC cells, suggesting H19 could be a prognostic marker and therapeutic target for ESCC patients [27]. Two years later, another group confirmed that downregulating H19 inhibited the growth, migration, invasion, metastasis, and EMT of EC cells by modulating the let-7c/STAT3/EZH2/β-catenin pathway (Fig. 1A) [28]. The study by Li et al. [19] investigated the clinicopathological parameters of H19 in ESCC patients, revealing that increased H19 expression was associated with larger tumor size, poor clinical stage, and shorter OS.
Data also suggest that H19 plays a role in the efficacy of radiotherapy in ESCC patients. A team from Shandong University revealed that knocking-down H19 decreased WNT1 expression, suppressing radioresistance of ESCC cells with regards to growth and migration by upregulating miR-22-3p (Fig. 1B) [29]. These data showed that H19 could be a prognostic marker and/or therapeutic target for EC.
H19 in gastric cancer (GC)
GC is an important malignant tumor of the digestive system, with China having one of the highest rates of GC in the world. Therefore, further studies of the molecular mechanisms involved in GC are urgently needed.
Nine years ago, it was first demonstrated that GC tissues showed an upregulation of H19 [30]. Overexpression of H19 was then found to promote cell growth via regulating p53 in GC. Subsequently, several groups found that H19 accelerated GC progression, demonstrating that H19 could serve as a biomarker for the early diagnosis and prognosis of GC [31-35]. Zhang et al. [36] revealed that H19 promoted cell proliferation by increasing NF-κB-involved inflammation in GC. Subsequent studies showed that upregulating H19 improved the sensitivity of GC cells to X-rays and chemotherapy, leading to greater tumor weights and larger tumor sizes [21].
H19 mediates mechanisms involved in digestive system tumors. (A). H19 could promote the expression of STAT3/EZH2/Catenin by targeting let-7c. (B). H19 could promote the expression of WNT1 by targeting miR-22-3p. (C1-C3). H19 could promote the expression of CALN1 (C1), RUNX1 (C2), and FADD (C3) by targeting miR-675. (D). H19 could promote the expression of ZEB1 by targeting miR-141. (E). H19 could promote the expression of HER2 by targeting let-7c. (F). H19 could promote the expression of Snail1 by targeting miR-22-3p. (G1-G2). H19 could promote the expression of RB (G1) and VDR (G2) by targeting miR-675. (H). H19 could promote the expression of ZEB1/ZEB2 by targeting miR-138/200a. (I). H19 could promote the expression of β-Catenin by targeting miR-200a. (J). H19 could promote the expression of PGRN by targeting miR-29b-3p. (K1-K2). H19 could promote the expression of FoxM1 (K1) and SIRT1 (K2) by targeting miR-194-5p. (L). H19 could promote the expression of MMP14 by targeting miR-22-3p. (M). H19 could promote the expression of HMGA1 by targeting miR-138. (N). H19 could promote the expression of β-Catenin by targeting miR-141. (O). H19 could promote the expression of TWIST1 by targeting miR-326. (P). H19 could promote the expression of CDC42/PAK1 by targeting miR-15b. (Q). H19 could promote the expression of MAPK1 by targeting miR-193b. (R). H19 could promote the expression of LIMK1 by targeting miR-520a-3p. (S). H19 could promote the expression of AKT2 by targeting miR-194-5p. (T). H19 could promote the expression of FOXM1 by targeting miR-342-3p. (U). H19 could promote the expression of CXCR4 by targeting miR-372/373. (V). H19 could promote the expression of IL-6 by targeting let-7a/b. (W). H19 could promote the expression of BCL-2 by targeting miR-612. (X). H19 could promote the expression of HMGA2 by targeting let-7. (Y1-Y2). H19 could promote the expression of E2F-1 (Y1) and SOCS5/STAT3 (Y2) by targeting miR-675. (Z). H19 could promote the expression of PFTK1 by targeting miR-194.
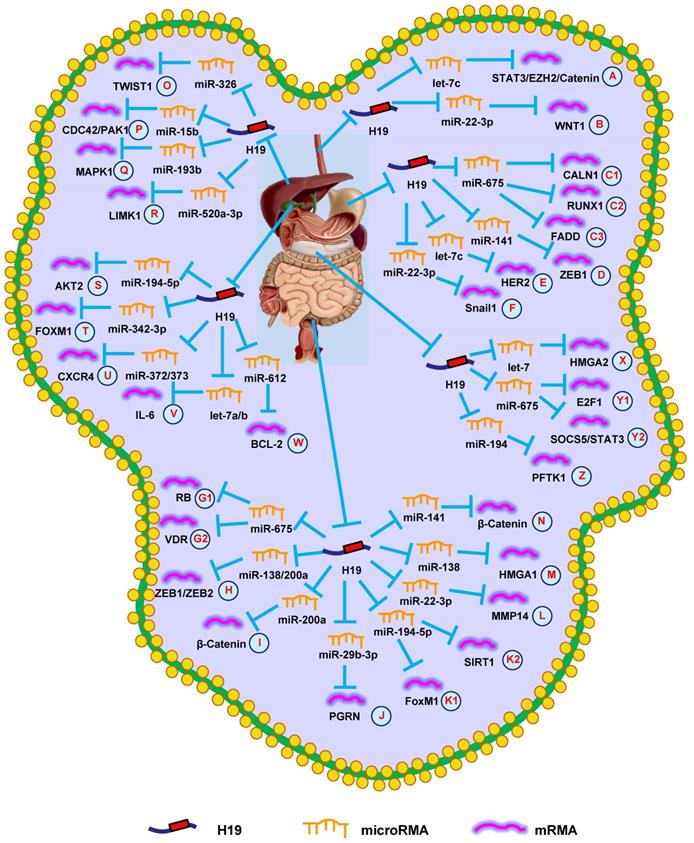
Recently, several research groups have provided novel insights into the active lncRNA-miRNA-mRNA network in GC. Li et al. [20] demonstrated that the effects of H19 were partially through directly upregulating ISM1 and indirectly downregulating CALN1 via miR-675 (Fig. 1C1). Similar results indicated that H19 could modulate GC progression through the miR-675/RUNX1 pathway (Fig. 1C2), which revealed a potential target for GC therapy [37]. Just a month later, another research group came to the same conclusion [38]. Additionally, the H19/miR-675 axis also participates in the development of GC through FADD/caspase 8 signaling (Fig. 1C3) [39]. Besides miR-675, H19 has also be shown to regulate ZEB1 expression via sequestering miR-141, suggesting an important role of the lncRNA-miRNA functional network in GC (Fig. 1D) [40]. The study by Wei et al. [41] indicated that H19 functioned as a competing endogenous RNA (ceRNA) to modulate HER2 expression by antagonizing let-7c in GC, providing other potential H19-based therapeutic strategies for GC (Fig. 1E). Subsequent studies uncovered that H19 controlled cell proliferation and metastasis through the miR-22-3p/Snail1 axis in GC (Fig. 1F) [42]. These findings showed that H19 could provide a diagnostic option for GC.
H19 in colorectal cancer (CRC)
CRC is the most common tumor of the digestive system and has the third highest morbidity and mortality rate of all malignancies in the United States [43]. Despite advances in early diagnosis over the last few years, the OS of CRC patients with metastases remains low. Therefore, a complete understanding of the pathogenic mechanisms of CRC represents key progress toward CRC treatments.
The study by Tsang et al. [44] revealed that H19 and its derivative, miR-675, promoted CRC cell cycle progression by targeting RB (Fig. 1G1). Other researchers reported that H19 accelerated cell growth and EMT by regulating ZEB1, ZEB2, and vimentin via functioning as a ceRNA of miR-200a and miR-138 (Fig. 1H) [45]. Additionally, H19 could bind eIF4A3 to promote cell growth and influence tumor differentiation and TNM stage in CRC patients [46]. H19 also promoted cell growth, migration, and EMT in CRC [47]. Subsequent studies found that H19 promoted the proliferation, invasion, and metastasis of CRC by activating important cancer-related signaling pathways, such as RAS/MAPK [48], Rb/E2F, CDK8/β-catenin [49], and Raf/ERK [50].
Recently, several groups have focused on the lncRNA-miRNA-mRNA axis in CRC. Studies on the role of H19 in CRC revealed that H19 repressed β-catenin expression via binding miR-200a to promote cell growth (Fig. 1I) [51]. Additionally, H19 was shown to be involved in EMT of CRC cells via the miR-29b-3p/PGRN/Wnt pathway (Fig. 1J) [52]. Another group revealed that the H19/miR-194-5p/FoxM1 axis influenced EMT and could serve as a therapeutic target in CRC (Fig. 1K1) [53]. Hu et al. [54] found that knocking-down HDAC2 increased H19 expression and induced EMT via the miR-22-3p/MMP14 pathway (Fig. 1L). Subsequent studies revealed that H19 increased cell migration and invasion by modulating the miR-138/HMGA1 axis, providing a novel insight for CRC treatment (Fig. 1M) [55].
H19 has also been proven to affect drug resistance during CRC treatment. The study by Ren et al. [56] indicated that carcinoma-associated fibroblasts increased the chemoresistance and stemness of CRC by transferring exosomal H19. Mechanistic investigations suggested that H19 can activate β-catenin signaling by functioning as a ceRNA for miR-141 (Fig. 1N). Another group indicated that abnormal overexpression of H19 facilitated resistance to 1,25(OH)2D3 treatment via the miR-675-5p/VDR axis (Fig. 1G2) [57]. Additionally, methotrexate (MTX) resistance has impeded its application in CRC therapy. A recent study revealed that knocking-down H19 inhibited MTX resistance and promoted apoptosis via suppressing Wnt/β-catenin signaling in CRC [58]. Subsequent studies revealed that H19 promoted 5-fluorouracil (5-Fu) resistance in CRC cells. Mechanistically, it was demonstrated that H19 led to 5-Fu resistance through miR-194-5p/SIRT1-mediated autophagy in CRC (Fig. 1K2) [11]. These findings confirmed that H19 provided an option for suppressing CRC progression.
H19 in hepatocellular carcinoma (HCC)
Although the incidence of HCC is relatively low, its mortality rate is high. Because HCC frequently metastasizes, patients often have a poor prognosis. Thus, it is essential to discover effective treatments for HCC patients.
The study by Lv et al. [59] reported that aflatoxin B1 promoted E2F1 and H19 expression, which increased the growth and invasion of HCC cells. Another lab revealed that H19 expression was higher in HCC than in normal hepatic tissues, and was positively correlated with lymphatic and distant metastasis. Increasing H19 expressing promoted the progression of HCC cells by targeting miR-22 [60].
Furthermore, it has been demonstrated that H19 promoted HCC progression through miRNA-mRNA pathways. Silencing H19 repressed the proliferation, migration, and invasion of HCC cells. Further investigations into the underlying mechanisms indicated that the H19/miR-326/TWIST1 axis was involved in HCC progression (Fig. 1O) [61]. Additionally, downregulating H19 suppressed growth, migration, and invasion by regulating the miR-15b/CDC42/PAK1 axis (Fig. 1P) [62]. Furthermore, bioinformatics analysis and in vitro experiments showed that H19 served as a miR-193b sponge to protect MAPK1 and promoted aggressive behaviors of HCC (Fig. 1Q) [63]. Similarly, H19 also promoted invasive behaviors through the miR-520a-3p/LIMK1 axis (Fig. 1R) [64].
H19 is also thought to be involved in chemotherapeutic resistance in HCC patients. Ding et al. [65] reported that silencing H19 reduced expression of the chemoresistance gene MDR1 by blocking MAPK/ERK signaling. Xu et al. [66] revealed that knocking-down H19 decreased miR-675 expression, which increased sorafenib sensitivity by inhibiting EMT in HCC cells. Together, these data revealed that H19 functioned as an oncogene in HCC.
H19 in gallbladder and bile duct tumors
Both gallbladder cancer (GBC) and cholangiocarcinoma (CCC) are low incidence tumors of the digestive system. Due to early metastasis, only a limited number of GBC or CCC cases can be resected, and the 5-year OS rate is only 20%-40% [67, 68]. Therefore, understanding the pathogenesis of GBC and CCC is vital to reveal therapeutic targets.
Wang et al. [69] reported that knocking-down H19 decreased GBC cell growth, causing them to arrest in the G0/G1 phase via regulating miR-194-5p/AKT2 signaling (Fig. 1S). Another group discovered that H19 expression was higher in GBC tissues than in normal bladder tissues, and was positively correlated with tumor size and lymphatic metastasis. Increasing H19 expression promoted invasion and EMT by regulating Twist1 expression in GBC cells [70]. Additionally, H19 functioned as a ceRNA of miR-342-3p to increase cell growth and invasion by enhancing FOXM1 expression in GBC cells (Fig. 1T) [71].
According to previous research, H19 promotes CCC cell migration and invasion by targeting CXCR4 and IL-6 via sponging miR-372/miR-373 and let-7a/b, respectively (Fig. 1U and 1V) [72]. Additionally, silencing H19 promoted apoptosis and inhibited growth, migration, and invasion by reversing EMT in CCC cells [73]. HIF1α-mediated H19 overexpression in CCC cells promoted proliferation, migration, and invasion via regulating the miR-612/Bcl-2 axis (Fig. 1W) [74]. Overall, these data confirmed that H19 had an oncogenic activity in GBC and CCC and represented a promising diagnostic target.
H19 in pancreatic cancer
Pancreatic cancer is the least common digestive tumor, but is the fourth leading cause of death from cancer in the United States [43]. Despite significant efforts of researchers to study the pathogenesis of pancreatic cancer, its five-year OS rate remains extremely low [75]. Therefore, understanding the pathogenesis of pancreatic cancer is crucial for the development of successful treatments.
A recent study revealed that H19 expression was increased in pancreatic cancer compared with normal pancreatic tissue. Silencing H19 in pancreatic cancer cells led to decrease HMGA2 expression and blocked cell migration and invasion by regulating let-7 (Fig. 1X) [76]. Another study showed that knocking-down H19 impaired the viability and growth of pancreatic cancer cells by decreasing E2F1 expression [77]. Two years later, the same group demonstrated that H19 regulated E2F1 expression by sponging miR-675, which served as an underlying biomarker for diagnosing pancreatic cancer (Fig. 1Y1) [78]. In situ hybridization rates of H19 also suggested it had an important role in pancreatic cancer metastasis, which implied that suppressing H19 could be a novel treatment for pancreatic cancer [79]. Recently, it was reported that H19 promoted pancreatic cancer cell invasion and metastasis via increasing cell adhesion and cancer stem cell self-renewal by regulating CD24 and integrin expression [80]. Another group found that knocking-down H19 inhibited the growth and migration of pancreatic cancer cells via altering the miR-194/PFTK1 signaling (Fig. 1Z) [81]. Studies to better understand these related molecular mechanisms showed that increased H19 expression promoted chemoresistance, EMT, migration, and invasion through the miR-675-3p/SOCS5 axis (Fig. 1Y2) [82]. Moreover, H19 increased the expression of VGF to activate the PI3K/AKT/CREB signaling pathway and promote aggressive phenotypes of pancreatic cancer [83]. In summary, H19 played a vital role in the prognosis of pancreatic cancer.
The role of H19 in respiratory system tumors
H19 in nasopharyngeal cancer (NPC)
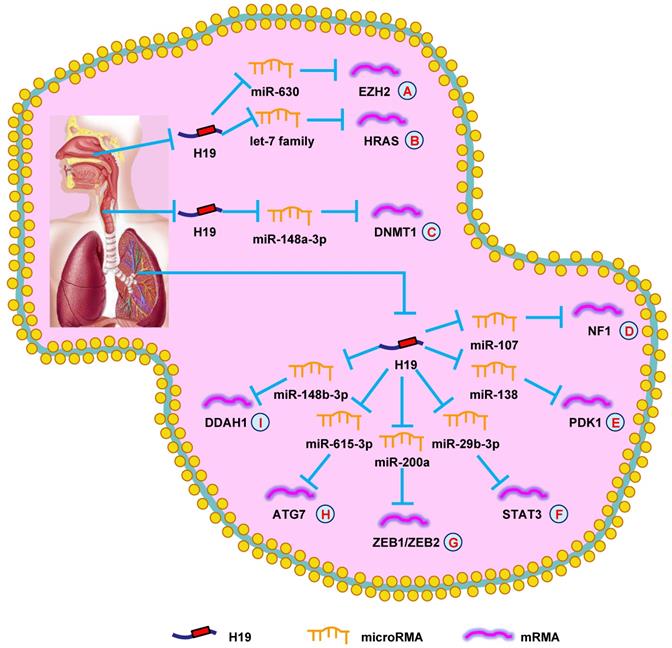
NPC is a malignant tumor of the respiratory system, and most NPC patients are diagnosed in advanced stages [84]. Because of its high sensitivity, radiotherapy is the primary treatment for NPC, but NPC often relapses after treatment [85]. Therefore, discovering novel biomarkers and therapeutic strategies could be pivotal for NPC.
In 2003, it was found that H19 was highly expressed in undifferentiated human NPC cell lines, but not in well-differentiated NPC cells. Additionally, it was demonstrated that hypomethylation of the CpG site in the H19 promoter region induced abnormal H19 expression in the well-differentiated NPC cells. Thus, hypermethylation of the H19 promoter region could be a significant epigenetic marker that played a vital role in the differentiation of NPC cells and the transcriptional silencing of imprinted genes [86]. Li et al. [87] found that H19 suppressed E-cadherin expression and promoted NPC cell invasion by regulating the miR-630/EZH2 pathway, which suggested a possible therapy for NPC (Fig. 2A). In another study, increased H19 expression was associated with poorer prognosis. Mechanistically, H19 showed oncogenic activity through the let-7/HRAS pathway and promoted NPC oncogenesis and metastasis (Fig. 2B) [88]. Additionally, upregulating H19 promoted the growth of NPC cells and decreased the chemosensitivity. Silencing H19 could be an effective method to suppress tumor growth [89]. Together, these data supported the conclusion that H19 functioned as an oncogene and promoted NPC progression.
H19 in laryngeal cancer
Laryngeal cancer is a common tumor of the respiratory system, among which laryngeal squamous cell carcinoma (LSCC) is the main subtype. In 2021, an estimated 12,620 new cases will be diagnosed, and approximately 3,770 patients will die from this LSCC [43]. Therefore, it is urgent to find new diagnostic biomarkers and novel therapies for LSCC.
Wu et al. [90] revealed that H19 expression was increased in LSCC. Silencing H19 in LSCC suppressed growth, migration, and invasion. H19 performed its biological activity in LSCC by targeting the miR-148a-3p/DNMT1 pathway (Fig. 2C). In summary, the data suggested that H19 played an important role in LSCC development and could be a therapeutic target.
H19 in lung cancer
Lung cancer is the malignancy with the highest mortality in the world. In 2021, approximately 235,760 patients will be diagnosed with lung and bronchial cancer in the United States, resulting in approximately 131,880 deaths [43]. The pathogenesis of lung cancer is not well understood, although great progress has been achieved in recent decades. Therefore, sufficient research into lung cancer will help us defeat it.
Since 2015, many scientists have focused their attention on the role of H19 in lung cancer. Cui et al. [91] discovered that H19 expression was higher in non-small cell lung cancer (NSCLC) compared with normal lung tissues. H19 expression was induced by c-Myc and promoted mitotic progression via regulating miR-107 in NSCLC cells. A year later, another group revealed a similar phenomenon, in that knocking-down H19 suppressed the growth of NSCLC cells by regulating c-Myc transcription. H19 could be a novel therapeutic target and diagnostic marker for NSCLC [92]. Additionally, the relationship between H19 and chemotherapeutic resistance was revealed in lung adenocarcinoma for the first time in 2017 [93]. Increased H19 expression was negatively correlated with cisplatin response in lung adenocarcinoma patients, which was associated with increased cell growth and metastasis and a cell-cycle arrest. Another study of NSCLC indicated that H19 functioned via exosomes in NSCLC cells. H19 was secreted into exosomes, mediated by hnRNPA2B1, and induced gefitinib resistance [94]. Moreover, FOXF2 was found to promote the progression of NSCLC cells by mediating decreased PTEN expression through H19 [95]. Moreover, H19 was involved in methylation-mediated lung cancer progression. Finally, it was demonstrated that silencing H19 suppressed growth and EMT while promoting apoptosis of lung cancer cells through suppressing the CDH1 promoter [96].
Recently, several researcher groups have shown that H19 promotes lung cancer progression through lncRNA-miRNA-mRNA network. H19 promoted NSCLC progression by modulating NF1 expression via competitively binding to miR-107 (Fig. 2D) [97]. Additionally, overexpressing H19 stimulated cell proliferation via the miR-138/PDK1 axis in NSCLC (Fig. 2E) [98]. H19 also promoted EMT and cell viability by modulating the miR-29b-3p/STAT3 axis (Fig. 2F) [99]. In another study, the H19/miR-200a/ZEB1/ZEB2 axis was shown to be involved in the growth and metastasis of lung cancer (Fig. 2G) [100].
Drug resistance is a major factor leading to chemotherapy failure in lung cancer patients. Pan et al. [101] validated that exosomal H19 expedited erlotinib resistance through the miR-615-3p/ATG7 axis, providing a new diagnostic and therapeutic target for NSCLC (Fig. 2H). Moreover, H19 facilitated resistance to gefitinib through the miR-148b-3p/DDAH1 axis in lung adenocarcinoma, offering a novel insight into resistance to EGFR inhibitors (Fig. 2I) [102]. Together, these studies demonstrated that H19 participated in lung cancer progression and functioned as a diagnostic biomarker and therapeutic target.
H19 mediates mechanisms involved in respiratory system tumors. (A). H19 could promote the expression of EZH2 by targeting miR-630. (B). H19 could promote the expression of HRAS by targeting let-7 family. (C). H19 could promote the expression of DNMT1 by targeting miR-148a-3p. (D). H19 could promote the expression of NF1 by targeting miR-107. (E). H19 could promote the expression of PDK1 by targeting miR-138. (F). H19 could promote the expression of STAT3 by targeting miR-29b-3p. (G). H19 could promote the expression of ZEB1/ZEB2 by targeting miR-200a. (H). H19 could promote the expression of ATG7 by targeting miR-615-3p. (I). H19 could promote the expression of DDAH1 by targeting miR-148b-3p.
The role of H19 in breast cancer (BC)
BC is the greatest threat to women's health. It is estimated that in 2021, BC will account for 30% of new diagnoses and 15% of deaths of all cancers in the United States [43]. Most BC cases occur in women >50-year-old; however, the incidence is rising in younger women. The prevalence of BC is 1.9% and 10.5% in women aged 20-34 and 35-44, respectively [103]. Therefore, it is necessary to explore the etiological mechanisms of BC.
A recent study showed that H19 was induced by estrogen, and had higher expression in estrogen receptor (ER)-positive BC than in ER-negative BC. Moreover, increasing H19 expression accelerated BC cell growth, which could serve as a predictive factor for BC [104]. Subsequently, multiple investigators have taken notice of the oncogenic role of H19; a study revealed that H19 expression was significantly correlated to ER, progesterone receptor, c-erbB-2, and lymph node metastasis in BC patients [105]. Furthermore, H19 expression in postoperative plasma was lower than in samples taken before surgery, which could be an early prognostic monitoring factor for BC.
Additionally, H19 also plays an important role in BC drug resistance. Si et al. [106] indicated that H19, functioning as a downstream target of ERα, restrained apoptosis in response to paclitaxel treatment by suppressing transcription of NOXA and BIK. Silencing H19 restored paclitaxel chemosensitivity through the AKT pathway in BC cells [107]. Furthermore, increasing H19 expression resulted in resistance to paclitaxel and anthracyclines. Silencing H19 increased drug sensitivity through the CUL4A-ABCB1/MDR1 pathway [108]. Tamoxifen is another drug commonly used to treat ER+ BC patients. However, tamoxifen resistance resulted in recurrence and reduced OS in BC patients. A report showed that knocking-down H19 helped overcome tamoxifen and fulvestrant resistance by blocking c-MET and Notch signaling [109]. Gao et al. [110] and Wang et al. [111] showed that silencing H19 elevated tamoxifen sensitivity by inhibiting cell growth or autophagy, which provided a novel option in fighting BC. Additionally, H19 knockdown restored trastuzumab sensitivity in BC [112] and restored doxorubicin (ADM) resistance by attenuating cell viability and colony-forming ability [113]. Also, another recent study showed that blocking H19 decreased cell proliferation, migration, invasion, EMT, and induced a cell cycle arrest by targeting the p53/TNFAIP8 axis in triple-negative BC [114]. Additionally, lncRNA PTCSC3 curbed cell growth by suppressing H19 in triple-negative BC [115].
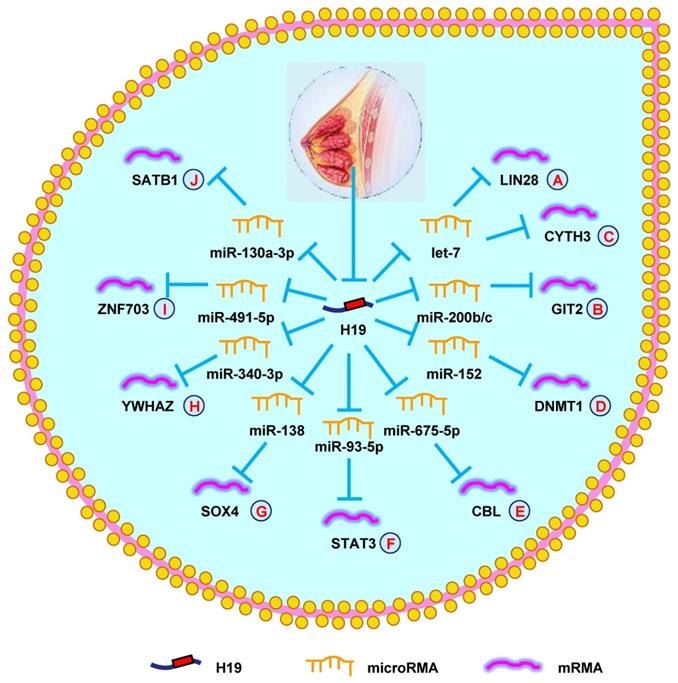
More recently, hundreds of investigators have further explored the lncRNA-miRNA-mRNA network in BC. In 2017, a study on BC showed that H19, let-7, and LIN 28 formed a double-negative feedback loop that played a vital role in BC formation. Further studies into the underlying mechanisms revealed that H19 functioned as a ceRNA of let-7, leading to increased LIN 28 expression (Fig. 3A) [116]. Another report demonstrated a similar result that the H19/let-7/LIN 28 network also increased autophagy by suppressing EMT in BC [117]. Moreover, H19 was found to mediate mesenchymal-to-epithelial transition and EMT via serving as a sponge for let-7 and miR-200b/c and regulating their targets Cyth3 and Git2 in BC (Fig. 3B and 3C) [118]. Li et al. [119] showed that H19 enhanced cell growth and invasion through the miR-152/DNMT1 axis, providing a new mechanism for BC development (Fig. 3D). Another group found that Huaier extract decreased the viability of BC cells by inducing apoptosis through the H19/miR-675-5p/CBL axis (Fig. 3E) [120]. Since 2019, myriad regulatory networks have been found to be involved in the occurrence and development of BC. The newly identified network H19/miR-93-5/STAT3 was shown to promote an aggressive phenotype of BC cells (Fig. 3F) [121]. Silencing H19 suppressed invasive behaviors by modulating the miR-138/SOX4 axis in BC (Fig. 3G) [122]. Yan et al. [123] uncovered that H19 functioned as a ceRNA to accelerate BC progression by regulating the miR-340-3p/YWHAZ axis, providing a potential therapeutic and prognostic biomarker for BC (Fig. 3H). Another report indicated that H19 increased growth, invasion, and migration in BC cells by sponging miR-491-5p to suppress ZNF703 (Fig. 3I) [124]. Finally, silencing H19 inhibited BC tumorigenesis by regulating the miR-130a-3p/SATB1 axis (Fig. 3J) [125]. These findings indicated that H19 was a novel oncogene that promoted BC progression.
The role of H19 in genitourinary system tumors
H19 in endometrial cancer
Endometrial cancer is a rare malignancy of the female reproductive system. Although the incidence of endometrial cancer is not very high, it can cause significant pain to patients. Therefore, it is essential to clarify the underlying mechanisms of this malignancy.
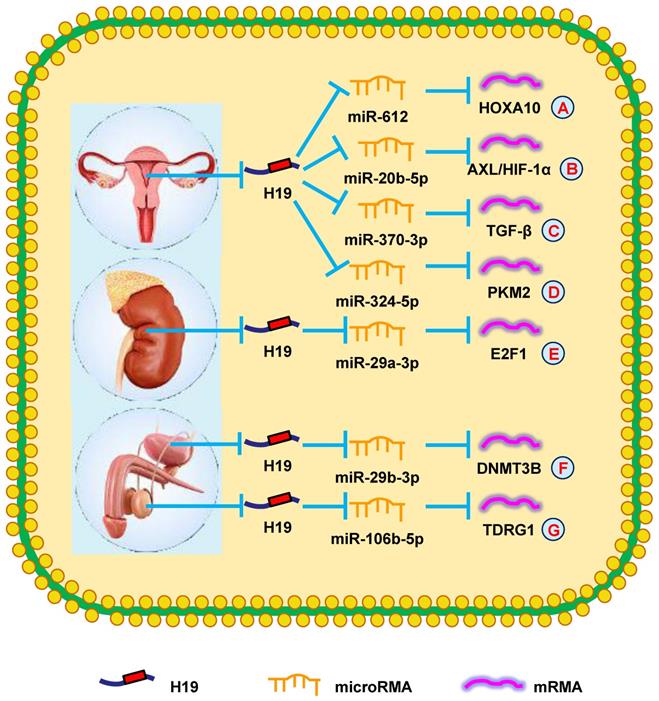
Zhang et al. [126] reported high H19 levels in endometrial cancer. Increased H19 levels promoted HOXA10 expression, which increased cell growth by targeting miR-612 (Fig. 4A). Another group showed that H19 induced the aggressive phenotype of endometrial cancer by targeting miR-20b-5p/AXL/HIF-1α signaling, providing a further target for treating endometrial cancer (Fig. 4B) [127]. These findings indicated that H19 participated in endometrial cancer progression.
H19 in ovarian cancer (OC)
OC is an important tumor of the female genitourinary system. In 2021, OC is expected to account for 5% of all deaths of women from cancer in the United States [43]. Therefore, there is an urgent need to uncover therapeutic targets in OC.
Silencing H19 was shown to inhibit cell proliferation by regulating certain cell cycle- and apoptosis-related proteins [128]. Moreover, increasing H19 expression led to higher TGF-β levels, which promoted EMT of OC cells via antagonizing miR-370-3p, which suggested this pathway could be a potential therapeutic target (Fig. 4C) [129]. Another report suggested that suppressing H19 with ginsenoside 20(S)-Rg3 increased the repression of PKM2 by miR-324-5p, thereby repressing OC tumorigenesis. Similarly, H19 mediates drug resistance in OC (Fig. 4D) [130]. Sajadpoor et al. [131] showed that valproic acid inhibited H19 expression and blocked cell growth and cisplatin resistance via the EZH2/p21/PTEN pathway. In summary, these studies provided novel insights into the mechanisms of H19 in OC and could be developed into OC therapies.
H19 in renal cell carcinoma (RCC)
RCC is a common malignancy of the urinary system. Early-stage RCC is difficult to detect due to a lack of typical clinical manifestations. Therefore, investigating the molecular mechanisms of RCC is critical.
Wang et al. [132] showed that increased H19 expression was associated with poorer prognosis and advanced clinical stage in RCC patients. Knocking-down H19 in RCC cells attenuated their proliferation, invasion, and migration. Additionally, silencing H19 suppressed E2F1 expression by sponging miR-29a-3p and restrained cell migration and invasion (Fig. 4E) [133]. These findings showed that H19 could be a therapeutic target in RCC.
H19 in bladder cancer
Bladder cancer is the most prevalent and fatal tumor of the urinary system [43]. Because there are no obvious clinical manifestations in early stages, many bladder cancer patients are diagnosed in advanced stages. Therefore, novel therapies to fight bladder cancer are urgently needed.
Studies to better understand the specific molecular mechanisms of bladder cancer have revealed that increased H19 expression promoted bladder cancer cell metastasis by suppressing E-cadherin [134, 135]. Furthermore, it was reported that increased H19 expression accelerated cell proliferation by modulating ID2 expression in bladder cancer [136]. Recently, another study revealed that YAP1-enhanced H19 overexpression was associated with poorer clinicopathological prognoses of bladder cancer patients [137]. Two years later, Lv et al. [138] showed that H19 attenuated the inhibitory effect of DNMT3B by functioning as a ceRNA for miR-29b-3p in bladder cancer (Fig. 4F). Additionally, Wang et al. [139] proposed that exosomal H19 expression was increased in bladder cancer patients and that these patients had reduced OS compared with other patients. In summary, H19 played a vital in bladder cancer prognosis and could be the target of novel bladder cancer treatments.
H19 mediates mechanisms involved in breast cancer. (A). H19 could promote the expression of LIN28 by targeting let-7. (B). H19 could promote the expression of GIT2 by targeting miR-200b/c. (C). H19 could promote the expression of CYTH3 by targeting let-7. (D). H19 could promote the expression of DNMT1 by targeting miR-152. (E). H19 could promote the expression of CBL by targeting miR-675-5p. (F). H19 could promote the expression of STAT3 by targeting miR-93-5p. (G). H19 could promote the expression of SOX4 by targeting miR-138. (H). H19 could promote the expression of YWHAZ by targeting miR-340-3p. (I). H19 could promote the expression of ZNF703 by targeting miR-491-5p. (J). H19 could promote the expression of SATB1 by targeting miR-130a-3p.
H19 mediates mechanisms involved in genitourinary system tumors. (A). H19 could promote the expression of HOXA10 by targeting miR-612. (B). H19 could promote the expression of AXL/HIF-1α by targeting miR-20b-5p. (C). H19 could promote the expression of TGF-β by targeting miR-370-3p. (D). H19 could promote the expression of PKM2 by targeting miR-324-5p. (E). H19 could promote the expression of E2F1 by targeting miR-29a-3p. (F). H19 could promote the expression of DNMT3B by targeting miR-29b-3p. (G). H19 could promote the expression of TDRG1 by targeting miR-106b-5p.
H19 in testicular tumors
Although testicular tumors account for a small percentage of malignancies in men, they are life-threatening to young men. Testicular neoplasms are difficult to detect in early stages because of their atypical clinical symptoms. Thus, it is essential to define more efficient diagnostic markers.
A new report revealed that H19 expression was elevated in cisplatin-resistant seminoma cells. H19 increased TDRG1 expression by sponging miR-106b-5p to stimulate cell survival in cisplatin-based chemotherapeutic conditions (Fig. 4G) [140]. These results demonstrated that H19 could be a novel therapeutic target for chemoresistant testicular tumors.
The role of H19 in nervous system tumors
Glioma is a common tumor of the nervous system. Due to its rapid progression and highly aggressive nature, the 5-year OS rate of glioma patients is only 15 months [141]. Therefore, discovering the pathogenesis of glioma and novel therapeutic methods is important for glioma patients.
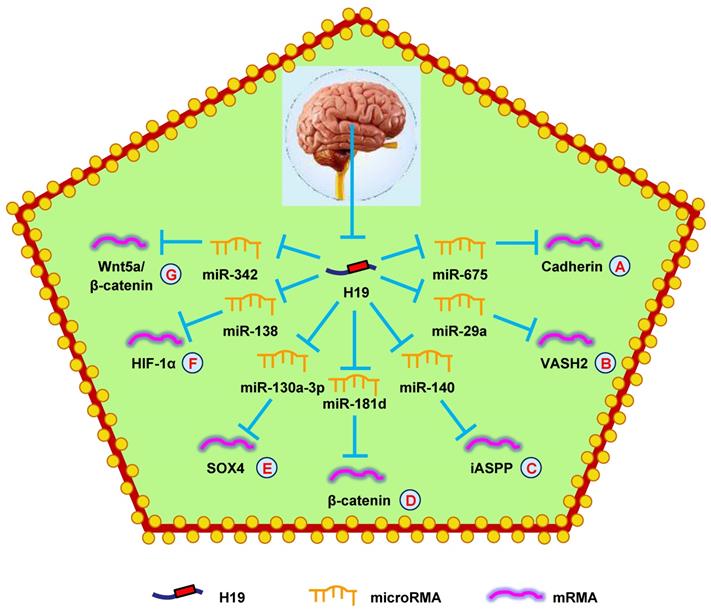
A recent report showed that H19 was elevated in glioma cells and contributed to maintain the stemness properties and malignant behaviors of glioma cells [142]. Another report showed that increased H19 expressed stimulated the tumorigenicity of glioma cells [143]. Subsequently, H19 was confirmed to bind to EZH2 and regulate glioma cell viability, migration, and invasion by repressing NKD1 [144]. Additionally, knocking-down H19 suppressed the growth, migration, invasion, and cell cycle progression of glioma cells and increased apoptosis by attenuating Wnt/β-catenin signaling [145]. In 2014, Shi et al. [146] first identified that miRNAs were involved in H19-mediated glioma progression. They reported that H19, mediated by miR-675, promoted cell proliferation in glioma. They found that H19 was upregulated in glioma cells and tissues, and was negatively correlated with patient survival (Fig. 5A). Another group came to a similar conclusion [147]. Not long after, a study reported that H19 promoted cell growth and invasion by repressing miR-152 [148].
H19 has also been found to regulate glioma progression through miRNA-mRNA network. Downregulation of H19 decreased VASH2 expression and inhibited tumor angiogenesis by upregulating miR-29a (Fig. 5B) [149]. Another study showed that H19 regulated cell proliferation and metastasis by controlling miR-140-mediated iASPP expression in glioma, which could be a new therapeutic biomarker for glioma (Fig. 5C) [150]. Another study found that hypoxia facilitated H19 expression, which relieved β-catenin suppression by binding miR-181d, enhancing the invasion and migration of glioma cells (Fig. 5D) [151]. Moreover, H19 also competed with SOX4 by sponging miR-130a-3p to influence EMT, migration, and invasion in glioma (Fig. 5E) [152]. Liu et al. [153] discovered that overexpressing H19 promoted the growth, invasion, and migration of glioma cells by serving as a ceRNA and regulating miR-138/HIF-1α signaling (Fig. 5F) [153]. Additionally, a mechanistic study revealed that H19 accelerated cell proliferation and metastasis by modulating Wnt5a/β-catenin signaling via miR-342, providing novel therapeutic targets in glioma (Fig. 5G) [154].
H19 mediates mechanisms involved in nervous system tumors. (A). H19 could promote the expression of Cadherin by targeting miR-675. (B). H19 could promote the expression of VASH2 by targeting miR-29a. (C). H19 could promote the expression of iASPP by targeting miR-140. (D). H19 could promote the expression of β-catenin by targeting miR-181d. (E). H19 could promote the expression of SOX4 by targeting miR-130a-3p. (F). H19 could promote the expression of HIF-1α by targeting miR-138. (G). H19 could promote the expression of Wnt5a/β-catenin by targeting miR-342.
H19 is also involved in glioma drug resistance. Jiang et al. [155] indicated significantly increased H19 levels in temozolomide (TMZ)-resistant glioma patients compared with TMZ-sensitive patients. H19 played a vital role in TMZ-resistance in glioma by altering the expression of drug resistance genes such as MRP, MDR, and ABCG2. Similarly, another study showed that H19 attenuated TMZ-resistance in glioma cells by inhibiting EMT via suppressing the Wnt/β-catenin pathway [156]. Finally, H19 induced TMZ resistance in glioma cells via activating NF-κB [157]. Together, these data demonstrated that H19 was an oncogene in glioma that could be a therapeutic target.
The role of H19 in tumors of other systems
The role of H19 in lymphoma
Lymphoma is a class of malignant tumors that derive from the lymphatic hematopoietic system. Hodgkin's lymphoma (HL) is a major lymphoma subtype that most often occurs in people over 55-years-old and between 15- and 35-years-old. Therefore, it is meaningful to look for diagnostic biomarkers of HL.
Wang et al. [158] reported that H19 was upregulated in HL tissues and inversely associated with OS in HL patients. H19 stimulated HL cell proliferation through AKT. These data confirmed that H19 promoted HL progression and functioned as an oncogene.
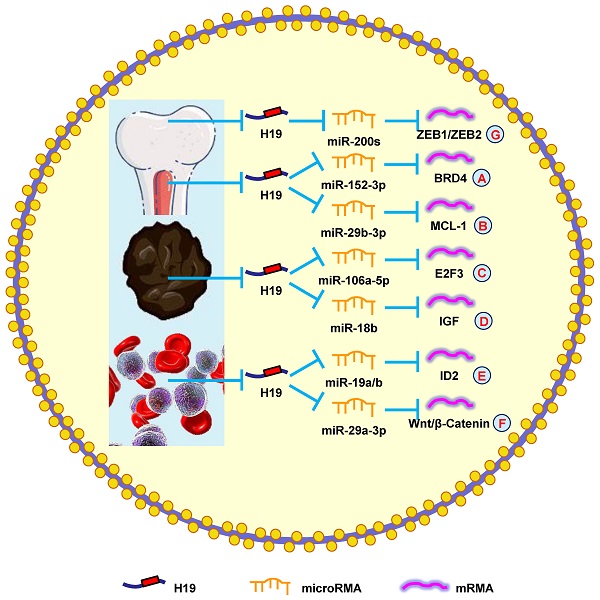
The role of H19 in myeloma
Myeloma is a malignant hematological tumor characterized by excessive proliferation of bone marrow plasma cells. Although there has been recent progress in treatments for myeloma, these are associated with adverse reactions such as severe infection, myelosuppression, neutropenia, and drug resistance. Therefore, improving myeloma treatment, including new drugs or combination therapies, is an urgent issue.
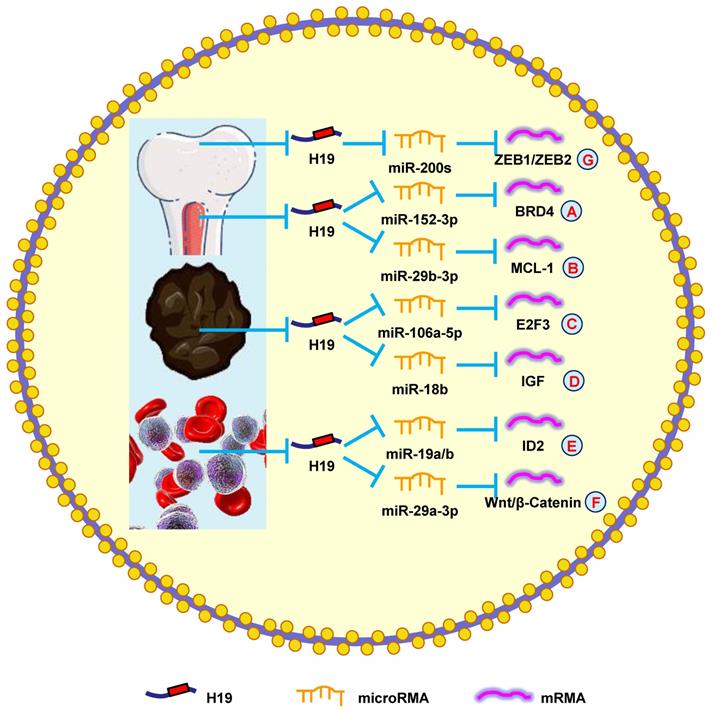
A previous study showed that H19 was elevated in myeloma patients and cell lines. The severity of myeloma was also shown to be associated with H19 levels in the serum of patients, suggesting that H19 could also be a therapeutic target [159]. Another study reported that blocking H19 along with reducing NF-κB expression blocked the growth of myeloma cells [160]. Silencing H19 attenuated the tumorigenesis of myeloma cells by blocking BRD4 expression through the miR-152-3p pathway (Fig. 6A) [161].
Moreover, H19 is involved in myeloma drug resistance. Pan et al. [8] revealed that H19 reduced chemosensitivity to bortezomib via upregulating MCL-1 by functioning as a miRNA sponge and sequestering miR-29b-3p (Fig. 6B). Additionally, upregulating H19 and AKT suppressed apoptosis, while silencing H19 and AKT accelerated apoptosis. Anti-H19 was a potential way to block drug resistance in myeloma [162]. These results confirmed that H19 was a therapeutic target in myeloma.
The role of H19 in melanoma
The World Health Organization estimates that 66,000 people die from skin cancer worldwide each year; melanoma is responsible for 80% of those deaths [163]. Therefore, understanding the pathogenesis of melanoma is vitally important for exploring new therapeutic targets.
It was previously reported that H19 accelerates the growth of melanoma cells by functioning as a miR-106a-5p sponge and increasing E2F3 expression (Fig. 6C) [9]. H19 is associated with poor prognoses, which means it could be a novel therapeutic target for melanoma. Zhu et al. [164] found that overexpressing H19 promoted the growth and invasion of melanoma cells by upregulating MMP2 and MMP9. Another study discovered that silencing H19 suppressed melanoma cell migration and invasion by deactivating NF‑κB expression via the PI3K/Akt pathway [165]. Furthermore, H19 overexpression in melanoma patients was correlated with poor clinical prognosis, such as lymph node metastasis, distant metastasis, and shorter OS. Silencing H19 inhibited the migration and invasion of melanoma cells and induced melanoma cell apoptosis after a G0/G1 arrest [166]. Finally, H19 promoted cisplatin-resistance by regulating miR-18b/IGF signaling, which suggests it could be a therapeutic target for melanoma (Fig. 6D) [22]. Overall, these data indicated that H19 had an oncogenic role in melanoma and represented a novel therapeutic target.
The role of H19 in leukemia
Leukemia is a malignant tumor with high morbidity and mortality in both men and women [43]. Recently, several groups have tried to explore the underlying mechanisms of leukemia, but there has not been a breakthrough. Hence, a full understanding of leukemia development would facilitate better disease management.
According to the study by Zhao et al. [167], silencing H19 decreased ID2 expression by competitive binding to miR-19a and miR-19b, which restrained cell growth (Fig. 6E). Expectedly, ectopic H19 expression was associated with shorter OS and lower complete remission, which was confirmed using the Gene Ontology Omnibus and The Cancer Genome Atlas datasets. It was also determined that H19 produced oncogenic effects through the downstream gene ID2 in leukemia [168]. Finally, H19 was shown to sequester miR-29a-3p to promote cell growth and inhibit apoptosis in leukemia through the Wnt/β-catenin pathway (Fig. 6F) [169]. Taken together, these results suggested that H19 played an oncogenic role in leukemia.
The role of H19 in osteosarcoma
Osteosarcoma is a rare malignant tumor that occurs in both adolescents and children. The highest incidence of osteosarcoma is between the ages of 10 and 20. With improved surgical techniques and chemotherapy regimens, the 5-year OS rate has increased from 20% to 70% [170]. However, it remains urgent to explore effective diagnostic and prognostic biomarkers for osteosarcoma to further improve OS.
Li et al. [171] reported that H19 increased metastasis via elevating ZEB1 and ZEB2 expression by combining with the miR-200 family (Fig. 6G). Another group revealed that patients with high H19 expression had a shorter OS compared with those with low H19 expression. Downregulating H19 attenuated cell invasion and migration by suppressing NF-κB signaling [172]. These findings indicated that H19 was a potential biomarker that could be used to diagnose and treat osteosarcoma.
H19 mediates mechanisms involved in tumors of other systems. (A). H19 could promote the expression of BRD4 by targeting miR-152-3p. (B). H19 could promote the expression of MCL-1 by targeting miR-29b-3p. (C). H19 could promote the expression of E2F3 by targeting miR-106a-5p. (D). H19 could promote the expression of IGF by targeting miR-18b. (E). H19 could promote the expression of ID2 by targeting miR-19a/b. (F). H19 could promote the expression of Wnt/β-Catenin by targeting miR-29a-3p. (G). H19 could promote the expression of ZEB1/ZEB2 by targeting miR-200s.
Conclusion and future perspectives
Cancer has been the leading cause of death in China since 2010, becoming a dominant public health issue in the country and worldwide [173]. Because early clinical symptoms are often not obvious and there is a lack of effective biological markers, many patients are diagnosed in advanced stages. The current lack of efficient therapeutic strategies for advanced tumors directly contributes to the high mortality rate for many malignancies. Therefore, studies to identify early-stage diagnostic markers and therapeutic targets have been performed by many researchers.
LncRNAs can function as oncogenes or tumor suppressor genes, and thus are involved in the occurrence and development of many different tumor types. Several lncRNAs are potential diagnostic and/or prognostic biomarkers including lncRNA HOTAIR [174], lncRNA MALAT1 [175], lncRNA MEG3 [176], lncRNA PVT1 [177], lncRNA XIST [178], and H19 [27]. Although H19 is one of the most studied lncRNAs, many molecular mechanisms remain unelucidated. Therefore, a comprehensive study of its downstream effectors and upstream regulatory mechanisms may provide a novel perspective to better counteract H19 in cancer.
In this review, we highlighted several examples of increased H19 expression and its role in cancer development, progression, prognosis, and treatment. H19 functions through a variety of mechanisms, such as interactions with miRNAs and/or target proteins to maintain cancer characteristics. Abnormal overexpression of H19 has been identified to be tightly associated with clinicopathological characteristics of the different cancers. H19 can also competitively combine with mRNAs by antagonizing miRNAs, revealing a regulatory network model of “H19-miRNAs-mRNAs.” Additionally, H19 overexpression is an important reason for chemotherapy resistance in malignant tumors.
Although further investigations are needed to expand our understanding of the molecular function of H19 in a more comprehensive way, the clinical application of H19 has aroused great interest. Recent studies have shown that H19 can be released from a variety of cancers and can be detected in patients' serum, which could be used for early detection and establishment of personalized treatments. For example, plasma H19 levels have been proposed by some scholars as a predictive biomarker for GC, BC, and lung cancer, and as an important tool for monitoring cancer development [32, 105, 179]. In addition, Sorin et al. [180] found that liver metastatic growth in treated animals was significantly reduced by using a plasmid approach to selectively kill H19-expressing cells with the diphtheria toxin A chain gene controlled by the H19 promoter (DTA-H19/BC-819). The method of BC-819 instillations to limit tumor recurrence is conducted in phase 1/2a trial for ovarian cancer and in phase 2b trial for BC [181, 182]. Finally, H19 increases resistance to ADM, 5-Fu, PTX, gefitinib, TMZ, sorafenib, tamoxifen, and MTX in almost all types of cancer, indicating the importance of designing anti-H19 therapy to improve the response of cancer patients to a broad range of treatment regimens.
Overall, several lines of evidence indicated that H19 plays important roles in tumor development and progression. H19 expression is proposed as a novel biomarker for many tumors. Antineoplastic drugs targeting H19 could be used to more accurately and safely treat malignant tumors.
Abbreviations
lncRNAs: Long non-coding RNAs; H19: LncRNA H19; ncRNAs: Non-coding RNAs; EMT: Epithelial-mesenchymal transition; OS: Overall survival; miRNA: MicroRNA; EC: Esophageal cancer; ESCC: Esophageal squamous cell carcinoma; GC: Gastric cancer; CRC: Colorectal cancer; MTX: Methotrexate; 5-Fu: 5-fluorouracil; HCC: Hepatocellular carcinoma; GBC: Gallbladder cancer; CCC: Cholangiocarcinoma; NPC: Nasopharyngeal cancer; LSCC: Laryngeal squamous cell carcinoma; NSCLC: Non-small cell lung cancer; BC: Breast cancer; ER: Estrogen receptor; PTX: Paclitaxel; ADM: Doxorubicin; OC: Ovarian cancer; RCC: Renal cell carcinoma; TMZ: Temozolomide; HL: Hodgkin's lymphoma; DTA: Diphtheria toxin A.
Acknowledgements
We thank Yun Cui from the Department of Urology, National Urological Cancer Center, Peking University First Hospital and Institute of Urology for helping us prepare the manuscript. We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Consent for publication
Written consent for publication was obtained from all the participants.
Funding
This work was supported by National Natural Science Foundation of China (Grant No. 82072835) to K Wang, Key Research and Development Joint Program of Liaoning Province (Grant No. 2020JH 2/10300139) to K Wang, Natural Science Foundation of Liaoning Province (Grant No. 2019-MS-360) to K Wang, Shenyang Science and Technology Bureau Plan Projects (Grant No. 20-205-4-076) to K Wang, 345 Talent Project of Shengjing Hospital of China Medical University (Grant No. M0366) to K Wang, and Natural Science Foundation of Liaoning Province (Grant No. 2019-MS-371) to M Qi.
Author Contributions
KW, JY and MQ conceived the review; KW, XF and XW wrote the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kapranov P, Cheng J, Dike S. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484-1488
2. Jarroux J, Morillon A, Pinskaya M. History, Discovery, and Classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1-46
3. Dahariya S, Paddibhatla I, Kumar S. et al. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol Immunol. 2019;112:82-92
4. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494-1504
5. Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097-1109
6. Smits G, Mungall AJ, Griffiths-Jones S. et al. Conservation of the H19 noncoding RNA and H19-IGF2imprinting mechanism in therians. Nat Genet. 2008;40:971-976
7. Yoshimura H, Matsuda Y, Yamamoto M. et al. Expression and role of long non-coding RNA H19 in carcinogenesis. Front Biosci (Landmark Ed). 2018;23:614-625
8. Pan Y, Zhang Y, Liu W. et al. LncRNA H19 overexpression induces bortezomib resistance in multiple myeloma by targeting MCL-1 via miR-29b-3p. Cell Death Dis. 2019;10:106
9. Luan W, Zhou Z, Ni X. et al. Long non-coding RNA H19 promotes glucose metabolism and cell growth in malignant melanoma via miR-106a-5p/E2F3 axis. J Cancer Res Clin Oncol. 2018;144:531-542
10. Peperstraete E, Lecerf C, Collette J. et al. Enhancement of Breast Cancer Cell Aggressiveness by lncRNA H19 and Its Mir-675 Derivative: Insight into Shared and Di_erent Actions. Cancers (Basel). 2020;12:1730
11. Wang M, Han D, Yuan Z. et al. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9:1149
12. Wu Z, Yan L, Liu Y. et al. Inhibition of mTORC1 by lncRNA H19 via disrupting 4E-BP1/Raptor interaction in pituitary tumours. Nat Commun. 2018;9:4624
13. Zhang Y, Liu Y, Tang H. et al. Exosome-Transmitted lncRNA H19 Inhibits the Growth of Pituitary Adenoma. J Clin Endocrinol Metab. 2019;104:6345-6356
14. Wu Z, Zheng Y, Xie W. et al. The long noncoding RNA-H19/miRNA-93a/ATG7 axis regulates the sensitivity of pituitary adenomas to dopamine agonists. Mol Cell Endocrinol. 2020;518:111033
15. Jiao X, Lu J, Huang Y. et al. Long non-coding RNA H19 may be a marker for prediction of prognosis in the follow-up of patients with papillary thyroid cancer. Cancer Biomark. 2019;26:203-207
16. Liu L, Yang J, Zhu X. et al. Long noncoding RNA H19 competitively binds miR-17-5p to regulate YES1 expression in thyroid cancer. FEBS J. 2016;283:2326-2339
17. Qi D, Wang M, Yu F. Knockdown of lncRNA-H19 inhibits cell viability, migration and invasion while promotes apoptosis via microRNA-143/RUNX2 axis in retinoblastoma. Biomed Pharmacother. 2019;109:798-805
18. Li L, Chen W, Wang Y. et al. Long non-coding RNA H19 regulates viability and metastasis, and is upregulated in retinoblastoma. Oncol Lett. 2018;15:8424-8432
19. Li X, Yang H, Wang J. et al. High level of lncRNA H19 expression is associated with shorter survival in esophageal squamous cell cancer patients. Pathol Res Pract. 2019;215:152638
20. Li H, Yu B, Li J. et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318-2329
21. Jia J, Zhang X, Zhan D. et al. LncRNA H19 interacted with miR-130a-3p and miR-17-5p to modify radio-resistance and chemo-sensitivity of cardiac carcinoma cells. Cancer Med. 2019;8:1604-1618
22. An L, Huang J, Han X. et al. Downregulation of lncRNA H19 sensitizes melanoma cells to cisplatin by regulating the miR-18b/IGF1 axis. Anticancer Drugs. 2020;31:473-482
23. Mao Y, Wang YM, Dong LX. et al. Hypoxic exosomes facilitate angiogenesis and metastasis in esophageal squamous cell carcinoma through altering the phenotype and transcriptome of endothelial cells. J Exp Clin Cancer Res. 2019;38:389
24. Roshandel G, Nourouzi A, Pourshams A. et al. Endoscopic Screening for Esophageal Squamous Cell Carcinoma. Arch Iran Med. 2013;16:351-357
25. Hibi K, Nakamura H, Hirai A. et al. Loss of H19 imprinting in esophageal cancer. Cancer Res. 1996;56:480-482
26. Huang C, Cao L, Qiu L. et al. Upregulation of H19 promotes invasion and induces epithelial-to-mesenchymal transition in esophageal cancer. Oncol Lett. 2015;10:291-296
27. Tan D, Wu Y, Hu L. et al. Long noncoding RNA H19 is up-regulated in esophageal squamous cell carcinoma and promotes cell proliferation and metastasis. Dis Esophagus. 2017;30:1-9
28. Chen M, Deng J, Chen C. et al. LncRNA H19 promotes epithelial mesenchymal transition and metastasis of esophageal cancer via STAT3/EZH2 axis. Int J Biochem Cell Biol. 2019;113:27-36
29. Luo W, Liu W, Yao J. et al. Downregulation of H19 decreases the radioresistance in esophageal squamous cell carcinoma cells. Onco Targets Ther. 2019;12:4779-4788
30. Yang F, Bi J, Xue X. et al. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159-3165
31. Zhang E, Han L, Yin D. et al. c-Myc-induced, long, noncoding H19 affects cell proliferation and predicts a poor prognosis in patients with gastric cancer. Med Oncol. 2014;31:914
32. Zhou X, Yin C, Dang Y. et al. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516
33. Chen J, Wang Y, Zhang X. et al. H19 serves as a diagnostic biomarker and up-regulation of H19 expression contributes to poor prognosis in patients with gastric cancer. Neoplasma. 2016;63:223-230
34. Hashad D, Elbanna A, Ibrahim A. et al. Evaluation of the Role of Circulating Long Non-Coding RNA H19 as a Promising Novel Biomarker in Plasma of Patients with Gastric Cancer. J Clin Lab Anal. 2016;30:1100-1105
35. Yang T, Zeng H, Chen W. et al. Helicobacter pylori infection, H19 and LINC00152 expression in serum and risk of gastric cancer in a Chinese population. Cancer Epidemiol. 2016;44:147-153
36. Zhang Y, Yan J, Li C. et al. LncRNA H19 induced by helicobacter pylori infection promotes gastric cancer cell growth via enhancing NF-κB-induced inflammation. J Inflamm (Lond). 2019;16:23
37. Zhuang M, Gao W, Xu J. et al. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448:315-322
38. Liu G, Xiang T, Wu Q. et al. Long Noncoding RNA H19-Derived miR-675 Enhances Proliferation and Invasion via RUNX1 in Gastric Cancer Cells. Oncol Res. 2016;23:99-107
39. Yan J, Zhang Y, She Q. et al. Long Noncoding RNA H19/miR-675 Axis Promotes Gastric Cancer via FADD/Caspase 8/Caspase 3 Signaling Pathway. Cell Physiol Biochem. 2017;42:2364-2376
40. Zhou X, Ye F, Yin C. et al. The Interaction Between MiR-141 and lncRNA-H19 in Regulating Cell Proliferation and Migration in Gastric Cancer. Cell Physiol Biochem. 2015;36:1440-1452
41. Wei Y, Liu Z, Fang J. H19 functions as a competing endogenous RNA to regulate human epidermal growth factor receptor expression by sequestering let-7c in gastric cancer. Mol Med Rep. 2018;17:2600-2606
42. Gan L, Lv L, Liao S. Long non-coding RNA H19 regulates cell growth and metastasis via the miR-22-3p/Snail1 axis in gastric cancer. Int J Oncol. 2019;54:2157-2168
43. Siegel RL, Miller KD, Fuchs HE. et al. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7-33
44. Tsang W, Enders K, Simon S. et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350-358
45. Liang W, Fu W, Wong C. et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513-22525
46. Han D, Gao X, Wang M. et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7:22159-22173
47. Chen S, Zhu J, Ma J. et al. Overexpression of long non-coding RNA H19 is associated with unfavorable prognosis in patients with colorectal cancer and increased proliferation and migration in colon cancer cells. Oncol Lett. 2017;14:2446-2452
48. Yang W, Redpath RE, Zhang C. et al. Long non-coding RNA H19 promotes the migration and invasion of colon cancer cells via MAPK signaling pathway. Oncol Lett. 2018;16:3365-3372
49. Ohtsuka M, Ling H, Ivan C. et al. H19 Noncoding RNA, an Independent Prognostic Factor, Regulates Essential Rb-E2F and CDK8-β-Catenin Signaling in Colorectal Cancer. EBioMedicine. 2016;13:113-124
50. Zhang Y, Huang W, Yuan Y. et al. Long non-coding RNA H19 promotes colorectal cancer metastasis via binding to hnRNPA2B1. J Exp Clin Cancer Res. 2020;39:141
51. Yang W, Ning N, Jin X. The lncRNA H19 Promotes Cell Proliferation by Competitively Binding to miR-200a and Derepressing β-Catenin Expression in Colorectal Cancer. Biomed Res Int. 2017;2017:2767484
52. Ding D, Li C, Zhao T. et al. LncRNA H19/miR-29b-3p/PGRN Axis Promoted Epithelial-Mesenchymal Transition of Colorectal Cancer Cells by Acting on Wnt Signaling. Mol Cells. 2018;41:423-435
53. Li C, Li Y, Wang Y. et al. The Effect of LncRNA H19/miR-194-5p Axis on the Epithelial-Mesenchymal Transition of Colorectal Adenocarcinoma. Cell Physiol Biochem. 2018;50:196-213
54. Hu X, Xing W, Zhao R. et al. HDAC2 inhibits EMT-mediated cancer metastasis by downregulating the long noncoding RNA H19 in colorectal cancer. J Exp Clin Cancer Res. 2020;39:270
55. Yang Q, Wang X, Tang C. et al. H19 promotes the migration and invasion of colon cancer by sponging miR-138 to upregulate the expression of HMGA1. Int J Oncol. 2017;50:1801-1809
56. Ren J, Ding L, Zhang D. et al. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8:3932-3948
57. Chen S, Bu D, Ma Y. et al. H19 Overexpression Induces Resistance to 1,25(OH)2D3 by Targeting VDR Through miR-675-5p in Colon Cancer Cells. Neoplasia. 2017;19:226-236
58. Wu K, Liang W, Feng L. et al. H19 mediates methotrexate resistance in colorectal cancer through activating Wnt/β-catenin pathway. Exp Cell Res. 2017;350:312-317
59. Lv J, Yu Y, Li S. et al. Aflatoxin B1 promotes cell growth and invasion in hepatocellular carcinoma HepG2 cells through H19 and E2F1. Asian Pac J Cancer Prev. 2014;15:2565-2570
60. Li L, Han T, Liu K. et al. LncRNA H19 promotes the development of hepatitis B related hepatocellular carcinoma through regulating microRNA-22 via EMT pathway. Eur Rev Med Pharmacol Sci. 2019;23:5392-5401
61. Wei L, Li L, Lu C. et al. Involvement of H19/miR-326 axis in hepatocellular carcinoma development through modulating TWIST1. J Cell Physiol. 2019;234:5153-5162
62. Zhou Y, Fan R, Qin C. et al. LncRNA-H19 activates CDC42/PAK1 pathway to promote cell proliferation, migration and invasion by targeting miR-15b in hepatocellular carcinoma. Genomics. 2019;111:1862-1872
63. Ye Y, Guo J, Xiao P. et al. Macrophages-induced long noncoding RNA H19 up-regulation triggers and activates the miR-193b/MAPK1 axis and promotes cell aggressiveness in hepatocellular carcinoma. Cancer Lett. 2020;469:310-322
64. Wang D, Xing N, Yang T. et al. Exosomal lncRNA H19 promotes the progression of hepatocellular carcinoma treated with Propofol via miR-520a-3p/LIMK1 axis. Cancer Med. 2020;9:7218-7230
65. Ding K, Liao Y, Gong D. et al. Effect of long non-coding RNA H19 on oxidative stress and chemotherapy resistance of CD133+ cancer stem cells via the MAPK/ERK signaling pathway in hepatocellular carcinoma. Biochem Biophys Res Commun. 2018;502:194-201
66. Xu Y, Liu Y, Li Z. et al. Long non-coding RNA H19 is involved in sorafenib resistance in hepatocellular carcinoma by upregulating miR-675. Oncol Rep. 2020;44:165-173
67. Jarnagin WR, Fong Y, DeMatteo RP. et al. Staging, resectability, and outcome in 225 patients with hilar cholangiocarcinoma. Ann Surg. 2001;234:507-517
68. Bridgewater J, Galle PR, Khan SA. et al. Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. J Hepatol. 2014;60:1268-1289
69. Wang S, Wu X, Zhang M. et al. Long noncoding RNA H19 contributes to gallbladder cancer cell proliferation by modulated miR-194-5p targeting AKT2. Tumour Biol. 2016;37:9721-9730
70. Wang S, Wu X, Zhang M. et al. Upregulation of H19 indicates a poor prognosis in gallbladder carcinoma and promotes epithelial-mesenchymal transition. Am J Cancer Res. 2015;6:15-26
71. Wang S, Ma F, Tang Z. et al. Long non-coding RNA H19 regulates FOXM1 expression by competitively binding endogenous miR-342-3p in gallbladder cancer. J Exp Clin Cancer Res. 2016;35:160
72. Wang W, Ye H, Wei P. et al. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J Hematol Oncol. 2016;9:117
73. Xu Y, Wang Z, Jiang X. et al. Overexpression of long noncoding RNA H19 indicates a poor prognosis for cholangiocarcinoma and promotes cell migration and invasion by affecting epithelial-mesenchymal transition. Biomed Pharmacother. 2017;92:17-23
74. Yu A, Zhao L, Kang Q. et al. Transcription factor HIF1α promotes proliferation, migration, and invasion of cholangiocarcinoma via long noncoding RNA H19/microRNA-612/Bcl-2 axis. Transl Res. 2020;224:26-39
75. Kleeff J, Korc M, Apte M. et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022
76. Ma C, Nong K, Zhu H. et al. H19 promotes pancreatic cancer metastasis by derepressing let-7's suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014;35:9163-9169
77. Ma L, Tian X, Wang F. et al. The long noncoding RNA H19 promotes cell proliferation via E2F-1 in pancreatic ductal adenocarcinoma. Cancer Biol Ther. 2016;17:1051-1061
78. Ma L, Tian X, Guo H. et al. Long noncoding RNA H19 derived miR-675 regulates cell proliferation by down-regulating E2F-1 in human pancreatic ductal adenocarcinoma. J Cancer. 2018;9:389-399
79. Yoshimura H, Matsuda Y, Yamamoto M. et al. Reduced expression of the H19 long non-coding RNA inhibits pancreatic cancer metastasis. Lab Invest. 2018;98:814-824
80. Sasaki N, Toyoda M, Yoshimura H. et al. H19 long non-coding RNA contributes to sphere formation and invasion through regulation of CD24 and integrin expression in pancreatic cancer cells. Oncotarget. 2018;9:34719-34734
81. Sun Y, Zhu Q, Yang W. et al. LncRNA H19/miR-194/PFTK1 axis modulates the cell proliferation and migration of pancreatic cancer. J Cell Biochem. 2019;120:3874-3886
82. Wang F, Rong L, Zhang Z. et al. LncRNA H19-Derived miR-675-3p Promotes Epithelial-Mesenchymal Transition and Stemness in Human Pancreatic Cancer Cells by targeting the STAT3 Pathway. J Cancer. 2020;11:4771-4782
83. Ji M, Yao Y, Liu A. et al. lncRNA H19 binds VGF and promotes pNEN progression via PI3K/AKT/CREB signaling. Endocr Relat Cancer. 2019;26:643-658
84. Zhao L, Fong AHW, Liu N. et al. Molecular subtyping of nasopharyngeal carcinoma (NPC) and a microRNA-based prognostic model for distant metastasis. J Biomed Sci. 2018;25:16
85. Kam MKM, Wong FCS, Kwong DLW. et al. Current controversies in radiotherapy for nasopharyngeal carcinoma (NPC). Oral Oncol. 2014;50:907-912
86. Aylwin N, Jing PT, Christopher HKG. et al. Regulation of the H19 imprinting gene expression in human nasopharyngeal carcinoma by methylation. Int J Cancer. 2003;104:179-187
87. Li X, Lin Y, Yang X. et al. Long noncoding RNA H19 regulates EZH2 expression by interacting with miR-630 and promotes cell invasion in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2016;473:913-919
88. Zhang Y, Zhu R, Wang J. et al. Upregulation of lncRNA H19 promotes nasopharyngeal carcinoma proliferation and metastasis in let-7 dependent manner. Artif Cells Nanomed Biotechnol. 2019;47:3854-3861
89. Zhu H. Silencing long non-coding RNA H19 combined with paclitaxel inhibits nasopharyngeal carcinoma progression. Int J Pediatr Otorhinolaryngol. 2020;138:110249
90. Wu T, Qu L, He G. et al. Regulation of laryngeal squamous cell cancer progression by the lncRNA H19/miR-148a-3p/DNMT1 axis. Oncotarget. 2016;7:11553-11566
91. Cui J, Mo J, Luo M. et al. c-Myc-activated long non-coding RNA H19 downregulates miR-107 and promotes cell cycle progression of non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:12400-12409
92. Zhang E, Li W, Yin D. et al. c-Myc-regulated long non-coding RNA H19 indicates a poor prognosis and affects cell proliferation in non-small-cell lung cancer. Tumour Biol. 2016;37:4007-4015
93. Wang Q, Cheng N, Li X. et al. Correlation of long non-coding RNA H19 expression with cisplatin-resistance and clinical outcome in lung adenocarcinoma. Oncotarget. 2017;8:2558-2567
94. Lei Y, Guo W, Chen B. et al. Tumor-released lncRNA H19 promotes gefitinib resistance via packaging into exosomes in non-small cell lung cancer. Oncol Rep. 2018;40:3438-3446
95. Xu J, Hua T, Ding J. et al. FOXF2 aggravates the progression of non-small cell lung cancer through targeting lncRNA H19 to downregulate PTEN. Eur Rev Med Pharmacol Sci. 2019;23:10796-10802
96. Gao L, Xu S, Zheng Y. et al. Long non-coding RNA H19 is responsible for the progression of lung adenocarcinoma by mediating methylation-dependent repression of CDH1 promoter. J Cell Mol Med. 2019;23:6411-6428
97. Qian B, Wang D, Gu X. et al. LncRNA H19 serves as a ceRNA and participates in non-small cell lung cancer development by regulating microRNA-107. Eur Rev Med Pharmacol Sci. 2018;22:5946-5953
98. Huang T, Wen Y, Peng B. et al. Upregulated lncRNA H19 promotes non-small cell lung cancer cell proliferation through miR-138/PDK1 axis. Int J Clin Exp Pathol. 2017;10:9012-9020
99. Liu L, Liu L, Lu S. lncRNA H19 promotes viability and epithelial-mesenchymal transition of lung adenocarcinoma cells by targeting miR-29b-3p and modifying STAT3. Int J Oncol. 2019;54:929-941
100. Zhao Y, Feng C, Li Y. et al. LncRNA H19 promotes lung cancer proliferation and metastasis by inhibiting miR-200a function. Mol Cell Biochem. 2019;460:1-8
101. Pan R, Zhou H. Exosomal Transfer of lncRNA H19 Promotes Erlotinib Resistance in Non-Small Cell Lung Cancer via miR-615-3p/ATG7 Axis. Cancer Manag Res. 2020;12:4283-4297
102. Huang Z, Ma Y, Zhang P. et al. Long non-coding RNA H19 confers resistance to gefitinib via miR-148b-3p/DDAH1 axis in lung adenocarcinoma. Anti-cancer Drugs. 2020;31:44-54
103. DeSantis C, Ma JM, Bryan L. et al. Breast Cancer Statistics, 2013. CA Cancer J Clin. 2014;64:52-62
104. Sun H, Wang G, Peng Y. et al. H19 lncRNA mediates 17β-estradiol-induced cell proliferation in MCF-7 breast cancer cells. Oncol Rep. 2015;33:3045-3052
105. Zhang K, Luo Z, Zhang Y. et al. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomark. 2016;17:187-194
106. Si X, Zang R, Zhang E. et al. LncRNA H19 confers chemoresistance in ERα-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget. 2016;7:81452-81462
107. Han J, Han B, Wu X. et al. Knockdown of lncRNA H19 restores chemo-sensitivity in paclitaxel-resistant triple-negative breast cancer through triggering apoptosis and regulating Akt signaling pathway. Toxicol Appl Pharmacol. 2018;359:55-61
108. Zhu Q, Wang G, Guo Y. et al. LncRNA H19 is a major mediator of doxorubicin chemoresistance in breast cancer cells through a cullin4A-MDR1 pathway. Oncotarget. 2017;8:91990-92003
109. Basak P, Chatterjee S, Bhat V. et al. Long Non-Coding RNA H19 Acts as an Estrogen Receptor Modulator that is Required for Endocrine Therapy Resistance in ER+ Breast Cancer Cells. Cell Physiol Biochem. 2018;51:1518-1532
110. Gao H, Hao G, Sun Y. et al. Long noncoding RNA H19 mediated the chemosensitivity of breast cancer cells via Wnt pathway and EMT process. Onco Targets Ther. 2018;11:8001-8012
111. Wang J, Xie S, Yang J. et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J Hematol Oncol. 2019;12:81
112. Sun Z, Zhang C, Wang T. et al. Correlation between long non-coding RNAs (lncRNAs) H19 expression and trastuzumab resistance in breast cancer. J Cancer Res Ther. 2019;15:933-940
113. Wang X, Pei X, Guo G. et al. Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J Cell Physiol. 2020;235:6896-6904
114. Li Y, Ma H, Hu X. et al. LncRNA H19 promotes triple-negative breast cancer cells invasion and metastasis through the p53/TNFAIP8 pathway. Cancer Cell Int. 2020;20:200
115. Wang N, Hou M, Zhan Y. et al. LncRNA PTCSC3 inhibits triple-negative breast cancer cell proliferation by downregulating lncRNA H19. J Cell Biochem. 2019;120:15083-15088
116. Peng F, Li T, Wang K. et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017;8:e2569
117. Xiong H, Shen J, Chen Z. et al. H19/let-7/Lin28 ceRNA network mediates autophagy inhibiting epithelial-mesenchymal transition in breast cancer. Int J Oncol. 2020;56:794-806
118. Zhou W, Ye X, Xu J. et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal. 2017;10:eaak9557
119. Li Z, Li Y, Li Y. et al. Long non-coding RNA H19 promotes the proliferation and invasion of breast cancer through upregulating DNMT1 expression by sponging miR-152. J Biochem Mol Toxicol. 2017 31
120. Wang J, Wang X, Chen T. et al. Huaier Extract Inhibits Breast Cancer Progression Through a LncRNA-H19/MiR-675-5p Pathway. Cell Physiol Biochem. 2017;44:581-593
121. Li J, Xiang Y, Fan L. et al. Long noncoding RNA H19 competitively binds miR-93-5p to regulate STAT3 expression in breast cancer. J Cell Biochem. 2019;120:3137-3148
122. Si H, Chen P, Li H. et al. Long non-coding RNA H19 regulates cell growth and metastasis via miR-138 in breast cancer. Am J Transl Res. 2019;11:3213-3225
123. Yan L, Yang S, Yue C. et al. Long noncoding RNA H19 acts as a miR-340-3p sponge to promote epithelial-mesenchymal transition by regulating YWHAZ expression in paclitaxel-resistant breast cancer cells. Environ Toxicol. 2020;35:1015-1028
124. Wang Y, Wu Z, Li Y. et al. Long Non-Coding RNA H19 Promotes Proliferation, Migration and Invasion and Inhibits Apoptosis of Breast Cancer Cells by Targeting miR-491-5p/ZNF703 Axis. Cancer Manag Res. 2020;12:9247-9258
125. Zhong G, Lin Y, Wang X. et al. H19 Knockdown Suppresses Proliferation and Induces Apoptosis by Regulating miR-130a-3p/SATB1 in Breast Cancer Cells. Onco Targets Ther. 2020;13:12501-12513
126. Zhang L, Wang D, Yu P. LncRNA H19 regulates the expression of its target gene HOXA10 in endometrial carcinoma through competing with miR-612. Eur Rev Med Pharmacol Sci. 2018;22:4820-4827
127. Zhu H, Jin Y, Lyu X. et al. Long noncoding RNA H19 regulates HIF-1α/AXL signaling through inhibiting miR-20b-5p in endometrial cancer. Cell Cycle. 2019;18:2454-2464
128. Zhu Z, Song L, He J. et al. Ectopic expressed long non-coding RNA H19 contributes to malignant cell behavior of ovarian cancer. Int J Clin Exp Pathol. 2015;8:10082-10091
129. Li J, Huang Y, Deng X. et al. Long noncoding RNA H19 promotes transforming growth factor-β-induced epithelial-mesenchymal transition by acting as a competing endogenous RNA of miR-370-3p in ovarian cancer cells. Onco Targets Ther. 2018;11:427-440
130. Zheng X, Zhou Y, Chen W. et al. Ginsenoside 20(S)-Rg3 Prevents PKM2-Targeting miR-324-5p from H19 Sponging to Antagonize the Warburg Effect in Ovarian Cancer Cells. Cell Physiol Biochem. 2018;51:1340-1353
131. Sajadpoor Z, Amini-Farsani Z, Teimori H. et al. Valproic Acid Promotes Apoptosis and Cisplatin Sensitivity Through Downregulation of H19 Noncoding RNA in Ovarian A2780 Cells. Appl Biochem Biotechnol. 2018;185:1132-1144
132. Wang L, Cai Y, Zhao X. et al. Down-regulated long non-coding RNA H19 inhibits carcinogenesis of renal cell carcinoma. Neoplasma. 2015;62:412-418
133. He H, Wang N, Yi X. et al. Long non-coding RNA H19 regulates E2F1 expression by competitively sponging endogenous miR-29a-3p in clear cell renal cell carcinoma. Cell Biosci. 2017;7:65
134. Luo M, Li Z, Wang W. et al. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213-221
135. Zhu Z, Xu L, Wan Y. et al. Inhibition of E-cadherin expression by lnc-RNA H19 to facilitate bladder cancer metastasis. Cancer Biomark. 2018;22:275-281
136. Luo M, Li Z, Wang W. et al. Upregulated H19 contributes to bladder cancer cell proliferation by regulating ID2 expression. FEBS J. 2013;280:1709-1716
137. Li S, Yu Z, Chen S. et al. The YAP1 oncogene contributes to bladder cancer cell proliferation and migration by regulating the H19 long noncoding RNA. Urol Oncol. 2015;33:427
138. Lv M, Zhong Z, Huang M. et al. lncRNA H19 regulates epithelial-mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim Biophys Acta Mol Cell Res. 2017;1864:1887-1899
139. Wang J, Yang K, Yuan W. et al. Determination of Serum Exosomal H19 as a Noninvasive Biomarker for Bladder Cancer Diagnosis and Prognosis. Med Sci Monit. 2018;24:9307-9316
140. Wei J, Gan Y, Peng D. et al. Long non-coding RNA H19 promotes TDRG1 expression and cisplatin resistance by sequestering miRNA-106b-5p in seminoma. Cancer Med. 2018;7:6247-6257
141. Ostrom Q, Cote D, Ascha M. et al. Adult Glioma Incidence and Survival by Race or Ethnicity in the United States From 2000 to 2014. JAMA Oncol. 2018;4:1254-1262
142. Li W, Jiang P, Sun X. et al. Suppressing H19 Modulates Tumorigenicity and Stemness in U251 and U87MG Glioma Cells. Cell Mol Neurobiol. 2016;36:1219-1227
143. Jiang X, Yan Y, Hu M. et al. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J Neurosurg. 2016;2016:129-136
144. Fazi B, Garbo S, Toschi N. et al. The lncRNA H19 positively affects the tumorigenic properties of glioblastoma cells and contributes to NKD1 repression through the recruitment of EZH2 on its promoter. Oncotarget. 2018;9:15512-15525
145. Guan N, Wang R, Guo W. et al. Long non-coding RNA H19 regulates the development of gliomas through the Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:4243-4253
146. Shi Y, Wang Y, Luan W. et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9:e86295
147. Zhang T, Wang Y, Zeng F. et al. LncRNA H19 is overexpressed in glioma tissue, is negatively associated with patient survival, and promotes tumor growth through its derivative miR-675. Eur Rev Med Pharmacol Sci. 2016;20:4891-4897
148. Chen L, Wang Y, He J. et al. Long Noncoding RNA H19 Promotes Proliferation and Invasion in Human Glioma Cells by Downregulating miR-152. Oncol Res. 2018;26:1419-1428
149. Jia P, Cai H, Liu X. et al. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016;381:359-369
150. Zhao H, Peng R, Liu Q. et al. The lncRNA H19 interacts with miR-140 to modulate glioma growth by targeting iASPP. Arch Biochem Biophys. 2016;610:1-7
151. Wu W, Hu Q, Nie E. et al. Hypoxia induces H19 expression through direct and indirect Hif-1α activity, promoting oncogenic effects in glioblastoma. Sci Rep. 2017;7:45029
152. Hu Q, Yin J, Zeng A. et al. H19 Functions as a Competing Endogenous RNA to Regulate EMT by Sponging miR-130a-3p in Glioma. Cell Physiol Biochem. 2018;50:233-245
153. Liu Z, Tian Y, Wu H. et al. LncRNA H19 promotes glioma angiogenesis through miR-138/HIF-1α/VEGF axis. Neoplasma. 2020;67:111-118
154. Zhou Q, Liu Z, Wu H. et al. LncRNA H19 Promotes Cell Proliferation, Migration, and Angiogenesis of Glioma by Regulating Wnt5a/β-Catenin Pathway via Targeting miR-342. Cell Mol Neurobiol. 2020 7
155. Jiang P, Wang P, Sun X. et al. Knockdown of long noncoding RNA H19 sensitizes human glioma cells to temozolomide therapy. Onco Targets Ther. 2016;9:3501-3509
156. Jia L, Tian Y, Chen Y. et al. The silencing of LncRNA-H19 decreases chemoresistance of human glioma cells to temozolomide by suppressing epithelial-mesenchymal transition via the Wnt/β-Catenin pathway. Onco Targets Ther. 2018;11:313-321
157. Duan S, Li M, Wang Z. et al. H19 induced by oxidative stress confers temozolomide resistance in human glioma cells via activating NF-κB signaling. Onco Targets Ther. 2018;11:6395-6404
158. Wang Y, Wang L, Sui M. Long non-coding RNA H19 promotes proliferation of Hodgkin's lymphoma via AKT pathway. J BUON. 2019;24:763-769
159. Pan Y, Chen H, Shen X. et al. Serum level of long noncoding RNA H19 as a diagnostic biomarker of multiple myeloma. Clin Chim Acta. 2018;480:199-205
160. Sun Y, Pan J, Zhang N. et al. Knockdown of long non-coding RNA H19 inhibits multiple myeloma cell growth via NF-κB pathway. Sci Rep. 2017;7:18079
161. Zheng J, Guo N, Zi F. et al. Long Noncoding RNA H19 Promotes Tumorigenesis of Multiple Myeloma by Activating BRD4 Signaling by Targeting MicroRNA 152-3p. Mol Cell Biol. 2020;40:e00382-19
162. Wang Y, Xu S, Wang L. et al. Long noncoding RNA H19 promotes vincristine resistance in multiple myeloma by targeting Akt. Cell Mol Biol (Noisy-le-grand). 2020;66:76-80
163. Lens MB, Dawes M. Global perspectives of contemporary epidemiological trends of cutaneous malignant melanoma. Br J Dermatol. 2004;150:179-185
164. Zhu X, Li W, Meng Q. LncRNA H19 promotes proliferation and invasion in A375 human melanoma cell line. Int J Clin Exp Pathol. 2018;11:1063-1073
165. Liao Z, Zhao J, Yang Y. Downregulation of lncRNA H19 inhibits the migration and invasion of melanoma cells by inactivating the NF-κB and PI3K/Akt signaling pathways. Mol Med Rep. 2018;17:7313-7318
166. Shi G, Li H, Gao F. et al. lncRNA H19 predicts poor prognosis in patients with melanoma and regulates cell growth, invasion, migration and epithelial-mesenchymal transition in melanoma cells. Onco Targets Ther. 2018;11:3583-3595
167. Zhao T, Jia H, Zhang Z. et al. LncRNA H19 regulates ID2 expression through competitive binding to hsa-miR-19a/b in acute myelocytic leukemia. Mol Med Rep. 2017;16:3687-3693
168. Zhang T, Zhou J, Zhang W. et al. H19 overexpression promotes leukemogenesis and predicts unfavorable prognosis in acute myeloid leukemia. Clin Epigenetics. 2018;10:47
169. Zhao T, Liu X. LncRNA-H19 inhibits apoptosis of acute myeloid leukemia cells via targeting miR-29a-3p. Eur Rev Med Pharmacol Sci. 2019;23:224-231
170. Bagcchi S. Osteosarcoma survivors' risk of second cancer. Lancet Oncol. 2014;15:e425
171. Li M, Chen H, Zhao Y. et al. H19 Functions as a ceRNA in Promoting Metastasis Through Decreasing miR-200s Activity in Osteosarcoma. DNA Cell Biol. 2016;35:235-40
172. Zhao J, Ma S. Downregulation of lncRNA H19 inhibits migration and invasion of human osteosarcoma through the NF-κB pathway. Mol Med Rep. 2018;17:7388-7394
173. Chen W, Zheng R, Baade P. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132
174. Zhang J, Qiu W, Zhu H. et al. HOTAIR contributes to the carcinogenesis of gastric cancer via modulating cellular and exosomal miRNAs level. Cell Death Dis. 2020;11:780
175. Shaath H, Vishnubalaji R, Elango R. et al. Single-cell long noncoding RNA (lncRNA) transcriptome implicates MALAT1 in triple-negative breast cancer (TNBC) resistance to neoadjuvant chemotherapy. Cell Death Discov. 2021;7:23
176. Ding C, Yi X, Xu J. et al. Long Non-Coding RNA MEG3 Modifies Cell-Cycle, Migration, Invasion, and Proliferation Through AKAP12 by Sponging miR-29c in Meningioma Cells. Front Oncol. 2020;10:537763
177. Shigeyasu K, Toden S, Ozawa T. et al. The PVT1 lncRNA is a novel epigenetic enhancer of MYC, and a promising risk-stratification biomarker in colorectal cancer. Mol Cancer. 2020;19:155
178. Zhao Y, Yu Z, Ma R. et al. lncRNA-Xist/miR-101-3p/KLF6/C/EBPα axis promotes TAM polarization to regulate cancer cell proliferation and migration. Mol Ther Nucleic Acids. 2020;23:536-551
179. Luo J, Li Q, Pan J. et al. Expression level of long noncoding RNA H19 in plasma of patients with nonsmall cell lung cancer and its clinical significance. J Cancer Res Ther. 2018;14:860-863
180. Sorin V, Ohana P, Mizrahi A. et al. Regional therapy with DTA-H19 vector suppresses growth of colon adenocarcinoma metastases in the rat liver. Int J Oncol. 2011;39:1407-1412
181. Lavie O, Edelman D, Levy T. et al. A phase 1/2a, dose-escalation, safety, pharmacokinetic, and preliminary efficacy study of intraperitoneal administration of BC-819 (H19-DTA) in subjects with recurrent ovarian/peritoneal cancer. Arch Gynecol Obstet. 2017;295:751-761
182. Gofrit O, Benjamin S, Halachmi S. et al. DNA based therapy with diphtheria toxin-A BC-819: a phase 2b marker lesion trial in patients with intermediate risk nonmuscle invasive bladder cancer. J Urol. 2014;191:1697-1702
Author contact
![]() Corresponding author: Kefeng Wang, address: #36 Sanhao Street, Heping District, Shenyang, Liaoning, China. Email: wang.kefengcom. Tel: +86 18940254849, Fax: +86 24 31939077.
Corresponding author: Kefeng Wang, address: #36 Sanhao Street, Heping District, Shenyang, Liaoning, China. Email: wang.kefengcom. Tel: +86 18940254849, Fax: +86 24 31939077.

 Global reach, higher impact
Global reach, higher impact