10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(13):3369-3380. doi:10.7150/ijbs.61439 This issue Cite
Review
The role of RNA m5C modification in cancer metastasis
1. Hepatic Surgery Center, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430030, China.
2. Hubei Province for the Clinical Medicine Research Center of Hepatic Surgery, Wuhan, Hubei 430030, China.
3. Hubei key laboratory of Hepato-Pancreato-Biliary Diseases, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430030, China.
† Qiaofeng Zhang and Furong Liu contributed equally to this work.
Received 2021-6-17; Accepted 2021-7-19; Published 2021-8-2
Abstract
Epigenetic modification plays a crucial regulatory role in the biological processes of eukaryotic cells. The recent characterization of DNA and RNA methylation is still ongoing. Tumor metastasis has long been an unconquerable feature in the fight against cancer. As an inevitable component of the epigenetic regulatory network, 5-methylcytosine is associated with multifarious cellular processes and systemic diseases, including cell migration and cancer metastasis. Recently, gratifying progress has been achieved in determining the molecular interactions between m5C writers (DNMTs and NSUNs), demethylases (TETs), readers (YTHDF2, ALYREF and YBX1) and RNAs. However, the underlying mechanism of RNA m5C methylation in cell mobility and metastasis remains unclear. The functions of m5C writers and readers are believed to regulate gene expression at the post-transcription level and are involved in cellular metabolism and movement. In this review, we emphatically summarize the recent updates on m5C components and related regulatory networks. The content will be focused on writers and readers of the RNA m5C modification and potential mechanisms in diseases. We will discuss relevant upstream and downstream interacting molecules and their associations with cell migration and metastasis.
Keywords: Metastasis, cell migration, 5-methylcytosine, ribosome acid, regulatory network
Background
In eukaryotes, a wide range of nucleotide modifications, including acetylation, methylation and glycosylation, are essential for cellular biological processes and carcinogenesis. Among the hundreds of chemical modifications, 5-methylcytosine (m5C) is a highly focused epigenetic modification that has been identified in DNA and various RNA families. 5-Methylcytosine in DNA (5mC) is well established as a regulator of gene expression and genome stability [1, 2]. Otherwise, the m5C modification is also present in transcript. The biological roles of this modification in RNA, including RNA export, translation, RNA fragmentation and ribosome composition, were not fully comprehended until recently [3-5]. Using metabolic or radioactive labeling detection, canonical bisulfite sequencing, methylated RNA immunoprecipitation sequencing (MeRIP-seq) and liquid chromatograph mass spectrometer (LC-MS), an increasing number of mapping approaches for m5C have been improved and employed to further elucidate the functions of the m5C modification [6-9]. For example, a newly developed solid-phase method for single RNA molecule sequencing may be used as a solution to the insufficient sensitivity of second-generation sequencing to detect RNA modification [10]. As researchers have increasingly recognized the RNA m5C modification, extensive target sites were discovered in multiple RNA classes and diverse organisms [11-17], revealing the crucial effect of the RNA m5C modification on eukaryotes.
An integrated m5C regulatory network has been revealed, and their writers (methyltransferase), demethylases and readers (proteins that specifically bind to methylation sites) have been identified. Earlier studies have detected DNA methylation induced by the human homologous DNA methyltransferase (DNMT) family, including DNMT1, DNMT2 and DNMT3A/3B. Exclusively, a multiomics study revealed that human DNMT2 not only methylates DNA but also a small RNA (aspartic acid transfer RNA) [18]. In recent years, a notable methyltransferase family, NOP2/Sun RNA methyltransferases (NSUNs), has been shown to methylate multiple sites in transfer RNA [13, 17, 19]. Additionally, some NSUN family members have target sites on mRNA, rRNA, mitochondrial RNA, noncoding RNA and even viral RNA [7, 20-22]. For m5C demethylation, ten-eleven translocation (TET) genes were initially implicated as tumor suppressors [23], but subsequently determined to mediate oxidation to 5-hydroxymethyl, 5-formyl, and 5-carboxylcytosine (hm5C, f5C and ca5C), followed by the excision of f5C or ca5C which is probably induced by thymine DNA glycosidase (TDG) in DNA [24]. Studies at m5C readers and subsequent physiological functions further indicate the complicacy of cascading regulation at the molecular level. The RNA m5C modification is involved in a large number of human cancers, including leukemia, lung cancer, gastric cancer, squamous cell carcinomas, hepatocellular carcinoma and gynecologic cancers [25-29]. The conceivable demethylase TET-mediated oxidation and absence of DNA 5mC are associated with cancer progression [30-34]. In summary, the m5C modification and its elimination are closely associated with molecular and cytological processes in diseases and various cancer types, and thus research on the mechanism and clinical application are hotspots.
In this review, we focus on the pivotal mechanisms of RNA m5C modifications involved in metastasis, cell migration and invasion, discussing their functions in cellular biological processes and tumor development. Classified by enzyme family, we review the recent progress in studies of m5C writers, demethylases and readers to summarize the current knowledge on methylation associated molecular regulation and pathway activation, as well as the reversion of demethylation. We then discuss the clinical advances in the evaluation and prognosis of human cancers.
Reversible methylation of nucleic acids
N6-methyladenosine
In earlier studies, the DNA N6-methyladenosine (m6A) modification was established as a pivotal modification in prokaryotes but was not detectable in eukaryotes. Importantly, m6A modifications have now been identified as closely related to many biological processes and diseases, such as RNA metabolism related gene expression regulation, DNA damage repair, cell development and differentiation, cell cycle, immune response and even cancer progression [5, 35-38]. For instance, research on DNA m6A in the human genome indicated that specific enzymes induce methylation and demethylation-mediated gene transcription by targeting the AGG motif [39]. The well-known m6A methyltransferase and demethylase, methyltransferase-like 3 (METTL3) and fat mass and obesity-associated protein (FTO), respectively, are involved in the DNA damage response to ultraviolet irradiation [40]. In contrast, as the most abundant RNA modification in eukaryotes, m6A plays a crucial role in epigenetic regulation at the transcriptional level. To date, various RNA classes have been verified to be regulated by m6A modification, including precursor mRNAs, long noncoding RNAs, ribosomal RNA and microRNAs [41-43]. The diversity of modification sites implies multiple biological functions that reveal an intricate regulatory network of m6A. Similarly, METTL3-induced m6A promotes the mRNA translation of several oncogenes in human acute myeloid leukemia cells, and then mediates cell proliferation, apoptosis and leukemia progression [44]. FTO functions as a regulator of poly(A) site and 3' UTR processing and exon splicing in nuclear mRNAs [45].
5-methylcytosine
Studies of the DNA 5mC modification have focused on the transformation of the second structure at first. As physiological and pathological effects are being revealed, DNA 5mC-associated methyltransferases are being identified, including DNMT1, DNMT2, DNMT3A and DNMT3B [46]. For instance, the structural and biochemical data of the DNMT1-DNA complex indicate the methylation maintenance function in methylase-induced DNA methylation [47]. Otherwise, DNMT1 translocates to the mitochondrial matrix and binds to mitochondrial DNA, which is associated with asymmetric regulation of the translation of heavy chains and light chains in mtDNA [48]. In the nucleus, a specific complex composed of DNMT3A or 3B and nucleosomes stabilizes free DNMT3A/3B enzymes, protects them from elimination and promotes further DNA methylation [49]. Interestingly, DNMT2 was confirmed to bind DNA in human eukaryotic cells [50]. However, according to whole-genome bisulfite sequencing, DNMT2-involved genome modification lacked detectable DNA methylation patterns in a large number of eukaryotes [51]. In addition, a human homologue of this methylase, tRNA aspartic acid methyltransferase 1 (TRDMT1), has been reported to share a sequence and function similar to DNMT2 in mediating tRNA m5C. We view these two molecules as one RNA m5C methylase, which is accepted by mainstream academics.[18, 52]. Meanwhile, the NSUN family has been revealed to mediate m5C modification in different RNA classes, including tRNA, mRNA, rRNA, vault RNA and enhancer RNA [14, 17, 53-55]. The main targets of NSUNs are the 3' untranslated region of mRNA and various cytosines of tRNA. As the most well-known NSUN family menber, NSUN2 (yeast homologue TRM4) participates in cell cycle progression and tumor growth, probably by targeting at known C34, 48, 49, 50 of tRNA in a range of tumors, such as human squamous cell carcinoma and breast cancer [56, 57]. While other NSUNs have a variety of functions at post-transcriptional level [11, 14, 22, 29, 58-60].
N1-methyladenosine
For the N1-methyladenosine (m1A) modification, related functional research has become more common in the last five years. The biological effect of the reversible m1A modification is to meditate RNA stability and RNA processing, and this modification has been verified to have a transcriptome-wide distribution in mammalian cells and tissues [61, 62]. According to recent studies, m1A is distributed in mRNAs and enriched at translation start sites (5' UTRs), which may promote translation [63, 64]. In addition to the many target sites in mRNAs, m1A also occurs in several regions of tRNAs and mitochondrial genes and regulates corresponding biological processes [65]. Nevertheless, as an essential component of epigenetic modifications, the biological functions of m1A remain ambiguous. The m1A level in mitochondrial tRNAs is significantly altered in various human cancers, and is associated with the clinical prognosis [66]. For example, by catalyzing the demethylation of m1A in tRNA, alkB homolog 1 (ALKBH1) facilitates cancer progression and migration in vivo, resulting in demethylation-induced tRNA fragmentation [67]. In summary, the clinical relevance of the RNA m1A modification remains unclear and requires further identification.
m5C writers, demethylases and readers
Methyltransferases mediating the RNA m5C modification
The formation of m5C indicates the installation of a methyl group on the fifth carbon of cytosine in CpG islands, which is regulated by m5C methyltransferases and demethylases. The so-called m5C “writer” typically possesses highly conserved functional regions that are present in other members of the enzyme family. Numerous m5C writers participate in the methylation of extensive RNAs and epigenetic regulation (Figure 1). DNMT2 proteins contain a DNMT family conserved region in the target recognition domain, which shares the DNA-MTase catalytic triad to methylate tRNA substrates [68]. An in vitro RNA electrophoretic mobility shift assay confirmed that DNMT2 preferentially binds specific mRNAs other than DNA [69]. A similar pattern of double-substrate targeting has been observed for other epigenetic modifications, such as m6A, hm5C and m1A [70-75]. To date, 7 human NSUN variants have been identified that recognize RNAs through aspartate side chains in motif VI and target at specific cytosines [76]. Despite sharing conserved functional sites with other family members, NSUN2 significantly upregulates the abundance of m5C in mRNAs rather than in total RNAs [7]. In addition to mRNA methylation, NSUN2-induced m5C installation in tRNAs and vault RNAs has also been verified [15, 21, 77]. In eukaryotic cells, NSUN1 (yeast homologue NOP2), NSUN4 and NSUN5 (yeast homologue RCM1) target 25S and 28S rRNAs [11, 14, 22]. NSUN3 and NSUN6 individually methylate cytosine 34 of mitochondrial tRNA and cytosine 72 of cytoplasmic tRNA, which improves their stability [60, 78]. These targeting sites at positions 2278 and 2870 in 25S rRNA and position 3782 in 28S rRNA are in highly conserved ribosome regions, suggesting the functional importance of rRNA m5C modification [79]. Unlike mRNA methylation, m5C enrichment in tRNAs is a structure-specific modification. Targeting sites are mostly located in the T stem loop at positions 48-50 and anticodon loop positions 34 and 38. Sites have also been reported at position 72 of the acceptor arm and at the D stem loop. NSUN7 interacts with promoter-derived enhancer RNAs to indirectly regulate gene expression indirectly [55]. The function of NSUN proteins requires an RNA-recognition motif and Rossman-fold catalytic core, which is slightly different from the catalytic site of DNMT2 [80]. Furthermore, a novel cofactor mechanism is essential for the NSUN-mediated m5C modification, such as the S-adenosylmethionine induced methyl transfer process [81].
m5C reversion and the TET family
The oxidation of m5C to hm5C, f5C and ca5C is an essential epigenetic modification in eukaryotes. TET protein-mediated demethylation of DNA 5mC was verified in early studies [82, 83]. Notably, the appellation of “5-methylcytosine eraser” for TETs is not widely accepted because the essence of TET induced demethylation is to replace the modification by facilitating subsequent oxidations. The oxidation cascade relies on the conserved catalytic core at the carboxyl terminus, which depends on a cofactor. A recent study revealed that reduced iron and α-ketoglutarate are recruited by the double-stranded β‑helix domain in the TET catalytic region for oxidation [84]. Interestingly, human TETs prefer hydroxyl and formyl depositions, suggesting a substrate-specific feature [85]. Three influential mechanisms for demethylation of the 5mC oxidized base have been proposed: direct removal of the oxidized methyl group, a passive replication-dependent dilution process, and DNA repair-associated excision of modified nucleotides. Active removal is mediated by TDG coupled with base excision repair, which is specific to the formation of f5C and ca5C [86, 87]. In addition, passive degradation of oxidized substrates occurs frequently during persistent DNA replication. This particular process was reported to be related to 5mC demethylation [88]. Finally, base excision repair-induced removal of 5mC residues has been widely accepted, although the detailed mechanism of accurate residue elimination has not been elucidated. In addition, non-canonical DNA mismatch repair-induced removal of alkylated and oxidized nucleotides is one of the probable pathways mediating active DNA demethylation [89].
Recently, TET protein-induced RNA hm5C modification has been reported in specific eukaryotic cells, including mouse embryonic stem cells, bone marrow mononuclear cells, Drosophila and mouse brain cells [70, 75, 90, 91]. For example, the distribution and enrichment of RNA hm5C in mouse brain tissues was verified by a dot blot analysis, and this modification is associated with Parkinson's disease. In human tissues and cells, a low level of RNA hm5C was detected using improved LC-MS/MS methods [92, 93]. Researches have not clearly determined whether hm5C modifications in RNA and DNA use the same mechanism and regulatory patterns. A recent study suggested that isocitrate dehydrogenases block TET-induced oxidative activity in both DNA and mRNA based on the results of an LC-MS/MS analysis [94]. Hence, further study at the molecular level is needed to assess the functional pattern of RNA hm5C. However, few mature sequencing methods at single-base resolution are available for oxidized m5C mapping, and thus the study of the mechanism of RNA m5C demethylation becomes quite challenging. Immunoprecipitation is an RNA-friendly sequencing method to further reveal the elusive biological functions of m5C oxidation and demethylation [90]. Moreover, a novel bisulfite-free and high-resolution method uses peroxotungstate to conduct the hm5C-to-T transition, following cDNA synthesis and base-resolution sequencing [9], which is helpful to comprehensively identify the hm5C distribution and function.
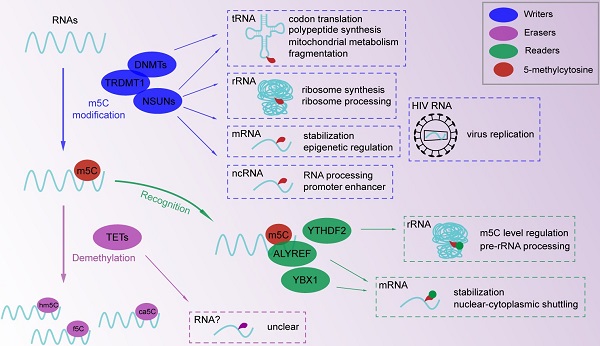
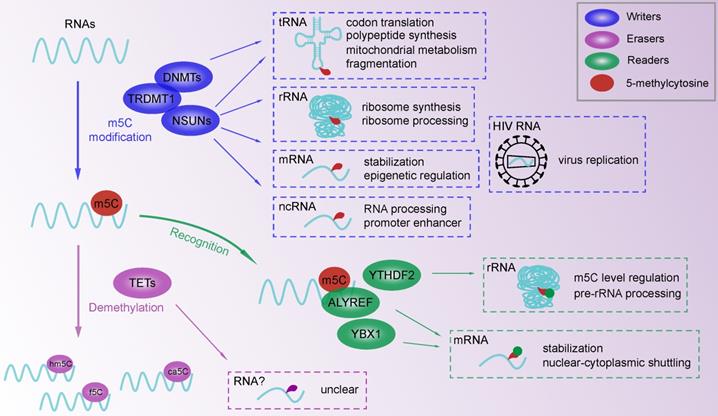
RNA m5C modifications with functions of m5C writers and readers. M5C methylation in tRNA, rRNA, mRNA and ncRNA is involved in RNA processing and metabolism. Binding proteins (YTHDF2, ALYREF, YBX1) for m5C modification take part in m5C modification in methylase-dependent or independent way. A multicomponent regulatory network is constructed to affect epigenetic regulation.
Proteins binding to RNA m5C sites
After the m5C modification of RNA, several proteins specifically bind to modified sites, leading to the subsequent regulation of biological processes (Figure 1). We named these particular proteins “readers” due to their capability of recognizing m5C-containing oligonucleotides. According to a recent study, the m6A binding protein, YTH domain-containing family 2 (YTHDF2), shares a conserved residue at the hydrophobic pocket for binding m5C-modified RNA [95]. Specifically, Aly/REF export factor (ALYREF) displayed a significantly enhanced binding ability to m5C-modified mRNAs across the cytoplasm and nucleus, which was confirmed by ALYREF protein sequencing [7]. Found in zebrafish early embryos, Y-box binding protein 1 (YBX1), a well-known multifunctional DNA and RNA binding protein , can recognize and bind m5C modified mRNAs by residue Trp45 in the cold shock domain of YBX1 [96, 97]. Overall, the m5C regulatory network mainly consists of functional readers and downstream effector, which needs further investigation of novel specific binding proteins.
Biological function of m5C in eukaryote and disease
The m5C modification is widely distributed in RNA involved in various biological processes. As a fundamental function of the RNA m5C modification, the regulation of RNA function and metabolism is crucial to the development of biological processes (Figure 1). With improved detection approaches for RNA methylation assays, more types of RNA have been identified as m5C targets. The RNA m5C modification induced by NSUN1 (yeast homologue Nop2) and NSUN5 (yeast homologue Rcm1) regulate ribosome synthesis and processing [11, 14]. NSUN4 even participates in mitochondrial ribosome biogenesis [29, 58]. The processing of mature RNA is also associated with m5C methylation in mRNAs and noncoding RNAs (ncRNAs). For example, DNMT2 is responsible for RNA processing by relocating and interacting with mRNA as a component of the processing complex [98]. The processing of noncoding vault RNA in human cells is induced by the m5C modification at C69, which maintains the cell cycle in progenitor cells [21]. The m5C modification promotes RNA stabilization, transportation and translation, but not translation termination induced by NSUN6 mediated mRNA methylation to allow RNAs to perform their regular functions [99]. For instance, NSUN2 targets the 3' untranslated region and stabilizes the mRNA of heparin binding growth factor, which regulates cancer progression [54]. The reader YBX1 recognizes and binds m5C-modified mRNAs at Trp45 to enhance mRNA stability and early embryogenesis in zebrafish [96]. For nucleus-to-cytoplasm transportation, the reader ALYREF interacts with NSUN2-methylated mRNA to mediate shuttling and subsequent translation in the cytoplasm [100]. In addition, protein translation and polypeptide synthesis involving tRNAs are regulated by DNMT2-induced m5C of the anticodon stem-loop [12, 77, 101]. The RNA m5C modification is involved in epigenetic regulation by affecting RNA metabolism. RNA fragmentation is a common metabolic pattern of tRNA that is related to epigenetic regulation and cancer progression [102]. DNMT2 and NSUN2 were reported to be associated with the biogenesis of tRNA-derived noncoding fragments by altering tRNA fragmentation [103, 104]. In addition, NSUN3 targets mitochondrial tRNA, which may be responsible for energy metabolism and protein synthesis [22, 59, 60].
Since RNA function and epigenetic regulation are strongly associated with the occurrence and development of diseases, the role of the RNA m5C modification in diseases and cancer is noteworthy. The disorder of tRNA fragmentation induced by DNMT2 leads to high-fat-diet-induced metabolic disturbance in sperm cells [103]. The accumulation of tRNA-derived small RNA fragments due to NSUN2 mutation and the absence of tRNA m5C leads to reduced protein translation rates, which activates stress pathways and promotes neuronal apoptosis [105]. NSUN2 also targets tRNAs in Purkinje cells and neuroepithelial stem cells, which regulate cell replication, differentiation, migration and neurocognitive development [19, 106]. NSUN5 was reported to be a promoter of radial glial cell migration and cerebral cortex development [107]. Based on these findings, the RNA m5C modification is strongly correlated with dysplasia and dysfunction of the nervous system. Diseases related to energy metabolism disorder are also associated with the mitochondrial RNA m5C modification. For example, NSUN3 may be associated with multisystem mitochondrial disease associated with a combined oxidative phosphorylation deficiency [108]. In leukemia cells, NSUN3/DNMT2 and NSUN1 regulate the formation of 5-azacitidine-sensitive chromatin structures in an antagonistic manners [25]. Additionally, the canonical DNA demethylase TET2 was reported to mediate mRNA m5C that leads to myelopoiesis in infections, such as sepsis and parasitosis, in the mammalian system [91].
The RNA m5C modification in malignant solid tumors has been a focus of cancer research at the epigenetic level. Various m5C writers and readers play crucial roles in cancer progression, including cell proliferation, differentiation, migration, invasion and drug sensitivity. Taking the typical m5C methylase NSUN2 as an example, NSUN2 has been shown to participate in multiple pan-cancer pathways: promoting cell progression, migration and invasion in breast cancer [57, 109]; promoting tumorigenesis and development in skin cancer [27]; promoting cell growth, tumor progression and metastasis in urothelial carcinoma of the bladder [54]; promoting tumorigenesis and cell proliferation in gall bladder cancer [110]; increasing the sensitivity of HeLa cells to 5-fluorouracil [28]; promoting cell migration, invasion and drug resistance in esophageal squamous cell carcinoma (ESCC) [111]; promoting proliferation in gastric cancer [112]; promoting tumorigenesis, migration and invasion in hepatocellular carcinoma (HCC) [113]. NSUN4, NSUN5, NSUN6 and NSUN7 are associated with cancer development in HCC, head and neck squamous cell carcinoma (HNSCC), pancreatic cancer and glioma [29, 114-116]. In addition, the tumor immune microenvironment is affected by NSUN2 and NSUN6 through multiple pathways, such as the regulation of RNA metabolism, the cell cycle and immune cell activation [117]. In addition to the NSUN family-induced RNA m5C modification in cancer, DNMT1 overexpression directly leads to hypermethylation of tumor suppressor genes, which results in lung tumorigenesis and a poor prognosis [26]. Regarding m5C readers, ALYREF is associated with tumor progression in patients with glioblastoma and HCC, but regulates glucose metabolism and tumorigenesis in individuals with bladder cancer [29, 118]. YBX1 promotes cell growth, tumor progression and metastasis in bladder urothelial carcinoma [54]. More importantly, m5C readers are associated with cancer development in collaboration with methylases [54, 119]. For the TET family, a downregulated RNA hm5C level was reported in cancerous tissues compared to adjacent normal tissues in human colorectal carcinoma and hepatocellular carcinoma, suggesting possible anticancer mechanisms [120]. In general, upregulated RNA m5C components and high levels of the m5C modification are significantly correlated with malignancy and a poor prognosis in patients with the cancers mentioned above and some other cancer types, such as head and neck squamous cell carcinoma (HSNCC) and ovarian cancer [27, 121, 122].
In addition to eukaryotes, in RNA virus HIV-1, the transcription and replication of TAR RNA are restricted by NSUN1 induced RNA methylation [20], while the splicing and translation of mRNA are related to NSUN2-induced RNA methylation [123]. The efficient prolongation of HIV latency in CD4+ T cells suggests a possible treatment for acquired immune deficiency syndrome.
Functional role of RNA m5C in facilitating metastasis
As the first beneficiary in the field of nucleic acid methylation, m6A in RNA possesses multiple epigenetic regulatory activities in various biological processes, diseases and especially cancer [124]. With a specific regulator, RNA m5C can mediates the activation of oncogenic pathways and forms a microenvironment suitable for the migration and metastasis of various cancer cell lines (Table 1). For instance, NSUN5 and NSUN6 were reported to be associated with metastasis in skin cancer and breast cancer. The former methylase and the specific reader ALYREF are overexpressed in metastatic stage of head and neck squamous cell carcinoma [119]. Although the latter participates in RNA-protein interactions, an MST1/2-antagonizing lncRNA for YAP activation inhibits the activity of macrophage stimulating 1 (a protein serine kinase) in an NSUN6-dependent manner, which facilitates bone metastasis in breast cancer [125]. In urothelial carcinoma of the bladder, NSUN2 targets the 3' untranslated region (3'-UTR) and stabilizes the mRNA of HGDF by generating the RNA m5C modification, while the reader YBX1 binds to the m5C region with the help of the partner protein ELAVL1 (an mRNA stability maintainer). The activation of the NSUN2/YBX1/HDGF axis was proven to promote cell growth, tumor progression and metastasis [54]. The expression levels of NSUN2 and NSUN3 are downregulated by knocking out proto-oncogenic isozymes (sphingosine kinases 1 and 2, SK1 and SK2) in metastatic prostate cancer and breast cancer. SK1 and SK2 silencing alter the activation of protumorigenic genes relevant to classic membrane signal transduction pathways, such as epidermal growth factor, the EMT, cell cycle, cell motility and DNA stability [126]. NSUN2 recognizes and binds to the canonical lncRNA H19 and NMR (NSUN2 methylated lncRNA). In ESCC, methylated NMR interacts with bromodomain PHD finger transcription factor (BPTF) and leads to the overexpression of oncogenes (MMP3 and MMP10) that are involved in cancer cell migration and invasion by activating ERK 1/2 [111]. Although H19 expression is elevated in various tumors, the lncRNA is stabilized by the NSUN2-induced m5C modification, which specifically binds to oncoprotein G3BP1. The mechanism was verified to be associated with tumorigenesis, malignancy and cell migration and invasion. Poor differentiation of cancer cells was observed in H19-overexpressing hepatocellular carcinoma [113]. The mRNA encoding autotaxin (ATX) is one target of NSUN2. ATX and its product are famous for exerting multiple membrane functions and mediating cancer development, and activation of the NSUN2/ATX/ALYREF axis promotes mRNA nucleus to cytoplasm transport and cell migration in glioma [127]. NSUN2 induced tRNA methylation in neuroepithelial stem cells also mediates differentiation and migration. The underlying mechanism is related to the activation of chemoattractant fibroblast growth factor 2 (FGF2), which facilitates cell maturation and cellular activities [106]. Moreover, following the upregulation of NSUN2, the proliferation and migration of HEK 293 cells is enhanced by the overexpression of oncogenes that are associated with the cell cycle, focal adhesion, TGF-β signaling pathway and Notch signaling pathway [52, 128]. Taken together, NSUN2 is a notable biomarker for predicting tumor metastasis and the prognosis and a promising therapeutic target in clinic applications. Although NSUN2 overexpression was reported to promote metastasis and cell invasion by promoter methylation [109], these studies explicitly indicate that NSUN family-induced RNA m5C modifications play a pivotal role in tumor metastasis and cell motility (Figure 2).
M5C RNA methylation induced cell migration and metastasis
| Cell type | M5C component | Role | Effects | Refs |
|---|---|---|---|---|
| Bladder urothelial carcinoma | NSUN2 & YBX1 | Writer & reader | NSUN2 stabilize the mRNA of HDGF, while YBX1 bind to m5C methylation with a partner protein ELAVL1 and promote cell growth, tumor progression and metastasis | [54] |
| Breast cancer | NSUN2 | Writer | NSUN2 overexpression promotes metastasis and invasion of breast cancer | [109] |
| Breast cancer | NSUN6 | Writer | The antagonizing lncRNA for YAP activation inhibits activity of the protein serine kinase MST1 in a NSUN6-dependent way, which facilitates bone metastasis in breast cancer | [125] |
| ESCC | NSUN2 | Writer | NSUN2 methylated lncRNA bind to chromatin regulator BPTF and promotes MMPs expression through ERK pathway | [111] |
| Glioma | NSUN2 | Writer | NSUN2 deficiency inhibits glioma cell migration by striking autotaxin and lysophosphatidic acid associated pathway | [100] |
| HCC | NSUN2 | Writer | NSUN2 methylated H19 bind to oncoprotein G3BP1, which may be associated with tumor genesis, migration and invasion | [113] |
| HEK 293 cell | NSUN2 | Writer | NSUN2 promote cell proliferation and migration by regulating oncogenes that are associated with cell cycle, focal adhesion, TGF-β signaling pathway and Notch signaling pathway | [52], [128] |
| HNSCC | NSUN5 & ALYREF | Writer & reader | ALYREF and NSUN5 are overexpressed in metastasis stage of head and neck squamous cell carcinoma | [119] |
| Neuroepithelial stem cells | NSUN2 | Writer | NSUN2 mediate FGF2 induced migration and differentiation of neuroepithelial stem cells | [106] |
| Prostate and breast cancer | NSUN2 | Writer | Expression level of NSUN2 and NSUN3 are regulated by proto-oncogenic isozymes (sphingosine kinases 1 and 2) in prostate metastasis cancer and breast cancer, by a possible mediation of epidermal growth factor associated cell migration | [126] |
| Prostate cancer | DNMTs | Writer | DNMT1, DNMT2, and DNMT3 are involeved in lymph node metastases of prostate cancer | [144] |
M5C writers and readers function as promoters of metastasis in pan-cancer. ESCC, esophageal squamous cell carcinoma. HCC, hepatocellular carcinoma. HNSCC, head and neck squamous cell carcinoma
Molecular interactions between m5C components and metastasis related genes. M5C writers promote metastasis in various cancers by regulate oncogenes or suppressor genes. Several writers and readers are associated with cancer metastasis by unclear mechanism. Methylase and binding protein for m5C can work synergistically and promote metastasis.
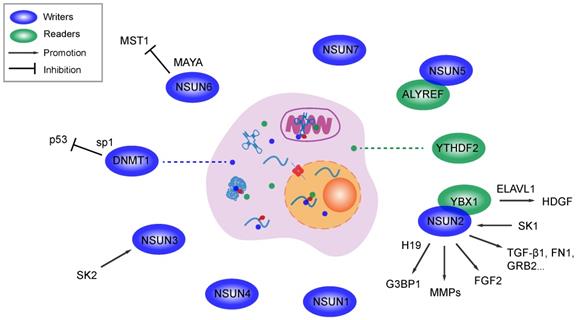
As an immature RNA methylase family, the relationship between the DNMT-induced m5C modification and cell mobility or metastasis remains unclear. However these proteins play a supporting role in pathways through which tumor suppressor genes function in a DNA methylation-dependent pattern. For example, DNMT1 activity is increased by high level of specificity protein 1 (SP1)-induced endogenous p53 inactivation in lung cancer, while p53 is now better known for its anticancer effect than as an oncoprotein in various tumors, as it is associated with cell survival, invasion, tumor growth, metastasis, the EMT and multiple pro-oncogenic signaling pathways [129]. Analogously, TETs-associated DNA demethylation mediates metastasis in malignancies such as hepatocellular carcinoma and lung cancer [130, 131]. The abovementioned effects of these writers, demethylases and readers on DNA 5mC do not refute their potential to mediate metastasis and cell migration. More research on m5C modification at the transcriptional level and related oncogenic pathways is needed to improve our knowledge of the epigenetic regulation of tumor metastasis.
Conclusions
Epigenetic modification is increasing in significance in biological processes. The elucidation of nucleic acid methylation has been reported in recent years. Our review focused on the RNA m5C modification in tumor cells and other diseases is inevitably deficient and has the potential for further exploration. The m5C modification may not be as popular as sibling methylation m6A, but it is still an integral part of epigenetic modification. Abundant writers and readers for RNA m5C have been identified, leading to a variety of interactions and regulation of biological processes. Typically, DNMT2 recognizes and methylates tRNA through a highly conserved region that was previously linked to DNA methylation. Along with NSUN2, NSUN3 and NSUN6, these enzymes target tRNAs and participate in tRNA fragmentation and energy metabolism. NSUN1, NSUN4 and NSUN5 preferentially bind rRNA and induce the process of ribosome metabolism. For mRNA, DNMT2 and NSUN2 are the main writers, since mRNA m5C is critical for numerous cellular biological processes, such as cell growth, differentiation, movement and RNA modification. Furthermore, NSUN2 and NSUN7 bind to noncoding RNAs that regulate cell metabolism [21, 55, 115]. Writers of RNA m5C usually work in partnership with readers. YBX1 and ALYREF tend to recognize NSUN-induced methylation, which promotes the transportation and improves the stability of target mRNAs. YTHDF2, on the other hand, binds to m5C-modified rRNA at the same conserved residue for the m6A modification. Unfortunately, erasers for RNA m5C have not been comprehensively identified. Members of the well-known DNA demethylase family TETs trigger cascade oxidations, which may activate the underlying mechanism of RNA demethylation in the m5C region. Since a duality of methylation in DNA and RNA was found in m6A, m5C, hm5C and m1A modifications, studies aiming to identify whether a co-function of same methylation component induces RNA and DNA methylation or demethylation exists and the related mechanisms and outcomes are needed. Nevertheless, different types of RNA methylation are able to collaborate to regulate downstream targets and modulate the development of diseases. For example, NSUN2-mediated m5C collaborates with METTL-mediated m6A promote p21 expression, and various types of RNA methylation participate in glioma genesis and progression [132, 133].
RNA m5C is relevant to numerous diseases, including energy metabolism disorders, hematopoietic system diseases, viral infection and the most remarkable pathological process, the development of malignancies. The expression of RNA m5C writers and readers is significantly correlated with a poor prognosis and the TNM classification of neoplasms. These writers and readers are mainly involved in pathways of tumorigenesis and metastasis in the brain, lung, breast, prostate and others tissue. Many cytokines, such as HDGF, TGF-β, FGF2 and G3BP1, which are validated tumor promoters, participate in cell migration and metastasis induced by the m5C modification, indicating a strong correlation between the RNA m5C level and mobility of cancer cells. Moreover, some studies discovered high RNA m5C levels in circulating tumor cells [134]. Since the global amount of m5C in blood DNA was identified as a promising marker for cancer prognosis, increased general RNA m5C levels may be a novel indicator of metastasis. Nevertheless, a more comprehensive regulatory network for cancer cell transfer requires many mechanistic investigations of epigenetic modifications. RNA m5C has the potential to be a unique target for clinical cancer assessment and treatment.
Regarding the existing targets of RNA m5C modification, gene therapy is a feasible treatment for RNA m5C-induced diseases and cancer development. By modifying the gene sequence of key RNA m5C methylases, such as NSUN2, reduced RNA methylation may prevent the occurrence of disease or reverse cancer progression. However, considering the non-specificity of gene therapy, a well-designed inhibitor based on molecular research may effectively reduce the function of a specific RNA m5C writer and reverse the development of diseases since inhibitors of m6A components are being used in cancer therapy. In addition, the upstream molecules that regulate the functional activity of the RNA m5C component may represent an optional treatment for diseases related to specific molecules. For example, aurora kinase B phosphorylates NSUN2 at Ser-139 and reduces RNA methylation levels, suggesting that regulation at the protein level is feasible [135]. More research focused on molecular structures and functional pathways is needed to completely understand the effect of the RNA m5C modification and exploit it in clinical applications.
However, barriers still exist to further RNA m5C research when the final goal is to fully understand the RNA m5C modification and translate the findings into the clinic. Regarding the detection and research trends of RNA m6A modification, detection methods based on homogeneous techniques are still evolving to obtain better resolution or sensitivity [136]. We anticipate that RNA methylation detection is both a limitation in research and a key to breakthrough. Because traditional high-throughput techniques for identifying m5C sites are inefficient, computer-based learning techniques that quickly and accurately predict RNA m5C modifications are urgently needed. For example, the novel m5C detection method, direct RNA sequencing revealed a more complex transcriptome characterization than expected in Arabidopsis [137]. New methods such as “iRNA-m5C” and “iRNA-m5C_SVM” for RNA m5C site detection with good predictive performance will be helpful in structural and functional research on the RNA m5C modification [138, 139]. However, the immediate application of these new approaches will be a further obstacle in research and clinical translation. Integration of broad data sets and establishment of databases are also effective for improving RNA m5C research. Database websites such as “RMBase” integrate a large number of RNA methylation data for review and verification while “RMDisease” and “RMVar” have been applied in RNA modification-related disease research [140-142]. Prediction tools such as “PEA-m5C” trained with the forest algorithm predict candidate m5C modification sites for functional research [143].
Abbreviations
M5C: RNA 5-methylcytosine; 5mC: DNA 5-methylcytosine; DNMT: DNA methyltransferase; TRDMT1: tRNA aspartic acid methyltransferase; NSUN: NOP2/Sun RNA methyltransferase; TET: ten-eleven translocation enzyme; YTHDF2: YTH domain-containing family 2; ALYREF: Aly/REF export factor; YBX1: Y-box binding protein 1; hm5C: 5-hydroxymethylcytosine; f5C: 5-formylcytosine; ca5C: 5-carboxylcytosine; HNSCC: head and neck squamous cell carcinoma; ESCC: esophageal squamous cell carcinoma; HCC: hepatocellular carcinoma; NMR: NSUN2 methylated lncRNA.
Authors' contributions
BXZ and ZGZ conceived this study and provided financial and administrative support. QFZ and FRL did most of the data generalization and summarization. WC, HRM and HFL help collecting supplement information. All authors further discussed about data and prepared the panels for tables and figures. QFZ wrote the manuscript. All authors read and approved the final manuscript.
Availability of data and materials
The researches that support the conclusion of this review have been referenced within this article.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Breiling A, Lyko F. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics Chromatin. 2015;8:24
2. Greenberg MVC, Bourc'his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 2019;20:590-607
3. Trixl L, Lusser A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip Rev RNA. 2019;10:e1510
4. García-Vílchez R, Sevilla A, Blanco S. Post-transcriptional regulation by cytosine-5 methylation of RNA. Biochim Biophys Acta Gene Regul Mech. 2019;1862:240-252
5. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell. 2017;169:1187-1200
6. Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ. et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023-5033
7. Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY. et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27:606-625
8. Cao J, Shu X, Feng XH, Liu J. Mapping messenger RNA methylations at single base resolution. Curr Opin Chem Biol. 2021;63:28-37
9. Yuan F, Bi Y, Siejka-Zielinska P, Zhou YL, Zhang XX, Song CX. Bisulfite-free and base-resolution analysis of 5-methylcytidine and 5-hydroxymethylcytidine in RNA with peroxotungstate. Chem Commun (Camb). 2019;55:2328-2331
10. Athapattu US, Amarasekara CA, Immel JR, Bloom S, Barany F, Nagel AC. et al. Solid-phase XRN1 reactions for RNA cleavage: application in single-molecule sequencing. Nucleic Acids Res. 2021;49:e41
11. Janin M, Ortiz-Barahona V, de Moura MC, Martínez-Cardús A, Llinàs-Arias P, Soler M. et al. Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol. 2019;138:1053-1074
12. Jeltsch A, Ehrenhofer-Murray A, Jurkowski TP, Lyko F, Reuter G, Ankri S. et al. Mechanism and biological role of Dnmt2 in Nucleic Acid Methylation. RNA Biol. 2017;14:1108-1123
13. Li J, Li H, Long T, Dong H, Wang ED, Liu RJ. Archaeal NSUN6 catalyzes m5C72 modification on a wide-range of specific tRNAs. Nucleic Acids Res. 2019;47:2041-2055
14. Sharma S, Yang J, Watzinger P, Kötter P, Entian KD. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 2013;41:9062-9076
15. Shinoda S, Kitagawa S, Nakagawa S, Wei FY, Tomizawa K, Araki K. et al. Mammalian NSUN2 introduces 5-methylcytidines into mitochondrial tRNAs. Nucleic Acids Res. 2019;47:8734-8745
16. Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G. et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. Embo j. 2015;34:2350-2362
17. Van Haute L, Lee SY, McCann BJ, Powell CA, Bansal D, Vasiliauskaitė L. et al. NSUN2 introduces 5-methylcytosines in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2019;47:8720-8733
18. Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X. et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395-398
19. Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V. et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90:856-863
20. Kong W, Biswas A, Zhou D, Fiches G, Fujinaga K, Santoso N. et al. Nucleolar protein NOP2/NSUN1 suppresses HIV-1 transcription and promotes viral latency by competing with Tat for TAR binding and methylation. PLoS Pathog. 2020;16:e1008430
21. Sajini AA, Choudhury NR, Wagner RE, Bornelöv S, Selmi T, Spanos C. et al. Loss of 5-methylcytosine alters the biogenesis of vault-derived small RNAs to coordinate epidermal differentiation. Nat Commun. 2019;10:2550
22. Spåhr H, Habermann B, Gustafsson CM, Larsson NG, Hallberg BM. Structure of the human MTERF4-NSUN4 protein complex that regulates mitochondrial ribosome biogenesis. Proc Natl Acad Sci U S A. 2012;109:15253-15258
23. Saint-Martin C, Leroy G, Delhommeau F, Panelatti G, Dupont S, James C. et al. Analysis of the ten-eleven translocation 2 (TET2) gene in familial myeloproliferative neoplasms. Blood. 2009;114:1628-1632
24. Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet. 2017;18:517-534
25. Cheng JX, Chen L, Li Y, Cloe A, Yue M, Wei J. et al. RNA cytosine methylation and methyltransferases mediate chromatin organization and 5-azacytidine response and resistance in leukaemia. Nat Commun. 2018;9:1163
26. Lin RK, Wu CY, Chang JW, Juan LJ, Hsu HS, Chen CY. et al. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res. 2010;70:5807-5817
27. Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J. et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534:335-340
28. Okamoto M, Fujiwara M, Hori M, Okada K, Yazama F, Konishi H. et al. tRNA modifying enzymes, NSUN2 and METTL1, determine sensitivity to 5-fluorouracil in HeLa cells. PLoS Genet. 2014;10:e1004639
29. He Y, Yu X, Li J, Zhang Q, Zheng Q, Guo W. Role of m(5)C-related regulatory genes in the diagnosis and prognosis of hepatocellular carcinoma. Am J Transl Res. 2020;12:912-922
30. Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J. et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32:663-669
31. Liu Z, Lin H, Gan Y, Cui C, Zhang B, Gu L. et al. P16 Methylation Leads to Paclitaxel Resistance of Advanced Non-Small Cell Lung Cancer. J Cancer. 2019;10:1726-1733
32. Frycz BA, Murawa D, Borejsza-Wysocki M, Marciniak R, Murawa P, Drews M. et al. Decreased expression of ten-eleven translocation 1 protein is associated with some clinicopathological features in gastric cancer. Biomed Pharmacother. 2014;68:209-212
33. Ma L, Qi T, Wang S, Hao M, Sakhawat A, Liang T. et al. Tet methylcytosine dioxygenase 1 promotes hypoxic gene induction and cell migration in colon cancer. J Cell Physiol. 2019;234:6286-6297
34. Hsu CH, Peng KL, Kang ML, Chen YR, Yang YC, Tsai CH. et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2012;2:568-579
35. Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol Cancer. 2020;19:88
36. Liu S, Zhuo L, Wang J, Zhang Q, Li Q, Li G. et al. METTL3 plays multiple functions in biological processes. Am J Cancer Res. 2020;10:1631-1646
37. Cao G, Li HB, Yin Z, Flavell RA. Recent advances in dynamic m6A RNA modification. Open Biol. 2016;6:160003
38. Zhang X, Blumenthal RM, Cheng X. A Role for N6-Methyladenine in DNA Damage Repair. Trends Biochem Sci. 2021;46:175-183
39. Xiao CL, Zhu S, He M, Chen D, Zhang Q, Chen Y. et al. N(6)-Methyladenine DNA Modification in the Human Genome. Mol Cell. 2018;71:306-318.e307
40. Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W. et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543:573-576
41. Coker H, Wei G, Brockdorff N. m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim Biophys Acta Gene Regul Mech. 2019;1862:310-318
42. Zhou Y, Kong Y, Fan W, Tao T, Xiao Q, Li N. et al. Principles of RNA methylation and their implications for biology and medicine. Biomed Pharmacother. 2020;131:110731
43. Ignatova VV, Stolz P, Kaiser S, Gustafsson TH, Lastres PR, Sanz-Moreno A. et al. The rRNA m(6)A methyltransferase METTL5 is involved in pluripotency and developmental programs. Genes Dev. 2020;34:715-729
44. Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G. et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369-1376
45. Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, Vanacova S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing. Nucleic Acids Res. 2017;45:11356-11370
46. Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018;19:81-92
47. Song J, Teplova M, Ishibe-Murakami S, Patel DJ. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science. 2012;335:709-712
48. Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci U S A. 2011;108:3630-3635
49. Sharma S, De Carvalho DD, Jeong S, Jones PA, Liang G. Nucleosomes containing methylated DNA stabilize DNA methyltransferases 3A/3B and ensure faithful epigenetic inheritance. PLoS Genet. 2011;7:e1001286
50. Dong A, Yoder JA, Zhang X, Zhou L, Bestor TH, Cheng X. Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res. 2001;29:439-448
51. Raddatz G, Guzzardo PM, Olova N, Fantappié MR, Rampp M, Schaefer M. et al. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc Natl Acad Sci U S A. 2013;110:8627-8631
52. Xue S, Xu H, Sun Z, Shen H, Chen S, Ouyang J. et al. Depletion of TRDMT1 affects 5-methylcytosine modification of mRNA and inhibits HEK293 cell proliferation and migration. Biochem Biophys Res Commun. 2019;520:60-66
53. Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y. et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4:255-261
54. Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X. et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21:978-990
55. Aguilo F, Li S, Balasubramaniyan N, Sancho A, Benko S, Zhang F. et al. Deposition of 5-Methylcytosine on Enhancer RNAs Enables the Coactivator Function of PGC-1α. Cell Rep. 2016;14:479-492
56. Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol. 2006;16:971-981
57. Frye M, Dragoni I, Chin SF, Spiteri I, Kurowski A, Provenzano E. et al. Genomic gain of 5p15 leads to over-expression of Misu (NSUN2) in breast cancer. Cancer Lett. 2010;289:71-80
58. Cámara Y, Asin-Cayuela J, Park CB, Metodiev MD, Shi Y, Ruzzenente B. et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527-539
59. Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell CA. et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun. 2016;7:12039
60. Nakano S, Suzuki T, Kawarada L, Iwata H, Asano K, Suzuki T. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNA(Met). Nat Chem Biol. 2016;12:546-551
61. Oerum S, Dégut C, Barraud P, Tisné C. m1A Post-Transcriptional Modification in tRNAs. Biomolecules. 2017 7
62. Xiong X, Li X, Yi C. N(1)-methyladenosine methylome in messenger RNA and non-coding RNA. Curr Opin Chem Biol. 2018;45:179-186
63. Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS. et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441-446
64. Li X, Xiong X, Wang K, Wang L, Shu X, Ma S. et al. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol. 2016;12:311-316
65. Zhang C, Jia G. Reversible RNA Modification N(1)-methyladenosine (m(1)A) in mRNA and tRNA. Genomics Proteomics Bioinformatics. 2018;16:155-161
66. Idaghdour Y, Hodgkinson A. Integrated genomic analysis of mitochondrial RNA processing in human cancers. Genome Med. 2017;9:36
67. Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J. et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019;47:2533-2545
68. Jurkowski TP, Meusburger M, Phalke S, Helm M, Nellen W, Reuter G. et al. Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism. Rna. 2008;14:1663-1670
69. Dev RR, Ganji R, Singh SP, Mahalingam S, Banerjee S, Khosla S. Cytosine methylation by DNMT2 facilitates stability and survival of HIV-1 RNA in the host cell during infection. Biochem J. 2017;474:2009-2026
70. Miao Z, Xin N, Wei B, Hua X, Zhang G, Leng C. et al. 5-hydroxymethylcytosine is detected in RNA from mouse brain tissues. Brain Res. 2016;1642:546-552
71. Wang X, Huang J, Zou T, Yin P. Human m(6)A writers: Two subunits, 2 roles. RNA Biol. 2017;14:300-304
72. Sathyamoorthy B, Shi H, Zhou H, Xue Y, Rangadurai A, Merriman DK. et al. Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A. Nucleic Acids Res. 2017;45:5586-5601
73. Wan L, Lam SL, Lee HK, Guo P. Effects of Adenine Methylation on the Structure and Thermodynamic Stability of a DNA Minidumbbell. Int J Mol Sci. 2021 22
74. DeNizio JE, Liu MY, Leddin EM, Cisneros GA, Kohli RM. Selectivity and Promiscuity in TET-Mediated Oxidation of 5-Methylcytosine in DNA and RNA. Biochemistry. 2019;58:411-421
75. Lan J, Rajan N, Bizet M, Penning A, Singh NK, Guallar D. et al. Functional role of Tet-mediated RNA hydroxymethylcytosine in mouse ES cells and during differentiation. Nat Commun. 2020;11:4956
76. Bohnsack KE, Höbartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m⁵C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes (Basel). 2019 10
77. Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S. et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900-905
78. Liu RJ, Long T, Li J, Li H, Wang ED. Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6. Nucleic Acids Res. 2017;45:6684-6697
79. Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK. et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303-d307
80. King MY, Redman KL. RNA methyltransferases utilize two cysteine residues in the formation of 5-methylcytosine. Biochemistry. 2002;41:11218-11225
81. Long T, Li J, Li H, Zhou M, Zhou XL, Liu RJ. et al. Sequence-specific and Shape-selective RNA Recognition by the Human RNA 5-Methylcytosine Methyltransferase NSun6. J Biol Chem. 2016;291:24293-24303
82. Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129-1133
83. Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472-479
84. Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341-356
85. Hu L, Lu J, Cheng J, Rao Q, Li Z, Hou H. et al. Structural insight into substrate preference for TET-mediated oxidation. Nature. 2015;527:118-122
86. Weber AR, Krawczyk C, Robertson AB, Kuśnierczyk A, Vågbø CB, Schuermann D. et al. Biochemical reconstitution of TET1-TDG-BER-dependent active DNA demethylation reveals a highly coordinated mechanism. Nat Commun. 2016;7:10806
87. Malik SS, Coey CT, Varney KM, Pozharski E, Drohat AC. Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA. Nucleic Acids Res. 2015;43:9541-9552
88. Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM. et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841-4849
89. Grin I, Ishchenko AA. An interplay of the base excision repair and mismatch repair pathways in active DNA demethylation. Nucleic Acids Res. 2016;44:3713-3727
90. Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R. et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351:282-285
91. Shen Q, Zhang Q, Shi Y, Shi Q, Jiang Y, Gu Y. et al. Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature. 2018;554:123-127
92. Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S. et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc. 2014;136:11582-11585
93. Zhang HY, Xiong J, Qi BL, Feng YQ, Yuan BF. The existence of 5-hydroxymethylcytosine and 5-formylcytosine in both DNA and RNA in mammals. Chem Commun (Camb). 2016;52:737-740
94. Xu Q, Wang K, Wang L, Zhu Y, Zhou G, Xie D. et al. IDH1/2 Mutants Inhibit TET-Promoted Oxidation of RNA 5mC to 5hmC. PLoS One. 2016;11:e0161261
95. Dai X, Gonzalez G, Li L, Li J, You C, Miao W. et al. YTHDF2 Binds to 5-Methylcytosine in RNA and Modulates the Maturation of Ribosomal RNA. Anal Chem. 2020;92:1346-1354
96. Yang Y, Wang L, Han X, Yang WL, Zhang M, Ma HL. et al. RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay. Mol Cell. 2019;75:1188-1202.e1111
97. Lyabin DN, Eliseeva IA, Ovchinnikov LP. YB-1 protein: functions and regulation. Wiley Interdiscip Rev RNA. 2014;5:95-110
98. Thiagarajan D, Dev RR, Khosla S. The DNA methyltranferase Dnmt2 participates in RNA processing during cellular stress. Epigenetics. 2011;6:103-113
99. Selmi T, Hussain S, Dietmann S, Heiß M, Borland K, Flad S. et al. Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res. 2021;49:1006-1022
100. Xu X, Zhang Y, Zhang J, Zhang X. NSun2 promotes cell migration through methylating autotaxin mRNA. J Biol Chem. 2020;295:18134-18147
101. Shanmugam R, Fierer J, Kaiser S, Helm M, Jurkowski TP, Jeltsch A. Cytosine methylation of tRNA-Asp by DNMT2 has a role in translation of proteins containing poly-Asp sequences. Cell Discov. 2015;1:15010
102. Yu M, Lu B, Zhang J, Ding J, Liu P, Lu Y. tRNA-derived RNA fragments in cancer: current status and future perspectives. J Hematol Oncol. 2020;13:121
103. Zhang Y, Zhang X, Shi J, Tuorto F, Li X, Liu Y. et al. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat Cell Biol. 2018;20:535-540
104. Gkatza NA, Castro C, Harvey RF, Heiß M, Popis MC, Blanco S. et al. Cytosine-5 RNA methylation links protein synthesis to cell metabolism. PLoS Biol. 2019;17:e3000297
105. Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P. et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. Embo j. 2014;33:2020-2039
106. Flores JV, Cordero-Espinoza L, Oeztuerk-Winder F, Andersson-Rolf A, Selmi T, Blanco S. et al. Cytosine-5 RNA Methylation Regulates Neural Stem Cell Differentiation and Motility. Stem Cell Reports. 2017;8:112-124
107. Chen P, Zhang T, Yuan Z, Shen B, Chen L. Expression of the RNA methyltransferase Nsun5 is essential for developing cerebral cortex. Mol Brain. 2019;12:74
108. Paramasivam A, Meena AK, Venkatapathi C, Pitceathly RDS, Thangaraj K. Novel Biallelic NSUN3 Variants Cause Early-Onset Mitochondrial Encephalomyopathy and Seizures. J Mol Neurosci. 2020;70:1962-1965
109. Yi J, Gao R, Chen Y, Yang Z, Han P, Zhang H. et al. Overexpression of NSUN2 by DNA hypomethylation is associated with metastatic progression in human breast cancer. Oncotarget. 2017;8:20751-20765
110. Gao Y, Wang Z, Zhu Y, Zhu Q, Yang Y, Jin Y. et al. NOP2/Sun RNA methyltransferase 2 promotes tumor progression via its interacting partner RPL6 in gallbladder carcinoma. Cancer Sci. 2019;110:3510-3519
111. Li Y, Li J, Luo M, Zhou C, Shi X, Yang W. et al. Novel long noncoding RNA NMR promotes tumor progression via NSUN2 and BPTF in esophageal squamous cell carcinoma. Cancer Lett. 2018;430:57-66
112. Mei L, Shen C, Miao R, Wang JZ, Cao MD, Zhang YS. et al. RNA methyltransferase NSUN2 promotes gastric cancer cell proliferation by repressing p57(Kip2) by an m(5)C-dependent manner. Cell Death Dis. 2020;11:270
113. Sun Z, Xue S, Zhang M, Xu H, Hu X, Chen S. et al. Aberrant NSUN2-mediated m(5)C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene. 2020;39:6906-6919
114. Jiang Z, Li S, Han MJ, Hu GM, Cheng P. High expression of NSUN5 promotes cell proliferation via cell cycle regulation in colorectal cancer. Am J Transl Res. 2020;12:3858-3870
115. Sato K, Tahata K, Akimoto K. Five Genes Associated With Survival in Patients With Lower-grade Gliomas Were Identified by Information-theoretical Analysis. Anticancer Res. 2020;40:2777-2785
116. Yang R, Liang X, Wang H, Guo M, Shen H, Shi Y. et al. The RNA methyltransferase NSUN6 suppresses pancreatic cancer development by regulating cell proliferation. EBioMedicine. 2021;63:103195
117. Huang Z, Pan J, Wang H, Du X, Xu Y, Wang Z. et al. Prognostic Significance and Tumor Immune Microenvironment Heterogenicity of m5C RNA Methylation Regulators in Triple-Negative Breast Cancer. Front Cell Dev Biol. 2021;9:657547
118. Wang JZ, Zhu W, Han J, Yang X, Zhou R, Lu HC. et al. The role of the HIF-1α/ALYREF/PKM2 axis in glycolysis and tumorigenesis of bladder cancer. Cancer Commun (Lond). 2021
119. Xue M, Shi Q, Zheng L, Li Q, Yang L, Zhang Y. Gene signatures of m5C regulators may predict prognoses of patients with head and neck squamous cell carcinoma. Am J Transl Res. 2020;12:6841-6852
120. Huang W, Lan MD, Qi CB, Zheng SJ, Wei SZ, Yuan BF. et al. Formation and determination of the oxidation products of 5-methylcytosine in RNA. Chem Sci. 2016;7:5495-5502
121. Yang JC, Risch E, Zhang M, Huang C, Huang H, Lu L. Association of tRNA methyltransferase NSUN2/IGF-II molecular signature with ovarian cancer survival. Future Oncol. 2017;13:1981-1990
122. Lu L, Gaffney SG, Cannataro VL, Townsend J. Transfer RNA methyltransferase gene NSUN2 mRNA expression modifies the effect of T cell activation score on patient survival in head and neck squamous carcinoma. Oral Oncol. 2020;101:104554
123. Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A. et al. Epitranscriptomic Addition of m(5)C to HIV-1 Transcripts Regulates Viral Gene Expression. Cell Host Microbe. 2019;26:217-227.e216
124. Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18:103
125. Li C, Wang S, Xing Z, Lin A, Liang K, Song J. et al. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat Cell Biol. 2017;19:106-119
126. Alshaker H, Wang Q, Brewer D, Pchejetski D. Transcriptome-Wide Effects of Sphingosine Kinases Knockdown in Metastatic Prostate and Breast Cancer Cells: Implications for Therapeutic Targeting. Front Pharmacol. 2019;10:303
127. Xu X, Zhang Y, Zhang J, Zhang X. NSun2 promotes cell migration through methylating autotaxin mRNA. J Biol Chem. 2020
128. Sun Z, Xue S, Xu H, Hu X, Chen S, Yang Z. et al. Effects of NSUN2 deficiency on the mRNA 5-methylcytosine modification and gene expression profile in HEK293 cells. Epigenomics. 2019;11:439-453
129. Roszkowska KA, Gizinski S, Sady M, Gajewski Z, Olszewski MB. Gain-of-Function Mutations in p53 in Cancer Invasiveness and Metastasis. Int J Mol Sci. 2020 21
130. Chen Q, Yin D, Zhang Y, Yu L, Li XD, Zhou ZJ. et al. MicroRNA-29a induces loss of 5-hydroxymethylcytosine and promotes metastasis of hepatocellular carcinoma through a TET-SOCS1-MMP9 signaling axis. Cell Death Dis. 2017;8:e2906
131. Zhao Y, Tan Y, Meng T, Liu X, Zhu Y, Hong Y. et al. Simultaneous targeting therapy for lung metastasis and breast tumor by blocking the NF-κB signaling pathway using Celastrol-loaded micelles. Drug Deliv. 2018;25:341-352
132. Li Q, Li X, Tang H, Jiang B, Dou Y, Gorospe M. et al. NSUN2-Mediated m5C Methylation and METTL3/METTL14-Mediated m6A Methylation Cooperatively Enhance p21 Translation. J Cell Biochem. 2017;118:2587-2598
133. Dong Z, Cui H. The Emerging Roles of RNA Modifications in Glioblastoma. Cancers (Basel). 2020 12
134. Huang W, Qi CB, Lv SW, Xie M, Feng YQ, Huang WH. et al. Determination of DNA and RNA Methylation in Circulating Tumor Cells by Mass Spectrometry. Anal Chem. 2016;88:1378-1384
135. Sakita-Suto S, Kanda A, Suzuki F, Sato S, Takata T, Tatsuka M. Aurora-B regulates RNA methyltransferase NSUN2. Mol Biol Cell. 2007;18:1107-1117
136. Zheng HX, Zhang XS, Sui N. Advances in the profiling of N(6)-methyladenosine (m(6)A) modifications. Biotechnol Adv. 2020;45:107656
137. Zhang S, Li R, Zhang L, Chen S, Xie M, Yang L. et al. New insights into Arabidopsis transcriptome complexity revealed by direct sequencing of native RNAs. Nucleic Acids Res. 2020;48:7700-7711
138. Dou L, Li X, Ding H, Xu L, Xiang H. Prediction of m5C Modifications in RNA Sequences by Combining Multiple Sequence Features. Mol Ther Nucleic Acids. 2020;21:332-342
139. Lv H, Zhang ZM, Li SH, Tan JX, Chen W, Lin H. Evaluation of different computational methods on 5-methylcytosine sites identification. Brief Bioinform. 2020;21:982-995
140. Xuan JJ, Sun WJ, Lin PH, Zhou KR, Liu S, Zheng LL. et al. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018;46:D327-d334
141. Chen K, Song B, Tang Y, Wei Z, Xu Q, Su J. et al. RMDisease: a database of genetic variants that affect RNA modifications, with implications for epitranscriptome pathogenesis. Nucleic Acids Res. 2021;49:D1396-d1404
142. Luo X, Li H, Liang J, Zhao Q, Xie Y, Ren J. et al. RMVar: an updated database of functional variants involved in RNA modifications. Nucleic Acids Res. 2021;49:D1405-d1412
143. Song J, Zhai J, Bian E, Song Y, Yu J, Ma C. Transcriptome-Wide Annotation of m(5)C RNA Modifications Using Machine Learning. Front Plant Sci. 2018;9:519
144. Tzelepi V, Logotheti S, Efstathiou E, Troncoso P, Aparicio A, Sakellakis M. et al. Epigenetics and prostate cancer: defining the timing of DNA methyltransferase deregulation during prostate cancer progression. Pathology. 2020;52:218-227
Author contact
![]() Corresponding authors: Prof. Zhanguo Zhang, or Bixiang Zhang; Hepatic Surgery Center, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Hubei Province for the Clinical Medicine Research Center of Hepatic Surgery, 1095 Jiefang Avenue, 430030 Wuhan, China. Tel.: +86 27 83663400; fax: +86 27 83662851; E-mail: zhanguo_tjhedu.cn, bixiangzhangcom
Corresponding authors: Prof. Zhanguo Zhang, or Bixiang Zhang; Hepatic Surgery Center, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Hubei Province for the Clinical Medicine Research Center of Hepatic Surgery, 1095 Jiefang Avenue, 430030 Wuhan, China. Tel.: +86 27 83663400; fax: +86 27 83662851; E-mail: zhanguo_tjhedu.cn, bixiangzhangcom

 Global reach, higher impact
Global reach, higher impact