10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(2):637-651. doi:10.7150/ijbs.66915 This issue Cite
Research Paper
CircCCNB1 silencing acting as a miR-106b-5p sponge inhibited GPM6A expression to promote HCC progression by enhancing DYNC1I1 expression and activating the AKT/ERK signaling pathway
1. Guangdong Provincial Key Laboratory of Medical Molecular Diagnostics, Institute of Aging Research, Guangdong Medical University, Dongguan, Guangdong, China.
2. Department of Clinical Laboratory, YueBei People's Hospital, Shaoguan, Guangdong, China.
3. The Third Clinical Medical College, Guangzhou University of Chinese Medicine, Guangzhou, Guangdong, China.
4. Department of Medical Technology, Medical College of Shaoguan University, Shaogguan, Guangdong, China.
5. Department of Anesthesiology, YueBei People's Hospital, Shaoguan, Guangdong, China.
6. Department of Hepatobiliary Surgery, YueBei People's Hospital, Shaoguan, Guangdong, China.
*These authors contributed equally to this work.
Abstract
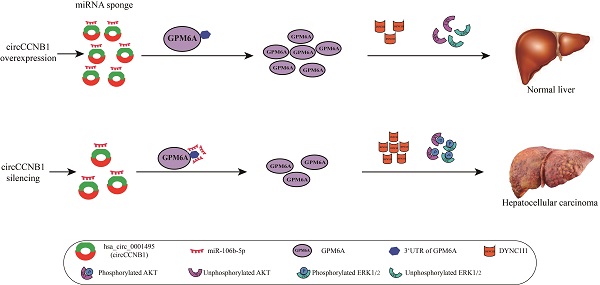
Background: Circular RNAs (circRNAs), which generally act as microRNA (miRNA) sponges to competitively regulate the downstream target genes of miRNA, play an essential role in cancer biology. However, few studies have been reported on the role of circRNA based competitive endogenous RNA (ceRNA) network in hepatocellular carcinoma (HCC). Herein, we aimed to screen and establish the circRNA/miRNA/mRNA networks related to the prognosis and progression of HCC and further explore the underlying mechanisms of tumorigenesis.
Methods: GEO datasets GSE97332, GSE108724, and GSE101728 were utilized to screen the differentially expressed circRNAs (DE-circRNAs), DE-miRNAs, and DEmRNAs between HCC and matched para-carcinoma tissues. After six RNA-RNA predictions and five intersections between DE-RNAs and predicted RNAs, the survival-related RNAs were screened by the ENCORI analysis tool. The ceRNA networks were constructed using Cytoscape software, based on two models of up-regulated circRNA/down-regulated miRNA/up-regulated mRNA and down-regulated circRNA/up-regulated miRNA/down-regulated mRNA. The qRT-PCR assay was utilized for detecting the RNA expression levels in HCC cells and tissues. The apoptosis, Edu, wound healing, and transwell assays were performed to evaluate the effect of miR-106b-5p productions on the proliferation, invasion, and metastasis of HCC cells. In addition, the clone formation, cell cycle, and nude mice xenograft tumor assays were used to investigate the influence of hsa_circ_0001495 (circCCNB1) silencing and overexpression on the proliferation of HCC cells in vitro and in vivo. Furthermore, the mechanism of downstream gene DYNC1I1 and AKT/ERK signaling pathway via the circCCNB1/miR-106b-5p/GPM6A network in regulating the cell cycle was also explored.
Results: Twenty DE-circRNAs with a genomic length less than 2000bp, 11 survival-related DE-miRNAs, and 61 survival-related DE-mRNAs were screened out and used to construct five HCC related ceRNA networks. Then, the circCCNB1/miR-106b-5p/GPM6A network was randomly selected for subsequent experimental verification and mechanism exploration at in vitro and in vivo levels. The expression of circCCNB1 and GPM6A were significantly down-regulated in HCC cells and cancer tissues, while miR-106b-5p expression was up-regulated. After transfections, miR-106b-5p mimics notably enhanced the proliferation, invasion, and metastasis of HCC cells, while the opposite was seen with miR-105b-5p inhibitor. In addition, circCCNB1 silencing promoted the clone formation ability, the cell cycle G1-S transition, and the growth of xenograft tumors of HCC cells via GPM6A downregulation. Subsequently, under-expression of GPM6A increased DYNC1I1 expression and activated the phosphorylation of the AKT/ERK pathway to regulate the HCC cell cycle.
Conclusions: We demonstrated that circCCNB1 silencing promoted cell proliferation and metastasis of HCC cells by weakening sponging of oncogenic miR-106b-5p to induce GPM6A underexpression. DYNC1I1 gene expression was up-regulated and further led to activation of the AKT/ERK signaling pathway.
Keywords: Circular RNA, hepatocellular carcinoma, circCCNB1, miR-106b-5p, GPM6A, DYNC1I1, AKT/ERK signal pathway

 Global reach, higher impact
Global reach, higher impact