10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(4):1724-1736. doi:10.7150/ijbs.66480 This issue Cite
Research Paper
RBMS2 Chemosensitizes Breast Cancer Cells to Doxorubicin by Regulating BMF Expression
1. Jiangsu Breast Disease Center, the First Affiliated Hospital with Nanjing Medical University, 300 Guangzhou Road, Nanjing, 210029, China.
2. Department of General Surgery, Comprehensive Breast Health Center, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine Shanghai, 200032, China.
3. Department of Breast Surgery, Fudan University Shanghai Cancer Center, No. 270, Dongan Road, Shanghai, 200032, China.
4. Department of Pharmacy, Jiangsu Cancer Hospital & Jiangsu Institute of Cancer Research & The Affiliated Cancer Hospital of Nanjing Medical University, Nanjing, 210029, China.
*These authors have contributed equally to this work.
Received 2021-8-25; Accepted 2022-1-20; Published 2022-2-7
Abstract
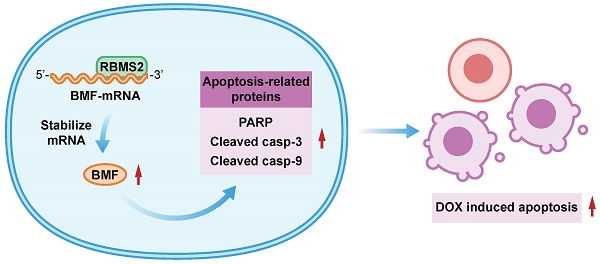
Chemoresistance is closely related to the therapeutic effect and prognosis in breast cancer patients. Increasing evidences demonstrated that RNA binding proteins (RBPs) have notable roles in regulating cancer cell proliferation, metastasis and chemotherapeutic sensitivity. RNA binding motif single stranded interacting protein 2 (RBMS2), an RBP, has been considered to be a tumor suppressor in several cancers. However, its role of doxorubicin sensitivity in breast cancer patients has not yet been fully revealed. Here, we performed doxorubicin cytotoxicity assay, flow cytometry and mouse xenograft model to examine the influence of RBMS2 on doxorubicin sensitization in vitro and in vivo. RIP assay and dual-luciferase reporter assay were performed to explore the relationship between RBMS2 and BMF. Our data demonstrated that upregulation of RBMS2 in breast cancer cells could enhance sensitivity to doxorubicin and promote apoptosis in the presence of doxorubicin, while inhibition of RBMS2 showed an opposite trend. Moreover, this chemosensitizing effect of RBMS2 could be reversed by the inhibition of Bcl-2 modifying factor (BMF). RBMS2 positively regulated BMF expression and increased BMF-induced expression of (cleaved) caspase 3, (cleaved) caspase 9 and poly (ADP-Ribose) polymerase (PARP). These results uncovered a novel mechanism for RBMS2 in the sensibilization of doxorubicin, suggesting that RBMS2 may act as a potential therapeutic target for drug-resistant breast cancer.
Keywords: RBMS2, BMF, apoptosis, chemosensitization, doxorubicin, breast cancer
Introduction
The incidence of breast cancer rises rapidly over the years, ranking as the first common cancer among females worldwide, and thus posing a considerable threat to female health [1, 2]. The current treatment strategies for breast cancer include surgery, chemotherapy, radiotherapy, targeted treatment and endocrine therapy [3]. Adjuvant systemic chemotherapy remains the preferred treatment for some patients of early stage and is suggested to be the major treatment for those with advanced stage [4]. The commonly used chemotherapeutic drugs includes doxorubicin (DOX), paclitaxel and cyclophosphamide [5]. However, the efficiency of chemotherapy is greatly limited by primary and acquired drug resistance [6, 7].
DOX is a cytotoxic anthracycline antibiotic used in anti-tumor therapy by inducing cancer cells apoptosis [8-10], which is regarded as the foundation of chemotherapy for breast cancer [11]. Cellular resistance to DOX often develops in breast cancer patients with recurrence and advanced stage. Its molecular mechanisms have not been fully revealed [12]. Various genetic and epigenetic factors can attribute to resistance of chemotherapy [13-15]. Most chemotherapeutic drugs kill tumor cells by inducing apoptosis, so the absence of proapoptotic protein or the overexpression of anti-apoptotic proteins leads to resistance to chemotherapeutic drugs [16]. BMF (Bcl-2 modifying factor) appertains to the Bcl-2 protein family and functions as a pro-apoptotic factor that is correlated with various cellular activities, including chemosensitivity. For instance, YAP/TEAD/SLUG axis suppressed apoptosis through transcriptional repression of BMF [17].
RBPs play a pivotal role at the post-transcriptional levels by several mechanisms, including mRNA stabilization, polyadenylation, transport, translation and RNA splicing [18, 19]. Previous studies showed that numerous RBPs are dysregulated in different cancers, causing change of drug sensitivity [20, 21]. Kim et al. found that non-POU domain containing octamer binding (NONO), an RBP, was highly expressed in triple-negative breast cancer and directly interacted with STAT3 protein by elevating its mRNA stability and transcriptional activity, which contributed to DOX resistance [22]. RBMS2, also known as SCR3, a member of RBPs family, was extracted via phenotypic complementation of cdc2 and cdc13 mutants of yeast [23]. Our previous study has illustrated that RBMS2 could act as a tumor suppressor and enhance the expression of P21 mRNA via directly binding to its AU-rich element (AREs) of 3′-untranslated region (3′-UTR) in breast cancer [24]. According to our results of RNA sequencing from the above study, we found BMF mRNA was significantly upregulated after overexpression of RBMS2. In addition, our data showed that RBMS2 could induce breast cancer cells apoptosis. Therefore, we hypothesized that RBMS2 could sensitize breast cancer cells to DOX via inducing apoptosis regulated by BMF expression.
Methods
Cell culture
Human breast cancer DOX resistance cell line MCF-7/DOX was kindly provided by Dr. Shui Wang (Nanjing Medical University, Nanjing, China); MCF-7 cell line was purchased from the American Type Culture Collection (ATCC, USA) and SUM 1315 provided by Dr. Stephen Ethier (University of Michigan, AnnArbor, MI, USA). These three cell lines were cultured in DMEM (Wisent, China) containing 10% FBS (Gibco, USA), 4.5 mg/ml glucose, 100 μg/ml streptomycin and 100 μg/ml penicillin (Hyclone, USA), in a humidified incubator containing 5% CO2 at 37 °C.
Lentivirus transfection and small interfering RNA
Lentiviral constructs to overexpress RBMS2 (RBMS2), the negative control (NC), knockdown RBMS2 (sh1, sh2) and the scramble control (SCR) were purchased from GenePharma (Shanghai, China) and generated following the manufacturer's instructions as previous described [24]. For recovery assays, cells were first transduced with overexpressed and control vectors of RBMS2, and then transduced with BMF-siRNA (GenePharma, Shanghai, China), p21-siRNA (GenePharma, Shanghai, China) and the nonspecific siRNA control. All sequences are listed in Table S1. Real-Time quantitative PCR (RT-qPCR) and western blot were used to detect the transfection efficiency.
Cytotoxicity assay
Cells were inoculated in 96-well plates (5×103 cells/well) and grew in complete medium containing different concentrations of DOX (MedChemExpress, USA), cisplatin (MedChemExpress, USA) and 5-fluorouracil (5-FU) (MedChemExpress, USA) for 24 h, respectively. The cell survival rate was further tested by CCK-8 kit (Dojindo, Japan) following the manufacturer's guidelines. For colony formation assay, cells were firstly seeded in six-well plates with 1×103 cells/well and incubated with DOX (0 or 1 μg/ml) for 24 h and then transferred to complete culture without DOX for 14 days. 1 μg/ml DOX was employed to keep the maximum inhibition rate below 50%. The resulting colonies were fixed in paraform for 20 min and stained with crystal violet solution for 20 min, then the colonies were counted and scanned.
Apoptosis assay
Apoptosis was tested by the apoptosis kit (Annexin V-APC/7-AAD Apoptosis Detection Kit, Multisciences, China) based on the manufacturer's protocols. Briefly, cells were digested by EDTA-free trypsin and washed twice with ice cold phosphate buffered saline (PBS) and centrifuged (1200 rpm,5 min). Then, 400 μl annexin binding buffer (1×106 cells/ml) was used to resuspend the cells. Finally, cells were dual stained using 5 μl APC-conjugated Annexin V and 10 μl 7-AAD and detected by flow cytometry (BD Biosciences, USA). Data were analyzed using FlowJo v10.0 (FlowJo, LLC).
RT-qPCR
Total RNAs were isolated by Trizol reagent (TaKaRa, Japan) and HiScript qRT SuperMix (Vazyme, China) was used for complementary DNA (cDNA) synthesis. Thirty pairs of snap-frozen breast cancer tumor and adjacent tissues were collected in the First Affiliated Hospital of Nanjing Medical University from 2006 to 2016, and approved by the ethics and research committee of the First Affiliated Hospital with Nanjing Medical University. All included patients provided written informed consents. RT-qPCR was conducted using AceQ qPCR SYBR Green Master Mix (Vazyme, China) in a real-time PCR instrument (Roche, USA) following the manufacturer's instructions. The 2-ΔΔCt method was calculated for relative mRNA expression and β-actin was utilized as the endogenous control. The sequences of primers used are listed in Table S2.
Western blot analysis
The total protein extracted from breast cancer cells was split with RIPA buffer (P0013C, Beyotime, China) containing 1% phosphatase inhibitor, 1% PMSF, and 0.1% protease inhibitor and separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), then electransferred onto an activated polyvinylidene fluoride (PVDF) membrane (Millipore, USA). Membranes were placed in Tris-buffered saline containing 0.1% Tween 20 (TBST) and 5% nonfat milk for 2 h and then incubated with primary antibodies against RBMS2 (1:1000, RayBiotech, USA, 102-12993), BMF (1:1000, Proteintech, China, 18298-1-AP), β-actin (1:1000, Proteintech, China, 20536-1-AP), Cleaved Caspase 3 (1:1000, Cell Signaling Technology, USA, 9664), Caspase 3 (1:1000, Cell Signaling Technology, USA, 14220), Cleaved Caspase 9 (1:1000, Cell Signaling Technology, USA, 9509), Caspase 9 (1:1000, Cell Signaling Technology, USA, 9504) , PARP (1:1000, Cell Signaling Technology, USA, 9532) overnight at 4 °C. Membranes were washed for 15 min for 3 times with TBST and then incubated for 2 h with secondary antibodies (1:5000, Cell Signaling Technology, USA, 7074P2). The expression levels of target protein were detected through Immobilob™ Western Chemiluminescent HRP Substrate (Millipore, USA).
RNA stability assays
MCF-7 and SUM 1315 cells were transferred into 6-well plates and added with actinomycin D (Act D, 5 μg/ml) at 0, 1, 2, 4, 6 and 8 h when the number of cells reached 3×105. Then total RNAs were extracted and the RNA expression levels of BMF were checked by RT-qPCR.
RNA immunoprecipitation (RIP)
MCF-7 and SUM 1315 cells (2×107) were washed with PBS and lysed by Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) following manufacturer's protocol. Then rabbit polyclonal anti-RBMS2 (5μg) and non-immunized rabbit IgG was incubated overnight at 4 °C. After RNA purification, protein A/G magnetic beads were used for the RNA protein immunocomplexes. Then RT-qPCR was conducted to examine the levels of BMF transcripts in the RBMS2 or IgG immunocomplexes.
Dual-luciferase reporter assay
MCF-7 and SUM 1315 cells were seeded evenly into a 24-well plate and co-transfected with 200ng of pGL3 reporter vectors carrying 3′-UTR or the corresponding AREs mutant region of BMF and 5 ng of pRL-TK (Promega, USA). The AREs mutant reporter converts the AUUUA motif in the BMF 3′-UTR into AGGGA. These cells were harvested after transfection for 48h, and dual luciferase reporter assay kit (Promega, USA) was performed to detect the luciferase activity in strict accordance with the manufacturer's instructions.
Xenograft tumors in nude mice
All animal investigations were performed according to the guidelines of Use Committee of the Nanjing Medical University and guidelines of Institutional Animal Care. Forty-eight female BALB/c nude mice (aged 4-6 weeks, 18-22 g) were purchased from the Experimental Animal Center of Nanjing Medical University (Nanjing, China). For subcutaneous inoculation, 1×106 SUM 1315 cells stably expressing RBMS2 and the negative control (NC) resuspended in 100 μl PBS were implanted subcutaneously into the right flank regions of the mice. When the subcutaneous tumors reached to an average size of 0.5-0.6 cm (approximately 2 weeks after cell injection), the nude mice were randomly divided into four groups, namely, the NC-DOX, RBMS2-DOX, NC+DOX and RBMS2+DOX group. Then, all nude mice received either 2 mg/kg of DOX or equal volume of PBS by tail vein injection every 3 days and the growth of tumors was recorded every week by a caliper, calculated as (length × width2)/2. The mice were sacrificed at the end of experiments and the subcutaneous tumors were dissected out and photographed.
Statistical analysis
GraphPad Prism (Version 8.0) was used to analyze and present the data. Significance of differences between groups was analyzed by Student′s t-test. Differenced expression profile of BMF in breast cancer and paired normal tissues was compared by the paired Student's t test. All experiments were independently repeated for three times, and p < 0.05 was used as statistically significant.
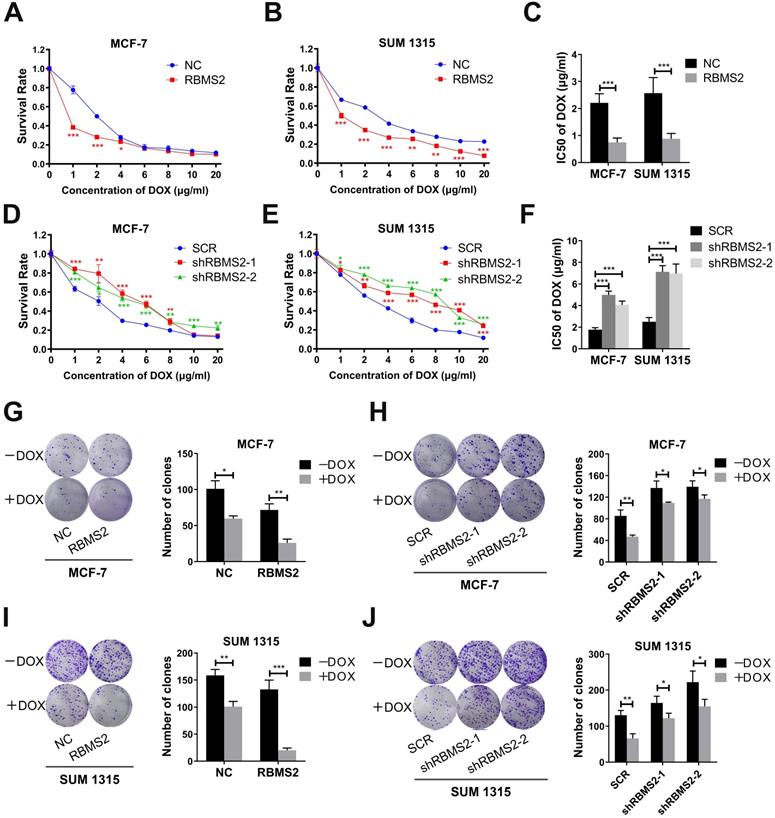
RBMS2 could sensitize breast cancer cells to DOX in vitro. RBMS2 overexpression and knockdown cell lines were treated with different dosages of DOX for 24 h, respectively. CCK-8 assay was used to examine the cell viability (A and D for MCF-7, B and E for SUM 1315) and IC50 value (C and F) of DOX. Transfected MCF-7 (G, H) and SUM 1315 (I, J) cells were treated with DOX (0 or 1 µg/ml) for 2 weeks. Representative images (left panel) and quantification (right panel) of colonies in the colony formation assay. Data were shown as mean ± SD. *p <0.05, **p <0.01, ***p<0.001.
Results
RBMS2 could sensitize breast cancer cells to DOX in vitro
Overexpression and knockdown of RBMS2 in MCF-7 and SUM 1315 stable cells constructed by lentivirus were validated via RT-qPCR and western blot (Fig. 4G-J). RBMS2 acted as a tumor suppressor and has relatively low expression in breast cancer, resulting in the minor knockdown effects. The half-maximal inhibitory concentration (IC50 value) of DOX in RBMS2 overexpressing cells was decreased by 66.3% in MCF-7 cells (Fig. 1A) and 65.7% in SUM 1315 cells (Fig. 1B). On the contrary, knockdown of RBMS2 showed an opposite trend, with the IC50 values increasing to 182% and 129% in MCF-7 cells (Fig. 1D) and 185% and 179% in SUM 1315 cells (Fig. 1E) respectively. IC50 values were shown in Fig. 1C and Fig. 1F. Although the proliferation of RBMS2 knockdown and overexpression cell lines were both inhibited by DOX, the IC50 values of overexpression group dropped more significantly than that of knockdown group. These data suggested that knockdown of RBMS2 could affect the sensitivity of DOX in breast cancer cell lines. The same effect was also observed in MCF-7/DOX cell lines after overexpression of RBMS2, with IC50 values decreasing by 58.8% (Fig. S1). As demonstrated in Fig. 1G and Fig. 1I, after DOX treatment, the ability of RBMS2 overexpressed breast cancer cells to form colonies was much worse than that of control cells, while inhibition of RBMS2 impaired the proliferation of breast cancer to a much lesser extent (Fig. 1H, J). Collectively, RBMS2 sensitized breast cancer cells to DOX in vitro.
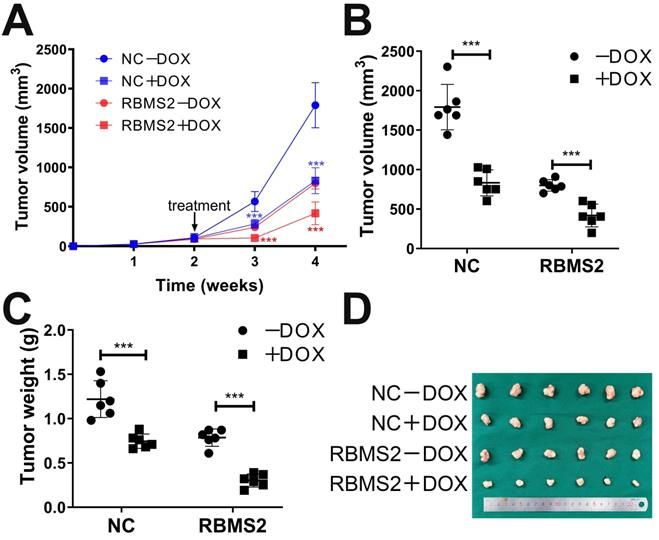
Overexpression of RBMS2 could enhance the therapeutic effect of DOX in vivo. Tumor volumes were measured in different treatment groups (A. B). The excised tumor lumps were weighed (C) and photographed (D). Data were shown as mean ± SD. *p <0.05, **p <0.01, ***p<0.001.
RBMS2 could sensitize breast cancer cells to DOX in vivo
To extend our results on the influence of RBMS2 on DOX sensitivity in breast cancer, a mouse xenograft model was carried out by subcutaneous injection with RBMS2 overexpression stable cells or control cells. As expected, with or without DOX treatment, tumors derived from RBMS2 group proliferate and expand significantly slower than that of control group (Fig. 2A). As demonstrated in Fig. 2B, tumors volume of RBMS2 and DOX combination group was relatively smaller than that of NC+DOX group. 4 weeks later, tumor weights in RBMS2+DOX were remarkably lighter than that of other groups, which indicated RBMS2 sensitized breast cancer cells to DOX in vivo (Fig. 2C-D).
RBMS2 could induce breast cancer cells apoptosis
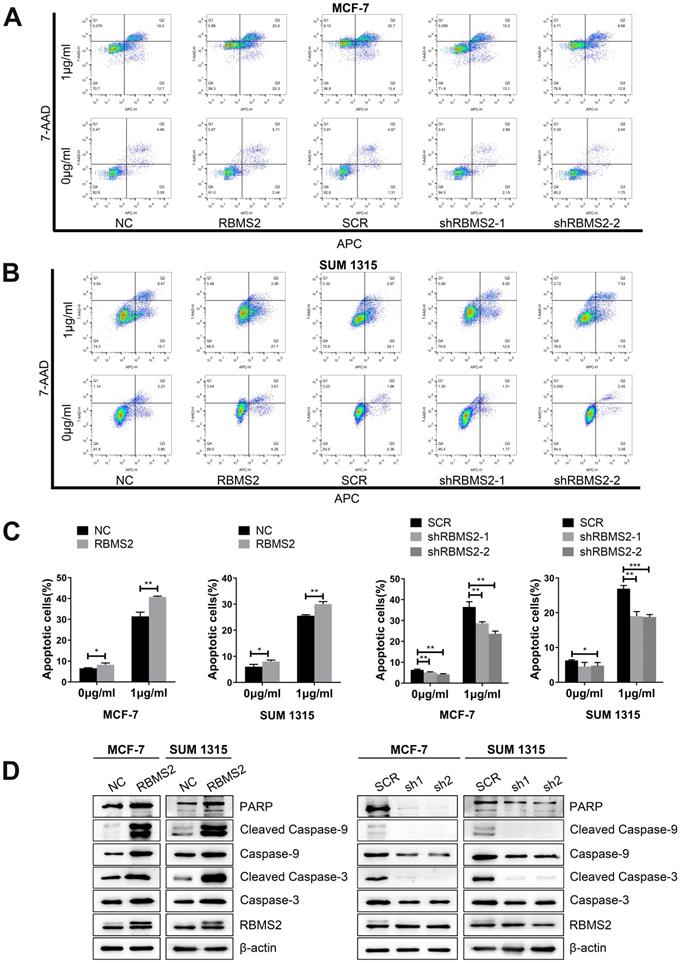
The effects of RBMS2 on apoptosis in MCF-7 and SUM 1315 cells were detected by flow cytometry. Overexpression of RBMS2 robustly strengthened DOX-induced apoptosis in breast cancer cells, while inhibition of RBMS2 abated DOX-induced apoptosis (Fig. 3A-B). Apoptosis ratios were shown in Fig. 3C. Then, western blotting was performed to investigate the expression of apoptosis-related proteins after the expression of RBMS2 was altered. The target protein expressions were consistent between NC and SCR groups in western blot analysis. The levels of the pro-apoptosis protein (cleaved) caspase 3, (cleaved) caspase 9 and PARP were increased compared to that of control group after the overexpression of RBMS2. In particular, cleaved caspase 3 and cleaved caspase 9 were more significantly upregulated than caspase 3 and caspase 9, indicating that RBMS2 could induce breast cancer cells apoptosis. In contrast, knockdown of RBMS2 showed an opposite trend (Fig. 3D). These data suggested RBMS2 could induce cell apoptosis and modulate apoptosis-related proteins in breast cancer cells.
RBMS2 could induce apoptosis and apoptosis related proteins in breast cancer cell lines. Flow cytometry analysis of apoptosis in RBMS2 overexpression and knockdown MCF-7 and SUM 1315 cells with or without DOX treatment (A, B). The statistical calculation of apoptotic rates was shown (C). The levels of (cleaved) caspase 3, (cleaved) caspase 9 and PARP in MCF-7 and SUM 1315 cells transfected with RBMS2 overexpression and knockdown were detected by Western blot (D). The top band of RBMS2 is the exogenously band in western blot. Data were shown as mean ± SD. *p <0.05, **p <0.01, ***p<0.001.
RBMS2 regulated the expression of pro-apoptotic protein BMF. Volcano plot represented the distribution of mapped transcripts (A). The size and color of the dots meant the number of enriched genes and the adjusted p values, respectively. BMF was found to be positively correlated with RBMS2 in breast cancer both from TIMER (Tumor Immune Estimation Resource, https://cistrome.shinyapps.io/timer) database (B), the patients' samples from our hospital (C). Correlation heatmap were used to analyze the correlation between RBMS2 and BMF (D). R1, R2, R3 and V5-1, V5-2 and V5-3 represented RBMS2-overexpressed group and the control group, respectively. The red arrow indicated that BMF was positively correlated with RBMS2. Expression of BMF in breast cancer tissues and normal tissues (E, F). Overexpression of RBMS2 significantly increased the expression of BMF at both mRNA and protein (G, I) levels. BMF was significantly down regulated after RBMS2 knock down at both mRNA and protein levels (H, J). The top band of RBMS2 is the exogenously band in western blot. Data were shown as mean ± SD. *p <0.05, **p <0.01, ***p<0.001.
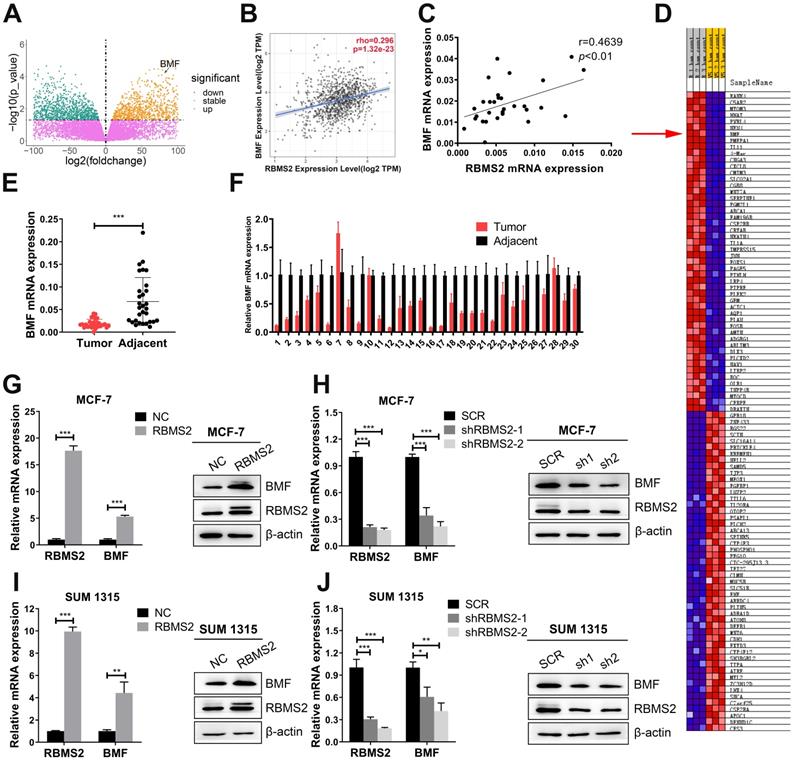
RBMS2 could elevate the expression of BMF in breast cancer cells
Previous RNA sequencing data of overexpressing RBMS2 and control group in SUM 1315 cells revealed that BMF might be a potential mRNA target mediated by RBMS2 [24]. BMF was dramatically upregulated as the expression of RBMS2 increased with log2 (foldchange) 2.35 and FDR < 0.05 (Fig. 4A). BMF was found to be positively correlated with RBMS2 in both TIMER (Tumor Immune Estimation Resource, https://cistrome.shinyapps.io/timer) database (Fig. 4B) and the breast cancer samples from our hospital (Fig. 4C). The same results were obtained for the correlation heatmap (Fig. 4D). Moreover, a remarkable decrease of BMF expression was reported in breast cancer tissues compared to that of paracancerous tissues (Fig. 4E-F). The expression of BMF was promoted after the MCF-7 (Fig. 4G) and SUM 1315 cells (Fig. 4I) were transfected with RBMS2 overexpressing lentivirus. Accordingly, the expression level of BMF in RBMS2 knockdown MCF-7 cells (Fig. 4H) and SUM 1315 cells (Fig. 4J) was significantly suppressed.
RBMS2 could increase BMF mRNA stability. In MCF-7 and SUM 1315 cell lines, RBMS2 overexpression prolonged the half-life of BMF mRNA after treated with Act D at a concentration of 5 µg/ml (A, B), while knockdown of RBMS2 shortened the half-life of BMF mRNA (C, D). MCF-7 and SUM 1315 cells lysates were immunoprecipitated with RBMS2 or IgG antibody and analyzed using PCR (E, G) and RT-qPCR (F, H) to detect transcript levels of BMF. Schematic diagram containing BMF 3'-UTR (upper) and AREs mutant region (lower) (I). The reporter containing BMF 3'-UTR was increased after overexpression of RBMS2 in MCF-7 (J) and SUM 1315 (K) cell lines. Firefly luciferase activity was detected and normalized to Renilla luciferase activity. Data were shown as mean ± SD. *p <0.05, **p <0.01, ***p<0.001.
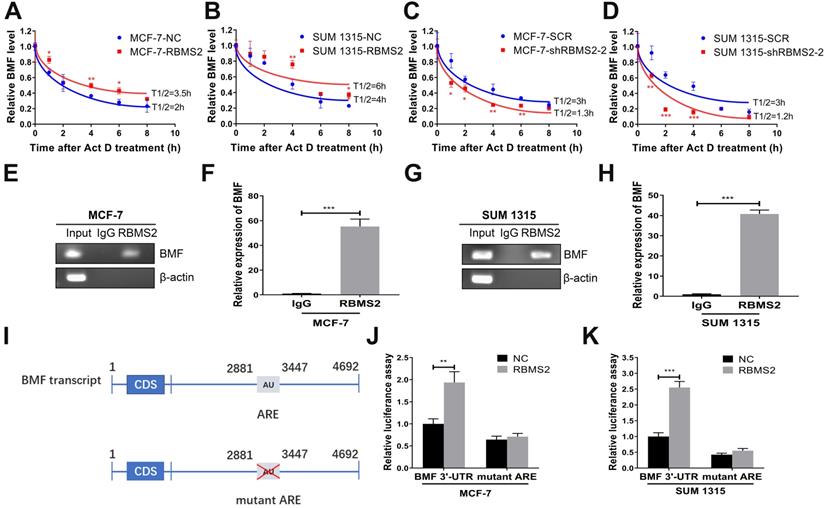
RBMS2 could increase BMF mRNA stability
After 5 μg/ml Act D treatment at different time points (0-8 h), the half-life of BMF mRNA in RBMS2 overexpressing cells increased from 2 to 3.5h in MCF-7 cells (Fig. 5A) and 4 to 6 h in SUM 1315 cells (Fig. 5B). In contrast, the opposite result was found when RBMS2 was knocked down, with the half-life decreasing from 3 to 1.3 h in MCF-7 cells (Fig. 5C) and 3 to 1.2 h in SUM 1315 cells (Fig. 5D). All these results indicated that RBMS2 could enhance BMF expression by elevating its mRNA stability. To further dissect whether RBMS2 binds directly to the BMF mRNA in breast cancer cells, RIP assay was used in MCF-7 (Fig. 5E-F) and SUM 1315 (Fig. 5G-H) cells followed by RT-PCR and RT-qPCR. The BMF mRNA was presented in RBMS2 and Input, whereas not in IgG. β-actin, which used as a negative control that could not bind to RBMS2 mRNA. The results showed that RBMS2 could physically bind to BMF mRNA. Then, luciferase reporter assay which contained the whole region of BMF 3′-UTR or AREs mutant BMF 3′-UTR was used to further explore whether RBMS2 specifically regulated BMF via binding to the AREs in 3′-UTR of BMF mRNA (Fig. 5I). Our results suggested that RBMS2 could enhance the luciferase activity of a reporter containing the 3′-UTR of the BMF mRNA in comparison with control. On the contrary, the luciferase activity of a reporter containing ARE sites mutant BMF 3′-UTR in RBMS2 and control group was similar (Fig. 5J-K). These findings indicated that RBMS2 could bind directly to the ARE sites in the 3′-UTR of BMF mRNA, enhancing its expression and stability.
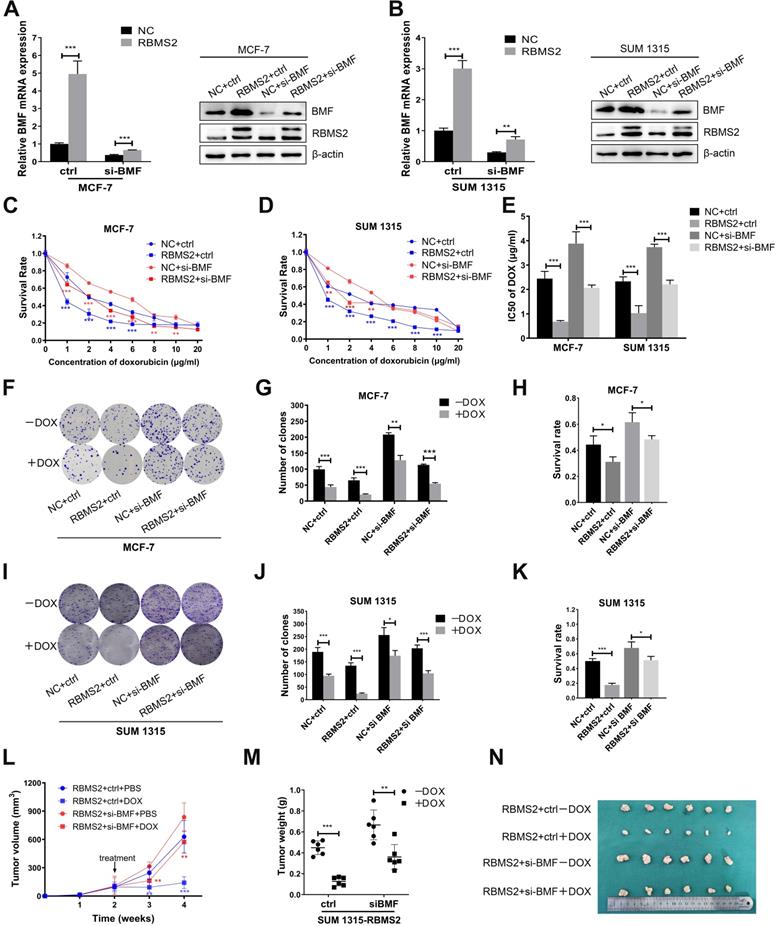
Inhibition of BMF could reverse the sensitization to DOX induced by RBMS2 both in vivo and in vitro
To explore whether the RBMS2 induced sensitization to DOX was depended on BMF expression, we performed rescue experiments. RBMS2-over-expressed MCF-7 and SUM 1315 cells were transfected with si-BMF or control. We set up four groups: NC+ctrl, NC+si-BMF, RBMS2+ctrl and RBMS2+si-BMF. RT-qPCR and western blot were performed to verify the transfection efficiency (Fig. 6A-B). IC50 value of DOX was reduced in the RBMS2-overexpressed groups compared with the NC group. However, on concurrent inhibition of BMF, this resistance to DOX reactivated significantly (Fig. 6C-E). Same trend was observed in colony formation assay (Fig. 6F-K) and tumor xenograft model (Fig. 6L-N).
BMF could reverse the sensitization to DOX induced by RBMS2 both in vivo and in vitro. Small interfering RNA of BMF was transfected into RBMS2 overexpression MCF- 7 and SUM 1315 cells. Transfection efficiency was confirmed via RT-qPCR and western blot (A, B). The top band of RBMS2 is the exogenously band in western blot. The sensitivity to DOX of MCF-7 and SUM 1315 cells mentioned above was examined using CCK-8 and colony formation. CCK-8 assay was used to examine cell viability (C for MCF-7, D for SUM 1315) and IC50 value (E) of DOX. Representative photographs (F, I), quantification (G, J) and cell survival after DOX treatment (H, K) were shown by colony formation assay. Tumor volumes were measured in different treatment groups (L). The excised tumor lumps were weighed (M) and photographed (N). Data were shown as mean ± SD. *p <0.05, **p <0.01, ***p<0.001.
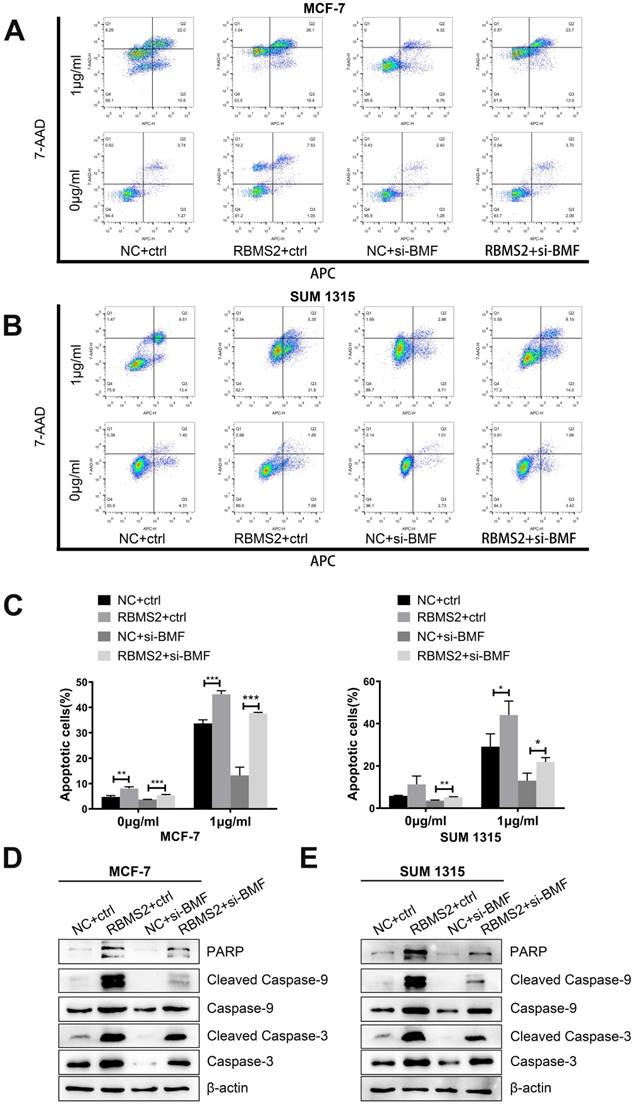
Inhibition of BMF could rescue the breast cancer cells apoptosis induced by RBMS2
To confirm whether the knockdown of BMF changes the increase in RBMS2-induced apoptosis rate in the presence of DOX, the apoptosis rate of all groups was measured by flow cytometry. RBMS2 overexpression dramatically increased the apoptosis under DOX treatment. But RBMS2+ si-BMF group dramatically decreased the apoptosis rate compared to the RBMS2+ctrl group (Fig. 7A-B). The apoptotic rates were shown in Fig. 7C. Western blotting showed the expression levels of (cleaved) caspase 3, (cleaved) caspase 9, (cleaved) caspase 3, (cleaved) caspase 9 and PARP were decreased after the attenuation of BMF. The increased ratio in cleaved caspase 3/caspase 3 and cleaved caspase 9/caspase 9 were partly reversed by the attenuation of BMF (Fig. 7D-E). Our previous study identified p21 as another key target gene mediating the tumor suppressing role of RBMS2 [24]. However, depletion of p21 could not reverse the sensitization to DOX induced by RBMS2 (Fig. S2). In summary, BMF could rescue the apoptosis of breast cancer cells induced by RBMS2.
Discussion
Our results revealed the unique mechanism on how RBMS2 could contribute to antineoplastic signaling. RBMS2 could promote BMF mRNA stability by binding AREs in the BMF mRNA 3′-UTR. In our study, we elucidated that RBMS2, together with BMF, could enhance apoptosis of breast cancer cells to promote the sensitivity to DOX.
Resistance to apoptosis is considered an important hallmark of cancer, and all chemical treatments are believed to achieve their function directly or indirectly through inducing cell apoptosis [25]. And inducing apoptosis has been used as a logical and realistic therapeutic strategy since the 1980s [26]. Our previous study illustrated that gene regulations mediated by RBPs are closely correlated with numerous cancer-related biological functions, such as proliferation and epithelial-mesenchymal transition (EMT) [27-29]. As overexpression of RBMS2 could enhance susceptibility to DOX in MCF-7/DOX cell lines, we hypothesized that, its ectopic expression might result in drug resistant phenotype by enhancing the cellular sensitivity to DOX. In this study, we found that overexpression of RBMS2 could improve DOX sensitivity and induce apoptosis in breast cancer cells, while inhibition of RBMS2 showed an opposite effect. We hypothesized that RBMS2 could enhance the sensitivity of DOX by inducing apoptosis, considering the resistance mechanism of DOX. Hence, apoptosis-related molecules were in the spotlight. According to our previous results of mRNA sequencing [24], we found that mRNA level of BMF was significantly upregulated in breast cancer cells. In our study, BMF was remarkably enhanced after RBMS2 overexpression. In general, BMF is associated with diverse cellular activities, including apoptosis and chemosensitivity [30, 31]. Various studies showed that overexpression of BMF could activate the release of BIM from BCL-XL to induce mitochondrial apoptosis mediated by (cleaved) caspase 3, (cleaved) caspase 9 and PARP [32, 33]. Then, we discovered that BMF expression was downregulated and positively correlated with RBMS2 expression in breast cancer and normal tissues from our hospital and TCGA databases. In our study, the expression of BMF and its effectors including (cleaved) caspase 3, (cleaved) caspase 9 and PARP were all enhanced by overexpression of RBMS2, resulting in the induction of apoptosis. And the sensibilization of DOX mediated by RBMS2 could be interfered by attenuating the expression of BMF, suggesting BMF was a primary target of RBMS2. Therefore, we concluded that RBMS2 regulated BMF, which could enhance the expression of (cleaved) caspase 3, (cleaved) caspase 9 and PARP, affecting the cellular sensitivity to DOX by promoting apoptosis. Knockdown of BMF alone in the absence of DOX could increase cell growth both in vitro and in vivo. The depletion of BMF may be a cause of resistance to DOX. Nevertheless, RBMS2-BMF axis could not sensitize breast cancer cells to 5-FU or cisplatin (Fig. S3). Further animal experiments confirmed the sensibilization to DOX of RBMS2 in breast cancer cells. Under the same dosage of DOX, tumors formed in nude mice with RBMS2-over-expressed SUM 1315 were more sensitive than the control group, consisting with the results in vitro. In our study, the same conclusions were confirmed both in ER+ and triple-negative breast cancer lines. However, whether there is a pervasive influence in different sub-types of breast cancer is unclear. Considering the few studies on RBMS2, further research will be conducted in other types of breast cancer.
In this study, overexpression of RBMS2 could increase BMF mRNA stability, as detected by RNA stability assays. RIP assay and dual-luciferase reporter assay also demonstrated that RBMS2 could bind directly to the AREs in 3′-UTR of BMF mRNA, leading to increased expression of (cleaved) caspase 3, (cleaved) caspase 9 and PARP, which were considered to be effectors of BMF and therefore contributed to higher apoptosis rate [34]. All the above data suggested RBMS2 could regulate the expression of BMF via a posttranscriptional regulation, which is the most common way for RBPs [35, 36]. Summarily, our study reveals a novel mechanism for RBMS2 as a regulator of chemotherapy response to DOX in breast cancer.
BMF could rescue the apoptosis and apoptosis related proteins induced by RBMS2 in breast cancer cell lines. Apoptosis and apoptosis related proteins were detected by flow cytometry analysis (A, B, C) and western blot (D, E). Data were shown as mean ± SD. *p <0.05, **p <0.01, ***p<0.001.
Conclusions
Here, our study first reported that RBMS2 could induce apoptosis and increase DOX sensitivity by elevating BMF expression in breast cancer cells. Mechanistically, RBMS2 could stabilize the mRNA of BMF by directly interacting with the AREs in the 3′-UTR. Activating RBMS2 expression may represent a novel strategy for drug-resistant breast cancer.
Abbreviations
RBPs: RNA binding proteins; RBMS2: RNA binding motif single stranded interacting protein 2; BMF: Bcl-2 Modifying Factor; PARP: poly (ADP-Ribose) polymerase; RT-qPCR: Real-time quantitative polymerase chain reaction; cDNA: Complementary DNA; 3′-UTR: 3-untranslated region; AREs: AU-rich element; RIP: RNA immunoprecipitation.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81972486, 81802644, 81802748), the Key Medical Talents of Jiangsu Province (ZDRCA2016029) and 333 High-level Talents Training Project of Jiangsu Province (BRA2016505). Meanwhile, the study was also funded by the International Cooperation Project of Jiangsu Provincial Science and Technology Department (BZ2018054) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) (JX10231801).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fan L, Strasser-Weippl K, Li J, St Louis J, Finkelstein D, Yu K. et al. Breast cancer in China. The Lancet Oncology. 2014;15:e279-89
2. Siegel R, Miller K, Jemal A. Cancer statistics, 2020. CA: a cancer journal for clinicians. 2020;70:7-30
3. Ma X, Wang M, Yin T, Zhao Y, Wei X. Myeloid-Derived Suppressor Cells Promote Metastasis in Breast Cancer After the Stress of Operative Removal of the Primary Cancer. Frontiers in oncology. 2019;9:855
4. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival. an overview of the randomised trials. Lancet (London, England). 2005;365:1687-717
5. Wang C, Jin H, Wang N, Fan S, Wang Y, Zhang Y. et al. Gas6/Axl Axis Contributes to Chemoresistance and Metastasis in Breast Cancer through Akt/GSK-3β/β-catenin Signaling. Theranostics. 2016;6:1205-19
6. Liu X, Zheng J, Sun W, Zhao X, Li Y, Gong N. et al. Ferrimagnetic Vortex Nanoring-Mediated Mild Magnetic Hyperthermia Imparts Potent Immunological Effect for Treating Cancer Metastasis. ACS Nano. 2019;13:8811-25
7. Ji X, Lu Y, Tian H, Meng X, Wei M, Cho W. Chemoresistance mechanisms of breast cancer and their countermeasures. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;114:108800
8. Venkatadri R, Muni T, Iyer A, Yakisich J, Azad N. Role of apoptosis-related miRNAs in resveratrol-induced breast cancer cell death. Cell death & disease. 2016;7:e2104
9. Pilco-Ferreto N, Calaf G. Influence of doxorubicin on apoptosis and oxidative stress in breast cancer cell lines. International journal of oncology. 2016;49:753-62
10. Nitiss J. Targeting DNA topoisomerase II in cancer chemotherapy. Nature reviews Cancer. 2009;9:338-50
11. Gradishar W, Anderson B, Abraham J, Aft R, Agnese D, Allison K. et al. Breast Cancer, Version 3.2020, NCCN Clinical Practice Guidelines in Oncology. Journal of the National Comprehensive Cancer Network: JNCCN. 2020;18:452-78
12. Zeng H, Wang L, Wang J, Chen T, Li H, Zhang K. et al. microRNA-129-5p suppresses Adriamycin resistance in breast cancer by targeting SOX2. Archives of biochemistry and biophysics. 2018;651:52-60
13. Liu J, Debnath J. The Evolving, Multifaceted Roles of Autophagy in Cancer. Advances in cancer research. 2016;130:1-53
14. Balaji S, Udupa N, Chamallamudi M, Gupta V, Rangarajan A. Role of the Drug Transporter ABCC3 in Breast Cancer Chemoresistance. PloS one. 2016;11:e0155013
15. Thomas S, Quinn B, Das S, Dash R, Emdad L, Dasgupta S. et al. Targeting the Bcl-2 family for cancer therapy. Expert opinion on therapeutic targets. 2013;17:61-75
16. Mortezaee K, Salehi E, Mirtavoos-Mahyari H, Motevaseli E, Najafi M, Farhood B. et al. Mechanisms of apoptosis modulation by curcumin: Implications for cancer therapy. Journal of cellular physiology. 2019;234:12537-50
17. Kurppa K, Liu Y, To C, Zhang T, Fan M, Vajdi A. et al. Treatment-Induced Tumor Dormancy through YAP-Mediated Transcriptional Reprogramming of the Apoptotic Pathway. Cancer cell. 2020;37:104-22.e12
18. Li X, Shi L, Zhou X, Wu J, Xia T, Zhou W. et al. The role of c-Myc-RBM38 loop in the growth suppression in breast cancer. Journal of experimental & clinical cancer research: CR. 2017;36:49
19. Pereira B, Billaud M, Almeida R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends in cancer. 2017;3:506-28
20. Panzeri V, Manni I, Capone A, Naro C, Sacconi A, Di Agostino S. et al. The RNA-binding protein MEX3A is a prognostic factor and regulator of resistance to gemcitabine in pancreatic ductal adenocarcinoma. Molecular oncology. 2021;15:579-95
21. Wang H, Huang R, Guo W, Qin X, Yang Z, Yuan Z. et al. RNA-binding protein CELF1 enhances cell migration, invasion, and chemoresistance by targeting ETS2 in colorectal cancer. Clinical science (London, England: 1979). 2020;134:1973-90
22. Kim S, Ju J, Kang M, Eun J, Kim Y, Raninga P. et al. RNA-binding protein NONO contributes to cancer cell growth and confers drug resistance as a theranostic target in TNBC. Theranostics. 2020;10:7974-92
23. Kanaoka Y, Nojima H. SCR: novel human suppressors of cdc2/cdc13 mutants of Schizosaccharomyces pombe harbour motifs for RNA binding proteins. Nucleic acids research. 1994;22:2687-93
24. Sun X, Hu Y, Wu J, Shi L, Zhu L, Xi P. et al. RBMS2 inhibits the proliferation by stabilizing P21 mRNA in breast cancer. Journal of experimental & clinical cancer research: CR. 2018;37:298
25. Burke P. Mitochondria, Bioenergetics and Apoptosis in Cancer. Trends in cancer. 2017;3:857-70
26. Carneiro B, El-Deiry W. Targeting apoptosis in cancer therapy. Nature reviews Clinical oncology. 2020;17:395-417
27. Zhu L, Xi P, Li X, Sun X, Zhou W, Xia T. et al. The RNA binding protein RBMS3 inhibits the metastasis of breast cancer by regulating Twist1 expression. Journal of experimental & clinical cancer research: CR. 2019;38:105
28. Xi P, Zhang X, Zhu L, Dai X, Cheng L, Hu Y. et al. Oncogenic action of the exosome cofactor RBM7 by stabilization of CDK1 mRNA in breast cancer. NPJ breast cancer. 2020;6:58
29. Wu J, Zhou X, Sun X, Xia T, Li X, Shi L. et al. RBM38 is involved in TGF-β-induced epithelial-to-mesenchymal transition by stabilising zonula occludens-1 mRNA in breast cancer. British journal of cancer. 2017;117:675-84
30. Tianhu Z, Shiguang Z, Xinghan L. Bmf is upregulated by PS-341-mediated cell death of glioma cells through JNK phosphorylation. Molecular biology reports. 2010;37:1211-9
31. Zhang Y, Adachi M, Kawamura R, Zou H, Imai K, Hareyama M. et al. Bmf contributes to histone deacetylase inhibitor-mediated enhancing effects on apoptosis after ionizing radiation. Apoptosis: an international journal on programmed cell death. 2006;11:1349-57
32. Hinds M, Smits C, Fredericks-Short R, Risk J, Bailey M, Huang D. et al. Bim, Bad and Bmf: intrinsically unstructured BH3-only proteins that undergo a localized conformational change upon binding to prosurvival Bcl-2 targets. Cell death and differentiation. 2007;14:128-36
33. Kutuk O, Letai A. Displacement of Bim by Bmf and Puma rather than increase in Bim level mediates paclitaxel-induced apoptosis in breast cancer cells. Cell death and differentiation. 2010;17:1624-35
34. Tan B, Tiong K, Choo H, Chung F, Hii L, Tan S. et al. Mutant p53-R273H mediates cancer cell survival and anoikis resistance through AKT-dependent suppression of BCL2-modifying factor (BMF). Cell death & disease. 2015;6:e1826
35. Xue J, Xia T, Liang X, Zhou W, Cheng L, Shi L. et al. RNA-binding protein RNPC1: acting as a tumor suppressor in breast cancer. BMC cancer. 2014;14:322
36. Zhou X, Wu J, Shi L, Li X, Zhu L, Sun X. et al. PTEN expression is upregulated by a RNA-binding protein RBM38 via enhancing its mRNA stability in breast cancer. Journal of experimental & clinical cancer research: CR. 2017;36:149
Author contact
![]() Corresponding authors: Prof. Qiang Ding, Jiangsu Breast Disease Center, The First Affiliated Hospital with Nanjing Medical University, 300 Guangzhou Road, 210029, Nanjing, China; E-mail: dingqiangedu.cn; Ji-Fu Wei, Department of Pharmacy, Jiangsu Cancer Hospital & Jiangsu Institute of Cancer Research & The Affiliated Cancer Hospital of Nanjing Medical University, Nanjing, China; Email: weijifuedu.cn; Xu-Jie Zhou, Jiangsu Breast Disease Center, The First Affiliated Hospital with Nanjing Medical University, 300 Guangzhou Road, 210029, Nanjing, China; Department of Breast Surgery, Fudan University Shanghai Cancer Center, No. 270, Dongan Road, Shanghai, 200032, China; E-mail: 1273474711com.
Corresponding authors: Prof. Qiang Ding, Jiangsu Breast Disease Center, The First Affiliated Hospital with Nanjing Medical University, 300 Guangzhou Road, 210029, Nanjing, China; E-mail: dingqiangedu.cn; Ji-Fu Wei, Department of Pharmacy, Jiangsu Cancer Hospital & Jiangsu Institute of Cancer Research & The Affiliated Cancer Hospital of Nanjing Medical University, Nanjing, China; Email: weijifuedu.cn; Xu-Jie Zhou, Jiangsu Breast Disease Center, The First Affiliated Hospital with Nanjing Medical University, 300 Guangzhou Road, 210029, Nanjing, China; Department of Breast Surgery, Fudan University Shanghai Cancer Center, No. 270, Dongan Road, Shanghai, 200032, China; E-mail: 1273474711com.

 Global reach, higher impact
Global reach, higher impact