10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(5):1844-1851. doi:10.7150/ijbs.66881 This issue Cite
Review
B.1.617.2 (Delta) Variant of SARS-CoV-2: features, transmission and potential strategies
1. Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410078, P. R. China; Institute of Clinical Pharmacology, Central South University; Hunan Key Laboratory of Pharmacogenetics, Changsha 410078, P. R. China.
2. Engineering Research Center of Applied Technology of Pharmacogenomics, Ministry of Education, 110 Xiangya Road, Changsha 410078, P. R. China.
3. National Clinical Research Center for Geriatric Disorders, 87 Xiangya Road, Changsha 410008, Hunan, P.R. China.
4. Hunan Key Laboratory of Precise Diagnosis and Treatment of Gastrointestinal Tumor, Changsha 410078, P. R. China.
#These authors contributed equally to this work.
Received 2021-9-7; Accepted 2022-1-16; Published 2022-2-14
Abstract
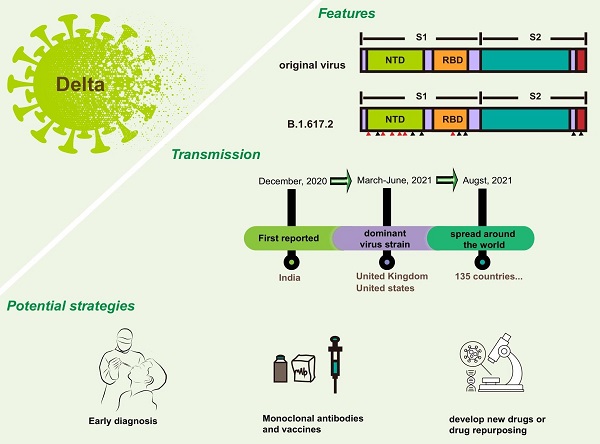
Coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a pandemic. With the continuous evolution of the viral genome, SARS-CoV-2 has evolved many variants. B.1.617.2, also called Delta, is one of the most concerned variants. The Delta variant was first reported in India at the end of 2020 but has spread globally, by now, to 135 countries and is not stand still. Delta shared some mutations with other variants, and owned its special mutations on spike proteins, which may be responsible for its strong transmission and increasing virulence. Under these circumstances, a systematic summary of Delta is necessary. This review will focus on the Delta variant. We will describe all the characteristics of Delta (including biological features and clinical characteristics), analyze potential reasons for its strong transmission, and provide potential protective ways for combating Delta.
Keywords: COVID-19, B.1.617.2, Delta variants, SARS-CoV-2, transmission, vaccines, diagnosis, structural biology, immune escape
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative pathogen of Coronavirus disease 2019 (COVID-19), emerged in late 2019 [1]. Because of the alarming levels of spread and severity, COVID-19 was characterized as a pandemic on March 11, 2020. Since the outbreak of the epidemic, a large number of lives have died in the process of fighting the virus. To date, the cumulative number of infection is nearly 212 million and the number of cumulative deaths is 4.4 million. COVID-19 has led to a serious public health crisis, and poses a huge threat to daily life.
The first whole genome sequence of SARS-CoV-2 was available within ten days after the occurrence of clustered cases [2]. Several months later, three million genome sequences were submitted. A great quantity of mutations obtained by analyzing these sequences reveal the variability of SARS-CoV-2 genome. Actually, it was not surprising that SARS-CoV-2 has new variants, because RNA virus evolves and changes easily. The mutations we can detect were those that can survive and spread successfully [3]. Since the first COVID-19 genome sequence was uploaded to the database, scientists began to monitor the evolution of SARS-CoV-2 in real time.
The emergence of B.1.1.7 has attracted attention [4, 5], because there is no previous evidence that variation will increase the adaptability of SARS-CoV-2 genome. In general, a new variant may increase circulation frequency or the change of clinical characteristics. B.1.1.7, also known as Alpha variant, was first reported in the United Kingdom. It exhibited a significant increase in transmission [6, 7] and was listed as variant of concern (VOC) by the World Health Organization (WHO) [7]. Currently, it has distributed all over the world. Mutations in Alpha variant make it easier for the virus to bind to the surface receptor of target cells, which greatly enhances the infectivity. Studies have shown that the infectivity of viral variant is increased by about 50% when compared with the original virus. In addition to B.1.1.7, there are some other VOCs, including B.1.351 (Beta) [8], P.1 (Gamma) [9], B.1.617.2 (Delta) and B.1.1.529 (Omicron). The possibility of SARS-CoV-2 variants with distinct characteristics to evolve was increased due to the immense number of current cases. B.1.351 starts in South Africa, and P.1 in Brazil, the outstanding feature of these two viruses is that they have the ability to escape immunity [10]. In other words, they may lead to decreased vaccine efficacy and ineffective drug treatment. Delta is also a VOC. Compared to the original virus, Delta has the following four features: (1) high viral load. In terms of viral load, Delta is 1,260 times that of last year's pandemic strain; (2) large exhaled virus concentration. It makes people more likely to be exposed to the virus; (3) highly contagious disease; (4) un-conspicuous symptoms. The two aspects mentioned later make Delta difficult to prevent. This review focused on the features and transmission of B.1.617.2 variant. The potential strategies against Delta will also be discussed.
Features of Delta variant
Recently, WHO announced that viruses within the lineage B.1.617 have been characterized as VOC or variant of interest (VOI). B.1.617 contains three sub-lineages, which included B.1.617.1 (also known as Kappa), B.1.617.2, and B.1.617.3 [11]. B.1.617.2 lineages have been divided into VOCs, which may affect the transmission ability of the virus, sensitivity to vaccine and its pathogenicity to humans. B.1.617.1 and B.1.617.3 lineage were VOIs that are significant variants requiring further observation [12].
Biological characteristics of Delta variant
In general, Delta virus is a variant of SARS-CoV-2, so their biological characteristics were similar to SARS-CoV-2, which is an enveloped, positive-sense single-stranded RNA virus, belonging to the beta coronavirus (β-CoV) [13, 14]. The genome of SARS-CoV-2 comprises fourteen open reading frames (ORFs) [15, 16], that encode sixteen non-structural proteins (NSP), nine accessory proteins and four structural proteins. NSPs participated in the formation of replicase complex and remaining parts are involved in viral entry, assembly and release [15, 17]. Spike protein (S) is critical for viral infection, which is an important target for combating SARS-CoV-2 [18]. S protein has a receptor-binding domain (RBD), an S1/S2 polybasic cleavage site and 3 O-linked glycans [19]. All specific functional structures are the product of natural evolution.
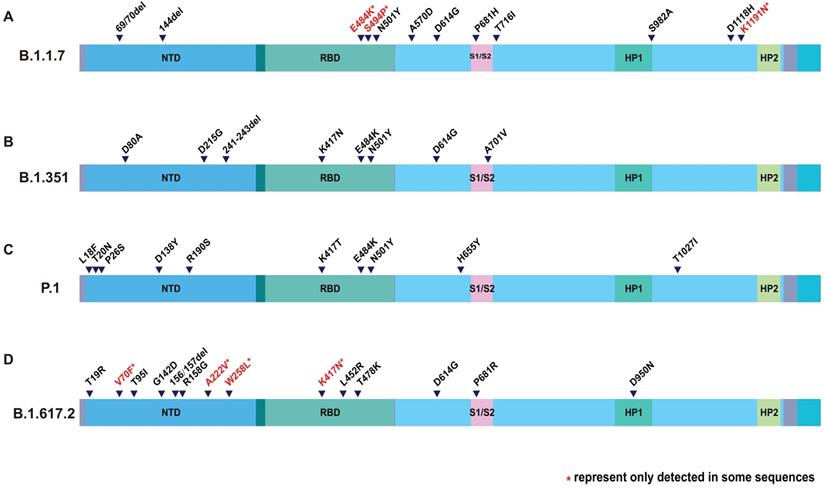
However, compared with the original virus, Delta is special. There are many mutations happened to Delta genome. Mutations in S protein are particularly abundant. According to the genome sequence analysis on GISAID, Delta lineage has 8 spike mutations (T19R, G142D, del157/158, L452R, T478K, D614G, P681R, and D950N) [20, 21]. T19R, G142D, and del157/158 are located in the N-terminal domain (NTD), and L452R and T478K are in the receptor binding domain (RBD). Together with the D614G, all of the mutations mentioned above occur on the S1 subunit. P681R and D950N occur on the S2 subunit (Figure 1). It is interesting that N501Y was not found in Delta, but can be found in Alpha, Beta and Gamma variant. [22, 23] Neither does the E484K [24]. Although Delta lacks these well-known mutations, that are common to multiple viral strains, it has produced many distinctive variants. For example, P681R, L452R and D950N. P681R resembled P681H of Alpha variant [25]. L452R and D950N are new ones (Table 1). Distinguishing features make Delta a new viral strain.
Clinical characteristics of Delta variant
The rapid spread of Delta virus is bringing a threat to us. Infection with Delta virus, causes flu-like symptoms [26]. Fever, dry cough, weakness, coughing with sputum, headache, short breath, and aching pain in muscle were considered to be the common symptoms [27]. Most patients had the first manifestation of hypoesthesia or loss of smell and taste [28]. Some patients with severe disease often have dyspnea or hypoxemia one week after onset, others can quickly develop into acute respiratory distress syndrome (ARDS), septic shock, metabolic acidosis, coagulation dysfunction and multiple organ failure [29]. Very few patients have the manifestations of central nervous system involvement and acral ischemic necrosis [30]. There was no significant difference between the clinical characteristics of Delta virus infection and the symptoms caused by other strains.
Transmission of Delta variant
According to the latest report issued by the WHO (Edition 51, published 3 August 2021), the cumulative number of patients with coronavirus infection reported globally is now nearly 197 million and the number of cumulative deaths is 4.2 million. Americas and Europe were the fastest growing regions in case incidence, and the Americas and South-East Asia Regions were reported to have the high mortality rate. Such scary number of infections is partly due to the prevalence of Delta virus.
Amino acid changes in S protein of VOCs. Description of SARS-CoV-2 spike mutation in different virus strain. (A) B.1.1.7; (B) B.1.351; (C) P.1; (D) B.1.617.2. The colored columns describe the structural domain of spike protein. NTD: N-terminal domain; RBD: receptor-binding domain; HP1: heat protein 1; HP2: heat protein 2.
Characteristics of SARS-CoV-2 Variants of Concern
| Variants of concern | Alpha | Beta | Gamma | Delta |
|---|---|---|---|---|
| Pango Lineage | B.1.1.7 | B.1.351 | P.1 | B.1.617 |
| First detected in | the United Kingdom | South Africa | Brazil | India |
| Earliest samples | September, 2020 | May, 2020 | November, 2020 | October, 2020 |
| Spike RBD mutations | E484K, S494P, N501Y | K417N, E484K, N501Y | K417T, E484K, N501Y | K417N, L452R, T478K |
| Spike non-RBD mutations | 69/70del, 144del, A570D, D614G, P681H, T716I, S982A, D118H | D80A, D215G, 241/243del, D614G, A701V | L18F, T20N, P26S, D138Y, R190S, H655Y, T1027I | T19R, V70F, T95I, G142D, del157/158, A222V, W258L, D614G, P681R, D950N |
| Transmissibility | ↑ | ↑ | ↑ | ↑ |
| Virulence | ↔ | ? | ? | ? |
Global spread of Delta variant
Delta was first reported in India at the end of 2020 but have spread globally. By detecting throat and nose swabs from people who aged 5 years and older in England, Paul et al. found that B.1.1.7, which first appeared in the United Kingdom, was gradually replaced by Delta [31]. And in the United States of America, since the first Delta infected person was detected in March, the dominant virus strain has also changed from B.1.1.7 to B.1.617 and P.1 [32]. Up to now, 135 countries have reported cases of the Delta variant [33, 34]. Delta variant has strong transmission and increasing virulence.
Possible reasons for rapid spread of Delta variant
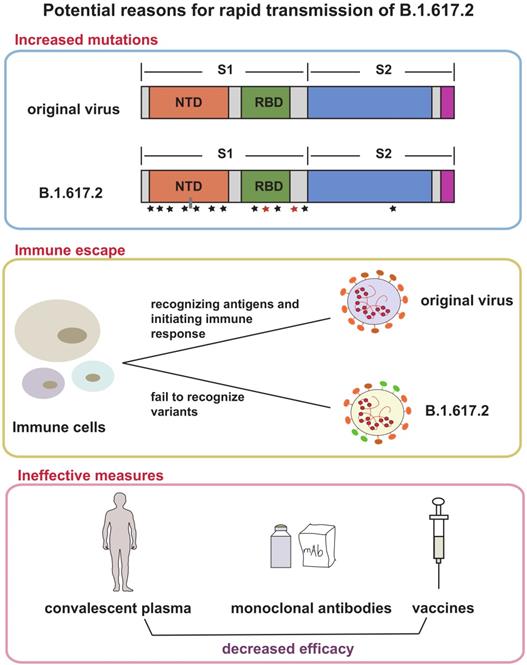
Based on current evidence, Delta variant is more transmissible than the Alpha variant. The ratio is about 40-60%, which may be related to greater risk of hospitalization. The factors driving the recent rapid growth of Delta-associated cases may be due to the followings (Figure 2): (1) More mutations and closer synergy. D614G exists in Delta genome. As the dominant viral strain in 2020, Delta is well known for its strong transmission and replication ability [35, 36]. P681R changed the furin cleavage sites of the virus, which accelerated cell-cell fusion [37, 38]. L452R increased the ability of virus to invade cells [39]. Although the role of many other mutations has not been studied clearly, the diversity of mutations does make Delta more susceptible; (2) Immune escape. A recent study found that SARS-CoV-2 spike L452R variant, a critical variant in Delta, evades cellular immunity and increases infectivity [40] The same conclusion was also confirmed in another study [41]; (3) Many protective measures are ineffective for Delta. A large number of experimental results suggested that current methods used for the prevention and treatment of infection do not work on Delta, including convalescent plasma [42], some monoclonal antibodies [43], and partial vaccines [43, 44]; (4) Insufficient prevention and control. Shorten generation intervals or high transmissibility are responsible for Delta pandemic [45]. Rigorous control could reduce these indicators.
Potential strategies for combating Delta variant
Variations of SARS-CoV-2 is threatening, but there are still many ways to deal with it. Some direct and indirect strategies are listed below.
Early diagnosis
Timely and effective diagnosis is necessary for the control of infectious diseases. Diagnosis of COVID-19 is the first step in the prevention and treatment. SARS-CoV-2 mutates frequently because of its unique genome structure and replication system. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) is a powerful molecular tool for detecting SARS-CoV-2 RNA, including VOCs and VOIs. Alves et.al conducted a clinical validation of colorimetric RT-LAMP, they found that RT-LAMP is a fast and sensitive diagnostic tool and can be used to detect SARS-CoV-2 variants [46]. Advances of CRISPR-Cas system indicates its significance in COVID-19 diagnosis, many excellent scientists build CRISPR-Cas-based system to identify infectious people. The most representative system are DETECTER [47] and SHERLOCK [48]. CRISPR-Cas system provides us with fast and reliable detection methods [49]. Early diagnosis is part and parcel of controlling Delta.
Possible reasons for Delta variant rapid transmission. This figure shows three reasons that explain the rapid spread of Delta variant and its emergence as the dominant mutant in many countries. The first frame suggested that the increased mutations in spike protein of Delta made contributions to transmission, the second frame depicts the occurrence of immune escape in Delta variant, and the third frame showed the decreased efficacy of convalescent plasma, monoclonal antibodies and vaccines. All above were potential reasons.
Effectiveness of various vaccines on Delta variant
| Name | Category | Country | Company | Effectiveness | Compared to other variants | Reference |
|---|---|---|---|---|---|---|
| BNT162b2 | mRNA | USA | Pfizer | 75%-82% | 2.5-fold↓(compared with B.1.1.7 and P.1) | [50, 67] |
| ChAdOx1(AZD1222) | Viral vector | UK | AstraZeneca | 53%-66% | 4.3-fold↓(compared with B.1.1.7 and P.1) | [67, 68] |
| mRNA-1273 | mRNA | USA | Moderna | 76% | 2.1 to 3.4 fold↓(compared with D614G) | [69, 70] |
| BBV152 /Covaxin | Inactivated | Indian | Bharat Biotech | 65.2% | 7-fold↓(compared with wildtype virus) | [71] |
| Sputnik V | Viral vector | Russia | Gamaleya Institute | 69.85% | 2.5-fold↓(compared with B.1.1.141 and B.1.1.317) | [72] |
| NVX-CoV2373 | Recombinant SARS‐CoV‐2 Spike protein nanoparticle | USA | Novovax | NA | NA | [73] |
| CoronaVac | Inactivated | China | SinoVac | 59% | NA | [74] |
| Ad26.COV2.S | Viral vector | USA | J&J/Janssen | 60% | NA | [75] |
Vaccines and monoclonal antibodies
Vaccines is a promising method to combating virus. Vaccination frequency is an important factor affecting the efficacy of mRNA vaccine. A study from NEJM shows that effectiveness after one dose of mRNA vaccine BNT162b2 was notably lower, only 30.7%. but the effectiveness of two doses has remarkable improvement, almost 88% [50]. At the same time, an article published in the Lancet disclosed this result. This article also believes that mRNA vaccine can fight against COVID-19, but it is best to vaccinate with two doses [51]. In addition to vaccination frequency, vaccination time and vaccination method will also have an impact on vaccine effectiveness. Zhang et al. found that vaccination in the morning produces a stronger protective immune response, which may be because the human immune system is affected by circadian rhythm [52]. Hassan et al. show that ChAd-SARS-CoV-2-S, an adenovirus vector vaccine, can protect against SARS-CoV-2 invasion, and intranasally administered ChAd-SARS-CoV-2-S induces durable protection in BALB/c mice [53]. The effectiveness of various vaccines on B.1.617.2 were summarized in Table 2. Monoclonal antibodies are equally important for curbing COVID-19. Although Bamlanivimab was proved ineffective, Etesivimab, Casirivimab and Imdevimab were proved to be useful [43].
Structural biology assists in identifying drug targets
Before the advent of high-resolution crystal structures, it was common to combine biology, chemistry and physics to uncover the feature of proteins or small molecule compounds. However, the resolution is low and error prone, and the combination of the two substances cannot be revealed. Technological innovation has removed the obstacles for scientific research. Structural biology plays an essential role in antiviral research. structural biologists have used the advanced technology, x-ray crystallography (X-ray) and cryo-electron microscopy (cryo-EM), to analyze critical information. Yao et al. reported the molecular assembly of the authentic SARS-CoV-2 virus using cryoelectron tomography (cryo-ET) and subtomogram averaging (STA), which revealed SARS-CoV-2 panorama in a delicate manner [54, 55]. Further refine the application of structural biology, receptor binding region [56], neutralizing antibody structure [57], spike mutation [58] are also within its scope. Using common viral strains, structural biology has helped people identify the key sites to break the virus and try to develop relevant inhibitors [59-61]. Jin et.al found more than 10,000 potentially valid compounds through structure-based drug design and screening, and six of these compounds exhibited promising M-pro inhibited activity in cell-based assays [59]. Beyond that, potential inhibitors are also identified by studying the complex structure of the drug binding to the virus [61-64]. Although the research on antiviral drugs is still making efforts to be closer to clinical transformation, some non-antiviral drugs, such as Branebrutinib, have been successfully entered into clinical trials based on the help of structural biology [65]. Therefore, we have reasons to trust structural biology to determine drug targets for Delta variant.
Epidemic prevention and control
To protect ourselves from infected by Delta, we should not only rely on external measures, but also improve self-protection awareness. A large amount of evidence shows that the strength of government prevention and control, and the degree of citizens' compliance are positively correlated with the infection. Delta strain is special, but its biological characteristics have not undergone subversive changes. For this reason, wear masks, wash hands frequently, keep social distance and avoid crowded gathering are still worth advocating.
Conclusions
After invading China, COVID-19 fights the world and defeats the defense lines of all countries, forcing WHO to declare that COVID-19 has reached the highest level of infectious disease. COVID-19 has spread worldwide over a year, and the extent and severity of the outbreak is deeply concerned. Many methods have been proposed to combat the virus, such as reduce exposure, drug treatment and vaccination, that can effectively delay the spread of the virus. While microorganisms are alive, they evolve in order to survive and reproduce, so is SARS-CoV-2. A lot of characteristic variations have been derived in the process of SARS-CoV-2 evolution. Delta is a SARS-CoV-2 variant originally found in India. It has spread to more than 135 countries in just half a year. At present, Delta has become the main variant in the world. The scientific community's understanding of Delta is still limited except its known infectious power. It is necessary to systematically expound the characteristics and global impact of Delta. In this article, we reviewed three major sections describing different aspects of Delta variant.
Firstly, we introduced features of Delta, including its biological features and clinical characteristics. Delta is a variant of SARS-CoV-2, similar to the original virus. But has its own distinguished feature. There are many remarkable mutations happened in S protein, for example, L452R and D950N. These mutations work together to make Delta virus become variant of concern.
Secondly, we described the dissemination of virus. Delta variant has become the leading variation and has involved in 135 countries. At the same time, a large number of studies show that the ultra-fast transmission speed not only increases the risk of infection, but also increases the hospitalization rate. Therefore, we summarized some reasons for this situation so that we can better understand and conquer Delta.
Finally, we proposed some possible measures for the treatment and prevention of Delta. We suggest that early diagnosis is important for control the infectious disease. Although there is no specific drug, vaccination is a reassuring treatment. There is a lot of evidence that the antibodies produced after vaccination can neutralize Delta variant. In addition, the latest research suggests that chimeric spike mRNA vaccines may be the direction to overcome the virus [66]. Using structural biology to find potential drug target is also suggested. Last but not least, improving self-protection awareness never go out of style.
In conclusion, understanding the relevant information of Delta will help us fight it better. Only by knowing ourselves and others can we defeat the virus.
Abbreviations
SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; COVID-19: coronavirus disease 2019; VOC: variant of concern; WHO: World Health Organization; VOI: variant of interest; β-CoV: beta coronavirus; ORFs: open reading frames; NSP: non-structural protein; S: Spike protein; RBD: receptor-binding domain; NTD: N-terminal domain; ARDS: acute respiratory distress syndrome; RT-LAMP: reverse transcription loop-mediated isothermal amplification; CRISPR: clustered regularly interspaced short palindromic repeats; X-ray: X-ray crystallography; cryo-ET: cryoelectron tomography; STA: subtomogram averaging.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81773823, 82073943), and Hunan Cancer Hospital Climb Plan (YF2020011).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J. et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727-33
2. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG. et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265-9
3. Otto SP, Day T, Arino J, Colijn C, Dushoff J, Li M. et al. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr Biol. 2021;31:R918-29
4. Washington NL, Gangavarapu K, Zeller M, Bolze A, Cirulli ET, Schiabor BK. et al. Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell. 2021;184:2587-94
5. Leung K, Shum MH, Leung GM, Lam TT, Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021 26
6. Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A. et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage - United States, December 29, 2020-January 12, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:95-9
7. Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L. et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266-9
8. Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J. et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438-43
9. Dejnirattisai W, Zhou D, Supasa P, Liu C, Mentzer AJ, Ginn HM. et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184:2939-54
10. Hoffmann M, Arora P, Gross R, Seidel A, Hornich BF, Hahn AS. et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384-93
11. Pascarella S, Ciccozzi M, Zella D, Bianchi M, Benedetti F, Benvenuto D, et al. SARS-CoV-2 B.1.617 Indian variants: Are electrostatic potential changes responsible for a higher transmission rate? J Med Virol. 2021
12. Ong S, Chiew CJ, Ang LW, Mak TM, Cui L, Toh M, et al. Clinical and virological features of SARS-CoV-2 variants of concern: a retrospective cohort study comparing B.1.1.7 (Alpha), B.1.315 (Beta), and B.1.617.2 (Delta). Clin Infect Dis. 2021
13. V'Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155-70
14. Wang MY, Zhao R, Gao LJ, Gao XF, Wang DP, Cao JM. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front Cell Infect Microbiol. 2020;10:587269
15. Flower TG, Buffalo CZ, Hooy RM, Allaire M, Ren X, Hurley JH. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc Natl Acad Sci U S A. 2021 118
16. Bianchi M, Borsetti A, Ciccozzi M, Pascarella S. SARS-Cov-2 ORF3a: Mutability and function. Int J Biol Macromol. 2021;170:820-6
17. Harrison AG, Lin T, Wang P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020;41:1100-15
18. Cai Y, Zhang J, Xiao T, Peng H, Sterling SM, Walsh RJ. et al. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586-92
19. Yang Q, Hughes TA, Kelkar A, Yu X, Cheng K, Park S. et al. Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration. Elife. 2020 9
20. Ong S, Chiew CJ, Ang LW, Mak TM, Cui L, Toh M, et al. Clinical and virological features of SARS-CoV-2 variants of concern: a retrospective cohort study comparing B.1.1.7 (Alpha), B.1.315 (Beta), and B.1.617.2 (Delta). Clin Infect Dis. 2021
21. Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017 22
22. Huang H, Zhu Y, Niu Z, Zhou L, Sun Q. SARS-CoV-2 N501Y variants of concern and their potential transmission by mouse. Cell Death Differ. 2021
23. Khan A, Zia T, Suleman M, Khan T, Ali SS, Abbasi AA. et al. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: An insight from structural data. J Cell Physiol. 2021;236:7045-57
24. Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM. et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:409-24
25. Peters MH, Bastidas O, Kokron DS, Henze CE. Transformations, Lineage Comparisons, and Analysis of Down-to-Up Protomer States of Variants of the SARS-CoV-2 Prefusion Spike Protein, Including the UK Variant B.1.1.7. Microbiol Spectr. 2021:e3021
26. Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y. et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382:1199-207
27. Islam KU, Iqbal J. An Update on Molecular Diagnostics for COVID-19. Front Cell Infect Microbiol. 2020;10:560616
28. Jiang F, Deng L, Zhang L, Cai Y, Cheung CW, Xia Z. Review of the Clinical Characteristics of Coronavirus Disease 2019 (COVID-19). J Gen Intern Med. 2020;35:1545-9
29. Li X, Ma X. Acute respiratory failure in COVID-19: is it "typical" ARDS? Crit Care. 2020;24:198
30. Meinhardt J, Radke J, Dittmayer C, Franz J, Thomas C, Mothes R. et al. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat Neurosci. 2021;24:168-75
31. Riley S, Eales O, Haw D. et al. REACT-1 round 13 interim report: acceleration of SARS-CoV-2 Delta epidemic in the community in England during late June and early July 2021. medRxiv. 2021
32. Alexandre Bolze ETCS, Dei Rossi TCSJ, Sandoval XWDW, Isaksson NLWW. Rapid displacement of SARS-CoV-2 variant B.1.1.7 by B.1.617.2 and P.1 in the United States. medRxiv. 2021
33. Zhang M, Xiao J, Deng A, Zhang Y, Zhuang Y, Hu T. et al. Transmission Dynamics of an Outbreak of the COVID-19 Delta Variant B.1.617.2-Guangdong Province, China, May-June 2021. CHINA CDC WEEKLY. 2021;3:584-6
34. Afrad MH, Khan MH, Rahman S, Bin MO, Hossain M, Alam AN. et al. Genome Sequences of 15 SARS-CoV-2 Sublineage B.1.617.2 Strains in Bangladesh. Microbiol Resour Announc. 2021;10:e56021
35. Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG. et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116-21
36. Volz E, Hill V, McCrone JT, Price A, Jorgensen D, O'Toole A. et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell. 2021;184:64-75
37. Lubinski B, Frazier LE, V TPM, Bugembe DL, Tang T, Daniel S, et al. Spike protein cleavage-activation mediated by the SARS-CoV-2 P681R mutation: a case-study from its first appearance in variant of interest (VOI) A.23.1 identified in Uganda. bioRxiv. 2021
38. Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M. et al. SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms. 2021 9
39. Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S. et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;182:1284-94
40. Motozono C, Toyoda M, Zahradnik J, Saito A, Nasser H, Tan TS. et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29:1124-36
41. Deng X, Garcia-Knight MA, Khalid MM, Servellita V, Wang C, Morris MK. et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell. 2021;184:3426-37
42. Liu C, Ginn HM, Dejnirattisai W, Supasa P, Wang B, Tuekprakhon A. et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220-36
43. Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM. et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276-80
44. Edara VV, Lai L, Sahoo MK, Floyd K, Sibai M, Solis D, et al. Infection and vaccine-induced neutralizing antibody responses to the SARS-CoV-2 B.1.617.1 variant. bioRxiv. 2021
45. Bandoy D, Weimer BC. Analysis of SARS-CoV-2 genomic epidemiology reveals disease transmission coupled to variant emergence and allelic variation. Sci Rep. 2021;11:7380
46. Dao TV, Herbst K, Boerner K, Meurer M, Kremer LP, Kirrmaier D. et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med. 2020 12
47. Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J. et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38:870-4
48. Patchsung M, Jantarug K, Pattama A, Aphicho K, Suraritdechachai S, Meesawat P. et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng. 2020;4:1140-9
49. Zhan Y, Li XP, Yin JY. COVID-19 one year later: a retrospect of CRISPR-Cas system in combating COVID-19. Int J Biol Sci. 2021;17:2080-8
50. Lopez BJ, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S. et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N Engl J Med. 2021;385:585-94
51. Wall EC, Wu M, Harvey R, Kelly G, Warchal S, Sawyer C. et al. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet. 2021;397:2331-3
52. Zhang H, Liu Y, Liu D, Zeng Q, Li L, Zhou Q. et al. Time of day influences immune response to an inactivated vaccine against SARS-CoV-2. Cell Res. 2021
53. Hassan AO, Shrihari S, Gorman MJ, Ying B, Yaun D, Raju S. et al. An intranasal vaccine durably protects against SARS-CoV-2 variants in mice. Cell Rep. 2021;36:109452
54. Arya R, Kumari S, Pandey B, Mistry H, Bihani SC, Das A. et al. Structural insights into SARS-CoV-2 proteins. J Mol Biol. 2021;433:166725
55. Yao H, Song Y, Chen Y, Wu N, Xu J, Sun C. et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell. 2020;183:730-8
56. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S. et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215-20
57. Barnes CO, Jette CA, Abernathy ME, Dam KA, Esswein SR, Gristick HB. et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682-7
58. Barcena M, Barnes CO, Beck M, Bjorkman PJ, Canard B, Gao GF. et al. Structural biology in the fight against COVID-19. Nat Struct Mol Biol. 2021;28:2-7
59. Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y. et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289-93
60. Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L. et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409-12
61. Rut W, Lv Z, Zmudzinski M, Patchett S, Nayak D, Snipas SJ. et al. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: A framework for anti-COVID-19 drug design. Sci Adv. 2020 6
62. Klemm T, Ebert G, Calleja DJ, Allison CC, Richardson LW, Bernardini JP. et al. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. Embo J. 2020;39:e106275
63. Fu Z, Huang B, Tang J, Liu S, Liu M, Ye Y. et al. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat Commun. 2021;12:488
64. Pathak N, Chen YT, Hsu YC, Hsu NY, Kuo CJ, Tsai HP. et al. Uncovering Flexible Active Site Conformations of SARS-CoV-2 3CL Proteases through Protease Pharmacophore Clusters and COVID-19 Drug Repurposing. Acs Nano. 2021;15:857-72
65. Watterson SH, Liu Q, Beaudoin BM, Batt DG, Li L, Pattoli MA. et al. Discovery of Branebrutinib (BMS-986195): A Strategy for Identifying a Highly Potent and Selective Covalent Inhibitor Providing Rapid in vivo Inactivation of Bruton's Tyrosine Kinase (BTK). J Med Chem. 2019;62:3228-50
66. Martinez DR, Schafer A, Leist SR, De la Cruz G, West A, Atochina-Vasserman EN. et al. Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science. 2021;373:991-8
67. Liu J, Liu Y, Xia H, Zou J, Weaver SC, Swanson KA. et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature. 2021;596:273-5
68. Sheikh A, McMenamin J, Taylor B, Robertson C. SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. 2021;397:2461-2
69. Choi A, Koch M, Wu K, Dixon G, Oestreicher J, Legault H. et al. Serum Neutralizing Activity of mRNA-1273 against SARS-CoV-2 Variants. J Virol. 2021;95:e131321
70. Puranik A, Lenehan PJ, Silvert E, Niesen M, Corchado-Garcia J, O'Horo JC. et al. Comparison of two highly-effective mRNA vaccines for COVID-19 during periods of Alpha and Delta variant prevalence. medRxiv. 2021
71. Thiagarajan K. What do we know about India's Covaxin vaccine? BMJ. 2021;373:n997
72. Jones I, Roy P. Sputnik V COVID-19 vaccine candidate appears safe and effective. Lancet. 2021;397:642-3
73. Keech C, Albert G, Cho I, Robertson A, Reed P, Neal S. et al. Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N Engl J Med. 2020;383:2320-32
74. Zhang Y, Zeng G, Pan H, Li C, Hu Y, Chu K. et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18-59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect Dis. 2021;21:181-92
75. Sadoff J, Le Gars M, Shukarev G, Heerwegh D, Truyers C, de Groot AM. et al. Interim Results of a Phase 1-2a Trial of Ad26.COV2.S Covid-19 Vaccine. N Engl J Med. 2021;384:1824-35
Author contact
![]() Corresponding author: Professor Ji-Ye Yin, Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410008; P. R. China. Tel: +86 731 84805380, Fax: +86 731 82354476, E-mail: yinjiyeedu.cn. ORCID: 0000-0002-1244-5045.
Corresponding author: Professor Ji-Ye Yin, Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410008; P. R. China. Tel: +86 731 84805380, Fax: +86 731 82354476, E-mail: yinjiyeedu.cn. ORCID: 0000-0002-1244-5045.

 Global reach, higher impact
Global reach, higher impact