10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(7):2744-2758. doi:10.7150/ijbs.70458 This issue Cite
Review
The role of Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) as m6A readers in cancer
1. College of Biological and Pharmaceutical Engineering, West Anhui University, Lu'an, China.
2. Department of Radiology, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Guangzhou, 510060, P.R. China.
# These authors contributed equally to this work.
Received 2021-12-25; Accepted 2022-3-14; Published 2022-3-28
Abstract
RNA can be modified by over 170 types of distinct chemical modifications, and the most abundant internal modification of mRNA in eukaryotes is N6-methyladenosine (m6A). The m6A modification accelerates mRNA process, including mRNA splicing, translation, transcript stability, export and decay. m6A RNA modification is installed by methyltransferase-like proteins (writers), and potentially removed by demethylases (erasers), and this process is recognized by m6A-binding proteins (readers). Notably, alterations of m6A-modified proteins (writers, erasers and readers) are involved in the tumorigenesis, progression and metastasis. Importantly, the fate of m6A-methylated mRNA is mediated mostly through m6A readers, and among these readers, insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) are unique RNA-binding proteins (RBPs) that stabilize their targets mRNA via m6A modification. In this review, we update the writers, erasers and readers, and their cross-talks in m6A modification, and briefly discuss the oncogenic role of IGF2BPs in cancer. Most importantly, we mainly review the up-to-date knowledges of IGF2BPs (IGF2BP1/2/3) as m6A readers in an m6A-modified manner in cancer progression.
Keywords: N6-methyladenosine (m6A), IGF2BPs, Reader, Stabilization, Cancer
Introduction
The insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs, IMPs), including IGF2BP1/2/3, are first identified in 1999 for their ability to bind to the name-giving 5'UTR of insulin-like growth factor Ⅱ (IGF-Ⅱ) leader 3 mRNA [1]. The first IGF2BPs family member described is IGF2BP1 that is identified as a 75kd polysome-associated protein to stabilize c-MYC mRNA by binding to its coding region determinant (CRD) [2]. IGF2BP2 is first described in 1999, and discovered as an intracellular antigen detected in 30-40% hepatocellular carcinoma patients [3, 4]. IGF2BP3, initially called KOC protein, is first demonstrated as a highly expressed gene in pancreatic cancer that encodes four K-homologous (KH) domains [5]. IGF2BPs are highly conserved RNA-binding proteins (RBPs) that are crucial players in regulating RNA processing, including mRNA splicing, translation, decay and stability [6]. IGF2BPs are expressed in most tissues during embryogenesis, and only IGF2BP2 is ubiquitously expressed in adult tissues [3, 7]. Importantly, IGF2BPs are characterized as oncofetal proteins, by which IGF2BPs are involved in carcinogenesis [8, 9]. Accumulating data have been demonstrated that IGF2BPs are highly expressed in a broad range of tumors and also associated with poor prognosis [8].
IGF2BPs uniquely contain two RNA recognition motifs (RRMs) and four KH domains. KH domains-containing proteins are first reported by Kiledjian to regulate mRNA stability in 1995 [10]. The underlying mechanism of mRNA stabilization by KH domains is revealed by Huang laboratory, by which IGF2BPs stabilize c-MYC by its KH domains (KH3-4) in m6A modification manner [11]. Since then, the function of IGF2BPs in m6A modification has been continuously reported, where the IGF2BPs participate in posttranscriptional process, including alternative splicing, metabolism, and stabilization [12].
In a manner similar with DNA and protein, RNAs can be modified by more than 170 chemically modifications [13]. Among them, N6-methyladenosine (m6A) is most abundant modification of messenger RNA (mRNAs) and non-coding RNA (ncRNAs) in mammals [14, 15]. Although first discovered in the 1970s, the m6A modification began to revive in 2012, when a next-generation sequencing method, MeRIP-Seq was described and used [16]. In mammalian cells, m6A modification can be catalyzed by a methyltransferase complex (“writers”) and removed by demethylases (“erasers”), thus m6A mRNA modification is a reversible and dynamic process [17]. Notably, the fate of m6A-modified RNA is mediated mostly through RNA-binding proteins (“readers”), including YTHDF, FMRP, CNBP, PRRC2A, HNRNPA2B1 and IGF2BPs [18-20]. The m6A modification by m6A readers affects mRNA fate by regulating RNA splicing, translation, stability, structure, export and decay of the modified RNA [21]. Recently, emerging evidence demonstrates that m6A modification also affects the mRNA fate by promoting the phase separation of m6A readers [22]. For example, m6A regulates the fate of cytosolic mRNA through scaffolding for binding YTHDF proteins, resulting in the formation of phase separation complex (YTHDF-m6A-mRNA) that partitions into phase-separated compartments, such as P-bodies and stress granules [23]. This effect is efficient for the polymethylated mRNAs that scaffold multiple YTHDF proteins. In addition, m6A modification is required for the liquid-liquid phase separation (LLPS) of YTHDC1 to form nuclear condensates that protect mRNAs from degradation, and regulate myeloid leukemic differentiation [24]. In addition to m6A readers, the roles of LLPS in m6A writers have been explored, and for instance, LLPS is an important player in regulating dynamic assembly of the mRNA m6A methyltransferase complex (METTL3/METTL14/WTAP) [25].
m6A modification governs mRNA fate and function in multiple biological processes, and m6A readers-mediated m6A modification is implicated in various human diseases, especially cancer [26]. Among m6A readers, IGF2BPs are very uniquely RNA-binding proteins that play critical roles in cancer progression through affecting mRNA fate in an m6A-dependent manner. In this review, we will briefly introduce the understanding of m6A modification by the writers, erasers and readers, and crosstalk between theses regulators. In addition, we also summarize the function of oncogenes, IGF2BPs in the tumorigenesis and cancer progression. Of note, we focus on how IGF2BPs promote tumorigenesis and progression via the m6A-dependent manner. Moreover, we further describe perspectives toward future questions and challenges in IGF2BPs-mediated m6A modification.
m6A writers, erasers, and readers
m6A, one of the most abundant chemical modifications in eukaryotic mRNA, has gained increasing attention as a mode of post-transcriptional gene regulation [27-29]. Genetic loss-of-function studies on m6A writers, erasers and readers highlight m6A modification as a dynamically and reversibly regulatory process in various biological processes [30, 31]. m6A methylation plays critical roles in regulating gene expression through mediating the mRNA stability, degradation and translation, and its disruption results in a series of diseases, including cancer [32-34]. The cross-talks among m6A writer, eraser, and reader are reported to determine the m6A levels of their targets and, consequently, the stability of these targets plays important roles in tumorigenesis, drug resistance, and metastasis [35-37].
m6A writers
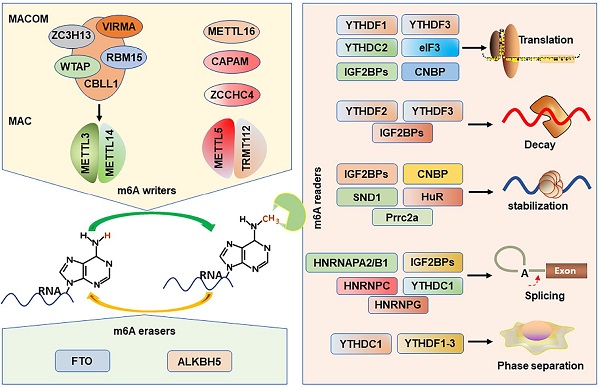
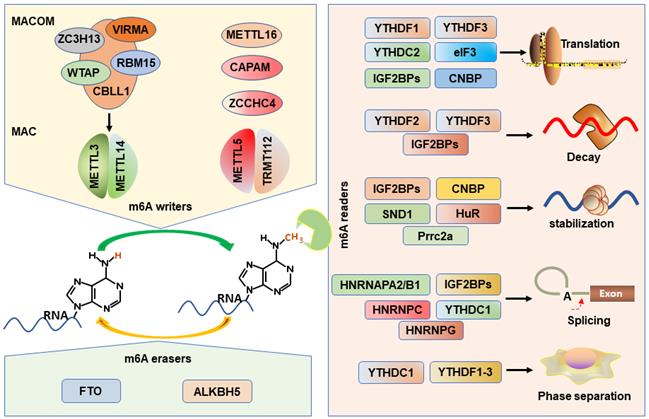
m6A is established by m6A methyltransferases complex (also called “writers”) that transfers a methyl group from s-adenosylmethionine (SAM) to the substrate adenosine in RNA [38, 39]. Methyltransferase-like protein 3 (METTL3) and METTL14 play as a catalytic core complex known as the m6A-METTL complex (MAC) that recognizes the DRACH motifs and promotes the m6A modification in the transcriptome [40-42]. Notably, METTL3 has catalytic activity, while METTL14 forms a heterodimer with METTL3 and then strengthens its catalytic action [39, 43, 44]. Interestingly, MAC interacts with m6A-METTL associated complex (MACOM) that composes of the wilms tumor 1 associated protein (WTAP), vir-like m6A methyltransferase-associated (VIRMA), RNA-binding motif 15 (RBM15), zinc-finger CCCH-type-containing 13 (ZC3H13), Cbl proto oncogene-like protein 1 (CBLL1) [45]. Although the MACOM itself lacks catalytic activity, its coordinated interaction with the MAC promotes it localization to nuclear speckles and modulates their recruitment to specific targets for m6A modification. METTL16 (a homologue of METTL3), a novel independent RNA methyltransferase, is a conserved U6 snRNA methyltransferase and regulates cellular SAM levels [46, 47]. CAPAM (also known as PCIF1) has been recently identified as an evolutionarily conserved methyltransferase, responsible for the m6A on the mRNA cap-adjacent Am-modified nucleotide [48-50]. The METTL5: TRMT112 complex has been recognized as a m6A rRNA methyltransferase that catalyzes the N6-methylation of A1832 (m6A1832) in human 18S rRNA [51]. In addition, ZCCHC4, another m6A rRNA MTase, facilitates the m6A4220 modification in all human 28S rRNA [52].
m6A erasers
The possible reversibility and dynamic of the m6A modification are identified by the m6A demethylases (erasers): fat mass and obesity-associated protein (FTO) and α-ketoglutarate-dependent dioxygenase alk B homolog 5 (ALKBH5) protein, which can selectively remove m6A-methylated groups from their targets RNA [53, 54]. The demethylase activity of ALKBH5 has been demonstrated to be preferential for m6A methylation in DRACH motif-dependent manner in RNA, whereas FTO demethylate a wide array of substrates including m6A [54]. Several recent studies suggest that FTO, however, preferentially demethylates the N6,2′-O-dimethyladenosine (m6Am), which suggests that ALKBH5 is more participated in global m6A demethylation than FTO [55].
m6A readers
m6A recognition proteins, known as “readers”, can decode m6A marks and perform diverse biological functions [56], and these m6A readers include YT521-B homology domain family proteins (YTHDF1/2/3), YT521-B homology domain containing 1 and 2 (YTHDC1/2) [57], eukaryotic translation initiation factor 3 (eIF3) [58], insulin like growth factor 2 mRNA-binding proteins (IGF2BP1/2/3) [11], heterogeneous nuclear ribonucleoproteins (HNRNPA2/B1, HNRNPC/G) [59], Proline rich coiled-coil 2 A (Prrc2a) [60], HuR (known as ELAVL1) [41], cellular nucleic acid binding protein (CNBP) [61], and SND1 [62]. YTHDF2 is first identified m6A reader that impairs the stability of targeted transcripts and promotes mRNAs degradation via recruiting the CCR4-NOT deadenylation complex [57, 63]. Conversely, YTHDF1 promotes mRNA translation by interacting with the translation initiation factor eIF3 [64]. YTHDF3 not only can interact cooperatively with YTHDF2 to promote mRNA decay, but also cooperates with YTHDF1 to promote the translation of the methylated RNAs [65, 66]. Interestingly, YTHDF3 loss decreases the binding of YTHDF1 and YTHDF2 to their transcripts, displaying the important role of YTHDF3 in the RNA binding specificity [65]. YTHDC1 mediates alternative splicing by recruiting the splicing factor serine and arginine-rich splicing factor 3 (SRSF3) and restricting the bind of exon-skipping factor SRSF10 [67]. YTHDC2 preferentially binds to m6A-containing RNA, and then promotes translation efficacy of its targets and reduce their mRNA abundance [68, 69]. eIF3 can directly bind to mRNAs-containing m6A in their 5'UTR, which is sufficient to stimulate the mRNA translation in the loss of the cap-binding factor eIF4E [58].
The HNRNPs family binds to m6A-containing mRNA through the mechanism of “m6A-switch”, in which m6A modification alters the mRNAs structures to expose the single stranded hnRNP binding motif [59, 70]. HNRNPA2B1 binds to pre-microRNA and accelerates its maturation [19, 71]. In addition, HNRNPA2B1 also functions as modulating the alternative splicing of transcripts [19]. HNRNPC participates in the processing of pre-mRNAs and intronless mRNAs [72]. HNRNPG selectively binds to RNAs and regulates alternative splicing by interacting with m6A-methylated mRNAs [73].
Recently, IGF2BPs are verified as a distinct and conserved family of m6A-readers, consisting of two RNA recognition motif (RRM) domains and four K homology (KH) domains [7]. Nevertheless, the third and fourth KH domains (KH3-4) are responsible for recognizing the m6A sites of targets mRNAs. IGF2BPs can promote the stability and facilitate the translation efficiency in an m6A-dependent fashion [11]. And the IGF2BPs-mediated mRNA stabilization can be enhanced by recruiting the co-factors of IGF2BPs, HuR and matrin 3 (MATR3), to prevent their targets from degradation [11]. In addition, IGF2BPs are also demonstrated to be involved in mRNA translation by modulating alternative splicing [74].
Prrc2a encodes a large proline-rich protein and is located at the MHC class III region [75]. A recent study has proved that Prrc2a functions as a novel m6A reader stabilizing the mRNA of Olig2 [60]. HuR, one of the members of the Hu family of RNA-binding proteins, is associated with the m6A bait and stabilizes m6A-containing mRNAs [41, 76]. In addition, the “Royal family” protein, SND1 also can read the m6A-modified mRNAs and promotes mRNA stabilization [62]. CNBP is recently identified a novel m6A reader that consists of seven highly conserved zinc finger domains involved in mRNA transcription, stabilization, and translation [20, 61, 77]. Interestingly, only CNBP protein is localized in the nucleoplasm while other readers are localized to the cytosol. However, all m6A writers are localized in the nucleus and most readers are localized to the cytosol, how writers-readers systems work remains poorly understood. The m6A writers, erasers, and readers have been characterized in Figure 1.
Working model of m6A writers, erasers and readers on m6A modification.
Crosstalk between m6A writers, erasers, and readers
First, the links between m6A writers and readers have been extensively studied. m6A readers are often required for m6A methyltransferases-catalyzed m6A methylation. For example, METTL3-dependent m6A hypermethylation up-regulates NLRP1 transcript, and knockdown of YTHDF1 reduces the NLRP1 mRNA [78]. In turn, m6A writers are also necessary for m6A readers-mediated m6A modification. For example, METTL14 depletion reduces Socs1 m6A methylation, and blunts YTHDF1 binds to the m6A sites [79]. Loss of METTL3 impairs YTHDF1-mediated translation of its target, SPRED2 in an m6A modification manner [80]. In addition, m6A writers and readers bind to the same target transcripts [81], and thus, m6A writers and readers combine together to regulate m6A methylation process. One reasonable possibility is that m6A writers and readers can form polymeric methyltransferase complex. Second, the relationships between the m6A erasers and readers are similar to the above links. m6A readers are also required for m6A demethylases-modified m6A methylation. For example, ALKBH5 suppresses pancreatic cancer progression through activation of PER1 in an YTHDF2-dependent m6A way [82]. Third, m6A writers and erasers regulate m6A modification in opposite direction. METTL3 and ALKBH5, for example, oppositely regulate the TFEB mRNA level in an m6A-modified manner [83]. In addition, m6A writers and erasers constitute positive feedback loops to control the stability of target transcripts [35]. Fourth, m6A writers and erasers can determine the m6A status of targets by controlling each other's expression and regulating m6A readers [35]. Thus, interplay among m6A writer, eraser and reader determines the m6A status and level.
The oncogenic role of IGF2BPs in cancer
All human IGF2BPs have been identified as oncofetal proteins, and among them, IGF2BP1 and IGF2BP3 are bona fide oncofetal proteins that are synthesized in many cancers [3]. In agreement, IGF2BP1 and IGF2BP3 display a high degree of amino acid identity (73%) with each other and show similar activity [84]. The oncogenic role of IGF2BP3 was first described due to its overexpression in pancreatic cancer in 1997 [5], and then IGF2BP3 modulates tumor cell fate, such as proliferation, migration, and chemo-resistance by controlling the translation and turnover of target transcripts, and regulating DNA methylation, and acetylation processes [3, 85]. IGF2BP1 is the most conserved oncogene of all three IGF2BPs that is required for the transport, stability, and localization of mRNAs in carcinogenesis, and chemo-resistance [84, 86]. Of note, IGF2BP1 is post-transcriptional driver of E2F1-driven hallmark in solid cancers [87]. IGF2BP2 is a unique member of IGF2BPs that is ubiquitously expressed in the adult organism, and IGF2BP2 is an important post-transcriptional regulator of RNAs via the ribonucleoprotein complex [7, 8]. Interestingly, IGF2BP2 can preferentially regulate glucose and lipid metabolism, and thus IGF2BP2 is considered as diabetes associated gene that impairs insulin secretion [88]. Notably, accumulating data demonstrates that IGF2BP2 promotes carcinogenesis by regulating cancer metabolism [8].
Nonetheless, all three IGF2BPs are highly expressed and function as independent prognostic factors in a variety of cancers, including lung cancer [89, 90], liver cancer [91-93], breast cancer [94, 95], colorectal cancer [96-98], pancreatic cancer [99-101], prostate cancer [102], bladder cancer [103-105], thyroid cancer [106-108], gastric cancer [109, 110], renal cell carcinoma [111], ovarian cancer [112, 113], esophageal squamous cell carcinoma [114, 115], acute myeloid leukemia [116]. In addition to the high expression of the IGF2BPs itself, intercellular communication factors, such as tumor-secreted extracellular vesicles (EVs), can maintain the stability of IGF2BPs in cells to promote the development of cancer. EVs are emerging as crucial messengers maintaining homeostasis in tumor progression, and metastasis [117]. Interestingly, the RBPs and their substrate RNAs are detected in EVs, and EVs can harbour sequence motifs to mirror the activity of RBPs [118]. circNEIL3 is packaged into exosomes and then transmitted to tumor associated macrophages (TAMs) that enable them to acquire immunosuppressive effect by stabilizing IGF2BP3, and promoting glioma progression [119]. EVs secreted by melanoma cells regulate the effect of IGF2BP1 on metastasis, and in turn, IGF2BP1 affects the cargo of the EVs [120]. Nevertheless, the connection of IGF2BPs' stability with EVs biology requires comparative and systematic studies. In addition, IGF2BPs also play important roles in regulating other cancer phenotypes, such as glycolysis, stemness and chemo-resistance [121-123]. In mechanism, the oncogenic roles of IGF2BPs are largely attributed to their m6A dependent mRNA stabilization of oncogenic targets [87]. Thus, in most cases, inhibition of IGF2BPs or their targets can suppress the proliferation and migration of cancer cells.
The role of IGF2BPs as m6A reader in cancer
The m6A modification is a dynamic process, and the biological function of m6A relies on m6A readers. These readers recognize m6A-containing RNAs either by directly binding to YT521-B homology (YTH) domain, or by binding to single-stranded RNA motifs [124]. Although YTH domains bind to RNA with low affinity between 100 and 300 nM, YTH domain-containing proteins recognize m6A to regulate mRNA splicing, metabolism, folding, and translation [17, 125]. Besides YTH domain, m6A-containing RNAs can be selectively recognized by K homology (KH) domain. In 2018, IGF2BPs are first recognized as a novel family of m6A readers that stabilize their targets in an m6A-dependent manner [11]. IGF2BPs have six characteristic RNA-binding domains, including four distinct KH domains at C-terminal region, and two RNA recognition motifs (RRMs) at N-terminal region [126]. IGF2BPs-recognized m6A by KH domains has been demonstrated by Huang's laboratory, by which mutations in the KH domains (KH3-4) completely abolish the function of IGF2BPs as m6A reader [11]. Importantly, the critical role of KH3-4 domains in cancer has been demonstrated before IGF2BPs are identified as new m6A reader [127]. Unlike the canonical YTH domain-containing proteins, IGF2BPs-recognized targets, such as c-MYC, display higher transcript level and longer half-life period [124]. As RNA stabilizers, IGF2BPs promote the stability of multiple mRNAs through m6A modification and IGF2BPs-modified m6A plays a crucial role in many pathological conditions, especially cancer.
IGF2BP1
Currently, m6A modification regulates the translation [128], splicing [129], maturation [130], stabilization [131], and decay [132] of noncoding RNAs, including miRNAs, lncRNAs and circRNAs in cancer. In turn, interestingly, noncoding RNAs regulate IGF2BPs mRNA stabilization or IGF2BPs-modificated m6A in cancer progression [133, 134]. LncRNA THAP7-AS1 is correlated with poor prognosis in gastric cancer patients, and mechanistically, METTL3 stabilizes THAP7-AS1 in IGF2BP1-mediated m6A modification manner [135]. The oncogene MYC (also known as c-MYC) is one of the most frequently activated in human cancers, and c-MYC overexpression causes tumorigenesis and maintains tumor growth [136]. Importantly, c-MYC is considered as a critical target of IGF2BPs, and lncRNAs recruits or binds to IGF2BP1 to stabilize or increase the mRNA of c-MYC, depending m6A modification in tumor progression. For example, a hypoxia-induced lncRNA KB-1980E6.3 maintains stemness of breast cancer stem cells (BCSC) under hypoxic microenvironment by recruiting IFG2BP1 to stabilize c-MYC mRNA in m6A-modified manner [137]. Before identified as the m6A reader, IGF2BPs are regulated by lncRNAs to mediate translation and mRNA stability of c-MYC [138]. Interestingly, lncRNAs regulate m6A modification on their targets' mRNA by IGF2BP1. LINC002661 encodes a 71-amino acid oncopeptide that binds to IGF2BP1 and then strengthens the m6A recognition of IGF2BP1 to increase mRNA stability of m6A-methylated c-MYC, which promotes tumorigenesis [139]. In addition to lncRNAs, a study reported that circPTPRA plays as a tumor suppressor in bladder cancer by interacting the KH domains of IGF2BP1 to block its m6A recognition of its targets, c-MYC and FSCN1 mRNA [105].
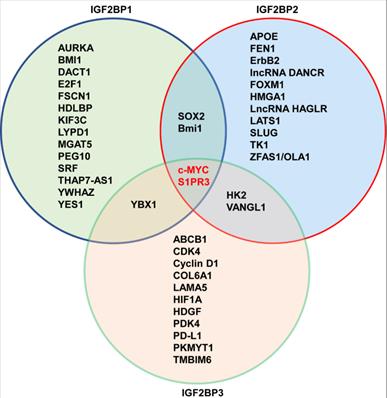
In addition to noncoding RNAs, m6A modification by IGF2BP1 also regulates the function and generation of mRNAs. High expression of IGF2BP1 is associated with poor prognosis in endometrial cancer patients, and mechanistically, IGF2BP1 recruits polyadenylate-binding protein 1 (PABPC1) to stabilize paternally expressed gene 10 (PEG10) mRNA in an m6A-dependent manner [140]. In endometrial cancer, IGF2BP1 is a direct downstream target of peptidylarginine deiminase II (PADI2) that is required for endometrial cancer progression, and IGF2BP1 binds to the m6A sites of oncogenic SOX2 and prevents its mRNA degradation [141]. In hepatocellular carcinoma (HCC), ALKBH5, an m6A demethylase, inhibits LY6/PLAUR Domain Containing 1 (LYPD1) that is recognized and then stabilized by IGF2BP1 [142]. In HCC, RNA-binding motif protein 15 (RBM15) facilitates cancer progression via promoting post-transcriptional activation of YES1 in IGF2BP1-mediated m6A manner [143]. In addition, ALKBH5 also blocks pancreatic cancer progression through activation of PER1 by another m6A reader, YTHDF2 [82]. In liver cancer stem cells (LCSC), IGF2BP1 facilitates LCSC phenotypes via promoting the stability of MGAT5 mRNA in an m6A modification manner [144]. In another study, IGF2BP1 is demonstrated as a post-transcriptional enhancer of serum response factor (SRF) in cancer with a 3' UTR and m6A-dependent manner [145]. A later study also shows that IGF2BP1 promotes tumorigenesis and metastasis of oral squamous cell carcinoma via enhancing Bmi1 mRNA translation in METTL3-mediated m6A modification manner [146]. A subsequent study shows that METTL3 promotes the mRNA stability of kinesin-like protein, KIP3C in IGF2BP1- modified m6A manner, accelerating prostate cancer progression [147]. Moreover, METTL3 methylates KRT7-AS to enhance the mRNA stability of keratin 7 (KRT7) depending on IGF2BP1-modified m6A, promoting breast cancer lung metastasis [148]. In addition to m6A methyltransferases, m6A demethylase FTO reduce the stability of DACT1 mRNA by IGF2BP1-modified m6A demethylation, facilitating osteosarcoma progression [149]. Recently, IGF2BP1 is reported to promote E2F1-3-driven G1/S transition in an m6A-dependent manner in cancer cells [87]. Thus, IGF2BP1, as an m6A-reader, plays as an important oncogene in cancer by stabilizing or enhancing mRNA of its oncogenic factors, and thus IFG2BP1 is druggable for cancer treatment. Interestingly, m6A readers (IGF2BPs) cooperatively interact with m6A writers or erasers to regulate cancer progression.
IGF2BP2
In similar with IGF2BP1, noncoding RNAs also interact with IGF2BP2 to stabilize or increase their targets mRNA, and IGF2BP2 can directly regulate noncoding RNAs in an m6A-dependent way. In colorectal cancer patients, high expressed LINC00460 is correlated with poor overall survival, and mechanistically, LINC00460 interacts with IGF2BP2 to bind to the 3'UTR of high-mobility group AT-hook 1 (HMGA1), and enhances the stability of HMGA1 mRNA [150]. In colorectal liver metastasis model, circNSUN2 binds to the KH3-4 domains of IGF2BP2 and stabilizes the high mobility group AT-hook 2 (HMGA2) [151]. In esophageal squamous cell carcinoma (ESCC), LncRNA CCAT2 increases IGF2BP2 expression, and then IGF2BP2 improves mRNA stability of thymidine kinase 1 (TK1) in m6A modification manner, which promotes tumor progression [152]. In prostate cancer, lncRNA PCAT6 interacts with IGF2BP2 to promote bone metastasis by stabilizing insulin-like growth factor1 receptor (IGF1R) mRNA [153]. In glioblastoma, IGF2BP2 recognizes the m6A site of lncRNA CASC9 and increases its stability, and CASC9 cooperates with IGF2BP2 to form a complex that stabilizes hexokinase 2 (HK2), promoting aerobic glycolysis [154]. In pancreatic cancer patients, IGF2BP2 is associated with poorer prognosis, and mechanistically, IGF2BP2 plays as m6A reader for modification of lncRNA DANCR and stabilizes its mRNA [155]. In thyroid cancer, lncRNA HAGLR increases IGF2BP2 expression, and IGF2BP2 recognizes the m6A modification of c-MYC and leads to increased c-MYC expression, which promotes cancer progression [156]. In addition, lncRNA LINRIS stabilizes IGF2BP2 by blocking its ubiquitination to promote the c-MYC-driven glycolysis in colorectal cancer [157]. In colorectal cancer, IGF2BP2 directly binds to the m6A sites of lncRNA ZFAS1 and increases its stability to activate the Warburg effect [158]. Moreover, LINC01021 promotes tumorigenesis and progression through enhancing the mRNA stability of target transcripts, MSX1 and JARID2 in IGF2BP2- mediated m6A modification [159]. Besides lncRNAs, circCD44 also directly interacts with IGF2BP2 to enhance the stability of c-MYC mRNA in m6A-modifed manner, promoting cancer progression in triple-negative breast cancer [160]. In addition, in cervical cancer, circARHGAP12 interacts with IGF2BP2 through m6A site in the exon-3 to increase the stability of forkhead box M1 (FOXM1) mRNA [161]. In turn, IGF2BP2 also directly stabilizes noncoding RNAs to promote cancer progression. For example, IGF2BP2 binds to lncRNA DUXAP9 and increases its stability in m6A manner, which facilitates proliferation and motility of renal cancer cells [162].
In addition to noncoding RNAs, the mechanism of IGF2BP2 activity also relies on the direct interaction with its protein partners. For example, higher expression of IGF2BP2 is associated with a poorer prognosis in HCC patients, and mechanistically, IGF2BP2 directly recognizes and binds to the m6A site of flap endonuclease-1 (FEN1), and maintains its mRNA expression [92]. Analogously, in colorectal cancer, IGF2BP2 recognizes and binds to m6A-modified YAP and enhances the stability of YAP mRNA, thereby facilitating tumorigenesis [163]. Similarly, IGF2BP2 recognizes the coding sequence (CDS) regions of transcription factor SOX2 and protects it from degradation in m6A- mediated manner, which facilitates tumorigenesis and metastasis in colorectal cancer [164]. In similar with c-MYC, oncogene SOX2 is highly susceptible to m6A modification [165, 166]. In thyroid cancer, m6A demethylase FTO inhibits cell growth and glycolysis by reducing the mRNA stability of target, APOE in IGF2BP2-mediated m6A-dependent manner [167]. IGF2BP2 also promotes lymphatic metastasis and epithelial-mesenchymal transition (EMT) of head and neck squamous carcinoma cells by stabilizing slug mRNA in an m6A-dependent manner [168]. In addition, human papillomavirus E6/E7 promotes aerobic glycolysis of cervical cancer by stabilizing MYC expression in IGF2BP2-mediated m6A-dependent way [169]. In breast cancer stem-like cells (BCSC), aurora kinase A (AURKA) binds to IGF2BP2 and strengthens IGF2BP2 to stabilize DROSHA mRNA in an m6A-modified way, thereby increasing BCSC stemness maintenance [170].
In addition to cancer, IGF2BP2-mediated m6A modification also plays crucial roles in other physiological and pathological contexts. Most recently, IGF2BP2 maintains mitochondrial homeostasis of hematopoietic stem cells (HSCs) through maintaining the mRNA stability of its downstream target Bmi1, indicating that IGF2BP2-mediated m6A modification is critical for HSCs maintenance and hematopoiesis [171]. In the immune process, IGF2BP2 regulates macrophage phenotypic activation by stabilizing TSC1 and PPARγ mRNA in an m6A-dependent manner [172]. Moreover, IGF2P2 also binds to CCAAT/enhancer binding proteins (C/EBPs) to enhance the mRNA half-life and expression of C/EBPs in an m6A-modified manner in autoimmune inflammation [173]. In addition, IGF2BP2-modified m6A also plays important roles in regulating cardiac hypertrophy and aging-associated disorders [174, 175].
IGF2BP3
A major function of IGF2BP3 is interaction with the mRNA machinery, and plays as a stabilizer of oncogene in cancer [3]. In colon cancer, high expression of IGF2BP3 is associated with poorer overall survival, and IGF2BP3 recognizes and binds to the CDS region of Cyclin D1 to regulate cycle, and IGF2BP3 also regulates angiogenesis through m6A modification of vascular endothelial growth factor (VEGF) [176]. In gastric cancer, IGF2BP3 directly binds to hypoxia inducible factor-1a (HIF1A) at a specific m6A site in the CDS region, and knockout of IGF2BP3 inhibits cell migration and angiogenesis induced by hypoxia [177]. In addition, IGF2BP3 can bind to the m6A-modified region of the ATP-binding cassette transporters subfamily B member 1 (ABCB1) and promotes its mRNA stabilization, thereby triggering chemoresistance of colorectal cancer cells [178]. m6A demethylase ALKBH5 inhibits metastasis of gastric cancer through modulating expression of downstream target, PKMYT, and IGF2BP3 stabilize the mRNA stability of PKMYT1 by recognizing its m6A modification sites [179]. m6A methyltransferase METTL3 post-transcriptionally mediates PD-L1 mRNA activation in breast cancer with m6A-IGF2BP3-dependent manner [180]. Thus, m6A readers play important roles in m6A writers or erasers-mediated the stability of targets mRNA. Moreover, MYC-activated IGF2BP3 increase m6A-modified level of KPNA2, thereby promoting cell proliferation and metastasis nasopharyngeal carcinoma [181]. Alternatively, IGF2BPs also can stabilize mRNAs in an m6A-independent manner. For example, IGF2BP3 specifically binds to pregenomic RNA (pgRNA) and increases its stability without m6A modification, and promotes the stemness and tumorigenicity of HCC cells [182]. Thus, in addition to m6A modification, IGF2BPs can stabilize their targets mRNA through other mechanisms, and in general, preferentially through the m6A-modified manner.
In line with IGF2BP1 and IGF2BP2, noncoding RNAs play as guide or scaffold to recruit or interact with IGF2BP3 to regulate the function of their targets. For instance, lncRNA DMDRMR binds and cooperates with IGF2BP3 to stabilize multiple targets, including CDK4, in an m6A-dependent manner, and thereby drives cancer progression of clear cell renal cell carcinoma [111]. In addition, circular RNA, circ-TNPO3 serves as a protein decoy to competitively interact with IGF2BP3, and the stabilization role of IGF2BP3 on c-MYC mRNA is weakened, leading to inhibition of metastasis in gastric cancer [183].
The m6A modification is established by m6A methyltransferases (also known as m6A writer) [56]. METTL3 and METTL14 are core subunits of the methyltransferase complex that efficiently catalyses m6A modification [184]. Accumulating evidences in recent years demonstrate that METTL3, in most cases, plays as an oncogene in cancer [185]. Importantly, in some instances, the function of METTL3 in cancer depends on m6A readers, such as YTHDF2 [186]. In gastric cancer, higher expression of METTL3 is associated with poor prognosis, and mechanistically, METTL3 mediates the m6A modification of HDGF mRNA in a manner with IGF2BP3-dependent HDGF mRNA stability [187]. In addition to METTL3, another m6A writer, RBM15 regulates the m6A modification of its downstream target, TMBIM6, and enhances TMBIM6 mRNA stability through IGF2BP3-dependent way, and thereby facilitating progression of laryngeal squamous cell carcinoma [188]. Interestingly, cross-talk among m6A writers, erasers, readers maintains the m6A level that regulates tumor growth and progression. For example, METTL14 and ALKBH5 (eraser) determine the m6A level of targets via controlling each other expression and by inhibiting YTHDF3 (reader) [189]. In some cases, the m6A modification requires different readers to regulate their targets mRNA. For example, m6A-modified pyruvate dehydrogenase kinase 4 (PDK4) participates in glycolysis of cancer cells, and specifically, m6A-modified PDK4 regulates translation and mRNA stability via binding to YTHDF1 and IGF2BP3, respectively [121].
IGF2BPs
In 2018, IGF2BPs family was first identified as new m6A reader that has unique KH domains different from classical YTH domains [11]. IGF2BPs promote the stability and storage of c-MYC, and the oncogenic role of IGF2BPs depends on their function as m6A readers [11]. In renal cell cancer, transcription factor early growth response 2 (EGR2) increases the expressions of IGF2BPs, and IGF2BPs enhance the stability of sphingosine-1-phosphate receptor 3 (S1PR3) mRNA in m6A-dependent manner, and S1PR3 drives tumorigenesis and metastasis [190]. In acute myeloid leukemia (AML), RNA-binding protein YBX1 is required for survival of AML, and mechanistically, YBX1 can cooperate with IGF2BP1 and IGF2BP3 to increase the stability of c-MYC and BCL2 mRNA in an m6A dependent manner [191]. In hepatocellular carcinoma (HCC), the cancer-testis lncRNA-CTHCC promotes HCC growth and metastasis, and mechanistically, lncRNA-CTHCC is modified by m6A methylation with METTL3 and IGF2BP1/IGF2BP3 to maintain its stability and increase its expression [192]. In colorectal cancer, METTL3 promotes glycolysis metabolism to drive tumorigenesis, mechanistically, METTL3 mediates m6A modification to enhance the expressions of HK2 and SLC2A1 through IGF2BP2/3-dependent mRNA stability function [193]. In lung cancer, IGF2BPs, in particular, the IGF2BP2/3 increase the mRNA stability of VANGL1, and VANGL1 is associated with radio-resistance [194]. Since IGF2BPs are oncofetal, interestingly, degradation of IGF2BPs can be used for cancer treatment. For example, tumor suppressor gene, circNDUFB2 interacts with the KH domains of IGF2BP1/2/3 in an m6A-dependent manner, and facilitates ubiquitination and degradation of IGF2BPs, thus leading to inhibition of tumor growth of lung cancer [195]. The detail functions of IGF2BPs- modified m6A in cancer is shown in Table 1 and Figure 2, Figure 3.
Conclusion and perspectives
IGF2BPs-mediated m6A modification that controls mRNA fate is emerging as a rising star in cancer through more mechanistic analyses since 2018. However, the oncogenic role of IGF2BPs in stabilizing theirs targets, such as c-MYC, has garnered interest in developing small-molecule inhibitors targeting IGF2BPs before 2018. As a result, a novel IGF2BP1 inhibitor, BTYNB, was identified to inhibit IGF2BP1 and destabilize c-MYC, providing a therapeutic option for cancer treatment [199]. Besides IGF2BPs, several small-molecule inhibitors targeting other m6A modification proteins (writers, erasers) are discovered using high-throughput screening [200]. Nevertheless, it is possible that inhibition of IGF2BPs may lead to feedback activation of other readers (such as YTHDF1), inevitably developing drug resistance.
IGF2BPs participate in posttranscriptional RNA processing, such as RNA splicing, translation, stability and decay, and in most cases, IGF2BPs stabilize their targets in m6A-dependent manner. Nevertheless, many questions remain to be resolved. (i) If IGF2BPs stabilize their downstream targets in an m6A-independent way, the detail mechanisms are what? (ii) Whether IGF2BPs increase the stability of targets mRNA through m6A modification before IGF2BPs are identified as new m6A readers? (iii) Do IGF2BPs and other m6A readers compete for the same targets mRNA, such as c-MYC? (ⅳ) How m6A writers (such as METTL3) and readers (IGF2BPs) cooperate through the “writers-readers system”? Thus, future structural studies are strongly warranted to understand how IGF2BPs binds to their targets, such as c-MYC, and investigate other KH domains-containing proteins as potential m6A readers.
IGF2BPs-modified m6A in human cancers.
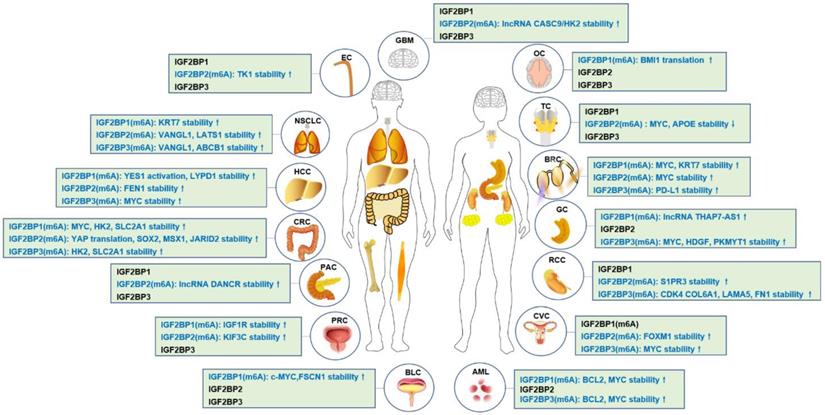
The functions of IGF2BPs as m6A readers in cancer
| Upstream target | m6A reader | Function | Cancer type | Reference |
|---|---|---|---|---|
| IGF2BPs | Enhances c-MYC mRNA stability and translation | Pan-cancer | [11] | |
| IGF2BPs | Stabilizes VANGL1 | Lung cancer | [194] | |
| IGF2BPs | Stabilizes HK2 and SLC2A1 | Colorectal cancer | [193] | |
| circNDUFB2 | IGF2BPs | Enhances IGF2BPs degradation | Lung cancer | [195] |
| EGR2 | IGF2BPs | Stabilizes S1PR3 mRNA | Renal cell carcinoma | [190] |
| YBX1 | IGF2BPs | Stabilizes BCL2, c-MYC mRNA | Myeloid leukemia | [191] |
| IGF2BPs | Stabilizes lncRNA-CTHCC | Hepatocellular carcinoma | [192] | |
| lncRNA KB-1980E6.3 | IGF2BP1 | Stabilizes c-MYC | Breast cancer | [137] |
| LINC00266-1 | IGF2BP1 | Stabilizes c-MYC | Colorectal cancer | [139] |
| CircPTPRA | IGF2BP1 | Stabilizes c-MYC and FSCN1 | Bladder cancer | [105] |
| IGF2BP1 | Stabilizes PEG10 | Endometrial cancer | [140] | |
| IGF2BP1 | Promotes SRF expression | Pan-cancer | [145] | |
| IGF2BP1 | Stabilizes MGAT5 | Liver cancer | [144] | |
| IGF2BP1 | Stabilizes AURKA, HDLBP and YWHAZ | Pan-cancer | [196] | |
| PADI2 | IGF2BP1 | Stabilizes SOX2 | Endometrial cancer | [141] |
| IGF2BP1 | Controls E2F1 turnover | Pan-cancer | [87] | |
| RBM15 | IGF2BP1 | Promotes post-transcriptional activation of YES1 | HCC | [143] |
| METTL3 | IGF2BP1 | Stabilizes KIF3C | Prostate cancer | [147] |
| METTL3 | IGF2BP1 | Stabilizes lncRNA THAP7-AS1 | Gastric cancer | [135] |
| ALKBH5 | IGF2BP1 | Stabilizes LYPD1 | HCC | [142] |
| METTL3 | IGF2BP1 | Promotes BMI1 translation | Squamous Cell Carcinoma | [146] |
| FTO | IGF2BP1 | Reduce mRNA stability of DACT1 | Osteosarcoma | [149] |
| IGF2BP2 | Reduces LncRNA HAGLR | Thyroid cancer | [197] | |
| IGF2BP2 | Stabilizes FEN1 mRNA | HCC | [92] | |
| IGF2BP2 | Promote YAP translation, and activates ErbB2 | Colorectal cancer | [163] | |
| IGF2BP2 | Stabilizes lncRNA DANCR | Pancreatic cancer | [155] | |
| CircARHGAP12 | IGF2BP2 | Stabilizes FOXM1 mRNA | Cervical cancer | [161] |
| HCG11 | IGF2BP2 | Stabilizes LATS1 mRNA | Lung cancer | [198] |
| lncRNA CCAT2 | IGF2BP2 | Stabilizes TK1 mRNA | Esophageal squamous cell carcinoma | [152] |
| LINC01021 | IGF2BP2 | Enhances mRNA stability of MSX1 and JARID2 | Colorectal cancer | [159] |
| IGF2BP2 | Stabilizes lncRNA CASC9/HK2 mRNA | Glioblastoma | [154] | |
| IGF2BP2 | Stabilizes the ZFAS1/OLA1 axis | Colorectal cancer | [158] | |
| METTL3 | IGF2BP2 | Prevents SOX2 mRNA degradation | Colorectal cancer | [164] |
| IGF2BP2 | Promotes Slug mRNA stability | Head and neck squamous carcinoma | [168] | |
| FTO | IGF2BP2 | Reduces APOE mRNA stability | Thyroid cancer | [167] |
| HPV E6/E7 | IGF2BP2 | Stabilize MYC expression | Cervical cancer | [169] |
| miR-204 | IGF2BP2 | Enhances c-MYC expression | Thyroid Cancer | [156] |
| LINC00460 | IGF2BP2 | Stabilizes HMGA1 mRNA | Colorectal cancer | [150] |
| IGF2BP3 | Stabilizes ABCB1 mRNA | Chemoresistance | [178] | |
| lncRNA (DMDRMR) | IGF2BP3 | Stabilizes CDK4 COL6A1, LAMA5, FN1 | Renal cell carcinoma | [111] |
| IGF2BP3 | Stabilizes HIF1A | Gastric cancer | [177] | |
| ALKBH5 | IGF2BP3 | Stabilize mRNA stability of PKMYT1 | Gastric cancer | [179] |
| METTL3 | IGF2BP3 | Promotes PD-L1 mRNA activation | Breast cancer | [180] |
| MYC | IGF2BP3 | Increases mRNA stability of KPNA2 | Nasopharyngeal carcinoma | [181] |
| RBM15 | IGF2BP3 | Stabilizes TMBIM6 | Laryngeal squamous cell carcinoma | [188] |
| IGF2BP3 | Reduces Cyclin D1 mRNA stability | Colon cancer | [176] | |
| IGF2BP3 | Stabilizes HDGF | Gastric cancer | [187] | |
| IGF2BP3 | Promotes translation and stability of PDK4 | Pan-cancer | [121] |
Overview of IGF2BPs-modified m6A on their downstream targets.
Abbreviations
IGF2BPs: insulin-like growth factor-2 mRNA-binding proteins; RBPs: RNA-binding proteins; m6A: N6-methyladenosine; KH: K-homologous; RRMs: RNA recognition motifs; SAM: s-adenosylmethionine; METTL3: methyltransferase-like protein 3; MAC: m6A-METTL complex; MACOM: m6A-METTL associated complex; WTAP: wilms tumor 1 associated protein; VIRMA: vir-like m6A methyltransferase-associated; RBM15: RNA-binding motif 15; ZC3H13: zinc-finger CCCH-type-containing 13; CBLL1: Cbl proto oncogene-like protein 1; FTO: fat mass and obesity-associated protein; ALKBH5: α-ketoglutarate-dependent dioxygenase alk B homolog 5; m6Am: N6,2′-O-dimethyladenosine; YTHDFs: YT521-B homology domain family proteins; YTHDC1: YT521-B homology domain containing 1; eIF3: eukaryotic translation initiation factor 3; HNRNPs: heterogeneous nuclear ribonucleoproteins; Prrc2a: Proline rich coiled-coil 2 A; CNBP: cellular nucleic acid binding protein; SRSF3: serine and arginine-rich splicing factor 3; BCSC: stemness of breast cancer stem cells; PABPC1: polyadenylate-binding protein 1; PADI2: peptidylarginine deiminase II; LYPD1: LY6/PLAUR Domain Containing 1; HMGA1: high-mobility group AT-hook 1; TK1: thymidine kinase 1; IGF1R: insulin-like growth factor1 receptor; HK2: hexokinase 2; FOXM1: forkhead box M1; FEN1: flap endonuclease-1; AURKA: aurora kinase A; C/EBPs: CCAAT/enhancer binding proteins; VEGF: vascular endothelial growth factor; HIF1A: hypoxia inducible factor-1a; ABCB1: ATP-binding cassette transporters subfamily B member 1; PDK4: pyruvate dehydrogenase kinase 4; EGR2: early growth response 2.
Acknowledgements
Funding
This study is supported by the grants from the Major Science and Technology Project of Anhui Province (No: 202003c08020004, 202103b06020004).
Author Contributions
SCY and CD drafted the manuscript, SCY and DBB revised the manuscript, CCW and LD approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Molecular and cellular biology. 1999;19(2):1262-1270
2. Bernstein PL, Herrick Dj Fau - Prokipcak RD, Prokipcak Rd Fau - Ross J, Ross J. Control of c-myc mRNA half-life in vitro by a protein capable of binding to a coding region stability determinant. (0890-9369 (Print)).
3. Lederer M, Bley N, Schleifer C, Huttelmaier S. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin Cancer Biol. 2014;29:3-12
4. Zhang JY, Chan Ek Fau - Peng XX, Peng Xx Fau - Tan EM, Tan EM. A novel cytoplasmic protein with RNA-binding motifs is an autoantigen in human hepatocellular carcinoma. (0022-1007 (Print)).
5. Müeller-Pillasch F, Lacher U Fau - Wallrapp C, Wallrapp C Fau - Micha A, Micha A Fau - Zimmerhackl F, Zimmerhackl F Fau - Hameister H, Hameister H Fau - Varga G, Varga G Fau - Friess H, Friess H Fau - Büchler M, Büchler M Fau - Beger HG, Beger Hg Fau - Vila MR. et al. Cloning of a gene highly overexpressed in cancer coding for a novel KH-domain containing protein. (0950-9232 (Print)).
6. Degrauwe N, Suvà ML, Janiszewska M, Riggi N, Stamenkovic I. IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. (1549-5477 (Electronic)).
7. Bell JL, Wachter K, Muhleck B, Pazaitis N, Kohn M, Lederer M, Huttelmaier S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70(15):2657-2675
8. Dai N. The Diverse Functions of IMP2/IGF2BP2 in Metabolism. Trends Endocrinol Metab. 2020;31(9):670-679
9. Yaniv K, Yisraeli JK. The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. (0378-1119 (Print)).
10. Kiledjian M, Wang X Fau - Liebhaber SA, Liebhaber SA. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. (0261-4189 (Print)).
11. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL. et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285-295
12. Dai N, Zhao L, Wrighting D, Kramer D, Majithia A, Wang Y, Cracan V, Borges-Rivera D, Mootha VK, Nahrendorf M. et al. IGF2BP2/IMP2-Deficient mice resist obesity through enhanced translation of Ucp1 mRNA and Other mRNAs encoding mitochondrial proteins. Cell Metab. 2015;21(4):609-621
13. Wiener D, Schwartz S. The epitranscriptome beyond m(6)A. Nat Rev Genet. 2021;22(2):119-131
14. Dierks D, Garcia-Campos MA, Uzonyi A, Safra M, Edelheit S, Rossi A, Sideri T, Varier RA, Brandis A, Stelzer Y. et al. Multiplexed profiling facilitates robust m6A quantification at site, gene and sample resolution. Nat Methods. 2021;18(9):1060-1067
15. Liu J, Gao M, He J, Wu K, Lin S, Jin L, Chen Y, Liu H, Shi J, Wang X. et al. The RNA m(6)A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature. 2021;591(7849):322-326
16. Meyer KD, Jaffrey SR. Rethinking m(6)A Readers, Writers, and Erasers. Annu Rev Cell Dev Biol. 2017;33:319-342
17. Liao S, Sun H, Xu C. YTH Domain: A Family of N(6)-methyladenosine (m(6)A) Readers. Genomics Proteomics Bioinformatics. 2018;16(2):99-107
18. Chang G, Shi L, Ye Y, Shi H, Zeng L, Tiwary S, Huse JT, Huo L, Ma L, Ma Y. et al. YTHDF3 Induces the Translation of m(6)A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell. 2020;38(6):857-871 e857
19. Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162(6):1299-1308
20. Deng J, Zhang J, Ye Y, Liu K, Zeng L, Huang J, Pan L, Li M, Bai R, Zhuang L. et al. N(6) -methyladenosine-Mediated Upregulation of WTAPP1 Promotes WTAP Translation and Wnt Signaling to Facilitate Pancreatic Cancer Progression. Cancer Res. 2021;81(20):5268-5283
21. Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, Yang C, Chen Y. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6(1):74
22. Gao Y, Pei G, Li D, Li R, Shao Y, Zhang QC, Li P. Multivalent m(6)A motifs promote phase separation of YTHDF proteins. Cell Res. 2019;29(9):767-769
23. Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, Patil DP, Kwak H, Lee JH, Jaffrey SR. m(6)A enhances the phase separation potential of mRNA. Nature. 2019;571(7765):424-428
24. Cheng Y, Xie W, Pickering BF, Chu KL, Savino AM, Yang X, Luo H, Nguyen DT, Mo S, Barin E. et al. N(6)-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell. 2021;39(7):958-972 e958
25. Han D, Longhini AP, Zhang X, Hoang V, Wilson MZ, Kosik KS. Dynamic assembly of the mRNA m6A methyltransferase complex is regulated by METTL3 phase separation. PLoS Biol. 2022;20(2):e3001535
26. Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18(1):103
27. Perry RP, Kelley DE, Friderici K, Rottman F. The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5' terminus. Cell. 1975;4(4):387-394
28. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71(10):3971-3975
29. Adams JM, Cory S. Modified nucleosides and bizarre 5'-termini in mouse myeloma mRNA. Nature. 1975;255(5503):28-33
30. Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18(1):31-42
31. Wang Y, Zhao JC. Update: Mechanisms Underlying N(6)-Methyladenosine Modification of Eukaryotic mRNA. Trends Genet. 2016;32(12):763-773
32. Wang X, He C. Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol. 2014;11(6):669-672
33. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet. 2014;15(5):293-306
34. Lin S, Liu Q, Lelyveld VS, Choe J, Szostak JW, Gregory RI. Mettl1/Wdr4-Mediated m(7)G tRNA Methylome Is Required for Normal mRNA Translation and Embryonic Stem Cell Self-Renewal and Differentiation. Mol Cell. 2018;71(2):244-255 e245
35. Panneerdoss S, Eedunuri VK, Yadav P, Timilsina S, Rajamanickam S, Viswanadhapalli S, Abdelfattah N, Onyeagucha BC, Cui X, Lai Z. et al. Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Science advances. 2018;4(10):eaar8263
36. Lin Z, Niu Y, Wan A, Chen D, Liang H, Chen X, Sun L, Zhan S, Chen L, Cheng C. et al. RNA m(6) A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020;39(12):e103181
37. Fukumoto T, Zhu H, Nacarelli T, Karakashev S, Fatkhutdinov N, Wu S, Liu P, Kossenkov AV, Showe LC, Jean S. et al. N(6)-Methylation of Adenosine of FZD10 mRNA Contributes to PARP Inhibitor Resistance. Cancer Res. 2019;79(11):2812-2820
38. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C. et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534(7608):575-578
39. Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. 2016;63(2):306-317
40. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149(7):1635-1646
41. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201-206
42. Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12(8):767-772
43. Scholler E, Weichmann F, Treiber T, Ringle S, Treiber N, Flatley A, Feederle R, Bruckmann A, Meister G. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA. 2018;24(4):499-512
44. Sledz P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife. 2016 5
45. Lence T, Paolantoni C, Worpenberg L, Roignant JY. Mechanistic insights into m(6)A RNA enzymes. Biochim Biophys Acta Gene Regul Mech. 2019;1862(3):222-229
46. Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, Conrad NK. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell. 2017;169(5):824-835 e814
47. Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Hobartner C, Sloan KE, Bohnsack MT. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18(11):2004-2014
48. Akichika S, Hirano S, Shichino Y, Suzuki T, Nishimasu H, Ishitani R, Sugita A, Hirose Y, Iwasaki S, Nureki O. et al. Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science. 2019 363(6423)
49. Sendinc E, Valle-Garcia D, Dhall A, Chen H, Henriques T, Navarrete-Perea J, Sheng W, Gygi SP, Adelman K, Shi Y. PCIF1 Catalyzes m6Am mRNA Methylation to Regulate Gene Expression. Mol Cell. 2019;75(3):620-630 e629
50. Boulias K, Toczydlowska-Socha D, Hawley BR, Liberman N, Takashima K, Zaccara S, Guez T, Vasseur JJ, Debart F, Aravind L. et al. Identification of the m(6)Am Methyltransferase PCIF1 Reveals the Location and Functions of m(6)Am in the Transcriptome. Mol Cell. 2019;75(3):631-643 e638
51. van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, Bohnsack KE, Bohnsack MT, Jaffrey SR, Graille M. et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47(15):7719-7733
52. Ren W, Lu J, Huang M, Gao L, Li D, Wang GG, Song J. Structure and regulation of ZCCHC4 in m(6)A-methylation of 28S rRNA. Nat Commun. 2019;10(1):5042
53. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885-887
54. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18-29
55. Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q. et al. Reversible methylation of m(6)Am in the 5' cap controls mRNA stability. Nature. 2017;541(7637):371-375
56. Zhao Y, Shi Y, Shen H, Xie W. m(6)A-binding proteins: the emerging crucial performers in epigenetics. J Hematol Oncol. 2020;13(1):35
57. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117-120
58. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5' UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015;163(4):999-1010
59. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518(7540):560-564
60. Wu R, Li A, Sun B, Sun JG, Zhang J, Zhang T, Chen Y, Xiao Y, Gao Y, Zhang Q. et al. A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29(1):23-41
61. Lee E, Lee TA, Kim JH, Park A, Ra EA, Kang S, Choi HJ, Choi JL, Huh HD, Lee JE. et al. CNBP acts as a key transcriptional regulator of sustained expression of interleukin-6. Nucleic Acids Res. 2017;45(6):3280-3296
62. Baquero-Perez B, Antanaviciute A, Yonchev ID, Carr IM, Wilson SA, Whitehouse A. The Tudor SND1 protein is an m(6)A RNA reader essential for replication of Kaposi's sarcoma-associated herpesvirus. Elife. 2019 8
63. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626
64. Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, Soller M. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540(7632):301-304
65. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27(3):315-328
66. Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, Sun HY, Zhu Q, Baidya P, Wang X. et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27(3):444-447
67. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY. et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61(4):507-519
68. Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, Qi M, Lu Z, Shi H, Wang J. et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27(9):1115-1127
69. Mao Y, Dong L, Liu XM, Guo J, Ma H, Shen B, Qian SB. m(6)A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat Commun. 2019;10(1):5332
70. Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR, Ma J. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun. 2018;9(1):420
71. Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482-485
72. McCloskey A, Taniguchi I, Shinmyozu K, Ohno M. hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science. 2012;335(6076):1643-1646
73. Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45(10):6051-6063
74. Zhou KI, Pan T. An additional class of m(6)A readers. Nat Cell Biol. 2018;20(3):230-232
75. Banerji J, Sands J, Strominger JL, Spies T. A gene pair from the human major histocompatibility complex encodes large proline-rich proteins with multiple repeated motifs and a single ubiquitin-like domain. Proc Natl Acad Sci U S A. 1990;87(6):2374-2378
76. Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, Zhao G. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18(1):142
77. Cao L, Zhang P, Li J, Wu M. LAST, a c-Myc-inducible long noncoding RNA, cooperates with CNBP to promote CCND1 mRNA stability in human cells. Elife. 2017 6
78. Chien CS, Li JY, Chien Y, Wang ML, Yarmishyn AA, Tsai PH, Juan CC, Nguyen P, Cheng HM, Huo TI. et al. METTL3-dependent N(6)-methyladenosine RNA modification mediates the atherogenic inflammatory cascades in vascular endothelium. Proc Natl Acad Sci U S A. 2021 118(7)
79. Du J, Liao W, Liu W, Deb DK, He L, Hsu PJ, Nguyen T, Zhang L, Bissonnette M, He C. et al. N(6)-Adenosine Methylation of Socs1 mRNA Is Required to Sustain the Negative Feedback Control of Macrophage Activation. Dev Cell. 2020;55(6):737-753 e737
80. Yin H, Zhang X, Yang P, Zhang X, Peng Y, Li D, Yu Y, Wu Y, Wang Y, Zhang J. et al. RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat Commun. 2021;12(1):1394
81. Yang Z, Yang S, Cui YH, Wei J, Shah P, Park G, Cui X, He C, He YY. METTL14 facilitates global genome repair and suppresses skin tumorigenesis. Proc Natl Acad Sci U S A. 2021 118(35)
82. Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, Feng Y, Pan Q, Wan R. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Molecular Cancer. 2020 19(1)
83. Song H, Feng X, Zhang H, Luo Y, Huang J, Lin M, Jin J, Ding X, Wu S, Huang H. et al. METTL3 and ALKBH5 oppositely regulate m(6)A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. 2019;15(8):1419-1437
84. Huang X, Zhang H, Guo X, Zhu Z, Cai H, Kong X. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in cancer. J Hematol Oncol. 2018;11(1):88
85. Mancarella C, Scotlandi K. IGF2BP3 From Physiology to Cancer: Novel Discoveries, Unsolved Issues, and Future Perspectives. Front Cell Dev Biol. 2019;7:363
86. Muller S, Bley N, Glass M, Busch B, Rousseau V, Misiak D, Fuchs T, Lederer M, Huttelmaier S. IGF2BP1 enhances an aggressive tumor cell phenotype by impairing miRNA-directed downregulation of oncogenic factors. Nucleic Acids Res. 2018;46(12):6285-6303
87. Muller S, Bley N, Busch B, Glass M, Lederer M, Misiak C, Fuchs T, Wedler A, Haase J, Bertoldo JB. et al. The oncofetal RNA-binding protein IGF2BP1 is a druggable, post-transcriptional super-enhancer of E2F-driven gene expression in cancer. Nucleic Acids Res. 2020;48(15):8576-8590
88. Regue L, Zhao L, Ji F, Wang H, Avruch J, Dai N. RNA m6A reader IMP2/IGF2BP2 promotes pancreatic beta-cell proliferation and insulin secretion by enhancing PDX1 expression. Mol Metab. 2021;48:101209
89. Kato T, Hayama S, Yamabuki T, Ishikawa N, Miyamoto M, Ito T, Tsuchiya E, Kondo S, Nakamura Y, Daigo Y. Increased expression of insulin-like growth factor-II messenger RNA-binding protein 1 is associated with tumor progression in patients with lung cancer. Clin Cancer Res. 2007;13(2 Pt 1):434-442
90. Guo W, Huai Q, Wan H, Guo L, Song P, Gao S, He J. Prognostic Impact of IGF2BP3 Expression in Patients with Surgically Resected Lung Adenocarcinoma. (1557-7430 (Electronic)).
91. Gutschner T, Hammerle M, Pazaitis N, Bley N, Fiskin E, Uckelmann H, Heim A, Grobeta M, Hofmann N, Geffers R. et al. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) is an important protumorigenic factor in hepatocellular carcinoma. Hepatology. 2014;59(5):1900-1911
92. Pu J, Wang J, Qin Z, Wang A, Zhang Y, Wu X, Wu Y, Li W, Xu Z, Lu Y. et al. IGF2BP2 Promotes Liver Cancer Growth Through an m6A-FEN1-Dependent Mechanism. Front Oncol. 2020;10:578816
93. Gao Y, Yang M, Jiang Z, Woda BA, Mercurio AM, Qin J, Huang X, Zhang F. IMP3 expression is associated with poor outcome and epigenetic deregulation in intrahepatic cholangiocarcinoma. Hum Pathol. 2014;45(6):1184-1191
94. Kim HY, Ha Thi HT, Hong S. IMP2 and IMP3 cooperate to promote the metastasis of triple-negative breast cancer through destabilization of progesterone receptor. Cancer Lett. 2018;415:30-39
95. Walter O, Prasad M, Lu S, Quinlan RM, Edmiston KL, Khan A. IMP3 is a novel biomarker for triple negative invasive mammary carcinoma associated with a more aggressive phenotype. Hum Pathol. 2009;40(11):1528-1533
96. Zhang XL, Li KJ, Feng JX, Liu GJ, Feng YL. Blocking the IGF2BP1-promoted glucose metabolism of colon cancer cells via direct de-stabilizing mRNA of the LDHA enhances anticancer effects. Mol Ther Nucleic Acids. 2021;23:835-846
97. Ye S, Song W, Xu X, Zhao X, Yang L. IGF2BP2 promotes colorectal cancer cell proliferation and survival through interfering with RAF-1 degradation by miR-195. FEBS Lett. 2016;590(11):1641-1650
98. Xu W, Sheng Y, Guo Y, Huang Z, Huang Y, Wen D, Liu CY, Cui L, Yang Y, Du P. Increased IGF2BP3 expression promotes the aggressive phenotypes of colorectal cancer cells in vitro and vivo. J Cell Physiol. 2019;234(10):18466-18479
99. Wan BS, Cheng M, Zhang L. Insulin-like growth factor 2 mRNA-binding protein 1 promotes cell proliferation via activation of AKT and is directly targeted by microRNA-494 in pancreatic cancer. World J Gastroenterol. 2019;25(40):6063-6076
100. Xu X, Yu Y, Zong K, Lv P, Gu Y. Up-regulation of IGF2BP2 by multiple mechanisms in pancreatic cancer promotes cancer proliferation by activating the PI3K/Akt signaling pathway. Journal of Experimental & Clinical Cancer Research. 2019 38(1)
101. Schaeffer DF, Owen DR, Lim HJ, Buczkowski AK, Chung SW, Scudamore CH, Huntsman DG, Ng SS, Owen DA. Insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) overexpression in pancreatic ductal adenocarcinoma correlates with poor survival. BMC Cancer. 2010;10:59
102. Zhang X, Wang D, Liu B, Jin X, Wang X, Pan J, Tu W, Shao Y. IMP3 accelerates the progression of prostate cancer through inhibiting PTEN expression in a SMURF1-dependent way. J Exp Clin Cancer Res. 2020;39(1):190
103. Wu Y, Liu Z, Wei X, Feng H, Hu B, Liu B, Luan Y, Ruan Y, Liu X, Liu Z. et al. Identification of the Functions and Prognostic Values of RNA Binding Proteins in Bladder Cancer. Front Genet. 2021;12:574196
104. Huang W, Li Y, Zhang C, Zha H, Zhou X, Fu B, Guo J, Wang G. IGF2BP3 facilitates cell proliferation and tumorigenesis via modulation of JAK/STAT signalling pathway in human bladder cancer. Journal of Cellular and Molecular Medicine. 2020;24(23):13949-13960
105. Xie F, Huang C, Liu F, Zhang H, Xiao X, Sun J, Zhang X, Jiang G. CircPTPRA blocks the recognition of RNA N(6)-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression. Mol Cancer. 2021;20(1):68
106. Haase J, Misiak D, Bauer M, Pazaitis N, Braun J, Potschke R, Mensch A, Bell JL, Dralle H, Siebolts U. et al. IGF2BP1 is the first positive marker for anaplastic thyroid carcinoma diagnosis. Mod Pathol. 2021;34(1):32-41
107. Wang X, Fu X, Zhang J, Xiong C, Zhang S, Lv Y. Identification and validation of m(6)A RNA methylation regulators with clinical prognostic value in Papillary thyroid cancer. Cancer Cell Int. 2020;20:203
108. Asioli S, Erickson LA, Righi A, Jin L, Volante M, Jenkins S, Papotti M, Bussolati G, Lloyd RV. Poorly differentiated carcinoma of the thyroid: validation of the Turin proposal and analysis of IMP3 expression. Mod Pathol. 2010;23(9):1269-1278
109. Wang X, Guan D, Wang D, Liu H, Wu Y, Gong W, Du M, Chu H, Qian J, Zhang Z. Genetic variants in m(6)A regulators are associated with gastric cancer risk. Arch Toxicol. 2021;95(3):1081-1088
110. Zhou Y, Huang T, Siu HL, Wong CC, Dong Y, Wu F, Zhang B, Wu WK, Cheng AS, Yu J. et al. IGF2BP3 functions as a potential oncogene and is a crucial target of miR-34a in gastric carcinogenesis. Mol Cancer. 2017;16(1):77
111. Gu Y, Niu S, Wang Y, Duan L, Pan Y, Tong Z, Zhang X, Yang Z, Peng B, Wang X. et al. DMDRMR-Mediated Regulation of m(6)A-Modified CDK4 by m(6)A Reader IGF2BP3 Drives ccRCC Progression. Cancer Res. 2021;81(4):923-934
112. Bley N, Schott A, Muller S, Misiak D, Lederer M, Fuchs T, Assmann C, Glass M, Ihling C, Sinz A. et al. IGF2BP1 is a targetable SRC/MAPK-dependent driver of invasive growth in ovarian cancer. RNA Biol. 2021;18(3):391-403
113. Hsu KF, Shen MR, Huang YF, Cheng YM, Lin SH, Chow NH, Cheng SW, Chou CY, Ho CL. Overexpression of the RNA-binding proteins Lin28B and IGF2BP3 (IMP3) is associated with chemoresistance and poor disease outcome in ovarian cancer. Br J Cancer. 2015;113(3):414-424
114. Wakita A, Motoyama S, Sato Y, Nagaki Y, Fujita H, Terata K, Imai K, Maeda E, Minamiya Y. IGF2BP3 Expression Correlates With Poor Prognosis in Esophageal Squamous Cell Carcinoma. J Surg Res. 2021;259:137-144
115. Barghash A, Golob-Schwarzl N, Helms V, Haybaeck J, Kessler SM. Elevated expression of the IGF2 mRNA binding protein 2 (IGF2BP2/IMP2) is linked to short survival and metastasis in esophageal adenocarcinoma. (1949-2553 (Electronic)).
116. He X, Li W, Liang X, Zhu X, Zhang L, Huang Y, Yu T, Li S, Chen Z. IGF2BP2 Overexpression Indicates Poor Survival in Patients with Acute Myelocytic Leukemia. Cell Physiol Biochem. 2018;51(4):1945-1956
117. Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell. 2016;30(6):836-848
118. Fabbiano F, Corsi J, Gurrieri E, Trevisan C, Notarangelo M, D'Agostino VG. RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins? J Extracell Vesicles. 2020;10(2):e12043
119. Pan Z, Zhao R, Li B, Qi Y, Qiu W, Guo Q, Zhang S, Zhao S, Xu H, Li M. et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol Cancer. 2022;21(1):16
120. Ghoshal A, Rodrigues LC, Gowda CP, Elcheva IA, Liu Z, Abraham T, Spiegelman VS. Extracellular vesicle-dependent effect of RNA-binding protein IGF2BP1 on melanoma metastasis. Oncogene. 2019;38(21):4182-4196
121. Li Z, Peng Y, Li J, Chen Z, Chen F, Tu J, Lin S, Wang H. N(6)-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat Commun. 2020;11(1):2578
122. Samanta S, Pursell B, Mercurio AM. IMP3 protein promotes chemoresistance in breast cancer cells by regulating breast cancer resistance protein (ABCG2) expression. J Biol Chem. 2013;288(18):12569-12573
123. Degrauwe N, Schlumpf TB, Janiszewska M, Martin P, Cauderay A, Provero P, Riggi N, Suva ML, Paro R, Stamenkovic I. The RNA Binding Protein IMP2 Preserves Glioblastoma Stem Cells by Preventing let-7 Target Gene Silencing. Cell Rep. 2016;15(8):1634-1647
124. Zhou KI, Pan T. An additional class of m(6)A readers. (1476-4679 (Electronic)).
125. Patil DP, Pickering BF, Jaffrey SR. Reading m(6)A in the Transcriptome: m(6)A-Binding Proteins. Trends Cell Biol. 2018;28(2):113-127
126. Korn SM, Ulshofer CJ, Schneider T, Schlundt A. Structures and target RNA preferences of the RNA-binding protein family of IGF2BPs: An overview. Structure. 2021;29(8):787-803
127. Wang G, Huang Z, Liu X, Huang W, Chen S, Zhou Y, Li D, Singer RH, Gu W. IMP1 suppresses breast tumor growth and metastasis through the regulation of its target mRNAs. (1949-2553 (Electronic)).
128. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y. et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27(5):626-641
129. Tang C, Xie Y, Yu T, Liu N, Wang Z, Woolsey RJ, Tang Y, Zhang X, Qin W, Zhang Y. et al. m(6)A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 2020;30(3):211-228
130. Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q. et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18(1):110
131. Qian X, Yang J, Qiu Q, Li X, Jiang C, Li J, Dong L, Ying K, Lu B, Chen E. et al. LCAT3, a novel m6A-regulated long non-coding RNA, plays an oncogenic role in lung cancer via binding with FUBP1 to activate c-MYC. J Hematol Oncol. 2021;14(1):112
132. Guo Y, Guo Y, Chen C, Fan D, Wu X, Zhao L, Shao B, Sun Z, Ji Z. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol Cancer. 2021;20(1):93
133. Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, Yuan W, Kan Q, Sun Z. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12(1):121
134. Hosono Y, Niknafs YS, Prensner JR, Iyer MK, Dhanasekaran SM, Mehra R, Pitchiaya S, Tien J, Escara-Wilke J, Poliakov A. et al. Oncogenic Role of THOR, a Conserved Cancer/Testis Long Non-coding RNA. Cell. 2017;171(7):1559-1572 e1520
135. Liu HT, Zou YX, Zhu WJ, Sen L, Zhang GH, Ma RR, Guo XY, Gao P. lncRNA THAP7-AS1, transcriptionally activated by SP1 and post-transcriptionally stabilized by METTL3-mediated m6A modification, exerts oncogenic properties by improving CUL4B entry into the nucleus. Cell Death Differ. 2021
136. Dhanasekaran R, Deutzmann A, Mahauad-Fernandez WD, Hansen AS, Gouw AM, Felsher DW. The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat Rev Clin Oncol. 2021
137. Zhu P, He F, Hou Y, Tu G, Li Q, Jin T, Zeng H, Qin Y, Wan X, Qiao Y. et al. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene. 2021;40(9):1609-1627
138. Gong C, Li Z, Ramanujan K, Clay I, Zhang Y, Lemire-Brachat S, Glass DJ. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev Cell. 2015;34(2):181-191
139. Zhu S, Wang JZ, Chen, He YT, Meng N, Chen M, Lu RX, Chen XH, Zhang XL, Yan GR. An oncopeptide regulates m(6)A recognition by the m(6)A reader IGF2BP1 and tumorigenesis. Nat Commun. 2020;11(1):1685
140. Zhang L, Wan Y, Zhang Z, Jiang Y, Gu Z, Ma X, Nie S, Yang J, Lang J, Cheng W. et al. IGF2BP1 overexpression stabilizes PEG10 mRNA in an m6A-dependent manner and promotes endometrial cancer progression. Theranostics. 2021;11(3):1100-1114
141. Xue T, Liu X, Zhang M, E Q, Liu S, Zou M, Li Y, Ma Z, Han Y, Thompson P. et al. PADI2-Catalyzed MEK1 Citrullination Activates ERK1/2 and Promotes IGF2BP1-Mediated SOX2 mRNA Stability in Endometrial Cancer. Adv Sci (Weinh). 2021;8(6):2002831
142. Chen Y, Zhao Y, Chen J, Peng C, Zhang Y, Tong R, Cheng Q, Yang B, Feng X, Lu Y. et al. ALKBH5 suppresses malignancy of hepatocellular carcinoma via m(6)A-guided epigenetic inhibition of LYPD1. Mol Cancer. 2020;19(1):123
143. Cai X, Chen Y, Man D, Yang B, Feng X, Zhang D, Chen J, Wu J. RBM15 promotes hepatocellular carcinoma progression by regulating N6-methyladenosine modification of YES1 mRNA in an IGF2BP1-dependent manner. Cell Death Discov. 2021;7(1):315
144. Yang Y, Wu J, Liu F, He J, Wu F, Chen J, Jiang Z. IGF2BP1 promotes the liver cancer stem cell phenotype by regulating MGAT5 mRNA stability via m6A RNA methylation. Stem Cells Dev. 2021
145. Muller S, Glass M, Singh AK, Haase J, Bley N, Fuchs T, Lederer M, Dahl A, Huang H, Chen J. et al. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 2019;47(1):375-390
146. Liu L, Wu Y, Li Q, Liang J, He Q, Zhao L, Chen J, Cheng M, Huang Z, Ren H. et al. METTL3 Promotes Tumorigenesis and Metastasis through BMI1 m(6)A Methylation in Oral Squamous Cell Carcinoma. Mol Ther. 2020;28(10):2177-2190
147. Ma H, Zhang F, Zhong Q, Hou J. METTL3-mediated m6A modification of KIF3C-mRNA promotes prostate cancer progression and is negatively regulated by miR-320d. (1945-4589 (Electronic)).
148. Chen F, Chen Z, Guan T, Zhou Y, Ge L, Zhang H, Wu Y, Jiang GM, He W, Li J. et al. N(6) -Methyladenosine Regulates mRNA Stability and Translation Efficiency of KRT7 to Promote Breast Cancer Lung Metastasis. Cancer Res. 2021;81(11):2847-2860
149. Lv D, Ding S, Zhong L, Tu J, Li H, Yao H, Zou Y, Zeng Z, Liao Y, Wan X. et al. M(6)A demethylase FTO-mediated downregulation of DACT1 mRNA stability promotes Wnt signaling to facilitate osteosarcoma progression. Oncogene. 2022
150. Hou P, Meng S, Li M, Lin T, Chu S, Li Z, Zheng J, Gu Y, Bai J. LINC00460/DHX9/IGF2BP2 complex promotes colorectal cancer proliferation and metastasis by mediating HMGA1 mRNA stability depending on m6A modification. J Exp Clin Cancer Res. 2021;40(1):52
151. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, Han K, Chen JW, Judde JG, Deas O. et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10(1):4695
152. Wu X, Fan Y, Liu Y, Shen B, Lu H, Ma H. Long Non-Coding RNA CCAT2 Promotes the Development of Esophageal Squamous Cell Carcinoma by Inhibiting miR-200b to Upregulate the IGF2BP2/TK1 Axis. Front Oncol. 2021;11:680642
153. Lang C, Yin C, Lin K, Li Y, Yang Q, Wu Z, Du H, Ren D, Dai Y, Peng X. m(6) A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. Clin Transl Med. 2021;11(6):e426
154. Liu H, Qin S, Liu C, Jiang L, Li C, Yang J, Zhang S, Yan Z, Liu X, Yang J. et al. m(6)A reader IGF2BP2-stabilized CASC9 accelerates glioblastoma aerobic glycolysis by enhancing HK2 mRNA stability. Cell Death Discov. 2021;7(1):292
155. Hu X, Peng WX, Zhou H, Jiang J, Zhou X, Huang D, Mo YY, Yang L. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. (1476-5403 (Electronic)).
156. Ye M, Dong S, Hou H, Zhang T, Shen M. Oncogenic Role of Long Noncoding RNAMALAT1 in Thyroid Cancer Progression through Regulation of the miR-204/IGF2BP2/m6A-MYC Signaling. Mol Ther Nucleic Acids. 2021;23:1-12
157. Wang Y, Lu J-H, Wu Q-N, Jin Y, Wang D-S, Chen Y-X, Liu J, Luo X-J, Meng Q, Pu H-Y. et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Molecular Cancer. 2019 18(1)
158. Lu S, Han L, Hu X, Sun T, Xu D, Li Y, Chen Q, Yao W, He M, Wang Z. et al. N6-methyladenosine reader IMP2 stabilizes the ZFAS1/OLA1 axis and activates the Warburg effect: implication in colorectal cancer. J Hematol Oncol. 2021;14(1):188
159. Wu H, Ding X, Hu X, Zhao Q, Chen Q, Sun T, Li Y, Guo H, Li M, Gao Z. et al. LINC01021 maintains tumorigenicity by enhancing N6-methyladenosine reader IMP2 dependent stabilization of MSX1 and JARID2: implication in colorectal cancer. Oncogene. 2022
160. Li J, Gao X, Zhang Z, Lai Y, Lin X, Lin B, Ma M, Liang X, Li X, Lv W. et al. CircCD44 plays oncogenic roles in triple-negative breast cancer by modulating the miR-502-5p/KRAS and IGF2BP2/Myc axes. Mol Cancer. 2021;20(1):138
161. Ji F, Lu Y, Chen S, Yu Y, Lin X, Zhu Y, Luo X. IGF2BP2-modified circular RNA circARHGAP12 promotes cervical cancer progression by interacting m(6)A/FOXM1 manner. Cell Death Discov. 2021;7(1):215
162. Tan L, Tang Y, Li H, Li P, Ye Y, Cen J, Gui C, Luo J, Cao J, Wei J. N6-Methyladenosine Modification of LncRNA DUXAP9 Promotes Renal Cancer Cells Proliferation and Motility by Activating the PI3K/AKT Signaling Pathway. Front Oncol. 2021;11:641833
163. Cui J, Tian J, Wang W, He T, Li X, Gu C, Wang L, Wu J, Shang A. IGF2BP2 promotes the progression of colorectal cancer through a YAP-dependent mechanism. Cancer Sci. 2021;112(10):4087-4099
164. Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, Chen ZH, Zeng ZL, Wang F, Zheng J. et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18(1):112
165. Zhang D, Ning J, Okon I, Zheng X, Satyanarayana G, Song P, Xu S, Zou MH. Suppression of m6A mRNA modification by DNA hypermethylated ALKBH5 aggravates the oncological behavior of KRAS mutation/LKB1 loss lung cancer. Cell Death Dis. 2021;12(6):518
166. Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, Somasundaram K. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37(4):522-533
167. Huang J, Sun W, Wang Z, Lv C, Zhang T, Zhang D, Dong W, Shao L, He L, Ji X. et al. FTO suppresses glycolysis and growth of papillary thyroid cancer via decreasing stability of APOE mRNA in an N6-methyladenosine-dependent manner. J Exp Clin Cancer Res. 2022;41(1):42
168. Yu D, Pan M, Li Y, Lu T, Wang Z, Liu C, Hu G. RNA N6-methyladenosine reader IGF2BP2 promotes lymphatic metastasis and epithelial-mesenchymal transition of head and neck squamous carcinoma cells via stabilizing slug mRNA in an m6A-dependent manner. J Exp Clin Cancer Res. 2022;41(1):6
169. Hu C, Liu T, Han C, Xuan Y, Jiang D, Sun Y, Zhang X, Zhang W, Xu Y, Liu Y. et al. HPV E6/E7 promotes aerobic glycolysis in cervical cancer by regulating IGF2BP2 to stabilize m(6)A-MYC expression. Int J Biol Sci. 2022;18(2):507-521
170. Peng F, Xu J, Cui B, Liang Q, Zeng S, He B, Zou H, Li M, Zhao H, Meng Y. et al. Oncogenic AURKA-enhanced N(6)-methyladenosine modification increases DROSHA mRNA stability to transactivate STC1 in breast cancer stem-like cells. Cell Res. 2021;31(3):345-361
171. Yin R, Chang J, Li Y, Gao Z, Qiu Q, Wang Q, Han G, Chai J, Feng M, Wang P. et al. Differential m(6)A RNA landscapes across hematopoiesis reveal a role for IGF2BP2 in preserving hematopoietic stem cell function. Cell Stem Cell. 2021
172. Wang X, Ji Y, Feng P, Liu R, Li G, Zheng J, Xue Y, Wei Y, Ji C, Chen D. et al. The m6A Reader IGF2BP2 Regulates Macrophage Phenotypic Activation and Inflammatory Diseases by Stabilizing TSC1 and PPARgamma. Adv Sci (Weinh). 2021;8(13):2100209
173. Bechara RA-O, Amatya NA-O, Bailey RA-O, Li YA-O, Aggor FA-O, Li DA-OX, Jawale CA-O, Coleman BA-O, Dai N, Gokhale NA-O. et al. The m(6)A reader IMP2 directs autoimmune inflammation through an IL-17- and TNFα-dependent C/EBP transcription factor axis. LID - 10.1126/sciimmunol.abd1287 [doi] LID - eabd1287. (2470-9468 (Electronic)).
174. Qian B, Wang P, Zhang D, Wu L. m6A modification promotes miR-133a repression during cardiac development and hypertrophy via IGF2BP2. Cell Death Discov. 2021;7(1):157
175. Wu Z, Shi Y, Lu M, Song M, Yu Z, Wang J, Wang S, Ren J, Yang YG, Liu GH. et al. METTL3 counteracts premature aging via m6A-dependent stabilization of MIS12 mRNA. Nucleic Acids Res. 2020;48(19):11083-11096
176. Yang Z, Wang T, Wu D, Min Z, Tan J, Yu B. RNA N6-methyladenosine reader IGF2BP3 regulates cell cycle and angiogenesis in colon cancer. J Exp Clin Cancer Res. 2020;39(1):203
177. Jiang L, Li Y, He Y, Wei D, Yan L, Wen H. Knockdown of m6A Reader IGF2BP3 Inhibited Hypoxia-Induced Cell Migration and Angiogenesis by Regulating Hypoxia Inducible Factor-1alpha in Stomach Cancer. Front Oncol. 2021;11:711207
178. Yang Z, Zhao F, Gu X, Feng L, Xu M, Li T, Liu X, Zhang X. Binding of RNA m6A by IGF2BP3 triggers chemoresistance of HCT8 cells via upregulation of ABCB1. (2156-6976 (Print)).
179. Hu Y, Gong C, Li Z, Liu J, Chen Y, Huang Y, Luo Q, Wang S, Hou Y, Yang S. et al. Demethylase ALKBH5 suppresses invasion of gastric cancer via PKMYT1 m6A modification. Mol Cancer. 2022;21(1):34
180. Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W, Guo W, Wu X, Pu C, Hu X. et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N(6)-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol Cancer. 2022;21(1):60
181. Du M, Peng Y, Li Y, Sun W, Zhu H, Wu J, Zong D, Wu L, He X. MYC-activated RNA N6-methyladenosine reader IGF2BP3 promotes cell proliferation and metastasis in nasopharyngeal carcinoma. Cell Death Discov. 2022;8(1):53
182. Ding WB, Wang MC, Yu J, Huang G, Sun DP, Liu L, Zhang JN, Yang Y, Liu H, Zhou WP. et al. HBV/Pregenomic RNA Increases the Stemness and Promotes the Development of HBV-Related HCC Through Reciprocal Regulation With Insulin-Like Growth Factor 2 mRNA-Binding Protein 3. Hepatology. 2021;74(3):1480-1495
183. Yu T, Ran L, Zhao H, Yin P, Li W, Lin J, Mao H, Cai D, Ma Q, Pan X. et al. Circular RNA circ-TNPO3 suppresses metastasis of GC by acting as a protein decoy for IGF2BP3 to regulate the expression of MYC and SNAIL. Mol Ther Nucleic Acids. 2021;26:649-664
184. Lee Y, Choe J, Park OH, Kim YK. Molecular Mechanisms Driving mRNA Degradation by m(6)A Modification. Trends Genet. 2020;36(3):177-188
185. Zeng C, Huang W, Li Y, Weng H. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol. 2020;13(1):117
186. Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM. et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254-2270
187. Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, Jiang Z, Zhang Y, Xu G, Zhang J. et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69(7):1193-1205
188. Wang X, Tian L, Li Y, Wang J, Yan B, Yang L, Li Q, Zhao R, Liu M, Wang P. et al. RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent. J Exp Clin Cancer Res. 2021;40(1):80
189. Panneerdoss SA-O, Eedunuri VK, Yadav P, Timilsina S, Rajamanickam S, Viswanadhapalli S, Abdelfattah NA-O, Onyeagucha BC, Cui XA-O, Lai Z. et al. Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. (2375-2548 (Electronic)).
190. Ying Y, Ma X, Fang J, Chen S, Wang W, Li J, Xie H, Wu J, Xie B, Liu B. et al. EGR2-mediated regulation of m(6)A reader IGF2BP proteins drive RCC tumorigenesis and metastasis via enhancing S1PR3 mRNA stabilization. Cell Death Dis. 2021;12(8):750
191. Feng M, Xie X, Han G, Zhang T, Li Y, Li Y, Yin R, Wang Q, Zhang T, Wang P. et al. YBX1 is required for maintaining myeloid leukemia cell survival by regulating BCL2 stability in an m6A-dependent manner. (1528-0020 (Electronic)).
192. Xia A, Yuan W, Wang Q, Xu J, Gu Y, Zhang L, Chen C, Wang Z, Wu D, He Q. et al. The cancer-testis lncRNA lnc-CTHCC promotes hepatocellular carcinogenesis by binding hnRNP K and activating YAP1 transcription. Nat Cancer. 2022;3(2):203-218
193. Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X, Zhang X, Cao Y, Ma D, Zhu X. et al. m(6)A-dependent glycolysis enhances colorectal cancer progression. Mol Cancer. 2020;19(1):72
194. Hao CC, Xu CY, Zhao XY, Luo JN, Wang G, Zhao LH, Ge X, Ge XF. Up-regulation of VANGL1 by IGF2BPs and miR-29b-3p attenuates the detrimental effect of irradiation on lung adenocarcinoma. J Exp Clin Cancer Res. 2020;39(1):256
195. Li B, Zhu L, Lu C, Wang C, Wang H, Jin H, Ma X, Cheng Z, Yu C, Wang S. et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun. 2021;12(1):295
196. Glass M, Misiak D, Bley N, Muller S, Hagemann S, Busch B, Rausch A, Huttelmaier S. IGF2BP1, a Conserved Regulator of RNA Turnover in Cancer. Front Mol Biosci. 2021;8:632219
197. Dong L, Geng Z, Liu Z, Tao M, Pan M, Lu X. IGF2BP2 knockdown suppresses thyroid cancer progression by reducing the expression of long non-coding RNA HAGLR. Pathol Res Pract. 2021;225:153550
198. Mao J, Qiu H, Guo L. LncRNA HCG11 mediated by METTL14 inhibits the growth of lung adenocarcinoma via IGF2BP2/LATS1. Biochem Biophys Res Commun. 2021;580:74-80
199. Mahapatra L, Andruska N, Mao C, Le J, Shapiro DJ. A Novel IMP1 Inhibitor, BTYNB, Targets c-Myc and Inhibits Melanoma and Ovarian Cancer Cell Proliferation. Transl Oncol. 2017;10(5):818-827
200. Lan Q, Liu PY, Bell JL, Wang JY, Huttelmaier S, Zhang XD, Zhang L, Liu T. The Emerging Roles of RNA m(6)A Methylation and Demethylation as Critical Regulators of Tumorigenesis, Drug Sensitivity, and Resistance. Cancer Res. 2021;81(13):3431-3440
Author contact
![]() Corresponding author: Prof. Dong Liu, College of Biological and Pharmaceutical Engineering, West Anhui University, Lu'an, P.R. China, E-mail: liudongcom.
Corresponding author: Prof. Dong Liu, College of Biological and Pharmaceutical Engineering, West Anhui University, Lu'an, P.R. China, E-mail: liudongcom.

 Global reach, higher impact
Global reach, higher impact