10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(13):4824-4836. doi:10.7150/ijbs.71234 This issue Cite
Research Paper
The m6A-induced lncRNA CASC8 promotes proliferation and chemoresistance via upregulation of hnRNPL in esophageal squamous cell carcinoma
1. Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Laboratory of Molecular Oncology, Peking University Cancer Hospital & Institute, Beijing 100142, China
2. Shenzhen Bay Laboratory, Shenzhen 518132, China
3. Research Unit of Molecular Cancer Research, Chinese Academy of Medical Sciences, Beijing 100021, China
*Those authors contributed equally to the work
Received 2022-1-20; Accepted 2022-6-12; Published 2022-7-18
Abstract
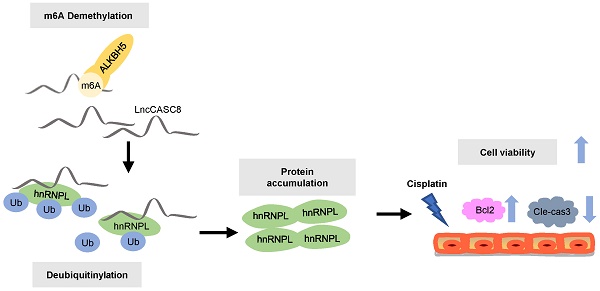
Long noncoding RNAs (lncRNAs) are dysregulated in many cancers. Here, we identified the molecular mechanisms of lncRNA Cancer Susceptibility Candidate 8 (CASC8) in promoting the malignancy of esophageal squamous cell carcinoma (ESCC). CASC8 was highly overexpressed in ESCC tissues and upregulation of CASC8 predicted poor prognosis in ESCC patients. Moreover, CASC8 decreased the cisplatin sensitivity of ESCC cells and promoted ESCC tumor growth in vivo. Mechanistically, CASC8 interacted with heterogeneous nuclear ribonucleoprotein L (hnRNPL) and inhibited its polyubiquitination and proteasomal degradation, thus stabilizing hnRNPL protein levels and activating the Bcl2/caspase3 pathway. Additionally, AlkB Homolog 5, RNA demethylase (ALKBH5)-mediated m6A demethylation stabilized the CASC8 transcript, resulting in CASC8 upregulation. Taken together, these findings identified an oncogenic function of CASC8 in the progression of ESCC, which suggest that CASC8 might become a potential prognostic biomarker in ESCC.
Keywords: lncRNA, esophageal squamous cell carcinoma, cell apoptosis, m6A demethylation, CASC8
Introduction
Esophageal carcinoma (EC) is one of the most frequent digestive cancers worldwide [1]. The majority subtype of EC is esophageal squamous cell carcinoma (ESCC), which occurs widely in China [2, 3]. The low five-year survival rate and the high mortality rate of ESCC are still prevalent, because of lacking effective methods for early diagnosis and therapy [4, 5]. Therefore, the clinical curative efficacy of ESCC treatment should be improved urgently. It has been found that plenty of risk factors are closely related with the formation and progression of ESCC, such as activated oncogene, inactivated tumor suppressor. Especially, the epigenetic abnormalities such as acetylation and methylation are reported to play essential roles in the malignant progress of ESCC [6, 7], however the underlying molecular mechanisms of ESCC tumorigenesis are still remained unknown.
In the last decade, the increasing transcriptome studies have displayed that most human genes encode for noncoding RNA (ncRNA), other than proteins [8, 9]. The ncRNA genes, particularly long ncRNA (lncRNA) genes, have been proven to link to various pathological development of cancers including ESCC. For example, lncRNA Linc01503 [10], POU3F3 [11], CCAT1 [12] and so on were highly upregulated in ESCC and act an promoting role in tumor progression. The function studies of these lncRNAs attracted new insight for the therapeutic strategies of ESCC.
Using whole-genome sequencing technology, our group has comprehensively reported genomic alterations and identified that the region 8q24.21 was significantly amplified in ESCC [13, 14]. The Cancer Susceptibility Candidate 8 (CASC8), which located at 8q24.21, showed significantly copy number amplification. The single-nucleotide polymorphisms of CASC8 have been found to be associated with the risk of EC [15], hepatocellular carcinoma [16] and colorectal cancer [17]. Moreover, CASC8 exhibited oncogenic role in promoting cell proliferation in bladder cancer [18] and retinoblastoma [19]. However, the molecular mechanism of CASC8 in tumorigenesis needs to be explored.
Here, we figured out an oncogenic role for lncRNA CASC8, which is frequently upregulated in ESCC tissues. Mechanistically, we demonstrated that the decrease in N6-methyladenosine (m6A)-modified RNA catalyzed by the m6A demethylase AlkB Homolog 5, RNA Demethylase (ALKBH5) induced the upregulation of CASC8, which subsequently promoted ESCC progression through the post-translational upregulation of hnRNPL.
Materials and Methods
Reagents
The primary antibodies of anti-hnRNPL (ab6106), IgG (ab171870) were from Abcam (Cambridge, MA, USA); the primary antibodies of anti-ALKBH5 (16837-1-AP), anti-β-actin (66009-1-g) and anti-METTL3 (15073-1-AP) were from Proteintech (Wuhan, China); the primary antibodies of anti-Bcl2 (3498S), anti-caspase3 (9665S), anti-caspase9 (9502S) and anti-Ubiquitin (3933S) were from Cell Signaling Technology (Danvers, MA, USA). Cisplatin, Actinomycin D, Cycloheximide and MG132 were purchased from MedChemExpress (Monmouth Junction, NJ, USA). Antisense LNATM GapmeRs for CASC8 were synthesized by QIAGEN (Frankfurt, Germany). siRNAs for CASC8, hnRNPL, and ALKBH5 were synthesized by Ribobio (Guangzhou, China).
Cell lines and transfection
The human ESCC cell lines: COLO-680N, KYSE-510, 450, 410, 180, 150, 30 and YES-2 were obtained from Dr. Y. Shimada of Kyoto University. The cells were cultured in RPMI 1640 (Lonza, Basel, Switzerland) with 10% fetal bovine serum (Gibco, Invitrogen, Carlsbad, USA) at 37 °C in a 5% CO2 humidified incubator. The Antisense LNATM GapmeRs were used to silence the expression of CASC8. The small interfering RNAs (siRNAs) are used to inhibit the expressions of hnRNPL and ALKKBH5. Lipofectamine 2000 transfection reagent (Invitrogen) was used to transfect plasmid and siRNA/GapmeR following the instruction. The detailed target sequences are showed below:
GapmeR/CASC8-1 sequence: CGGTAACCTAGATTGG;
GapmeR/CASC8-2 sequence: TCGTTGGTAGGCTAGT.
si/hnRNPL-1 target sequence: GACGGGTCTTGCAGTTACA;
si/hnRNPL-2 target sequence: GTCAGTCATACGGGTTGGA.
si/ALKBH5-1 target sequence: GATCGCCTGTCAGGAAACA;
si/ALKBH5-2 target sequence: GTCCTTCTTTAGCGACTCT.
Lentivirus-mediated transfection with pLenti-U6-CASC8-shRNA-GFP-Puro and its empty vector were used for stably knockdown of CASC8. Lentivirus-mediated transfection with pCDH-CMV-CASC8-EF1-CopGFP-P2A-Puro and its empty vector were used for stably overexpression of CASC8.
Tissue specimens
The tissue microarrays of ESCC were provided by Shanghai Outdo Biotech (HEsoS180Su09, Shanghai, China). All the patients who provides the tissues has signed the written informed consent. The study of these clinical specimens was permitted by Peking University Cancer Hospital Institutional Research Ethics Committee.
In situ hybridization
The DNA probes complementary to CASC8 were labeled with digoxigenin and synthesized by random primer labeling (Boster, Wuhan, China). The proportion of positive staining tumor cells was graded: < 5%, 0; 5-25%, 1; 26-50%, 2; 51-75%, 3 and > 75%, 4. The intensity of positive staining tumor cells was graded as follows: 0, negative; 1, weak; 2, moderate; and 3, strong. Final scores were the result of the multiplication of these two primary scores. Final scores of 0-6 were defined as low expression and scores of 6-12 were defined as high expression [20].
RNA isolation and qRT-PCR analyses
TRIzol reagent (Invitrogen) was used to isolated the total RNA from the cells and tissues following the manufacturer's protocol. The real-time quantitative polymerase chain reaction (qRT-RCR) assay was used to evaluate the gene expression. The reverse transcription kit (Takara, Dalian, China) was used to reverse the isolated RNA into cDNA and conducted in an ABI 7500 apparatus (Applied Biosystems, Foster City, CA, USA). The relative expression levels were normalized to the GAPDH expression. The PCR primers were as follows:
CASC8 F, 5′-CTGCTGATCATGTGCATAAGCC-3′; R, 5′-TGCTACATAACAGCCACCCAA-3′;
hnRNPL F, 5′-GTGTGGTGGAAGCAGACCTTGT-3′; R, 5′-CAAACTCCACCAGTGCTTGTCTC-3′;
18s F, 5′-CAGCCACCCGAGATTGAGCA-3′; R, 5′-TAGTAGCGACGGGCGGTGTG-3′;
U6 F, 5′-CTCGCTTCGGCAGCACA-3′; R, 5′-AACGCTTCACGAATTTGCGT-3′;
GAPDH F, 5′-CAATGACCCCTTCATTGACC-3′; R, 5′-TGGAAGATGGTGATGGGATT-3′.
Isolation of cytoplasmic and nuclear RNAs
The PARIS™ Kit (AM1921, Thermo Fisher Scientific, Scotts Valley, USA) was used to isolated cytoplasmic and nuclear RNAs according to the manufacturer's instructions. The cytoplasmic and nuclear RNAs were analyzed by qRT-RCR assay.
RNA fluorescence in situ hybridization (FISH)
The CASC8 probes were conjugated with fluorescence (Ribobio, Guangzhou, China) and applied for RNA-FISH as previously described [21]. The cells were seeded on the thick glass and fixed with ice-cold 4% paraformaldehyde. Hybridization was performed using DNA probe sets according to the manufacturer's instructions. Images were captured and visualized by confocal microscope (ST2, Leica, Wetzlar, Germany).
Cell viability assay
The cell viability was measured by Cell Titer 96® AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA). Briefly, 5 × 103 treated cells were seeded in the 96-well plate, 10% MTS in PBS was added and incubated at 37 °C for 1 h. The MTS solution absorbance was collected at 490 nm by infinite M200pro (TECAN, Hombrechtikon, Swiss).
Bromodeoxyuridine (BrdU) incorporation
5 × 103 transfected cells per well were plated into 96-well plates and incubated with BrdU for 24 h. Then the cells were fixed using ice-cold 70% ethanol for 5 min and washed with PBS. The immunostaining steps was performed according to the manufacturer's protocol. Finally, the plate was read at dual wavelengths of 450/550 nm by infinite M200pro (TECAN).
Western blot analysis
RIPA buffer (Beyotime, Shanghai, China) with protease inhibitors and phosphatase inhibitor was used to extracted the total protein. The sample was incubated on ice and vortexed for 5 min. The lysates were centrifuged at 12,000 × g for 15 min and collected the supernatant. Protein concentrations were determined by the Pierce BCA protein assay kit (Thermo). 20 μg of total protein were subjected to an SDS-polyacrylamide gel and transferred to PVDF membranes. The membranes were blocked in 5% non-fat dry milk blocking buffer for 1 h and incubated with antibodies at 4°C overnight. The Amersham Imager 600 (Amersham, Chalfont, UK) was used to detect chemiluminescence signals.
Colony formation assay
2 × 103 of transfected cells were placed in 6-well plates. After 10 days, the cell colonies were washed with PBS and fixed with methyl alcohol for 5 min. Then the 6-well plates were stained with Crystal Violet.
The tumor formation assay in nude mice
The male BALB/c nude mice (Vital River Laboratories, Beijing, China) were subcutaneously injected 1 × 106 cells/mL of transfected ESCC cells at the right flanks. The tumor volumes were evaluated by the equation, V = 1/2× length × width2. Tumor growth was measured weekly, and mice were euthanized one month after injection. The tumor growth was determined by Ki67 staining and Hematoxylin and eosin staining. The animal experiment was approved by the experimental animal department of Peking university cancer hospital.
Protein stability and mRNA stability assay
Cells were treated with 100 μg/ml cycloheximide (CHX) for indicated time to study protein stability by western blot. Cells were treated with 5 μg/ml actinomycin D (actD) for indicated time to study mRNA stability by qRT-PCR.
RNA pull-down assay
The Magnetic RNA-Protein Pull-Down kit was used for RNA pull-down assay following the manufacturer's instructions (Pierce, Waltham, MA, USA). The RiboMAX™ Large Scale RNA Production System was used to synthesize the full-length CASC8 (Promega, Madison, WI, USA). Then the biotin-labelled CASC8 was pre-incubated with the beads and the cell protein lysate was added for incubating overnight. The beads were washed three times and denaturalized in loading buffer. The CACS8 binding protein was analyzed by western blot.
RNA immunoprecipitation (RIP) assay
The Magna RIP RNA-Binding Protein Immunoprecipitation kit (Merck Millipore, Oakville, Ontario, Canada) was used for RIP assay following the manufacturer's instructions [22]. The hnRNPL antibodies were used to pull down the target RNA and and IgG was used as negative control. The qRT-PCR assay was performed to show the fold enrichment of CASC8.
Measurement of Total m6A in cells
Total m6A content was measured in 200 ng aliquots of total RNA extracted from Het-1A and KYSE-450 cells using an m6A RNA methylation quantification kit (Epigentek) according to the manufacturer's instructions.
Methylated RNA immune‑precipitation (MeRIP) assay
Total RNA was isolated from KYSE-450 cells. According to the instruction of Magna methylated RNA immune-precipitation (MeRIP) m6A Kit (Merck Millipore), the fragmented RNAs were immunoprecipitated by the m6A antibody. qRT-PCR assay was used to analyze enrichment of m6A containing RNA.
Statistical analysis
The correlation of CASC8 protein levels between cancer and adjacent normal tissue, or various clinical parameters was analyzed by chi-square test. Kaplan-Meier method and compared log-rank test were used to analyze overall survival. Statistical analysis was performed with one-way ANOVA or the 2-tailed Student's t-test. All data were obtained from at three independent experiments and showed as the mean ± SEM. All the statistical analysis was performed by SPSS software (version 25.0). A value of P < 0.05 was considered statistically significant.
Results
The upregulated CASC8 in ESCC predicts a poor prognosis
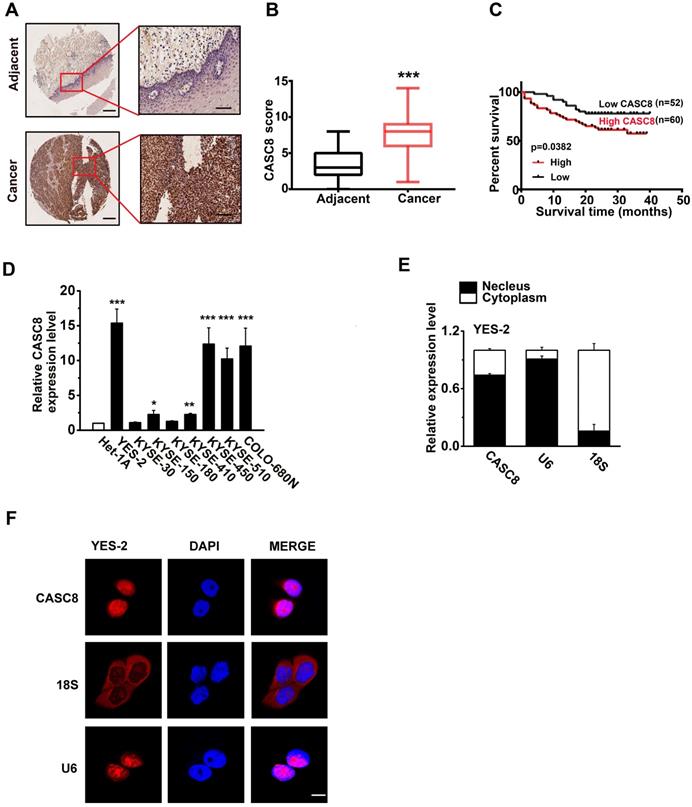
We previously analyzed the copy number alterations in ESCC and identified that the region 8q24.21 was significantly amplified [13, 14]. Intriguingly, we found that the lncRNA CASC8, which located in 8q24.21, had remarkable high frequency amplification (40%) in ESCC after analysis of our previously published data. Then, we analyzed the CASC8 in public database CCLE (https://sites.broadinstitute.org/ccle) and confirmed that CASC8 was highly expressed in esophageal carcinoma cell lines (Figure S1). For further validation, we examined the CASC8 expression in ESCC tissues by in situ hybridization assay. The ESCC tissue microarrays contained 112 cases of ESCC and 68 cases of normal esophageal tissue. CASC8 staining was dark brown in the ESCC whereas it was nearly undetectable in most normal tissues (Figure 1A). These results indicated that CASC8 was significantly upregulated in ESCC tissues, comparing to the adjacent normal tissues (Figure 1B). Moreover, ESCC patients with high expression level of CASC8 were related with poorer prognosis (P=0.0382; Figure 1C). Next, we explored the relationship between CASC8 expression levels and a various of clinicopathological parameters of ESCC patients. CASC8 expression was positively correlated with pathological grade (Table 1). In addition, the eight ESCC cell lines displayed high CASC8 expression, while the normal esophageal epithelial cell line Het-1A displayed the lowest CASC8 expression (Figure 1D). We then analyzed the subcellular localization of CASC8 in YES-2 cells by performed cellular fractionation assay. CASC8 was found mostly in the nucleus of YES-2 cells (Figure 1E). Further confirmation through RNA-FISH assay displayed that CASC8 expression was largely located in the nucleus in ESCC cells (Figure 1F).
Association of CASC8 Expression with clinicopathological features of 112 ESCC patients
| Clinicopathological Features | Total cases (%) | CASC8 expression | P value | |||
|---|---|---|---|---|---|---|
| High (%) | Low (%) | |||||
| Gender | Male | 94(83.9) | 49(52.1) | 45(47.9) | 0.6082 | |
| Female | 18(16.1) | 11(61.1) | 7(38.9) | |||
| Age | ≤ 65 | 66(58.9) | 35(53) | 31(47) | 1 | |
| > 65 | 46(44.1) | 25(54.3) | 21(45.7) | |||
| Lymph Node Metastasis | Yes | 51(45.5) | 27(52.9) | 24(47.1) | 1 | |
| No | 61(54.5) | 33(54.1) | 28(45.9) | |||
| Pathological Stage | < Ⅱ | 36(32.1) | 14(38.9) | 22(61.1) | 0.0426 | |
| ≥ Ⅱ | 76(67.9) | 46(60.5) | 30(39.5) | |||
| T Stage | T1+T2 | 27(24.1) | 16(59.3) | 11(40.7) | 0.4963 | |
| T3+T4 | 85(75.9) | 44(51.8) | 41(48.2) | |||
| AJCC Stage | 1 | 2(1.8) | 2(100) | 0(0) | 0.6011 | |
| 2 | 64(57.1) | 33(51.6) | 31(48.4) | |||
| 3 | 44(39.3) | 24(54.5) | 20(45.6) | |||
| 4 | 2(1.8) | 1(50.0) | 1(50.0) | |||
CASC8 regulates cell proliferation and cisplatin sensitive in ESCC
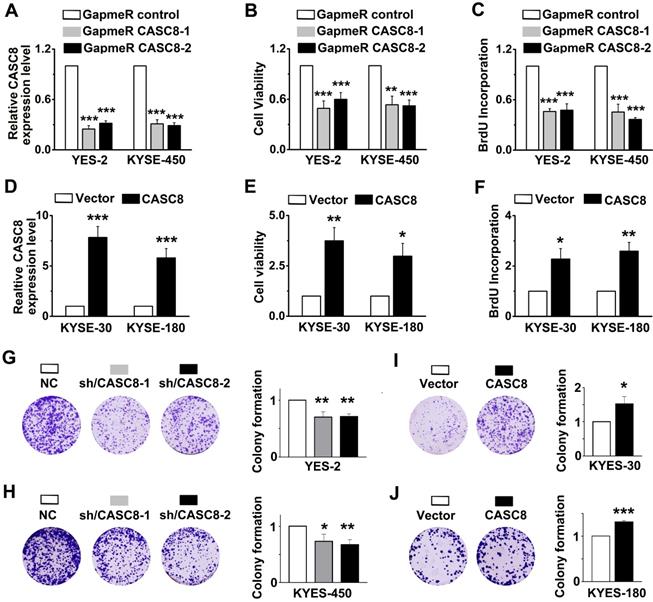
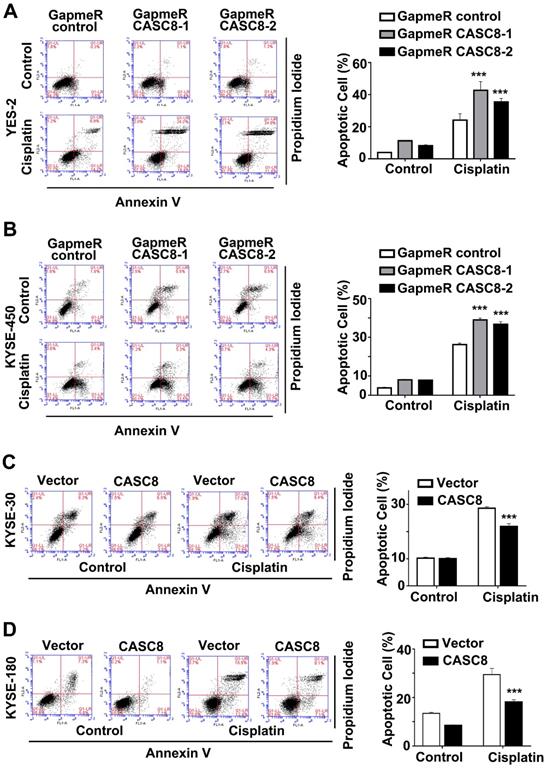
As CASC8 was highly expressed in ESCC, we further explored the function of CASC8 in ESCC cells by silencing its expression with locked nucleic acid GapmeRs. GapmeRs are antisense oligonucleotides that consist of deoxynucleotide monomers, which induce sufficiently RNase H-dependent cleavage of the targeted lncRNA [23]. As shown in figure 2A, transfection with CASC8-antisense GapmeRs led to a 70% reduction in transcript levels in both YES-2 and KYSE-450 cells. The cell viability and proliferation were inhibited when CASC8 was silenced in both YES-2 and KYSE-450 cells (Figure 2B and 2C). In contrast, overexpressing CASC8 in KYSE-30 and KYSE-180 cells improved cell viability and proliferation (Figure 2D-2F). We also used two independent siRNAs to knock down CASC8 expression in both YES-2 and KYSE-450 cells. These data suggested that siRNA treatment successfully inhibited the expression of nuclear-localized CASC8 (Figure S2). To further investigate the function of CASC8 during ESCC tumorigenesis, we established stable CASC8-knockdown ESCC cells using two small hairpin RNAs (shRNA), with shRNA-NC as negative control. The colony formation was decreased following CASC8 knockdown in both YES-2 and KYSE-450 cells (Figure 2G and 2H), whereas the colony formation was increased following CASC8 overexpression in both KYSE-30 and KYSE-180 cells (Figure 2I and 2J). Furthermore, CASC8 knockdown increased cisplatin-induced apoptosis in YES-2 and KYSE-450 cells (Figures 3A and 3B). On the contrary, CASC8 overexpression decreased cisplatin-induced apoptosis in both KYSE-30 and KYSE-180 cells (Figures 3C and 3D). Together, these observations indicated that CASC8 promoted cell proliferation and cisplatin resistance in ESCC.
The expression of CASC8 in esophageal squamous cell carcinoma. (A) The in suit hybridization showed the CASC8 RNA expression in tissues. Scale bar, left, 100μm; Right, 200μm. (B) The CASC8 staining scores in ESCC tissue and adjacent normal tissues. (C) The survival time of patients with high levels of CASC8 expression was shorter than patients with low levels of CASC8 expression (P=0.0382, log-rank test). (D) Expression of CASC8 in eight ESCC cell lines and in the normal esophageal epithelial cell Het-1A cell. (E) Representative analysis of the CASC8 cellular distribution in YES-2 cells by qRT-PCR. U6 mRNA was control for nuclear RNAs and 18s mRNA was control for cytoplasmic RNAs. (F) The expression of CASC8 in YES-2 cells detected by RNA-FISH. DAPI staining represented the nuclei. Scale bar, 10μm. *P<0.05, **P<0.01, and ***P<0.001. All of the values are expressed as the mean ± SEM.
CASC8 regulates cell proliferation in ESCC. (A) CASC8 silencing in YES-2 and KYSE-450 cells after transfection with GapmeR CASC8. (B) The cell viability was determined by MTS assay in YES-2 and KYSE-450 cells. (C) The synthesis of DNA was showed by BrdU incorporation assay in YES-2 and KYSE-450 cells. (D) The overexpression efficiency of CASC8 in KYSE-180 and KYSE-30 cells. (E) The cell viability of KYSE-30 and KYSE-180 cells measured by MTS assay. (F) The synthesis of DNA in were showed by BrdU incorporation assay KYSE-30 and KYSE-180 cells. (G-H) The colony forming assay in YES-2 and KYSE-450 cells. (I-J) The colony forming assay in KYSE-30 and KYSE-180 cells. GapmeR CASC8, GapmeR targeting CASC8; GapmeR control, GapmeR targeting scrambled control. Vector, cells were infected with empty lentiviruses; CASC8, cells were infected with lentiviruses expressing CASC8. Sh/CASC8, small hairpin RNA for CASC8; shRNA/NC, small hairpin RNA for negative control. ***P<0.001. All values are expressed as the mean ± SEM.
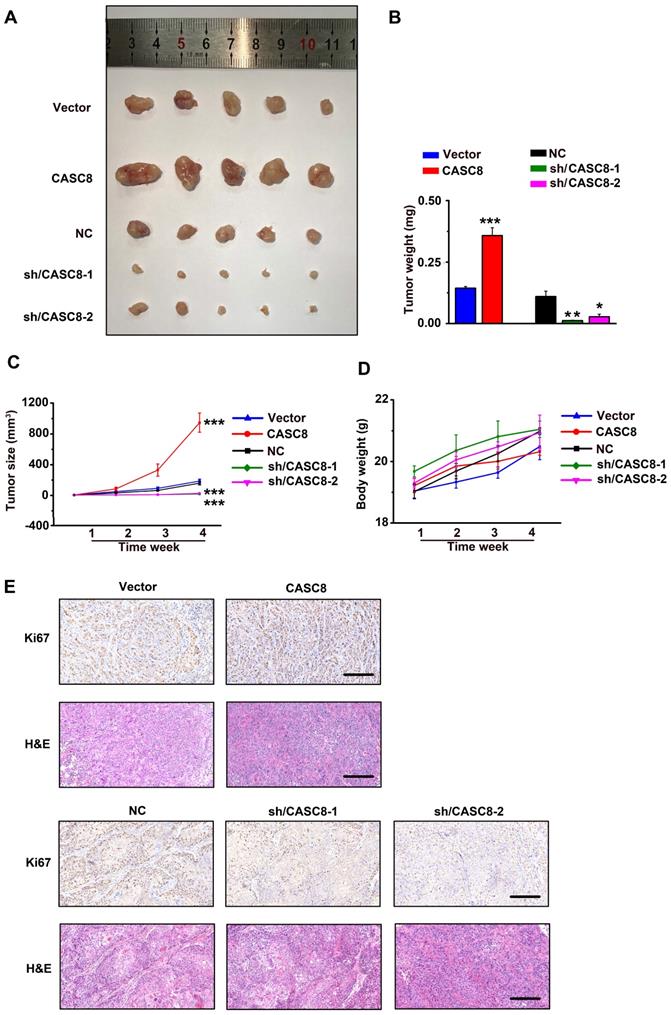
CASC8 promotes tumor growth in vivo.
Next, YES-2 cells transfected with shRNA/NC or shRNA/CASC8 and KYSE-30 cells transfected with empty vector (Vector) or vectors expressing CASC8 were subcutaneously injected into nude mice. Tumors from the shRNA-CASC8 group showed significant lower weights and smaller volumes compared with control mice (Figure 4A, 4B and 4C). On the contrary, we observed higher tumor weights and larger tumor volumes in CASC8 group compared with the vector group. Moreover, to exclude the side effect of CASC8 on physiological activities of the mice, we found no obvious body weight decrease in the mice after CASC8 knockdown or overexpression, indicating a low toxicity of CASC8 (Figure 4D). After CASC8 overexpression, the expression of Ki67, which act as a marker of proliferation, was significantly increased while the opposite results were observed after knockdown of CASC8, and the similar changes were observed in H&E staining assay (Figure 4E).
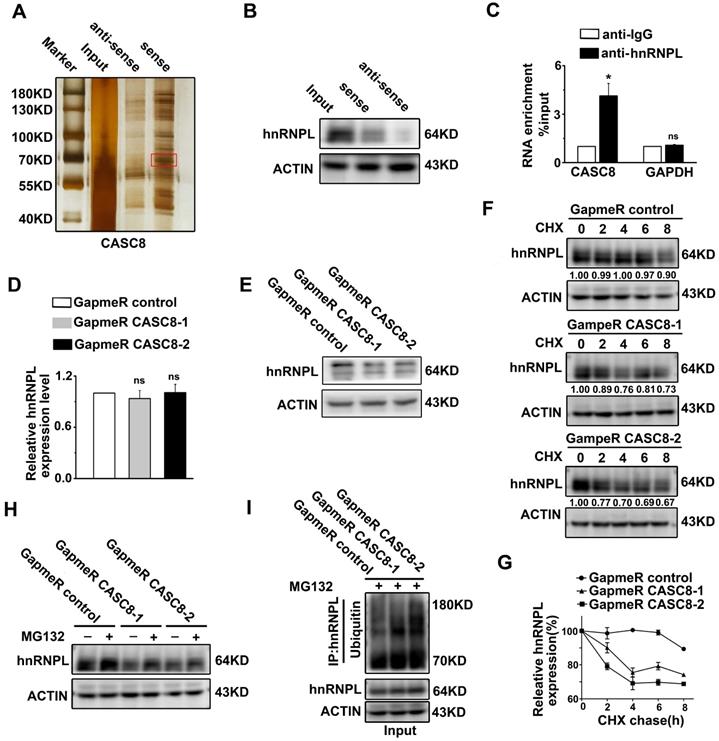
CASC8 interacts with hnRNPL and stabilizes hnRNPL
To determine the mechanism of how CASC8 promotes ESCC, we performed RNA pull-down assay combined with silver staining to identify the CASC8-binding proteins. There was one specific band in the CASC8 pull-down samples which was excised and analyzed by mass spectrometry (Figure 5A). In the list of the mass spectrometry annotated proteins, hnRNPL was selected as a potential CASC8 interacting protein. Further validation of the binding of CASC8 to hnRNPL was confirm by western blot using the retrieved proteins from the RNA pull-down assays (Figure 5B). Moreover, RNA immunoprecipitation in KYSE-450 cells exhibited that CASC8 interacted with hnRNPL protein (Figure 5C).
CASC8 promotes cisplatin resistance in ESCC. (A) The apoptotic cells in YES-2 was represented by Annexin V and PI staining using flow cytometry in left panel. Quantification of the apoptotic cells in right panel. (B) The apoptotic cells in YES-2 was represented by Annexin V and PI staining using flow cytometry in left panel. Quantification of the apoptotic cells in right panel. (C) The apoptotic cells in KYES-30 was represented by Annexin V and PI staining using flow cytometry in left panel. Quantification of the apoptotic cells in right panel. (D) The apoptotic cells in KYSE-180 cells was represented by Annexin V and PI staining using flow cytometry. Cells were treated with 16 μM cisplatin for 24 h. *P < 0.05, **P < 0.01, and ***P < 0.001. All values are expressed as the mean ± SEM.
To address the biological function of the interactions between CASC8 and hnRNPL, we explored the effects of CASC8 on protein and mRNA levels of hnRNPL. As shown in Figure 5D and E, CASC8 silencing decreased hnRNPL protein levels, but had not effect on hnRNPL mRNA levels, indicating that CASC8 upregulated hnRNPL protein levels at the post-transcriptional level. Considering that the ubiquitination/proteasome degradation is the common and quickest mechanism that regulated proteins stability in post-transcriptional way [24-26], we hypothesized that CASC8 upregulated hnRNPL by preventing its ubiquitination. To test whether CASC8 stabilized hnRNPL protein, we treated KYSE-450 cells with CHX, an inhibitor of protein biosynthesis, and found that CASC8 knockdown dramatically accelerated hnRNPL protein degradation (Figure 5F and 5G). To further confirm that CASC8 hinders hnRNPL protein degradation, we treated CASC8-knockdown KYSE-450 cells and control cells with MG132, a proteasome inhibitor. The result showed that cells treated with MG132 inhibited the decrease in the hnRNPL protein levels caused by knockdown of CASC8 (Figure 5H). We also performed an in vivo ubiquitination assay, which showed that silencing CASC8 increased hnRNPL polyubiquitination levels upon MG132 treatment (Figure 5I). Taken together, these results strongly suggested that CASC8 inhibited hnRNPL protein polyubiquitination and proteasomal degradation, thus led to increase of hnRNPL protein levels.
CASC8 promotes tumor growth in vivo. (A) The xenograft tumors from nude mice in different group. (B) Tumor weight of the tumors in different mice group. (C) Tumor growth curve and the tumor volumes of mice (n=5). (D) The body weight of the nude mice. (E) Hematoxylin and eosin staining and Ki67 staining for tumor tissues from mice. Scale bar, 50μm. *P<0.05, **P<0.01, and ***P<0.001. All values are expressed as the mean ± SEM.
CASC8 interacts with hnRNPL. (A) The silver staining showed specific bands in sense CASC8 through RNA pull-down assay in KYSE-450 cells. (B) Western blot showed the interaction of CASC8 and hnRNPL in KYSE-450 cells. (C) RNA immunoprecipitation and qRT-PCR assays indicated the CASC8 enrichment with hnRNPL antibody and negative controls in KYSE-450 cells. (D) qRT-PCR showed the unchanged mRNA expression of hnRNPL after CASC8 knockdown in KYSE-450 cells. (E) Western blot analyses showed the decreased protein expression of hnRNPL after CASC8 knockdown in KYSE-450 cells. (F) CASC8-knockdown KYSE-450 cells were treated with 100 μg/ml CHX for 8 h and then collected at the indicated times for western blotting. (G) The relative levels of hnRNPL. (H) KYSE-450 cells transfected with GapmeR targeting CASC8 were treated with or without 10 μM MG132 for 6 h and subjected to western blot analysis. (I) Lysates from KYSE-450 cells that had been transfected with GapmeR targeting CASC8 and treated with MG132 were subjected to immunoprecipitation with anti-hnRNPL antibody followed by immunoblotting analysis with anti-ubiquitin or anti-hnRNPL antibody. *P<0.05. All values are expressed as the mean ± SEM.
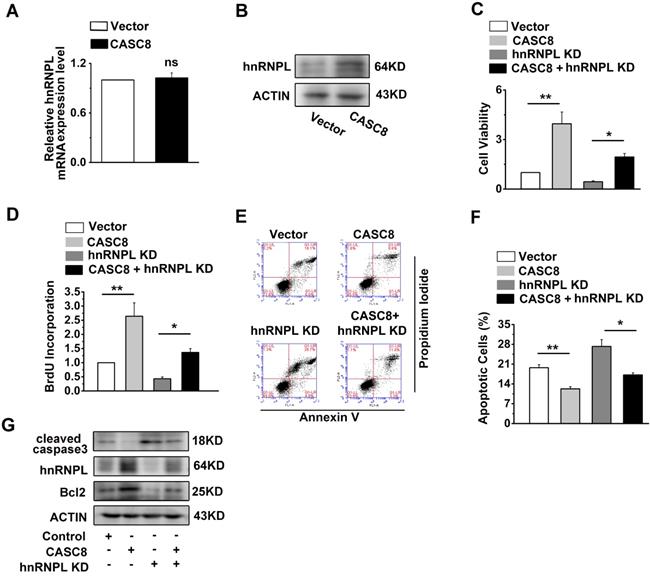
CASC8 promotes ESCC progression by upregulating hnRNPL
We observed that CASC8 overexpression increased the protein levels of hnRNPL, while the mRNA of hnRNPL displayed little changed (Figure 6A and 6B). The improvement of CASC8 overexpression on ESCC cell proliferation was inhibited by the depletion of hnRNPL (Figure 6C and D). Furthermore, the decreased percentage of apoptotic cells caused by CASC8 overexpression was successfully rescued by si-hnRNPL (Figure 6E and F). Considering that hnRNPL is linked to the Bcl2 and caspase family, which are necessary for the initiation and execution of cell apoptosis [27-29], we also detected the expression of cleaved caspase3 and Bcl2. CASC8 overexpression was found to increase the protein levels of Bcl2 and decreased the protein levels of cleaved caspase3, while silencing hnRNPL abrogated the stimulatory effect of CASC8 overexpression (Figure 6G). Collectively, these results suggested that CASC8 promoted ESCC progression probably by enhancing hnRNPL expression.
CASC8 promotes ESCC progression by upregulated hnRNPL. (A) Expression of hnRNPL in CASC8-overexpressed KYSE-450 cells. (B) The protein expression levels of hnRNPL in CASC8-overexpressed KYSE-450 cells. (C) The cell viability was determined by MTS assay in KYSE-450 cells. (D) The BrdU incorporation assay in KYSE-450 cells. (E) The percentage of apoptosis cells in KYSE-450 cells were determined by flow cytometry. (F) Quantification of the apoptotic cells. (G) Western blot showed the protein expression levels of Bcl2, hnRNPL, caspase3 and caspase9. *P<0.05, and **P<0.01. All values are expressed as the mean ± SEM.
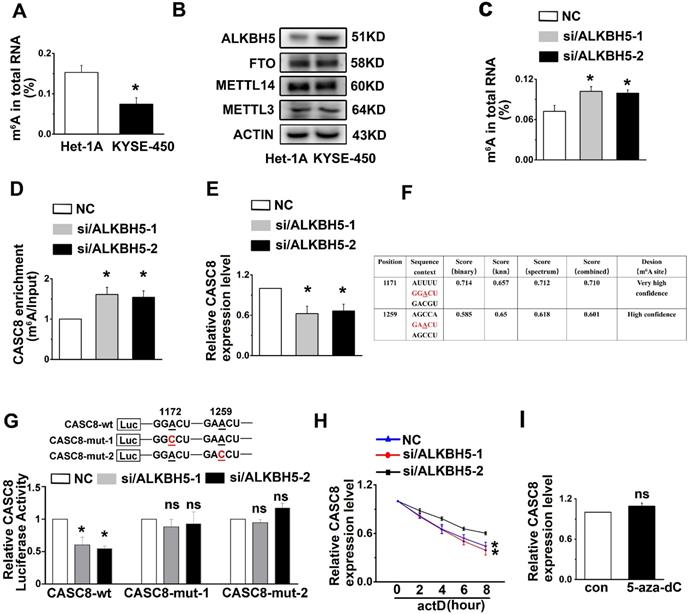
The ALKBH5-mediated m6A demethylation is involved in the upregulation of CASC8
A number of reports in tumor epigenetic regulation have demonstrated that m6A is largely involved in modification of lncRNA [30, 31]. We therefore wondered if m6A was associated with CASC8 upregulation in ESCC. First, Het-1A and KYSE-450 cells were used to detect the percentage of m6A residues among total adenosine residues in RNA. We found that the total m6A levels in RNA from Het-1A cells was higher than the levels in KYSE-450 cells (Figure 7A). We then examined the crucial m6A methyltransferase METTL3 and the m6A demethylase ALKBH5 in Het-1A and KYSE-450 cells and found that ALKBH5 was highly expressed in KYSE-450 compared to Het-1A cells, while the METTL3 expression showed no obvious changes between Het-1A and KYSE-450 cells (Figure 7B). To confirm that increased ALKBH5 expression led to decreased m6A modification of RNA in ESCC cells, we silenced the expression of ALKBH5 in KYSE-450 cells and found that ALKBH5 knockdown increased RNA demethylation in ESCC cells (Figure 7C). The m6A level in CASC8, as well as the CASC8 expression level, was also raised in ALKBH5-knockdown KYSE-450 cells compared to the control group (Figure 7D and 7E). We further identified two m6A modification sequence motifs in the 3'UTR of CASC8 using computation software called SRAMP (Figure 7F). To validate the m6A modification, we constructed a luciferase reporter inserting CASC8 wild type (CASC8-wt) or two site mutant sequence (CASC8-mut-1 and CASC8-mut-2), whose putative m6A sites were mutated. As expected, the luciferase activity of CASC8-wt cells tended to decrease when ALKBH5 was silenced, while the 1171 and 1259 site mutant groups seemed to be unchanged (Figure 7G). Considering that the m6A modification of the 3'UTR may affect RNA stability, an actinomycin D treatment was employed in the KYSE-450 cells to block transcription. The result displayed that the half-life of CASC8 became significantly shorter after ALKBH5 depletion (Figure 7H). Besides, we treated cells with 5-zaz-dc, a DNA methyltransferase inhibitor, to confirm whether CASC8 expression was affect by DNA methylation in ESCC. However, the CASC8 expression did no significantly changed after 5-zaz-dc treatment (Figure 7I). These findings revealed that ALKBH5-mediated m6A demethylation was associated with the upregulation of CASC8 in ESCC, probably by regulating the stability of its transcript.
Discussion
LncRNA dysregulation have been identified as an important pathogenic factor, especially in malignancies [32-35]. Several studies have reported that CASC8 exhibits high expression levels in EC which is correlated with overall survival of EC patients [36], but its function and mechanism are barely reported. In the present work, we demonstrated that lncRNA CASC8 played an important role in ESCC malignant progression. CASC8 was found to localized in the nucleus, and both gain and loss of function experiments confirmed that CASC8 could promote ESCC cell proliferation and inhibit cisplatin-induced apoptosis in vivo and in vitro.
The RNA-binding protein hnRNPL, a type of RNA binding protein, is likely to play a major role in various biological process including transcription, protein translation processing, alternative splicing and chromatin remodeling [37-40]. hnRNPL has also participated in many carcinogenic processes such as proliferation, apoptosis resistance, DNA damage response and repair [37, 38, 41-43], but its mechanism is unknown. We showed here that CASC8 was an oncogene and was able to regulate the ESCC progression by directly binding to hnRNPL to protect it from ubiquitin-mediated degradation. To our knowledge, we firstly demonstrated that hnRNPL exerts an important role in the cancer development via post-transcriptional modifications.
m6A modification is associated with CASC8 upregulation in ESCC cells. (A) Total RNA was extracted and m6A levels were determined as a percentage of all adenosine residues in RNA in KYSE-450 and Het-1A. (B) Western blot showed the protein expression levels of METTL3, METTL14, FTO and ALKBH5 in Het-1A and KYAE-450 cells. (C) Total RNA was extracted, and m6A content was determined as a percentage of all adenosine residues after ALKBH5 knockdown in KYSE-450. (D) The m6A level of CASC8 was measured in KYSE-450 cells with ALKBH5 knockdown in KYAE-450 cells. (E) CASC8 expression level was unchanged with ALKBH5 knockdown in KYSE-450 cells. (F) SRAMP was used to predict the m6A modification sequence motifs of CASC8. (G) The relative CASC8 luciferases activity under ALKBH5 knockdown. (H) KYSE-450 cells were treated with actinomycin D (5 µg/mL) and the CASC8 the expression level was measured by RT-qPCR. H. KYSE-450 cells were treated with or without 5-aza-dC (a DNA methyltransferase inhibitor), and CASC8 expression level was examined by RT-qPCR.
We also tried to determine if the E3 ubiquitin ligase MDM2 interacted with hnRNPL to regulate its ubiquitination, but unfortunately the results were not significant, so we speculate that there might be other ubiquitin ligases regulating hnRNPL expression which we will explore in the future study.
Total RNA m6A modification plays different roles in multiple types of tumors, including glioma [44], breast cancer [45] and endometrial cancer [46], in which total RNA m6A hypomethylation are shown to cancer progression [47]. However, two studies have shown very different results in hepatocellular carcinoma. It was reported that METTL14 was significantly downregulated in hepatocellular carcinoma, in turn reduce the total RNA m6A modifications and led to metastasis and invasion of hepatocellular carcinoma [48]. In contrast, other researchers reported that the overexpression of METTL3 was related to the poor prognosis of hepatocellular carcinoma [49]. In this study, we found that total m6A levels in RNA from KYSE-450 cells was lower than in Het-1A cells and that ALKBH5 was highly expressed in KYSE-450 compared to Het-1A cells. Furthermore, ALKBH5 knockdown increased RNA m6A modifications in ESCC cells. This is in line with the previous report that ALKBH5 is upregulated in ESCC and promotes cell proliferation.
In the recent years, the remarkable advancements have been obtained in our understanding of m6A modifications in influencing the RNAs [47, 50]. The m6A modification participates in the progression of many diseases by targeting the normal functions of mRNA or miRNA. Nevertheless, there are a lack of studies on m6A modification in lncRNAs in ESCC. One recent study revealed that smoking decreased m6A modification of LINC00278 in ESCC [51]. Herein, we demonstrated that ALKBH5 inhibited the m6A modification of CASC8, resulting regulate CASC8 RNA stability. We also confirmed that m6A methylation, but not DNA methylation in general, was involved in the upregulation of CASC8 in ESCC cells. In conclusion, our findings demonstrated the oncogenic role of lncRNA CASC8 in the progression of ESCC. We have discovered that CASC8 post-translationally regulates hnRNPL expression by protecting it from ubiquitin-mediated degradation. Furthermore, m6A modification may regulate CASC8 expression in ESCC cells. These results uncovered the molecular mechanism of CASC8 in the progression of ESCC, which would promote the development of novel clinical therapies.
Abbreviations
ESCC: Esophageal Squamous Cell Carcinoma; CASC8: Cancer Susceptibility Candidate 8; Lnc RNA: Long noncoding RNA; HnRNPL: Heterogeneous Nuclear Ribonucleoprotein L; ALKBH5: AlkB Homolog 5; m6A: N6-methyladenosine; BrdU: Bromodeoxyuridine; FISH: Fluorescence In-Situ Hybridization; EC: Esophageal Carcinoma.
Supplementary Material
Supplementary figures.
Acknowledgements
This study was supported by National Natural Science Foundation of China (No. 81988101, 81830086, 81902835 and 82003007), National key research and development program of China (2021YFC2501001), CAMS Innovation Fund for Medical Sciences (No. 2019-I2M-5-081), Guangdong Basic and Applied Basic Research Foundation (No. 2019B030302012).
Author contributions
Conceptualization: Qimin Zhan, Qingnan Wu, Hongyue Zhang, and Di Yang; Experiments design: Hongyue Zhang and Qingnan Wu; Methodology: Hongyue Zhang and Di Yang; Formal analysis and investigation: Qingnan Wu and Qingjie Min; Writing original draft preparation: Qingnan Wu and Hongyue Zhang; Review and editing: Qimin Zhan., Qingnna Wu and Weimin Zhang; Material preparation and data collection: Qingnan Wu, Hongyue Zhang and Yan Wang; Supervision: Qimin Zhan.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chen Y, Hao D, Wu X, Xing W, Yang Y, He C. et al. Neoadjuvant versus adjuvant chemoradiation for stage II-III esophageal squamous cell carcinoma: a single institution experience. Diseases of the esophagus: official journal of the International Society for Diseases of the Esophagus. 2017;30:1-7
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68:394-424
3. Wang LD, Zhou FY, Li XM, Sun LD, Song X, Jin Y. et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nature genetics. 2010;42:759-63
4. Lin DC, Dinh HQ, Xie JJ, Mayakonda A, Silva TC, Jiang YY. et al. Identification of distinct mutational patterns and new driver genes in oesophageal squamous cell carcinomas and adenocarcinomas. Gut. 2018;67:1769-79
5. Wu C, Hu Z, He Z, Jia W, Wang F, Zhou Y. et al. Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nature genetics. 2011;43:679-84
6. Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun ZM. et al. Genetic landscape of esophageal squamous cell carcinoma. Nature genetics. 2014;46:1097-102
7. Lin DC, Hao JJ, Nagata Y, Xu L, Shang L, Meng X. et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nature genetics. 2014;46:467-73
8. Matsui M, Corey DR. Non-coding RNAs as drug targets. Nature reviews Drug discovery. 2017;16:167-79
9. Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nature reviews Molecular cell biology. 2016;17:756-70
10. Xie JJ, Jiang YY, Jiang Y, Li CQ, Lim MC, An O. et al. Super-Enhancer-Driven Long Non-Coding RNA LINC01503, Regulated by TP63, Is Over-Expressed and Oncogenic in Squamous Cell Carcinoma. Gastroenterology. 2018;154:2137-51 e1
11. Li W, Zheng J, Deng J, You Y, Wu H, Li N. et al. Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 2014;146:1714-26 e5
12. Zhang E, Han L, Yin D, He X, Hong L, Si X. et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic acids research. 2017;45:3086-101
13. Song Y, Li L, Ou Y, Gao Z, Li E, Li X. et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91-5
14. Cui Y, Chen H, Xi R, Cui H, Zhao Y, Xu E. et al. Whole-genome sequencing of 508 patients identifies key molecular features associated with poor prognosis in esophageal squamous cell carcinoma. Cell Res. 2020;30:902-13
15. Sang Y, Gu H, Chen Y, Shi Y, Liu C, Lv L. et al. Long non-coding RNA CASC8 polymorphisms are associated with the risk of esophageal cancer in a Chinese population. Thorac Cancer. 2020;11:2852-7
16. Wu ER, Hsieh MJ, Chiang WL, Hsueh KC, Yang SF, Su SC. Association of lncRNA CCAT2 and CASC8 Gene Polymorphisms with Hepatocellular Carcinoma. International journal of environmental research and public health. 2019 16
17. Haerian MS, Haerian BS, Molanaei S, Kosari F, Sabeti S, Bidari-Zerehpoosh F. et al. Lack of association of CASC8 rs1447295 with colorectal cancer in Iranian population: A multicenter case-control study. Gene. 2017;634:74-6
18. Hu R, Zhong P, Xiong L, Duan L. Long Noncoding RNA Cancer Susceptibility Candidate 8 Suppresses the Proliferation of Bladder Cancer Cells via Regulating Glycolysis. DNA Cell Biol. 2017;36:767-74
19. Yang B, Gu B, Zhang J, Xu L, Sun Y. CASC8 lncRNA Promotes the Proliferation of Retinoblastoma Cells Through Downregulating miR34a Methylation. Cancer Manag Res. 2020;12:13461-7
20. Zhang H, Wang Y, Zhang W, Wu Q, Fan J, Zhan Q. BAALC-AS1/G3BP2/c-Myc feedback loop promotes cell proliferation in esophageal squamous cell carcinoma. Cancer Commun (Lond). 2021;41:240-57
21. Zhang H, Liu Y, Yan L, Zhang M, Yu X, Du W. et al. Increased levels of the long noncoding RNA, HOXA-AS3, promote proliferation of A549 cells. Cell death & disease. 2018;9:707
22. Kim KK, Kim YC, Adelstein RS, Kawamoto S. Fox-3 and PSF interact to activate neural cell-specific alternative splicing. Nucleic acids research. 2011;39:3064-78
23. Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T. et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circulation research. 2014;114:1389-97
24. Scott DC, Rhee DY, Duda DM, Kelsall IR, Olszewski JL, Paulo JA. et al. Two Distinct Types of E3 Ligases Work in Unison to Regulate Substrate Ubiquitylation. Cell. 2016;166:1198-214 e24
25. Zhu W, Li Z, Xiong L, Yu X, Chen X, Lin Q. FKBP3 Promotes Proliferation of Non-Small Cell Lung Cancer Cells through Regulating Sp1/HDAC2/p27. Theranostics. 2017;7:3078-89
26. Zhi X, Zhao D, Wang Z, Zhou Z, Wang C, Chen W. et al. E3 ubiquitin ligase RNF126 promotes cancer cell proliferation by targeting the tumor suppressor p21 for ubiquitin-mediated degradation. Cancer research. 2013;73:385-94
27. Goehe RW, Shultz JC, Murudkar C, Usanovic S, Lamour NF, Massey DH. et al. hnRNP L regulates the tumorigenic capacity of lung cancer xenografts in mice via caspase-9 pre-mRNA processing. The Journal of clinical investigation. 2010;120:3923-39
28. Shankarling G, Lynch KW. Living or dying by RNA processing: caspase expression in NSCLC. The Journal of clinical investigation. 2010;120:3798-801
29. Fiandalo MV, Kyprianou N. Caspase control: protagonists of cancer cell apoptosis. Experimental oncology. 2012;34:165-75
30. Dossin F, Pinheiro I, Zylicz JJ, Roensch J, Collombet S, Le Saux A. et al. SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature. 2020;578:455-60
31. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M. et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369-73
32. Mendell JT. Targeting a Long Noncoding RNA in Breast Cancer. The New England journal of medicine. 2016;374:2287-9
33. Munschauer M, Nguyen CT, Sirokman K, Hartigan CR, Hogstrom L, Engreitz JM. et al. Publisher Correction: The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature. 2018;563:E32
34. Yin Y, Lu JY, Zhang X, Shao W, Xu Y, Li P. et al. U1 snRNP regulates chromatin retention of noncoding RNAs. Nature. 2020;580:147-50
35. Tuvia J, Patel S, Patel M, Moskowitz GW, Levy L. Visualization of a pseudomeningocele on isotope cisternography. Clinical nuclear medicine. 1988;13:889-91
36. Liu H, Zhang Q, Lou Q, Zhang X, Cui Y, Wang P. et al. Differential Analysis of lncRNA, miRNA and mRNA Expression Profiles and the Prognostic Value of lncRNA in Esophageal Cancer. Pathology oncology research: POR. 2020;26:1029-39
37. Ji J, Xu R, Ding K, Bao G, Zhang X, Huang B. et al. Long Noncoding RNA SChLAP1 Forms a Growth-Promoting Complex with HNRNPL in Human Glioblastoma through Stabilization of ACTN4 and Activation of NF-kappaB Signaling. Clinical cancer research: an official journal of the American Association for Cancer Research. 2019;25:6868-81
38. Fei T, Chen Y, Xiao T, Li W, Cato L, Zhang P. et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E5207-E15
39. Xiu B, Chi Y, Liu L, Chi W, Zhang Q, Chen J. et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Molecular cancer. 2019;18:187
40. Klingenberg M, Gross M, Goyal A, Polycarpou-Schwarz M, Miersch T, Ernst AS. et al. The Long Noncoding RNA Cancer Susceptibility 9 and RNA Binding Protein Heterogeneous Nuclear Ribonucleoprotein L Form a Complex and Coregulate Genes Linked to AKT Signaling. Hepatology. 2018;68:1817-32
41. Chen C, He W, Huang J, Wang B, Li H, Cai Q. et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nature communications. 2018;9:3826
42. Listerman I, Sun J, Gazzaniga FS, Lukas JL, Blackburn EH. The major reverse transcriptase-incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis. Cancer research. 2013;73:2817-28
43. Asai A, Yano K, Mizukami T, Nakano H. Characterization of a duocarmycin-DNA adduct-recognizing protein in cancer cells. Cancer research. 1999;59:5417-20
44. Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z. et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer cell. 2017;31:591-606 e6
45. Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I. et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E2047-56
46. Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K. et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nature cell biology. 2018;20:1074-83
47. Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nature reviews Molecular cell biology. 2019;20:608-24
48. Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F. et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529-43
49. Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL. et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254-70
50. Clyde D. New tools for transcriptome-wide mapping of m(6)A. Nature reviews Genetics. 2020;21:387
51. Wu S, Zhang L, Deng J, Guo B, Li F, Wang Y. et al. A Novel Micropeptide Encoded by Y-Linked LINC00278 Links Cigarette Smoking and AR Signaling in Male Esophageal Squamous Cell Carcinoma. Cancer research. 2020;80:2790-803
Author contact
![]() Corresponding author: Qimin Zhan, 1Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Laboratory of Molecular Oncology, Peking University Cancer Hospital & Institute, Beijing 100142, China. E-mail: Zhanqiminedu.cn Phone: (+86) 010-88196735
Corresponding author: Qimin Zhan, 1Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Laboratory of Molecular Oncology, Peking University Cancer Hospital & Institute, Beijing 100142, China. E-mail: Zhanqiminedu.cn Phone: (+86) 010-88196735

 Global reach, higher impact
Global reach, higher impact