10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(15):5607-5623. doi:10.7150/ijbs.76281 This issue Cite
Review
Engineering neoantigen vaccines to improve cancer personalized immunotherapy
1. Department of Interventional Radiology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450052, China.
2. Interventional Institute of Zhengzhou University, Zhengzhou, Henan 450052, China.
3. Interventional Treatment and Clinical Research Center of Henan Province, Zhengzhou, Henan 450052, China.
4. Department of Gastroenterology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450052, China.
5. Department of Colorectal Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450052, China.
6. Department of Hepatobiliary and Pancreatic Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450052, China.
7. Department of Pediatric Urology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 40052, China.
†These authors have contributed equally to this work and share the first authorship
Received 2022-6-18; Accepted 2022-8-25; Published 2022-9-1
Abstract
Immunotherapy treatments harnessing the immune system herald a new era of personalized medicine, offering considerable benefits for cancer patients. Over the past years, tumor neoantigens emerged as a rising star in immunotherapy. Neoantigens are tumor-specific antigens arising from somatic mutations, which are proceeded and presented by the major histocompatibility complex on the cell surface. With the advancement of sequencing technology and bioinformatics engineering, the recognition of neoantigens has accelerated and is expected to be incorporated into the clinical routine. Currently, tumor vaccines against neoantigens mainly encompass peptides, DNA, RNA, and dendritic cells, which are extremely specific to individual patients. Due to the high immunogenicity of neoantigens, tumor vaccines could activate and expand antigen-specific CD4+ and CD8+ T cells to intensify anti-tumor immunity. Herein, we introduce the origin and prediction of neoantigens and compare the advantages and disadvantages of multiple types of neoantigen vaccines. Besides, we review the immunizations and the current clinical research status in neoantigen vaccines, and outline strategies for enhancing the efficacy of neoantigen vaccines. Finally, we present the challenges facing the application of neoantigens.
Keywords: Neoantigen, Cancer, Vaccine, Immunotherapy, Personalized therapy
Introduction
Malignant tumors are the leading cause of mortality and remain a primary stumbling block to improving life expectancy worldwide [1]. According to the 2022 Cancer Statistics, the number of new cancer cases and deaths from cancer in the United States is projected to be 1,918,030 and 609,360, respectively [2]. Traditional cancer treatments, including surgery, radiotherapy, chemotherapy, and targeted therapy, have drawbacks. Surgical treatment is limited due to the possibilities of postoperative infections, tumor metastasis, and recurrence. Repopulation of cancer cells, resistance, and high recurrence rate are considered as main factors limiting the efficacy of radiotherapy [3-5]. Chemotherapy outcomes are restricted by resistance, safe dosage, and unspecific cytotoxicity of chemotherapeutic drugs [6-8]. Drug resistance demonstrates a dominant problem for the failure of targeted therapy [9]. Besides, targeted therapy has certain requirements on the patient population, such as the compatibility of targets and the economic situation of patients [10]. Furthermore, not all tumors have suitable targeted drugs. For example, developing targeted drugs for triple-negative breast cancer (TNBC) is problematic [11].
A major contribution to cancer immunotherapy has been made by immune checkpoint inhibitors (ICIs). ICIs activate the immune system and stimulate the production of antitumor immune cells once they release the inhibitory brakes of T cells [12]. Presentation and recognition of neoantigens are crucial to the efficacy of ICIs [13]. Neoantigens, known as tumor-specific antigens, differ from tumor-associated antigens. Neoantigens are only expressed in cancer cells but absent in normal cells, however, tumor-associated antigens exist both in cancer cells and normal cells. Thereby, neoantigens prevent “off-target” damage to normal tissues and are not subject to central or peripheral tolerance, making neoantigens attractive for personalized vaccines [14, 15]. Neoantigen-specific T cells could be activated by sophisticated sequencing, recognization of individual mutations, computational forecasts of neoantigens, and vaccination of neoantigen vaccines [16]. Cancer patients with the most abundant CD8+ T cell infiltration and the highest neoantigen number, but not either alone, have the longest survival. Meanwhile, long-term survivors exhibited sustained T cell reactivity to high-quality neoantigens. It is identified that neoantigen quality rather than quantity might guide the application of immunotherapies [17]. In recent years, cancer vaccines have focused on neoantigens. Neoantigen vaccines furnish cancer patients with options and hopes, and show a bright future in different categories of cancers, including melanoma [18], non-small cell lung cancer (NSCLC) [19], glioma [20], and ovarian cancer [21].
In this review, we discussed the origin and identification of neoantigens, the immunological mechanism and the clinical application of neoantigen vaccines. Finally, we posed challenges to the utilization of neoantigens.
The origin of neoantigen
Single nucleotide variations
The somatic mutations that generated genetic variation within multicellular individuals were crucial to cancer occurrence and development [22]. Parkhurst et al. [23] performed whole exon sequencing (WES) on tumors of patients with gastrointestinal cancer, defecting mutations ranging from 22 to 928 with a median of 114. They found that single nucleotide variations (SNVs) accounted for the majority of mutations and minority gene products encoded by somatic non-synonymous SNVs (nsSNVs) were immunogenic [23]. Besides, 83% of patients possessed tumor-infiltrating lymphocytes (TILs) reactivity to neoantigens [23]. Point mutation in the arginine 132 (R132) residue of IDH1 was known as “hot spots” [24], serving as a potential immunotherapeutic target. IDH1 (R132H) was combined with the major histocompatibility complex (MHC) II allele human leukocyte antigen (HLA), whereby stimulated specific CD4+ T-helper-1 (Th1) cells to generate interferon-γ (IFN-γ). Further, a patient with IDH1 (R132H) glioma was detected to have IgG1 subclass-specific antibodies [25], anti-IDH vaccines might be a viable new treatment strategy. Additionally, the majority of diffuse gliomas showed a mutation at position 27 of histone H3.3 (H3.3K27M mutation) that changed lysine (K) to methionine (M) [26]. Long peptide vaccines induced the expansion of specific T cell lines in progressive midline glioma with an H3.3K27M mutation. The H3.3K27M peptide bound HLA-A*02:01 with high affinity, subsequently connected with CD8+ T cells. Moreover, in HLA-A2+H3.3K27M+ glioma cells, T cell receptor (TCR)-transduced cells killed tumor cells in an antigen- and HLA-specific manner, which provided a powerful method for adoptive T cell therapy (ACT) of neoantigens [27]. In addition, hepatocellular carcinoma (HCC) was a solid tumor with a low/moderate mutation load, and HCC neoantigens were rarely reported. Underlying neoantigens produced by nsSNVs in HCC were identified by WES and RNA sequencing. Most predictive peptides bound to HLA-A*02.01 and HLA-DRB1*01 representative class I and class II alleles and enjoyed immunogenicity, providing a novel strategy for HCC treatment [28]. It was noteworthy that nsSNVs and neoantigen loads were associated with checkpoint inhibitor responses. ICIs were highly efficient in tumors with high nsSNVs load, such as melanoma, lung cancer, head and neck squamous cell carcinoma, and bladder cancer [29].
Insertions and deletions
Insertions and deletions (Indels) gave rise to frame-shifting, formed a new open reading framework, and generated alien neoantigens. Across a pan-cancer cohort, renal cell carcinoma had the highest proportion and number of indels. Compared to nsSNVs, indels generated three times more predicted neoantigens as well as nine times more powerful mutant-binding neoantigens [30]. Indels were ideal sources of tumor-derived neoantigens and capable of inducing multiple neoantigen responsive T cells while decreasing susceptibility to the tolerance due to the increased number of mutant peptides. Proverbially, overexpression of the epidermal growth factor receptor (EGFR) was connected with various cancers [31]. The most frequent EGFR mutation was EGFR variant III (EGFRvIII), which arose from the deletion of 267 amino acids in exons 2-7 of the EGFR gene [32]. Cancer patients expressing EGFRvIII produced specific humoral and cellular immune responses to EGFRvIII [33], suggesting that EGFRvIII was an immunogenic neoantigen. Furthermore, the microsatellite instability-high (MSI-H) tumor phenotype was attributable to defects in the DNA repair mechanism caused by a loss of mismatch repair activity. Variations in DNA length resulting from nucleotide repetition in microsatellite site units were a cardinal characteristic of MSI-H. Indels in MSI-H cancer formed some shared frameshift peptides that could combine with MHC I and activate CD8+ T cells, producing IFN-γ and tumor necrosis factor-α (TNF-α). Moreover, the frameshift mutations were “further from self” [34]. The shared neoantigens resulting from frameshift mutations of MSI-H cancer patients extensively existed and owned strong immunogenicity, suitable for “off-the-shelf” design of cancer vaccines. Particularly, indel load was associated with increased cytotoxic T cell infiltration and superior ICI responses. The presence of indels was explicitly conjoined with significantly longer progression-free survival (PFS) and made a difference in overall response rates and disease control rates in NSCLC patients treated with ICIs, suggesting indels might play a predictive role in ICIs responses [35].
Gene fusions
Gene fusions played a crucial part in tumorigenesis, which were related to chromosomal structural variations [36]. In general, tumors with high somatic mutation burden exhibit a greater response to ICIs. However, it has been found that ICIs can also be effective in tumors with a low somatic mutation burden. Through further studies, researchers found the emergence of gene fusions led to the stimulation of cytotoxic T cell responses [37]. Peptides that spanned two gene breakpoints were excellent sources of neoantigens. For instance, Yang et al.[36] discovered the DEK-AFF2 gene fusion produced the mutant peptide DKESEEEVS, which bound to MHC I and stimulated T cell activation, generating IFN-γ and CD137 positive staining of CD8+ T cells. Additionally, peptides derived from MYB-NFIB or MYBL1-NFIB fusion gene could be treated as neoantigens in a group of adenoid cystic cancers of the head and neck. The fusion between CBFB and MYH11 genes arising from inv (16) and t (16;16) was momentous in taking acute myeloid leukemia (AML) forward. CBFB-MYH11+ HLA-B*40:01+ AML cell lines and primary human samples were killed by clones of high-affinity CD8+ T cells segregated from healthy donors. High-affinity CBFB-MYH11 epitope-specific TCR transduced into CD8+ T cells performing anti-leukemia activity [38]. These manifested that CBFB-MYH11 fusion took the shape of a neoantigen, which had potential as a target for immunotherapy and provided evidence of principle for fusion-directed T cell immunotherapy of AML. Similarly, the amino acid sequence at the break/fusion point of the EWS/FLI1 fusion gene was changed, forming a neoantigen. Functionally rejuvenated CTLs induced from pluripotent stem cells targeting the neoantigen encoded by EWS/FLI1 fusion gene may be a promising approach for treating Ewing sarcoma [39]. Nevertheless, the relationship between fusion protein and immunotherapy responses was indistinct, especially when the tumor mutational burden (TMB) of fusion-positive cancers was low [40].
Others
DNA mutations were a perfect source of neoantigens, whereas changes in RNA splicing led to the generation of new epitopes in the same way. Alternative splicing was a regulatory mechanism whereby a single gene produced multiple mRNA transcripts, significantly expanding proteome diversity [41]. Disruption of splicing mechanisms affected transcription and was capable of resulting in disease. Pharmacological modulation of RNA splicing could produce true neoantigens that inhibited tumor growth and augmented checkpoint blocking in a manner reliant on peptides present on host T cells and tumor MHC I [42]. Intron retention (IR) was the failure of the spliceosome to remove specific introns from the former mRNA molecule, allowing them to remain in the mature polyadenylated mRNA. IR was a significant source of multiple myeloma (MM) neoantigens, and newly diagnosed MM samples showed more IR events in comparison with normal plasma cells [43]. Patients with MM had poor overall survival when they had high IR neoantigen loads. Besides, endogenous retroelements, HLA-somatic mutation-derived antigens, post-translational TSAs, and viral-derived cancer antigens (for example, HPV and EBV) are classes of surrogate neoantigens [44]. For instance, dysregulation of protein citrullination was connected with autoimmune diseases. Citrullinated peptides could bind to MHC II, promoting the autoimmunity of rheumatoid arthritis and inducing a B cell response [45, 46]. Recently, citrullinated peptides targeting vimentin and α-enolase have been certified to have anti-tumor immune effects by triggering a CD4+ T cell response, building evidence for the potential immunogenicity of protein citrullinated in tumors [47, 48]. Likewise, Hiroyuki et al. [49] demonstrated that the protein citrulline was a source of cancer neoantigens.
Identification and prediction of neoantigens
Although neoantigens were weakly expressed, recent advances in mass spectrometry (MS), exosome, and bioinformatics offered powerful tools for mining sparse samples [50]. Somatic mutations were recognized using WES of tumor and normal cell DNA, followed by prioritization of mutated alleles using RNA sequencing, and the binding affinity between the putative neoantigen and MHC was predicted on an electronic computer subsequently[18, 51]. Though RNA sequencing had high false-positive and false-negative rates, WES lacked coverage and variant allele reads in the tumor. Nevertheless, combining the two was an effective and economical approach to predicting neoantigens, detecting about 70% of neoepitope candidates with high expression and rich mutant transcripts [52]. Next, the IC50 value or percentile rank score was extensively used for affinity prediction. IC50 values reflected predictions of direct binding affinity, and peptide thresholds <500 nM were applied to identify compounds most likely to bind HLA. While the percentile rank represented relative neoantigen binding affinity, a rank score ≤2 was accustomed to selecting potential neoantigen binders [53]. Unlike HLA I, the peptide-binding groove of HLA II was open to accommodate longer peptides. The flanking amino acids of the core binding sequence could affect the binding affinity [54]. In addition, HLA II peptide processing was extraordinarily intricate and obscure, posing a challenge to the development of algorithms [55]. Moreover, the prediction of neoantigens included other biological features other than gene expression and MHC affinities, such as mutation clonality, dissimilarity to self, and TCR recognition [56]. A powerful way to discover neoantigens was a combined genomic, proteomic, and immunopeptidomics approach [57]. Currently, various types of software applications are capable of discerning neoantigens. The positive predictive value of HLA antigens was increased up to ninefold through the EDGE model which used genomic dataset and HLA peptide MS data rather than HLA-peptide binding affinity data [58]. Moreover, the features based on immunopeptidomics MS increased neoantigen prioritization up to 50% [59]. Notably, MS provided posttranslational modification information, for example, glycosylation and phosphorylation [60, 61]. Structural parameters could improve the prediction of immunogenic neoantigens [62]. The convolutional neural network (CNN) was a particular type of deep neural network, more opportune for studying peptides whose amino acid spatial positions were critical for binding [63]. A deep CNN named Antigen Presentation Prediction Model outperformed the means proposed by the immune epitope database on specificity and positive predictive value [64]. Beyond that, the binding affinity to the same register was reduced by nearly 70 times in consideration of the proximal variation. Thereby, some weakly binding candidate peptides could be unsuitable [65]. It was essential to achieve accuracy in predicting neoantigens during the development of neoantigen vaccines. However, neoantigen prediction tools are currently limited to SNVs, and few tools are available to predict mutations other than SNVs in neoantigens [66]. Therefore, the development of bioinformatics is indispensable to improving the identification and evaluation of neoantigens.
Neoantigen vaccines
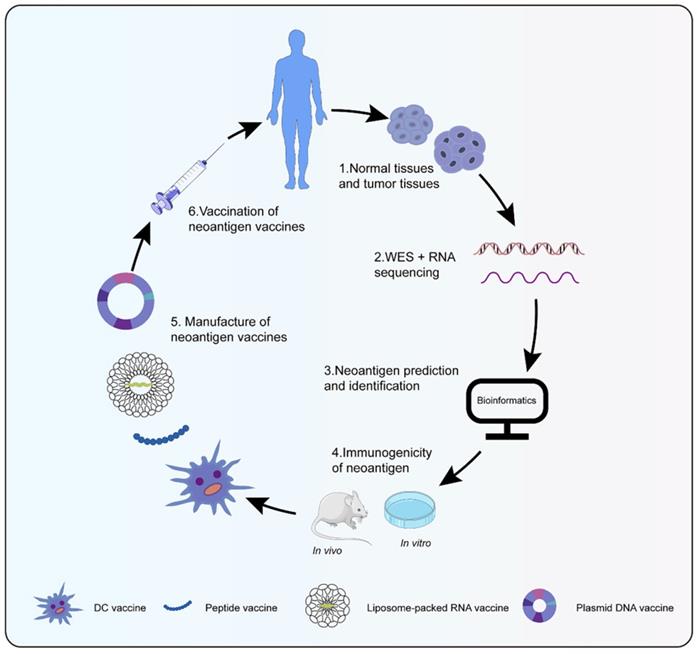
Neoantigen vaccines were highly personalized, whose process from tumor specimen collection, sequencing, and bioinformatics analysis to vaccine preparation generally took 3 to 5 months (Figure 1) [67]. Cancer vaccines were designed in four broad types: peptides, DNA, RNA, and dendritic cell (DC) (Table 1). Peptide vaccines, the most pervasive form of the vaccine, consisted of recombinant or purified proteins. Peptide vaccines composed of 20-30 amino acids might be processed and presented by antigen presentation cells (APCs) as a matter of priority [68]. After being cleaved by the immunoproteasome and antigen processing, short peptides (typically 9-11 amino acids in length) were affixed to MHC I, while long peptides (typically 14-16 amino acids in length) were attached to MHC II [69]. Id22, a peptide vaccine, was efficient enough to protect mice from the growth of MM, which induced antigen-specific CD4+ Th activity and anti-tumor immunity [70]. Meanwhile, an IDH1(R132H)-specific peptide vaccine (IDH1-vac) was equipped to engender specific Th cell responses arresting the growth of IDH1 (R132H) + tumors in mice [71]. Nonetheless, peptide vaccines were restricted as a result of their inimitable peptide epitopes, low molecular weight, simple degradation, and short half-life [72, 73]. Plasmid DNA vaccines represented another attractive approach for personalized vaccination because of fabrication easiness and low cost [74]. The probability of integration between host genome and DNA vaccines was extremely low, even lower than spontaneous mutations [75]. Robust antigen expression, processing, and presentation were in demand for the success of DNA vaccines. Tondini et al. [76] evaluated an optimized polyepitope DNA vaccine in murine melanoma. Vaccination with DNA vaccine was capable of eliciting T cell responses. Moreover, the combination of DNA vaccination and anti-PD-1 treatment suppressed tumor growth. RNA-based vaccines became a hotspot in the context of the coronavirus disease-2019 pandemic. Unlike plasmid DNA, RNA avoided possible insertion mutations and aberrant transcription. Additionally, the risk of side effects was reduced for fewer constituents to transform RNA into protein and biodegradation of RNA [77]. Compared with peptide vaccines, RNA also had distinct advantages such as low synthesis expenses, short synthesis cycle, simultaneous encoding of multiple antigen sequences, and lack of MHC haplotype restriction. Effective delivery of RNA into cells was essential to RNA vaccines. DP7-C could convey antigens into cells to its advantage by caveolin- and clathrin-dependent pathways [78, 79]. As a carrier of mRNA, DP7-C-modified DOTAP liposomes were more efficient in transferring mRNA into different types of DCs, which stimulated the maturation of DCs, production of CD103+ DCs (contributing to antigen presentation), and secretion of proinflammatory cytokine [80]. The principle behind making DC vaccines was simple. DCs were transferred back into the patient after isolation and cultivation of patient DC progenitors and loading of tumor antigens. Henceforth, specific T cells stimulated by DCs could engage in anti-tumor responses. Due to their low toxicity, lack of invasive procedures, and potential long-term effects, DC vaccines were a particularly attractive immunotherapy option [81]. However, the efficacy of DC vaccines was confined to advanced recurrent patients [82]. Sipuleucel-T was the first DC vaccine for prostate cancer approved by the United States Food and Drug Administration [83]. Different immune modes may produce distinct therapeutic effects and immune responses despite the same antigen. Neoantigen-pulsed DC vaccines were superior in immune and anti-tumor effects in contrast with the neoantigen-adjuvant vaccines [84]. In summary, a new era of neoantigen vaccines was coming; additional experiments were in need for further application of neoantigen vaccines.
Preparation of neoantigen vaccines. Obtain normal tissue and tumor tissue from cancer patients, then perform whole exons and RNA sequencing to detect mutations. Bioinformatics technology screen out candidate neoantigens. Immunogenic neoantigens are identified by in vivo and in vitro experiments. At last, various types of neoantigen vaccines are prepared to treat cancer patients.
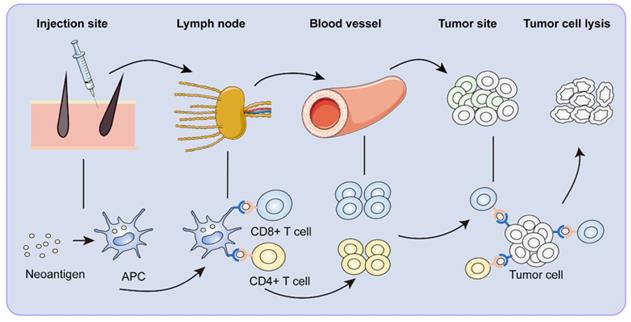
Immune response to neoantigen vaccines. After vaccination, neoantigen vaccines are first recognized by APCs around the injection site. The APCs reach the draining lymph nodes through lymphatic vessels. In the lymph node, neoantigen-specific T cells are activated. Activated CD8+ and CD4+ T cells clone and arrive at the tumor site with the circulatory system, specifically killing tumor cells.
Advantages and disadvantages of different forms of vaccines
| Vaccines type | Advantages | Disadvantages |
|---|---|---|
| Peptide | Stable, Safe, Low-complexity antigens, Low toxicity, Easy to transport | Low immunogenicity in the absence of adjuvants, Unrelated immune response due to degradation, Short half-life |
| DNA | Easy fabrication, Low cost, Polyepitope | Theoretical integration risk, Potential risk of abnormal transcription, Discomfort electroporation |
| RNA | Less side effects, Low synthesis expenses, Short synthesis cycle, Simultaneous encoding of multiple antigen sequences, Inadequate restriction of MHC haplotypes | Unstable, Innate immunogenicity, Easy degradation by RNases, Inefficient delivery, Harsh storage condition |
| DC | Low toxicity, Lack of invasive procedures, Potential long-term effects, Conventional antigen presentation pathways | Time-consuming, High cost, Less feasible to produce at large scale |
Neoantigen vaccines and immune
Immune responses
ICIs have revolutionized cancer therapy. However, microsatellite stable colorectal cancer was not sensitive to ICIs because of its low mutation rate and stable microsatellites. In contrast, neoantigens appeared to offer promising immunotherapeutic potential [85]. Therapeutic cancer vaccination was designed to produce a robust and sustained anti-tumor immune response [86, 87]. For the perceived advantages of neoantigen, neoantigen vaccines have developed expeditiously in recent years. Neoantigen vaccines underwent a series of complex maturation processes to become functional (Figure 2).
As generally believed, CD8+ T cells were a central participant in killing tumor cells. CD8+ T cells either took place directly via synaptic exocytosis of cytotoxic granules that contained perforin and granzymes into the target cells or caused the indirect destruction of cancer cells through secreting cytokines, such as IFN-γ and TNF. CD40, CD70, and CD80/86 were of great importance in the activation and persistence of T cells mediated by DCs. Chemokine receptor CX3CR1 was a marker of effector T cell differentiation [88, 89]. However, a recent study found the anti-tumor efficacy of neoantigen/toll-like receptor 3 (TLR3)/CD40 agonist vaccine and neoantigen-specific CX3CR1+ CD8+ T cell generation relied on CD40 and CD80/86, rather than the CD70 signaling pathway [90]. To induce cytotoxicity and neoepitope-specific CD8+ T cell responses, therapeutic vaccines required a conjoined helper epitope, such as P30 [91]. Besides, B cells and CD4+ T follicular helper cells synergically promoted the anti-tumor CD8+ T cell responses [92]. Nevertheless, without CD8+ T cells targeting the same antigen, CD4+ T cells have been proven to protect against tumor growth. The new epitope-specific TCR transgenic mice rejected MM cells. In the absence of CD8+ T and B cells, transgenic mice still had protection [93], demonstrating that neoepitope-specific CD4+ T cells offered sufficient production. These studies were the first to show that neoantigen-specific CD4+ T cells could repel tumor cells. Subsequent studies found that the transgenic mice also rejected B lymphoma cells [94]. Tumor cells were killed indirectly by CD4+ T cells via cytotoxic macrophages depending on the inducible nitric oxide synthetase way [95]. Furthermore, neoantigen vaccine-induced CD4+ T cells impacted the function of CD8+ T cells. On the one hand, they activated CD8+ T cells responses against non-vaccine, tumor-associated antigens. On the other hand, they promoted effector memory CD8+ T cells migrate to the tumor microenvironment (TME) [70, 96]. CD4+ Th cells could facilitate activation of CD8+ T cells by licensing DCs through CD40/CD40L interactions [97]. DCs' interaction with CD4+ T cells stimulated them to produce cytokines like IL-15 and increased CD80/86 and CD70 expression, which provoked a potent response from CD8+ T cells [98]. Besides, CD4+ T cells could eliminate cancer via a cytotoxic phenotype mediated by perforin and granzyme B [99].
In addition, the analysis of immunogenicity and immunological characteristics in clinical trials provided a theoretical basis for the specific T cell responses of neoantigen vaccines. Keskin et al. [100] administered personalized neoantigen vaccination to 10 patients with glioblastoma. CD4+ and CD8+ T cells expressed cytotoxic markers (PRF1, GZMA, and GZMK), and a few CD4+ T cells were Tregs, co-expressing IL2RA and FOXP3. Most CD4+ and CD8+ T cells expressed at least one effector cytokine (IFNG, IL2, or TNF) [100]. Co-inhibitory molecules (TIM-3, TIGIT, PD1, CTLA4, and LAG3) were also discriminatively expressed in CD4+ and CD8+ T cells. Moreover, peripheral blood neoantigen-specific T cells could migrate to intracranial glioblastoma [100]. In another clinical trial, eight patients with surgically resected stage IIIB/C or IVM1A/B melanoma were treated with neoantigen vaccine. Transcriptional status analysis at diverse vaccination stages demonstrated that the differentiation state associated with T cells underwent a marked shift from infantile to effector, apoptosis, and memory [101]. TCR sequence became abundant after inoculation. Furthermore, neoantigen vaccination expanded pre-existing T cell populations specific to neoantigens and resulted in a broader repertoire of novel T cell specificities, tipping the intra-tumoral balance in favor of better tumor control [18]. Taken together, these studies provided valuable insights into the immunogenicity of neoantigen vaccines and their therapeutic potential.
Immune escape
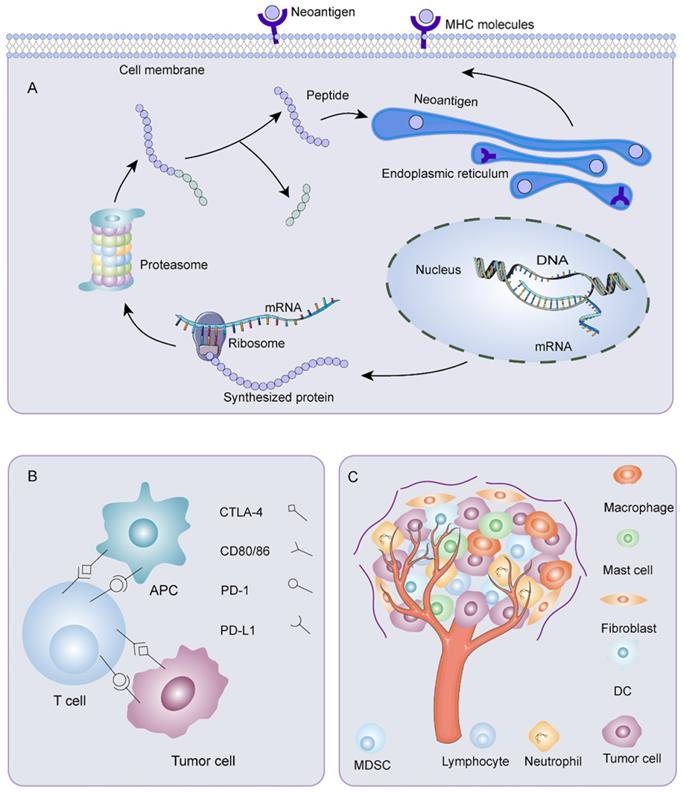
Immune escape was a crucial problem affecting the effectiveness of neoantigen vaccines (Figure 3). The reduced neoantigen expression was an immune escape mechanism [102]. DNA copy number variation induced by chromosomal instability may incite loss of neoantigens. Promoter hypermethylation caused preferential inhibition of genes containing neoantigen mutations, a latent mechanism for neoantigen deletion in the transcriptome [103, 104]. Post-translational mechanisms and the silencing of genomic segments encoding neoantigens at the epigenetic level may also be instrumental in deleting neoantigens [103]. Before being recognized by specific immune cells, neoantigens passed through a complicated intra-cellular mechanism involving the proteasome. Non-synonymous somatic mutations altered the chemical composition of amino acid sequences. As a result, the proteasomal cleavage properties varied on account of different cleavage preferences for basic, acidic, or hydrophobic amino acids. These may bring on ineffective neoantigen-specific T cell activation owing to a loss of binding affinity between the neoantigen and the MHC I complex or alteration of the complex's affinity for binding to the TCR. Lung cancer [105] and advanced bladder cancer [106] had a latent immune escape mechanism by altering proteasome antigen processsing. Meanwhile, HLA loss or mutation affected the stability of MHC and production of HLA enhancers, thus, disrupting antigen presentation, which provided an alternative mechanism for immune evasion [103, 107, 108]. HLA I deficiency was more frequent in advanced tumors than early tumors, and the degree of infiltration of CD8+ T cells was noticeably lower in HLA I deficient tumor areas than in HLA I preserved tumor areas [109]. Proteins were crucial for the maturation and exposure of antigens, such as antigen peptide transporter-2 (TAP2), beta2-microglobulin, and chaperone molecules, including calreticulin, ERp57 calnexin, and tapasin [110]. Alterations in TAP1/2 interfered with antigen-presenting. The deletion of the beta2-microglobulin gene hindered immune surveillance of tumors [111]. For example, in tumors with extrachromosomal DNA (ecDNA), immune cell infiltration and cytotoxic T cell activity were reduced. Through gene set enrichment analysis, expression of MHC-related genes decreased, which may be a dormant immune evasion mechanism of cancer with ecDNA [112]. Moreover, impaired DC cross-priming resulted in site-dependent immune escape. CD40 agonist enhanced cross-presentation and restored immune control by promoting T cell initiation and expanding response through epitope diffusion [113]. Besides, the exhaustion of T cells posed a significant challenge to antitumor immunity [114]. Depleted T cells exhibited a progressive loss of effector functions, multiple inhibitory receptor expression, dysregulated metabolism, unsatisfactory memory recall response, and dysfunctional homeostasis, contributing to ineffective cancer control [115]. Furthermore, immune checkpoints such as CTLA-4, PD-1, PD-L1, and PD-L2 were related to physiological self-tolerance. However, overexpression of immune checkpoints restrained anti-tumor immune responses. Meanwhile, TME, consisting of tumor cells, immune cells, interstitial tissue, and extracellular matrix, was inextricably linked with the development, invasion, and metastasis of tumors. The presence of tumor-associated fibroblasts [116], tumor-associated macrophages [117], tumor-associated mast cells [118], Tregs [119], and bone marrow-derived suppressor cells (MDSC) [120] helped tumor cells escape immune killing. In recent years, it has been found that tumor-associated neutrophils also had several connections to immune escape. In ovarian cancer, tumor-associated neutrophils coordinated intra-tumoral IL-8-driven immune escape through activation of Jagged2 [121]. In breast cancer, tumor-associated neutrophils advanced T cell immunosuppression through PD-L1 [122]. Metabolic enzymes like indoleamine 2, 3-dioxygenase 1 [123], serine/threonine-protein kinase 1 [124], and arginase 1 [125] created a tolerant microenvironment. In addition, immunosuppressive cytokines such as IL-6 [126], IL-4 [127], and transforming growth factor-β (TGF-β) [128] acted analogously.
Mechanisms of immune escape. A) Mutant gene sequences are transcribed and translated into mutated proteins. After processing, the mutant peptides enter the endoplasmic reticulum. They bind MHC I or MHC II to form peptide-MHC molecular complexes, which are transported to the surface of tumor cells. Any abnormality in the process of neoantigen processing and presentation could result in immune escape. B) Overexpression of immune checkpoints restrains anti-tumor immune responses. C) Various components of tumor microenvironment are associated with the occurrence of immune escape.
Clinical trials of personalized neoantigen-based vaccines
| ClinicalTrials.gov identifier | Phase | Tumor types | Number of patients | Treatment | Form of vaccine | Status |
|---|---|---|---|---|---|---|
| NCT03359239 | I | Urothelial/Bladder Cancer | 10 | PGV001 with atezolizumab; adjuvant Poly-ICLC | Peptide | Completed |
| NCT03633110 | I/II | Melanoma, NSCLC, SCCHN, RCC or urothelial carcinoma | 24 | GEN-009 alone or combined with nivolumab or pembrolizumab; adjuvant Poly-ICLC | Peptide | Completed |
| NCT03645148 | I | Pancreatic cancer | 7 | iNeo-Vac-P01; adjuvant GM-CSF | Peptide | Completed |
| NCT02454634 | I | Grade 3 and 4 IDH1(R132H) + astrocytomas | 32 | IDH1-vac | Peptide | Completed |
| NCT01970358 | I | Stage IIIB/C or stage IVM1b high-risk melanoma | 8 | NeoVax; adjuvant Poly-ICLC | Peptide | Completed |
| NCT02897765 | Ib | Advanced melanoma, NSCLC, or bladder cancer | 82 | NEO-PV-01 plus anti-PD-1; adjuvant Poly-ICLC | Peptide | Completed |
| NCT03662815 | I | Advanced solid tumors | 22 | iNeo-Vac-P01; adjuvant GM-CSF | Peptide | Active, not recruiting |
| NCT04799431 | I | Pancreatic cancer, colorectal cancer | 12 | Vaccine and retifanlimab; adjuvant Poly-ICLC | Peptide | Not yet recruiting |
| NCT05111353 | I | Pancreas cancer | 30 | Vaccine; adjuvant Poly-ICLC | Peptide | Not yet recruiting |
| NCT04487093 | I | NSCLC | 20 | Vaccine combined with targeted drug | Peptide | Recruiting |
| NCT03122106 | I | Pancreatic cancer | 15 | Vaccine alone | DNA | Active, not recruiting |
| NCT03199040 | I | TNBC | 18 | Vaccine alone or combined with durvalumab | DNA | Active, not recruiting |
| NCT03532217 | I | Metastatic hormone-sensitive prostate cancer | 19 | Vaccine in combination with nivolumab/ipilimumab and PROSTVAC | DNA | Active, not recruiting |
| NCT03988283 | I | Pediatric recurrent brain tumor | 10 | Vaccine alone | DNA | Not yet recruiting |
| NCT04015700 | I | Glioblastoma | 12 | Vaccine alone | DNA | Recruiting |
| NCT04397003 | II | Extensive-stage small cell lung cancer | 27 | Vaccine in combination with durvalumab | DNA | Recruiting |
| NCT03480152 | I/II | Metastatic gastrointestinal cancer | 4 | mRNA-4650 | RNA | Terminated, has results |
| NCT02035956 | I | Stage III-IV melanoma | 13 | mRNA vaccine alone or combined with PD-1 blockade | RNA | Completed |
| NCT03289962 | Ib | NSCLC, colorectal cancer, melanoma, and breast cancer | 29 | RO7198457 alone or combined with atezolizumab | RNA | Active, not recruiting |
| NCT05198752 | I | Solid tumor | 36 | SW1115C3 | RNA | Not yet recruiting |
| NCT00683670 | I | Stage III melanoma | 3 | Vaccine alone | DC | Completed |
| NCT02956551 | I | Advanced lung cancer | 12 | Vaccine alone or combine with PD-1 inhibitor | DC | Unknown |
| NCT03171220 | I | Solid tumors | 6 | Vaccine alone | DC | Unknown |
| NCT03871205 | I | Lung cancer | 30 | Vaccine alone | DC | Unknown |
| NCT04912765 | II | HCC | 60 | Vaccine and nivolumab | DC | Recruiting |
GM-CSF, granulocyte macrophage colony-stimulating factor; NSCLC, non-small cell lung cancer; SCCHN, squamous cell carcinoma of the head and neck; RCC, renal cell carcinoma; TNBC, triple-negative breast cancer; HCC, hepatocellular carcinoma
Neoantigen vaccine and clinical application
Monotherapy
Given the promising results from preclinical studies, numerous clinical trials of neoantigen vaccines are in progress (Table 2). The high recurrence rate after radical resection was the primary reason for the unfavorable prognosis of HCC patients. Transcatheter arterial chemoembolization (TACE) was an effective strategy for preventing recurrence. Cai et al. [129] subcutaneously injected neoantigen peptide vaccine into ten patients undergoing radical surgical resection and prophylactic TACE therapy. The median recurrence-free survival of the five patients with responsive neoantigens was considerably longer than those with non-responsive neoantigens or with only the initial vaccine. Notably, two patients maintained a relatively powerful neoantigen-specific immune response for ten months. In a clinical trial, 22 patients with advanced malignant tumors were treated with iNeo-Vac-P01, containing 5 to 20 peptides with 15 to 35 amino acids. Twenty participants experienced no or mild adverse reactions, while two had severe acute allergic reactions, showing a 71.4% disease control rate and a median PFS of 4.6 months. This trial also acknowledged that the mutations of genes and variations of copy number were predictive of the immune responses [130].
Nucleic acid-based vaccines could convey DNA or RNA encoding targeted epitopes to prevent and treat infectious diseases and cancer. Rather than targeting cancer neoantigens, most DNA vaccines focused on tumor-associated antigens, such as mammaglobin-A in breast cancer [131], HPV E6, and E7 in cervical cancer [132], and HER2/CEA in solid cancer [133]. Clinical trials of neoantigen DNA vaccines in glioblastoma, TNBC, and pancreatic cancer are ongoing. In TNBC patients, neoantigen DNA vaccination was conducive to a robust immune response and prolonged PFS [134]. Cafri et al. [135] recognized specific immunogenic mutations expressed in tumors, constructing a neoantigen vaccine called mRNA-4650, including up to 15 HLA I candidate neoantigens. Four patients with metastatic gastrointestinal cancer received the vaccine. Although no clinical response was observed, vaccine-induced CD4+ and CD8+ T cells were detected in three patients. RO7198457, an RNA-lipoplex vaccine, encoded up to twenty neoantigens. In phase Ib trial, it was single-tested in 29 patients with advanced solid tumors, including NSCLC, colorectal cancer, melanoma, and breast cancer. Achieve an objective response rate of 4% and a disease stabilization rate of 40%, slightly lower than the combination of RO7198457 and anti-PD-L1 antibody atezolizumab [136].
DCs were the dominant focus of cancer vaccines as antigen delivery carriers. In addition to acting as vaccines alone, various DNA, RNA, and peptides could be loaded onto DCs. The first neoantigen DC vaccine embarked on testing in 2015 [137]. In this trial, genomic analysis and computer simulation of neoantigen prediction were performed on three patients' surgically removed tumor tissues. Seven neoantigens appending to HLA-A*02:01 were screened out and incorporated into DC vaccine formulations together with melanoma gp100-derived peptides. T cell-triggered immune responses were enhanced, and all three patients survived without autoimmune adverse reactions. Furthermore, DC vaccines facilitated the expansion of the TCR repertoire of highly diverse neoantigens. Ding et al. [138] tested personalized neoantigen pulsed DC vaccine in 12 advanced lung cancer patients. After treatment, all adverse events were grade 1-2, and no delays due to toxic reactions to drugs occurred. Besides, they reached a 25% objective effectiveness rate, a 75% control rate for disease, a 5.5-month PFS, and a 7.9-month overall survival. Remarkably, a patient with extensively metastatic lung adenocarcinoma who had failed three treatments, including a PD-1 inhibitor, was treated with the vaccine with almost no metastatic lymph nodes, partial metastatic lesions, and a reduction in tumor target lesions.
In general, initial results suggested that peptide-, nucleic acid-, DC-based neoantigen vaccines were safe and immunogenic, revealing encouraging prospects as a feasible approach to cancer treatments and bringing prospective benefits to patients.
Combine with other therapies
Neoantigen vaccines could stimulate an autoimmune response. However, the effectiveness of neoantigen vaccines was confined, which was attributable to the lack of appropriate immune stimulation and the impact of the immunosuppressive TME. ICIs boosted T cell responses and conduced to prolonged survival in patients with previously untreatable cancers [139-141]. Although ICIs have been successful in numerous advanced malignancies, they were only effective in a small number of treated patients [142-144]. Preclinical studies have shown that combining neoantigen vaccines and ICIs exhibited different T cell phenotypic characteristics, enhanced immune responses, and appreciably increased the total number of TCR clones [145-147]. The combination therapy also upregulated the expression of genes linked to T cell activation and effector function. Neoantigen vaccines were expected to work synergistically with ICIs to reverse tumor-induced immunosuppression, contributing to clinical benefits. The first combination of an individualized neoantigen vaccine (NEO-PV-01) with PD-1 inhibition was reported in 60 patients with high TMB metastatic tumors, including melanoma, NSCLC, and urothelial carcinoma of the bladder. The objective response rates were 59%, 39%, and 27%, with median PFS of 23.5 months, 8.5 months, and 5.8 months. Meanwhile, the response to combination therapy was influenced by TMB and epitope quality [148]. Ott et al. [18] administered neoantigen vaccines to six melanoma patients, four free of recurrence at 25 months. Two sufferers received anti-PD-1 therapy after recurrence and had complete tumor regression. Some researchers treated twenty patients with unresectable advanced tumors with neoantigen vaccine in combination with pembrolizumab. Six clinical responses have been reported, including two of the 12 individuals who previously received ICIs [149]. These studies provided forceful support for the combined use of neoantigen vaccines and ICIs.
ACT targeted and killed tumor cells with the patients' T cells, which could promote the active invasion of T cells into tumor tissues and delay tumor progression. Therapeutic neoantigen vaccines and ACT transfer have shown propitious initial results. Tanaka et al. [150] sequenced the whole-exome and transcriptome of patients with the myelodysplastic syndrome to identify neoantigen candidates, then prepared T cells for specific neoantigen and injected them into the patients. No dose-limited toxicity or cytokine release syndrome was observed, and no apparent autoimmune response occurred. Kristensen et al. [151] performed ACT in patients with melanoma, finding that the number and frequency of new epitope-specific T cells in TILs infusion products were associated with improved clinical outcomes.
Furthermore, some preclinical studies have shown that neoantigen vaccines worked better when combined with other therapies. Tumor clearance after radiotherapy and chemotherapy was the induction of immunogenic cell death and the release of damage-associated molecular patterns [152]. Chemotherapy or radiotherapy could promote the release of neoantigens [153, 154] and reduce immunosuppression in TME, thus improving the effect of immunotherapy [155, 156]. Local immunochemotherapy significantly activated neoantigen-specific CD8+ T cells response, which recognized and lysed tumor cells [157]. In a mouse tumor model with lousy immunogenicity received radiation, immunogenic gene mutations increased [158]. CD4+ and CD8+ T cells could be induced by vaccination with new epitopes encoded by these genes, thus improving the efficacy of radiation therapy, disregarding their ineffectiveness in preventing tumor growth. Additionally, photothermal therapy (PTT) used photothermal agents to produce local hyperthermia, afterward, eliminated tumors. The combination of PTT and cancer vaccines effectively eradicated the sizeable primary tumor, and tumors wholly subsided [159]. What's more, the combination therapy exerted a powerful abscopal effect on distant metastases.
How can the efficacy of neoantigen vaccines be enhanced?
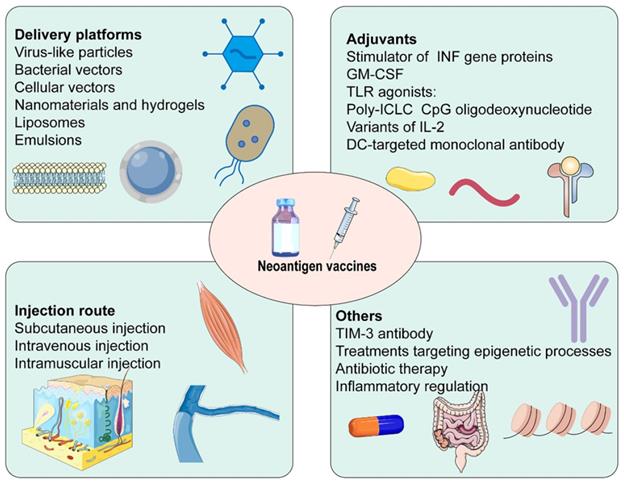
The efficacy of cancer vaccines was connected with generating robust T cell responses (Figure 4) [14]. Firstly, several neoantigen vaccine platforms have been established in recent years. Virus-like particles increased antibody titers, the efficiency of cross-presentation, as well as the proportion of CD4+ T cells, CD8+ T cells, and effector memory T cells. They could also decrease the proportion of MDSC [160]. Similarly, neoantigens expressed by engineered bacterial vectors infiltrated the tumor, resulting in a relative increment in CD4+ and CD8+ T cells and cytokine release [161]. Besides, nanomaterials and hydrogels delivered neoantigen vaccines and adjuvants to lymph nodes and APCs in coordination. They promoted the maturation of APCs, and cross-presentation of antigens, and were retained in lymph nodes for a long time, playing a robust and durable anti-tumor effect [162-164]. Secondly, adjuvants were critical to successful cancer vaccines. IFN-γ treatment not only increased HLA I and HLA II presenting peptides but also facilitated the expression of IFN-γ induction genes and immune proteasome genes [165]. Some adjuvants based on the stimulator of INF genes, such as cGAMP [166], ADU-V16 [167], and ADU-V19 [168], could trigger INF production, stimulate DCs maturation and antigen cross-presentation. IL-2, the first cytokine approved for clinical cancer treatment, was restricted by the proliferation of Tregs expressing high-affinity IL-2Rα (CD25), which hindered anti-tumor immunity. Several structural variants of IL-2 without binding to IL2Rα have been developed, for example, Bempegaldesleukin (BEMPEG: NKTR-214) [169] and Neoleukin-2 (Neo2/15) [170]. A study found that neoantigen vaccination alone was ineffective, BEMPEG monotherapy cured 20% of the mice, and the combination led to tumor regression in half mice. These may be related to the increased level of vaccine-induced T cells and CD8+ T cells infiltration, and the reduced population of Tregs caused by BEMPEG [169]. Furthermore, TLR played an essential role in anti-infective immune responses. CPG oligodeoxynucleotides, TLR9 agonists, were efficient vaccine adjuvants, promoting the secretion of IFN-γ, TNF-α, and IL-6, simultaneously activating the expression of T cells [171]. Thirdly, the immunosuppressive TME was a significant factor in the efficacy of neoantigen vaccines. TIM-3 antibody noticeably reduced Tregs in tumor tissue and enhanced levels of IFN-γ and IL-12P70. It promoted infiltration of CD8+ T cells and effectively suppressed the progression of HCC in situ with the combination of neoantigen vaccines [172]. Fourthly, treatment targeting epigenetic processes upregulated the expression of neoantigen and induced antigen-driven immune response, which provided a promising and appealing strategy for the efficacy enhancement of vaccines [173]. Fifthly, the efficacy of the vaccine response was influenced by the injection route. Subcutaneous injection significantly enhanced the ability of neoantigen vaccine delivery to lymph nodes and improved uptake of neoantigen. It stimulated neoantigen-specific T cell responses 7 times and 20 times as much as intramuscular and intravenous injection, respectively [174, 175]. Furthermore, the vaccination pathway affected the differentiation of neoantigen-specific CD8+ T cells. Tetramer staining showed that most neoantigen-specific CD8+ T cells were short-lived effector cells after subcutaneous injection, whereas a high proportion was primarily memory precursors effector cells after intravenous injection. Single-cell RNA sequencing showed that intravenous injection induced stem cell-like genes, while subcutaneous injection enriched effect genes. Therefore, intravenous injection had a better anti-tumor response after checkpoint blockade [175]. Besides, the development and differentiation of immune cells were linked to the intestinal flora. The decrease in the diversity of the microbiome on account of long-term antibiotic therapy produced a higher tumor-specific immune response, thereafter, performing a greater anti-tumor induced by neoantigen vaccines [176]. Finally, inflammatory regulation was a prospective strategy for improving the efficacy of neoantigen immunotherapy [177].
Some factors affect the efficacy of neoantigen vaccines.
Challenges
Despite the solid and enduring anti-tumor effect of personalized neoantigen vaccines in some patients, a series of open questions restricted their wide clinical application [134]. Firstly, thousands of heterologous gene mutations typically existed in tumor samples. However, only a few fulfilled the eligibility criteria of neoantigen. At present, effective screening methods are still lacking. Secondly, it was widely believed that neoantigen expression led to adaptive immunity and disease suppression. Surprisingly, neoantigen expression contributed to the deterioration of the fibroinflammatory microenvironment that resulted in progression and metastasis in pancreatic ductal adenocarcinoma, which was associated with pathogenic TH17 responses [178]. Thirdly, for patients with multifocal tumors, analysis of a single sample might inaccurately capture the complexity of the neoantigen pool. Intrahepatic metastasis (IM) or multicentric occurrence (MO) was a significant feature of HCC. Nonetheless, IM and MO had different genetic susceptibility, clonal structural, and mutation profiles [179]. Multi-region sequencing may provide more comprehensive information for neoantigen vaccine design. Fourthly, serious consideration of shared neoantigens was in need. Accumulation of mutations promoted tumorigenesis and ultimately led to tumor invasion and metastasis. Once a malignant tumor developed, a single cancer cell continued to acquire mutations, causing multiple different genomic maps within the tumor. Studies have found that neoantigens in both primary and metastatic tumors were similar in terms of the number and type. Nevertheless, the proportion of shared neoantigens remained modest [180]. Optimal therapeutic outcome was not provided by vaccines solely targeting primary tumor neoantigens. Fifthly, in vaccine development, various biometric assay technologies, from genome sequencing to the preparation of personalized neoantigen vaccines, were challenging and costly. These may limit the use of neoantigen vaccines in the population, and some patients may not receive ultimate treatment due to the long vaccine preparation cycle. Further, the frequency of vaccination and the interval of administration of neoantigen vaccines require clinical practice.
Conclusion
The immune system could attack cancer cells specifically and adapt to the evolving tumor and memory, making immunotherapy the fourth potent weapon in tumor control after surgery, radiotherapy, and chemotherapy. Neoantigens were mutated antigens expressed in tumor tissue specifically but did not exist in normal cells. Sequencing technologies improved the accuracy of neoantigen prediction and identification. Neoantigen vaccines effectively induced the production of tumor-specific T cells without killing normal cells. Syntactic peptides, DNA vaccines, RNA vaccines, and DC vaccines have shown reliable safety, tolerability, and immunogenicity in clinical trials. Whether used alone or in combination with other immunotherapies and conventional therapies, they have shown perfect anti-tumor effects, representing the frontier progress and prospect of cancer treatment. However, due to the loss of neoantigen, the barrier to neoantigen processing and presentation, and the influence of TME, some patients fail to achieve the expected effect of neoantigen vaccine therapy, which is a primary challenge for the application of neoantigen vaccines. Nevertheless, with a better understanding of neoantigens and tumor immunity, we have abundant reasons to believe that the future of neoantigen vaccines is bright in tumor immunotherapy.
Abbreviations
ACT: adoptive T cell therapy; AML: acute myeloid leukemia; APC: antigen presentation cell; CNN: convolutional neural network; DC: dendritic cell; ecDNA: extrachromosomal DNA; EGFR: epidermal growth factor receptor; EGFRvIII: EGFR variant III; HCC: hepatocellular carcinoma; HLA: human leukocyte antigen; ICIs: immune checkpoint inhibitors; IFN-γ: interferon-γ; IM: intrahepatic metastasis; Indels: insertions and deletions; IR: intron retention; K: lysine; MDSC: marrow-derived suppressor cells; M: methionine; MHC: major histocompatibility complex; MM: multiple myeloma; MO: multicentric occurrence; MS: mass spectrometry; MSI-H: microsatellite instability-high; NSCLC: non-small cell lung cancer; nsSNVs: non-synonymous SNVs; PFS: progression-free survival; PTT: photothermal therapy; R132: arginine 132; SNVs: single nucleotide variations; TACE: transcatheter arterial chemoembolization; TAP2: antigen peptide transporter-2; TCR: T cell receptor; Th1: T-helper-1; TILs: tumor-infiltrating lymphocytes; TIM-3: T-cell immunoglobulin and mucin-containing molecule 3; TLR: toll-like receptor; TMB: tumor mutational burden; TME: tumor microenvironment; TNBC: triple-negative breast cancer; TNF: tumor necrosis factor; Tregs: regulatory T cells; WES: whole exon sequencing.
Author contributions
ZQL, XWH, and QD provided direction and guidance throughout the preparation of this manuscript. JXL, ZQL, and QD wrote and edited the manuscript. QD reviewed and made significant revisions to the manuscript. LL, SYW, LBW, XYG, QD, ZKZ, YK, HYL, YLH, and ZQL collected and prepared the related papers. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Laversanne M, Weiderpass E, Soerjomataram I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer. 2021;127:3029-30
2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33
3. Penninckx S, Heuskin AC, Michiels C, Lucas S. Gold Nanoparticles as a Potent Radiosensitizer: A Transdisciplinary Approach from Physics to Patient. Cancers (Basel). 2020 12
4. Zhang J, Jin X, Zhou C, Zhao H, He P, Hao Y. et al. Resveratrol Suppresses Human Nasopharyngeal Carcinoma Cell Growth Via Inhibiting Differentiation Antagonizing Non-Protein Coding RNA (DANCR) Expression. Med Sci Monit. 2020;26:e923622
5. Li S, Huang XT, Wang MY, Chen DP, Li MY, Zhu YY. et al. FSCN1 Promotes Radiation Resistance in Patients With PIK3CA Gene Alteration. Front Oncol. 2021;11:653005
6. Caiado F, Maia-Silva D, Jardim C, Schmolka N, Carvalho T, Reforco C. et al. Lineage tracing of acute myeloid leukemia reveals the impact of hypomethylating agents on chemoresistance selection. Nat Commun. 2019;10:4986
7. Satoh Y, Kotani H, Iida Y, Taniura T, Notsu Y, Harada M. Supplementation of l-arginine boosts the therapeutic efficacy of anticancer chemoimmunotherapy. Cancer Sci. 2020;111:2248-58
8. Mateu-Sanz M, Tornin J, Brulin B, Khlyustova A, Ginebra MP, Layrolle P. et al. Cold Plasma-Treated Ringer's Saline: A Weapon to Target Osteosarcoma. Cancers (Basel). 2020 12
9. Baumann C, Ullrich A, Torka R. GAS6-expressing and self-sustaining cancer cells in 3D spheroids activate the PDK-RSK-mTOR pathway for survival and drug resistance. Mol Oncol. 2017;11:1430-47
10. Huang CW, Hsieh WC, Hsu ST, Lin YW, Chung YH, Chang WC. et al. The Use of PET Imaging for Prognostic Integrin alpha2beta1 Phenotyping to Detect Non-Small Cell Lung Cancer and Monitor Drug Resistance Responses. Theranostics. 2017;7:4013-28
11. Guo Z, Primeau T, Luo J, Zhang C, Sun H, Hoog J. et al. Proteomic Resistance Biomarkers for PI3K Inhibitor in Triple Negative Breast Cancer Patient-Derived Xenograft Models. Cancers (Basel). 2020 12
12. Bagchi S, Yuan R, Engleman EG. Immune Checkpoint Inhibitors for the Treatment of Cancer: Clinical Impact and Mechanisms of Response and Resistance. Annu Rev Pathol. 2021;16:223-49
13. Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1-10
14. Schumacher TN, Scheper W, Kvistborg P. Cancer Neoantigens. Annu Rev Immunol. 2019;37:173-200
15. Yarchoan M, Johnson BA 3rd, Lutz ER, Laheru DA, Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer. 2017;17:209-22
16. Wang G, Kang X, Chen KS, Jehng T, Jones L, Chen J. et al. An engineered oncolytic virus expressing PD-L1 inhibitors activates tumor neoantigen-specific T cell responses. Nat Commun. 2020;11:1395
17. Balachandran VP, Luksza M, Zhao JN, Makarov V, Moral JA, Remark R. et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551:512-6
18. Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ. et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547:217-21
19. Zhang W, Yin Q, Huang H, Lu J, Qin H, Chen S. et al. Personal Neoantigens From Patients With NSCLC Induce Efficient Antitumor Responses. Front Oncol. 2021;11:628456
20. Zhong H, Liu S, Cao F, Zhao Y, Zhou J, Tang F. et al. Dissecting Tumor Antigens and Immune Subtypes of Glioma to Develop mRNA Vaccine. Front Immunol. 2021;12:709986
21. Tanyi JL, Bobisse S, Ophir E, Tuyaerts S, Roberti A, Genolet R. et al. Personalized cancer vaccine effectively mobilizes antitumor T cell immunity in ovarian cancer. Sci Transl Med. 2018 10
22. Duan C, Huan Q, Chen X, Wu S, Carey LB, He X. et al. Reduced intrinsic DNA curvature leads to increased mutation rate. Genome Biol. 2018;19:132
23. Parkhurst MR, Robbins PF, Tran E, Prickett TD, Gartner JJ, Jia L. et al. Unique Neoantigens Arise from Somatic Mutations in Patients with Gastrointestinal Cancers. Cancer Discov. 2019;9:1022-35
24. Kelley RK, Bridgewater J, Gores GJ, Zhu AX. Systemic therapies for intrahepatic cholangiocarcinoma. J Hepatol. 2020;72:353-63
25. Schumacher T, Bunse L, Pusch S, Sahm F, Wiestler B, Quandt J. et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512:324-7
26. Tatavosian R, Duc HN, Huynh TN, Fang D, Schmitt B, Shi X. et al. Live-cell single-molecule dynamics of PcG proteins imposed by the DIPG H3.3K27M mutation. Nat Commun. 2018;9:2080
27. Chheda ZS, Kohanbash G, Okada K, Jahan N, Sidney J, Pecoraro M. et al. Novel and shared neoantigen derived from histone 3 variant H3.3K27M mutation for glioma T cell therapy. J Exp Med. 2018;215:141-57
28. Reparaz D, Ruiz M, Llopiz D, Silva L, Vercher E, Aparicio B. et al. Neoantigens as potential vaccines in hepatocellular carcinoma. J Immunother Cancer. 2022 10
29. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV. et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415-21
30. Turajlic S, Litchfield K, Xu H, Rosenthal R, McGranahan N, Reading JL. et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 2017;18:1009-21
31. Sigismund S, Avanzato D, Lanzetti L. Emerging functions of the EGFR in cancer. Mol Oncol. 2018;12:3-20
32. Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci U S A. 1990;87:8602-6
33. Purev E, Cai D, Miller E, Swoboda R, Mayer T, Klein-Szanto A. et al. Immune responses of breast cancer patients to mutated epidermal growth factor receptor (EGF-RvIII, Delta EGF-R, and de2-7 EGF-R). J Immunol. 2004;173:6472-80
34. Roudko V, Bozkus CC, Orfanelli T, McClain CB, Carr C, O'Donnell T. et al. Shared Immunogenic Poly-Epitope Frameshift Mutations in Microsatellite Unstable Tumors. Cell. 2020;183:1634-49 e17
35. Chae YK, Viveiros P, Lopes G, Sukhadia B, Sheikh MM, Saravia D. et al. Clinical and Immunological Implications of Frameshift Mutations in Lung Cancer. J Thorac Oncol. 2019;14:1807-17
36. Yang W, Lee KW, Srivastava RM, Kuo F, Krishna C, Chowell D. et al. Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nat Med. 2019;25:767-75
37. Hindson J. Gene-fusion neoantigens stimulate T cells. Nat Rev Cancer. 2019;19:364
38. Biernacki MA, Foster KA, Woodward KB, Coon ME, Cummings C, Cunningham TM. et al. CBFB-MYH11 fusion neoantigen enables T cell recognition and killing of acute myeloid leukemia. J Clin Invest. 2020;130:5127-41
39. Ishii M, Ando J, Yamazaki S, Toyota T, Ohara K, Furukawa Y. et al. iPSC-Derived Neoantigen-Specific CTL Therapy for Ewing Sarcoma. Cancer Immunol Res. 2021;9:1175-86
40. Gao Q, Liang WW, Foltz SM, Mutharasu G, Jayasinghe RG, Cao S. et al. Driver Fusions and Their Implications in the Development and Treatment of Human Cancers. Cell Rep. 2018;23:227-38 e3
41. Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457-63
42. Lu SX, De Neef E, Thomas JD, Sabio E, Rousseau B, Gigoux M. et al. Pharmacologic modulation of RNA splicing enhances anti-tumor immunity. Cell. 2021;184:4032-47 e31
43. Dong C, Cesarano A, Bombaci G, Reiter JL, Yu CY, Wang Y. et al. Intron retention-induced neoantigen load correlates with unfavorable prognosis in multiple myeloma. Oncogene. 2021;40:6130-8
44. Smith CC, Selitsky SR, Chai S, Armistead PM, Vincent BG, Serody JS. Alternative tumour-specific antigens. Nat Rev Cancer. 2019;19:465-78
45. Scherer HU, Huizinga TWJ, Kronke G, Schett G, Toes REM. The B cell response to citrullinated antigens in the development of rheumatoid arthritis. Nat Rev Rheumatol. 2018;14:157-69
46. Margulies DH. How MHC molecules grab citrullinated peptides to foster rheumatoid arthritis. J Biol Chem. 2018;293:3252-3
47. Brentville VA, Metheringham RL, Gunn B, Symonds P, Daniels I, Gijon M. et al. Citrullinated Vimentin Presented on MHC-II in Tumor Cells Is a Target for CD4+ T-Cell-Mediated Antitumor Immunity. Cancer Res. 2016;76:548-60
48. Cook K, Daniels I, Symonds P, Pitt T, Gijon M, Xue W. et al. Citrullinated alpha-enolase is an effective target for anti-cancer immunity. Oncoimmunology. 2018;7:e1390642
49. Katayama H, Kobayashi M, Irajizad E, Sevillarno A, Patel N, Mao X. et al. Protein citrullination as a source of cancer neoantigens. J Immunother Cancer. 2021 9
50. Zhu G, Zhang F, Ni Q, Niu G, Chen X. Efficient Nanovaccine Delivery in Cancer Immunotherapy. ACS Nano. 2017;11:2387-92
51. Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Lower M. et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222-6
52. Hashimoto S, Noguchi E, Bando H, Miyadera H, Morii W, Nakamura T. et al. Neoantigen prediction in human breast cancer using RNA sequencing data. Cancer Sci. 2021;112:465-75
53. Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J Immunol. 2017;199:3360-8
54. Lundegaard C, Lund O, Kesmir C, Brunak S, Nielsen M. Modeling the adaptive immune system: predictions and simulations. Bioinformatics. 2007;23:3265-75
55. Nielsen M, Lund O, Buus S, Lundegaard C. MHC class II epitope predictive algorithms. Immunology. 2010;130:319-28
56. Lang F, Schrors B, Lower M, Tureci O, Sahin U. Identification of neoantigens for individualized therapeutic cancer vaccines. Nat Rev Drug Discov. 2022;21:261-82
57. Olsson N, Heberling ML, Zhang L, Jhunjhunwala S, Phung QT, Lin S. et al. An Integrated Genomic, Proteomic, and Immunopeptidomic Approach to Discover Treatment-Induced Neoantigens. Front Immunol. 2021;12:662443
58. Bulik-Sullivan B, Busby J, Palmer CD, Davis MJ, Murphy T, Clark A. et al. Deep learning using tumor HLA peptide mass spectrometry datasets improves neoantigen identification. Nat Biotechnol. 2018
59. Muller M, Gfeller D, Coukos G, Bassani-Sternberg M. 'Hotspots' of Antigen Presentation Revealed by Human Leukocyte Antigen Ligandomics for Neoantigen Prioritization. Front Immunol. 2017;8:1367
60. Cobbold M, De La Pena H, Norris A, Polefrone JM, Qian J, English AM. et al. MHC class I-associated phosphopeptides are the targets of memory-like immunity in leukemia. Sci Transl Med. 2013;5:203ra125
61. Malaker SA, Ferracane MJ, Depontieu FR, Zarling AL, Shabanowitz J, Bai DL. et al. Identification and Characterization of Complex Glycosylated Peptides Presented by the MHC Class II Processing Pathway in Melanoma. J Proteome Res. 2017;16:228-37
62. Zaidi N, Soban M, Chen F, Kinkead H, Mathew J, Yarchoan M. et al. Role of in silico structural modeling in predicting immunogenic neoepitopes for cancer vaccine development. JCI Insight. 2020 5
63. Vang YS, Xie X. HLA class I binding prediction via convolutional neural networks. Bioinformatics. 2017;33:2658-65
64. Hao Q, Wei P, Shu Y, Zhang YG, Xu H, Zhao JN. Improvement of Neoantigen Identification Through Convolution Neural Network. Front Immunol. 2021;12:682103
65. Hundal J, Kiwala S, Feng YY, Liu CJ, Govindan R, Chapman WC. et al. Accounting for proximal variants improves neoantigen prediction. Nat Genet. 2019;51:175-9
66. Lancaster EM, Jablons D, Kratz JR. Applications of Next-Generation Sequencing in Neoantigen Prediction and Cancer Vaccine Development. Genet Test Mol Biomarkers. 2020;24:59-66
67. Li L, Goedegebuure SP, Gillanders WE. Preclinical and clinical development of neoantigen vaccines. Ann Oncol. 2017;28:xii11-xii7
68. Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443-73
69. Cruz FM, Colbert JD, Merino E, Kriegsman BA, Rock KL. The Biology and Underlying Mechanisms of Cross-Presentation of Exogenous Antigens on MHC-I Molecules. Annu Rev Immunol. 2017;35:149-76
70. Bekri S, Rodney-Sandy R, Gruenstein D, Mei A, Bogen B, Castle J. et al. Neoantigen vaccine-induced CD4 T cells confer protective immunity in a mouse model of multiple myeloma through activation of CD8 T cells against non-vaccine, tumor-associated antigens. J Immunother Cancer. 2022 10
71. Bunse L, Pusch S, Bunse T, Sahm F, Sanghvi K, Friedrich M. et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat Med. 2018;24:1192-203
72. Vergati M, Intrivici C, Huen NY, Schlom J, Tsang KY. Strategies for cancer vaccine development. J Biomed Biotechnol. 2010. 2010
73. Wang X, Li X, Yoshiyuki K, Watanabe Y, Sogo Y, Ohno T. et al. Cancer Immunotherapy: Comprehensive Mechanism Analysis of Mesoporous-Silica-Nanoparticle-Induced Cancer Immunotherapy (Adv. Healthcare Mater. 10/2016). Adv Healthc Mater. 2016;5:1246
74. Lopes A, Vandermeulen G, Preat V. Cancer DNA vaccines: current preclinical and clinical developments and future perspectives. J Exp Clin Cancer Res. 2019;38:146
75. Faurez F, Dory D, Le Moigne V, Gravier R, Jestin A. Biosafety of DNA vaccines: New generation of DNA vectors and current knowledge on the fate of plasmids after injection. Vaccine. 2010;28:3888-95
76. Tondini E, Arakelian T, Oosterhuis K, Camps M, van Duikeren S, Han W. et al. A poly-neoantigen DNA vaccine synergizes with PD-1 blockade to induce T cell-mediated tumor control. Oncoimmunology. 2019;8:1652539
77. Maruggi G, Zhang C, Li J, Ulmer JB, Yu D. mRNA as a Transformative Technology for Vaccine Development to Control Infectious Diseases. Mol Ther. 2019;27:757-72
78. Zhang R, Wu F, Wu L, Tian Y, Zhou B, Zhang X. et al. Novel Self-Assembled Micelles Based on Cholesterol-Modified Antimicrobial Peptide (DP7) for Safe and Effective Systemic Administration in Animal Models of Bacterial Infection. Antimicrob Agents Chemother. 2018 62
79. Zhang R, Tang L, Tian Y, Ji X, Hu Q, Zhou B. et al. Cholesterol-modified DP7 enhances the effect of individualized cancer immunotherapy based on neoantigens. Biomaterials. 2020;241:119852
80. Zhang R, Tang L, Tian Y, Ji X, Hu Q, Zhou B. et al. DP7-C-modified liposomes enhance immune responses and the antitumor effect of a neoantigen-based mRNA vaccine. J Control Release. 2020;328:210-21
81. Bol KF, Schreibelt G, Gerritsen WR, de Vries IJ, Figdor CG. Dendritic Cell-Based Immunotherapy: State of the Art and Beyond. Clin Cancer Res. 2016;22:1897-906
82. Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccines. Nat Med. 2004;10:909-15
83. Goldman B, DeFrancesco L. The cancer vaccine roller coaster. Nat Biotechnol. 2009;27:129-39
84. Zhang R, Yuan F, Shu Y, Tian Y, Zhou B, Yi L. et al. Personalized neoantigen-pulsed dendritic cell vaccines show superior immunogenicity to neoantigen-adjuvant vaccines in mouse tumor models. Cancer Immunol Immunother. 2020;69:135-45
85. Zheng Y, Fu Y, Wang PP, Ding ZY. Neoantigen: A Promising Target for the Immunotherapy of Colorectal Cancer. Dis Markers. 2022;2022:8270305
86. Lee CH, Yelensky R, Jooss K, Chan TA. Update on Tumor Neoantigens and Their Utility: Why It Is Good to Be Different. Trends Immunol. 2018;39:536-48
87. Carretero-Iglesia L, Couturaud B, Baumgaertner P, Schmidt J, Maby-El Hajjami H, Speiser DE. et al. High Peptide Dose Vaccination Promotes the Early Selection of Tumor Antigen-Specific CD8 T-Cells of Enhanced Functional Competence. Front Immunol. 2019;10:3016
88. Gordon CL, Lee LN, Swadling L, Hutchings C, Zinser M, Highton AJ. et al. Induction and Maintenance of CX3CR1-Intermediate Peripheral Memory CD8(+) T Cells by Persistent Viruses and Vaccines. Cell Rep. 2018;23:768-82
89. Gerlach C, Moseman EA, Loughhead SM, Alvarez D, Zwijnenburg AJ, Waanders L. et al. The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity. 2016;45:1270-84
90. Yamauchi T, Hoki T, Oba T, Kajihara R, Attwood K, Cao X. et al. CD40 and CD80/86 signaling in cDC1s mediate effective neoantigen vaccination and generation of antigen-specific CX3CR1(+) CD8(+) T cells. Cancer Immunol Immunother. 2022;71:137-51
91. Swartz AM, Congdon KL, Nair SK, Li QJ, Herndon JE 2nd, Suryadevara CM. et al. A conjoined universal helper epitope can unveil antitumor effects of a neoantigen vaccine targeting an MHC class I-restricted neoepitope. NPJ Vaccines. 2021;6:12
92. Cui C, Wang J, Fagerberg E, Chen PM, Connolly KA, Damo M. et al. Neoantigen-driven B cell and CD4 T follicular helper cell collaboration promotes anti-tumor CD8 T cell responses. Cell. 2021;184:6101-18 e13
93. Bogen B, Munthe L, Sollien A, Hofgaard P, Omholt H, Dagnaes F. et al. Naive CD4+ T cells confer idiotype-specific tumor resistance in the absence of antibodies. Eur J Immunol. 1995;25:3079-86
94. Lauritzsen GF, Weiss S, Dembic Z, Bogen B. Naive idiotype-specific CD4+ T cells and immunosurveillance of B-cell tumors. Proc Natl Acad Sci U S A. 1994;91:5700-4
95. Bogen B, Fauskanger M, Haabeth OA, Tveita A. CD4(+) T cells indirectly kill tumor cells via induction of cytotoxic macrophages in mouse models. Cancer Immunol Immunother. 2019;68:1865-73
96. Ahrends T, Busselaar J, Severson TM, Babala N, de Vries E, Bovens A. et al. CD4(+) T cell help creates memory CD8(+) T cells with innate and help-independent recall capacities. Nat Commun. 2019;10:5531
97. Atif SM, Gibbings SL, Redente EF, Camp FA, Torres RM, Kedl RM. et al. Immune Surveillance by Natural IgM Is Required for Early Neoantigen Recognition and Initiation of Adaptive Immunity. Am J Respir Cell Mol Biol. 2018;59:580-91
98. Ahrends T, Spanjaard A, Pilzecker B, Babala N, Bovens A, Xiao Y. et al. CD4(+) T Cell Help Confers a Cytotoxic T Cell Effector Program Including Coinhibitory Receptor Downregulation and Increased Tissue Invasiveness. Immunity. 2017;47:848-61 e5
99. Hartl CA, Bertschi A, Puerto RB, Andresen C, Cheney EM, Mittendorf EA. et al. Combination therapy targeting both innate and adaptive immunity improves survival in a pre-clinical model of ovarian cancer. J Immunother Cancer. 2019;7:199
100. Keskin DB, Anandappa AJ, Sun J, Tirosh I, Mathewson ND, Li S. et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565:234-9
101. Hu Z, Leet DE, Allesoe RL, Oliveira G, Li S, Luoma AM. et al. Personal neoantigen vaccines induce persistent memory T cell responses and epitope spreading in patients with melanoma. Nat Med. 2021;27:515-25
102. Nejo T, Matsushita H, Karasaki T, Nomura M, Saito K, Tanaka S. et al. Reduced Neoantigen Expression Revealed by Longitudinal Multiomics as a Possible Immune Evasion Mechanism in Glioma. Cancer Immunol Res. 2019;7:1148-61
103. Rosenthal R, Cadieux EL, Salgado R, Bakir MA, Moore DA, Hiley CT. et al. Neoantigen-directed immune escape in lung cancer evolution. Nature. 2019;567:479-85
104. Yi M, Dong B, Chu Q, Wu K. Immune pressures drive the promoter hypermethylation of neoantigen genes. Exp Hematol Oncol. 2019;8:32
105. Wessolly M, Stephan-Falkenau S, Streubel A, Werner R, Borchert S, Griff S. et al. A Novel Epitope Quality-Based Immune Escape Mechanism Reveals Patient's Suitability for Immune Checkpoint Inhibition. Cancer Manag Res. 2020;12:7881-90
106. Wessolly M, Mairinger FD, Herold T, Hadaschik B, Szarvas T, Reis H. Proteasomal Processing Immune Escape Mechanisms in Platinum-Treated Advanced Bladder Cancer. Genes (Basel). 2022 13
107. Montesion M, Murugesan K, Jin DX, Sharaf R, Sanchez N, Guria A. et al. Somatic HLA Class I Loss Is a Widespread Mechanism of Immune Evasion Which Refines the Use of Tumor Mutational Burden as a Biomarker of Checkpoint Inhibitor Response. Cancer Discov. 2021;11:282-92
108. Schrors B, Lubcke S, Lennerz V, Fatho M, Bicker A, Wolfel C. et al. HLA class I loss in metachronous metastases prevents continuous T cell recognition of mutated neoantigens in a human melanoma model. Oncotarget. 2017;8:28312-27
109. Iwasaki A, Shinozaki-Ushiku A, Kunita A, Yamazawa S, Sato Y, Yamashita H. et al. Human Leukocyte Antigen Class I Deficiency in Gastric Carcinoma: An Adaptive Immune Evasion Strategy Most Common in Microsatellite Instable Tumors. Am J Surg Pathol. 2021;45:1213-20
110. Amodio V, Mauri G, Reilly NM, Sartore-Bianchi A, Siena S, Bardelli A. et al. Mechanisms of Immune Escape and Resistance to Checkpoint Inhibitor Therapies in Mismatch Repair Deficient Metastatic Colorectal Cancers. Cancers (Basel). 2021 13
111. Germano G, Lu S, Rospo G, Lamba S, Rousseau B, Fanelli S. et al. CD4 T Cell-Dependent Rejection of Beta-2 Microglobulin Null Mismatch Repair-Deficient Tumors. Cancer Discov. 2021;11:1844-59
112. Wu T, Wu C, Zhao X, Wang G, Ning W, Tao Z. et al. Extrachromosomal DNA formation enables tumor immune escape potentially through regulating antigen presentation gene expression. Sci Rep. 2022;12:3590
113. Diamond MS, Lin JH, Vonderheide RH. Site-Dependent Immune Escape Due to Impaired Dendritic Cell Cross-Priming. Cancer Immunol Res. 2021;9:877-90
114. McLane LM, Abdel-Hakeem MS, Wherry EJ. CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu Rev Immunol. 2019;37:457-95
115. Thommen DS, Schumacher TN. T Cell Dysfunction in Cancer. Cancer Cell. 2018;33:547-62
116. Kieffer Y, Hocine HR, Gentric G, Pelon F, Bernard C, Bourachot B. et al. Single-Cell Analysis Reveals Fibroblast Clusters Linked to Immunotherapy Resistance in Cancer. Cancer Discov. 2020;10:1330-51
117. Wang QW, Sun LH, Zhang Y, Wang Z, Zhao Z, Wang ZL. et al. MET overexpression contributes to STAT4-PD-L1 signaling activation associated with tumor-associated, macrophages-mediated immunosuppression in primary glioblastomas. J Immunother Cancer. 2021 9
118. Galdiero MR, Varricchi G, Seaf M, Marone G, Levi-Schaffer F, Marone G. Bidirectional Mast Cell-Eosinophil Interactions in Inflammatory Disorders and Cancer. Front Med (Lausanne). 2017;4:103
119. Wang Z, He L, Li W, Xu C, Zhang J, Wang D. et al. GDF15 induces immunosuppression via CD48 on regulatory T cells in hepatocellular carcinoma. J Immunother Cancer. 2021 9
120. Yang Z, Guo J, Weng L, Tang W, Jin S, Ma W. Myeloid-derived suppressor cells-new and exciting players in lung cancer. J Hematol Oncol. 2020;13:10
121. Yang M, Zhang G, Wang Y, He M, Xu Q, Lu J. et al. Tumour-associated neutrophils orchestrate intratumoural IL-8-driven immune evasion through Jagged2 activation in ovarian cancer. Br J Cancer. 2020;123:1404-16
122. Kwantwi LB, Wang S, Zhang W, Peng W, Cai Z, Sheng Y. et al. Tumor-associated neutrophils activated by tumor-derived CCL20 (C-C motif chemokine ligand 20) promote T cell immunosuppression via programmed death-ligand 1 (PD-L1) in breast cancer. Bioengineered. 2021;12:6996-7006
123. Prendergast GC, Malachowski WJ, Mondal A, Scherle P, Muller AJ. Indoleamine 2,3-Dioxygenase and Its Therapeutic Inhibition in Cancer. Int Rev Cell Mol Biol. 2018;336:175-203
124. Wang W, Marinis JM, Beal AM, Savadkar S, Wu Y, Khan M. et al. RIP1 Kinase Drives Macrophage-Mediated Adaptive Immune Tolerance in Pancreatic Cancer. Cancer Cell. 2020;38:585-90
125. Arlauckas SP, Garren SB, Garris CS, Kohler RH, Oh J, Pittet MJ. et al. Arg1 expression defines immunosuppressive subsets of tumor-associated macrophages. Theranostics. 2018;8:5842-54
126. Jing B, Wang T, Sun B, Xu J, Xu D, Liao Y. et al. IL6/STAT3 Signaling Orchestrates Premetastatic Niche Formation and Immunosuppressive Traits in Lung. Cancer Res. 2020;80:784-97
127. Wang Y, Tan J, Hu P, Pei Q, Wen Y, Ma W. et al. Traditional Chinese medicine compound, Bu Sheng Hui Yang Fang, promotes the proliferation of lymphocytes in the immunosuppressed mice potentially by upregulating IL-4 signaling. Biomed Pharmacother. 2021;134:111107
128. Kondo Y, Suzuki S, Takahara T, Ono S, Goto M, Miyabe S. et al. Improving function of cytotoxic T-lymphocytes by transforming growth factor-beta inhibitor in oral squamous cell carcinoma. Cancer Sci. 2021;112:4037-49
129. Cai Z, Su X, Qiu L, Li Z, Li X, Dong X. et al. Personalized neoantigen vaccine prevents postoperative recurrence in hepatocellular carcinoma patients with vascular invasion. Mol Cancer. 2021;20:164
130. Fang Y, Mo F, Shou J, Wang H, Luo K, Zhang S. et al. A Pan-cancer Clinical Study of Personalized Neoantigen Vaccine Monotherapy in Treating Patients with Various Types of Advanced Solid Tumors. Clin Cancer Res. 2020;26:4511-20
131. Tiriveedhi V, Tucker N, Herndon J, Li L, Sturmoski M, Ellis M. et al. Safety and preliminary evidence of biologic efficacy of a mammaglobin-a DNA vaccine in patients with stable metastatic breast cancer. Clin Cancer Res. 2014;20:5964-75
132. Trimble CL, Morrow MP, Kraynyak KA, Shen X, Dallas M, Yan J. et al. Safety, efficacy, and immunogenicity of VGX-3100, a therapeutic synthetic DNA vaccine targeting human papillomavirus 16 and 18 E6 and E7 proteins for cervical intraepithelial neoplasia 2/3: a randomised, double-blind, placebo-controlled phase 2b trial. Lancet. 2015;386:2078-88
133. Diaz CM, Chiappori A, Aurisicchio L, Bagchi A, Clark J, Dubey S. et al. Phase 1 studies of the safety and immunogenicity of electroporated HER2/CEA DNA vaccine followed by adenoviral boost immunization in patients with solid tumors. J Transl Med. 2013;11:62
134. Supabphol S, Li L, Goedegebuure SP, Gillanders WE. Neoantigen vaccine platforms in clinical development: understanding the future of personalized immunotherapy. Expert Opin Investig Drugs. 2021;30:529-41
135. Cafri G, Gartner JJ, Zaks T, Hopson K, Levin N, Paria BC. et al. mRNA vaccine-induced neoantigen-specific T cell immunity in patients with gastrointestinal cancer. J Clin Invest. 2020;130:5976-88
136. Zhao X, Pan X, Wang Y, Zhang Y. Targeting neoantigens for cancer immunotherapy. Biomark Res. 2021;9:61
137. Carreno BM, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, Petti AA. et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348:803-8
138. Ding Z, Li Q, Zhang R, Xie L, Shu Y, Gao S. et al. Personalized neoantigen pulsed dendritic cell vaccine for advanced lung cancer. Signal Transduct Target Ther. 2021;6:26
139. Angelicola S, Ruzzi F, Landuzzi L, Scalambra L, Gelsomino F, Ardizzoni A. et al. IFN-gamma and CD38 in Hyperprogressive Cancer Development. Cancers (Basel). 2021 13
140. Oh S, Lee JH, Kwack K, Choi SW. Natural Killer Cell Therapy: A New Treatment Paradigm for Solid Tumors. Cancers (Basel). 2019 11
141. Shaheen S, Mirshahidi H, Nagaraj G, Hsueh CT. Conservative management of nivolumab-induced pericardial effusion: a case report and review of literature. Exp Hematol Oncol. 2018;7:11
142. Podojil JR, Glaser AP, Baker D, Courtois ET, Fantini D, Yu Y. et al. Antibody targeting of B7-H4 enhances the immune response in urothelial carcinoma. Oncoimmunology. 2020;9:1744897
143. Kolb R, De U, Khan S, Luo Y, Kim MC, Yu H. et al. Proteolysis-targeting chimera against BCL-XL destroys tumor-infiltrating regulatory T cells. Nat Commun. 2021;12:1281
144. Bertolini F. Desperately seeking..Models to find the right partner and the best use for checkpoint inhibitors. Br J Cancer. 2019;120:139-40
145. D'Alise AM, Leoni G, Cotugno G, Troise F, Langone F, Fichera I. et al. Adenoviral vaccine targeting multiple neoantigens as strategy to eradicate large tumors combined with checkpoint blockade. Nat Commun. 2019;10:2688
146. Fehlings M, Simoni Y, Penny HL, Becht E, Loh CY, Gubin MM. et al. Checkpoint blockade immunotherapy reshapes the high-dimensional phenotypic heterogeneity of murine intratumoural neoantigen-specific CD8(+) T cells. Nat Commun. 2017;8:562
147. Salvatori E, Lione L, Compagnone M, Pinto E, Conforti A, Ciliberto G. et al. Neoantigen cancer vaccine augments anti-CTLA-4 efficacy. NPJ Vaccines. 2022;7:15
148. Ott PA, Hu-Lieskovan S, Chmielowski B, Govindan R, Naing A, Bhardwaj N. et al. A Phase Ib Trial of Personalized Neoantigen Therapy Plus Anti-PD-1 in Patients with Advanced Melanoma, Non-small Cell Lung Cancer, or Bladder Cancer. Cell. 2020;183:347-62 e24
149. Blass E, Ott PA. Advances in the development of personalized neoantigen-based therapeutic cancer vaccines. Nat Rev Clin Oncol. 2021;18:215-29
150. Tanaka TN, Ferrari V, Tarke A, Fields H, Ferrari L, Ferrari F. et al. Adoptive transfer of neoantigen-specific T-cell therapy is feasible in older patients with higher-risk myelodysplastic syndrome. Cytotherapy. 2021;23:236-41
151. Kristensen NP, Heeke C, Tvingsholm SA, Borch A, Draghi A, Crowther MD. et al. Neoantigen-reactive CD8+ T cells affect clinical outcome of adoptive cell therapy with tumor-infiltrating lymphocytes in melanoma. J Clin Invest. 2022 132
152. Galluzzi L, Buque A, Kepp O, Zitvogel L, Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol. 2017;17:97-111
153. O'Donnell T, Christie EL, Ahuja A, Buros J, Aksoy BA, Bowtell DDL. et al. Chemotherapy weakly contributes to predicted neoantigen expression in ovarian cancer. BMC Cancer. 2018;18:87
154. Lussier DM, Alspach E, Ward JP, Miceli AP, Runci D, White JM. et al. Radiation-induced neoantigens broaden the immunotherapeutic window of cancers with low mutational loads. Proc Natl Acad Sci U S A. 2021 118
155. Mahmood J, Shukla HD, Soman S, Samanta S, Singh P, Kamlapurkar S. et al. Immunotherapy, Radiotherapy, and Hyperthermia: A Combined Therapeutic Approach in Pancreatic Cancer Treatment. Cancers (Basel). 2018 10
156. Shi J, Li J, Xu Z, Chen L, Luo R, Zhang C. et al. Celastrol: A Review of Useful Strategies Overcoming its Limitation in Anticancer Application. Front Pharmacol. 2020;11:558741
157. Gao J, Yuan X, Yuan J, Li L. Complete rejection of large established breast cancer by local immunochemotherapy with T cell activation against neoantigens. Cancer Immunol Immunother. 2021;70:3291-302
158. Lhuillier C, Rudqvist NP, Yamazaki T, Zhang T, Charpentier M, Galluzzi L. et al. Radiotherapy-exposed CD8+ and CD4+ neoantigens enhance tumor control. J Clin Invest. 2021 131
159. Nam J, Son S, Park KS, Moon JJ. Photothermal therapy combined with neoantigen cancer vaccination for effective immunotherapy against large established tumors and distant metastasis. Adv Ther (Weinh). 2021 4
160. Li W, Jing Z, Wang S, Li Q, Xing Y, Shi H. et al. P22 virus-like particles as an effective antigen delivery nanoplatform for cancer immunotherapy. Biomaterials. 2021;271:120726
161. Hyun J, Jun S, Lim H, Cho H, You SH, Ha SJ. et al. Engineered Attenuated Salmonella typhimurium Expressing Neoantigen Has Anticancer Effects. ACS Synth Biol. 2021;10:2478-87
162. Liang Z, Cui X, Yang L, Hu Q, Li D, Zhang X. et al. Co-assembled nanocomplexes of peptide neoantigen Adpgk and Toll-like receptor 9 agonist CpG ODN for efficient colorectal cancer immunotherapy. Int J Pharm. 2021;608:121091
163. Ni Q, Zhang F, Liu Y, Wang Z, Yu G, Liang B. et al. A bi-adjuvant nanovaccine that potentiates immunogenicity of neoantigen for combination immunotherapy of colorectal cancer. Sci Adv. 2020;6:eaaw6071
164. Delitto D, Zabransky DJ, Chen F, Thompson ED, Zimmerman JW, Armstrong TD. et al. Implantation of a neoantigen-targeted hydrogel vaccine prevents recurrence of pancreatic adenocarcinoma after incomplete resection. Oncoimmunology. 2021;10:2001159
165. Newey A, Griffiths B, Michaux J, Pak HS, Stevenson BJ, Woolston A. et al. Immunopeptidomics of colorectal cancer organoids reveals a sparse HLA class I neoantigen landscape and no increase in neoantigens with interferon or MEK-inhibitor treatment. J Immunother Cancer. 2019;7:309
166. Shae D, Baljon JJ, Wehbe M, Christov PP, Becker KW, Kumar A. et al. Co-delivery of Peptide Neoantigens and Stimulator of Interferon Genes Agonists Enhances Response to Cancer Vaccines. ACS Nano. 2020;14:9904-16
167. Kinkead HL, Hopkins A, Lutz E, Wu AA, Yarchoan M, Cruz K. et al. Combining STING-based neoantigen-targeted vaccine with checkpoint modulators enhances antitumor immunity in murine pancreatic cancer. JCI Insight. 2018 3
168. Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE. et al. Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell Rep. 2015;11:1018-30
169. D'Alise AM, Leoni G, De Lucia M, Langone F, Nocchi L, Tucci FG. et al. Maximizing cancer therapy via complementary mechanisms of immune activation: PD-1 blockade, neoantigen vaccination, and Tregs depletion. J Immunother Cancer. 2021 9
170. Silva DA, Yu S, Ulge UY, Spangler JB, Jude KM, Labao-Almeida C. et al. De novo design of potent and selective mimics of IL-2 and IL-15. Nature. 2019;565:186-91
171. Li Q, Ren J, Liu W, Jiang G, Hu R. CpG Oligodeoxynucleotide Developed to Activate Primate Immune Responses Promotes Antitumoral Effects in Combination with a Neoantigen-Based mRNA Cancer Vaccine. Drug Des Devel Ther. 2021;15:3953-63
172. Zhao Q, Wang Y, Zhao B, Chen H, Cai Z, Zheng Y. et al. Neoantigen Immunotherapeutic-Gel Combined with TIM-3 Blockade Effectively Restrains Orthotopic Hepatocellular Carcinoma Progression. Nano Lett. 2022;22:2048-58
173. Truong AS, Zhou M, Krishnan B, Utsumi T, Manocha U, Stewart KG. et al. Entinostat induces antitumor immune responses through immune editing of tumor neoantigens. J Clin Invest. 2021 131
174. Kuai R, Sun X, Yuan W, Xu Y, Schwendeman A, Moon JJ. Subcutaneous Nanodisc Vaccination with Neoantigens for Combination Cancer Immunotherapy. Bioconjug Chem. 2018;29:771-5
175. Baharom F, Ramirez-Valdez RA, Tobin KKS, Yamane H, Dutertre CA, Khalilnezhad A. et al. Intravenous nanoparticle vaccination generates stem-like TCF1(+) neoantigen-specific CD8(+) T cells. Nat Immunol. 2021;22:41-52
176. Lione L, Salvatori E, Petrazzuolo A, Massacci A, Maggio R, Confroti A. et al. Antitumor efficacy of a neoantigen cancer vaccine delivered by electroporation is influenced by microbiota composition. Oncoimmunology. 2021;10:1898832
177. Wang C, Ding Y, Liu Y, Zhang Q, Xu S, Xia L. et al. Identification of Mutated Peptides in Bladder Cancer From Exomic Sequencing Data Reveals Negative Correlation Between Mutation-Specific Immunoreactivity and Inflammation. Front Immunol. 2020;11:576603
178. Hegde S, Krisnawan VE, Herzog BH, Zuo C, Breden MA, Knolhoff BL. et al. Dendritic Cell Paucity Leads to Dysfunctional Immune Surveillance in Pancreatic Cancer. Cancer Cell. 2020;37:289-307 e9
179. Dong LQ, Peng LH, Ma LJ, Liu DB, Zhang S, Luo SZ. et al. Heterogeneous immunogenomic features and distinct escape mechanisms in multifocal hepatocellular carcinoma. J Hepatol. 2020;72:896-908
180. Jiang T, Cheng R, Pan Y, Zhang H, He Y, Su C. et al. Heterogeneity of neoantigen landscape between primary lesions and their matched metastases in lung cancer. Transl Lung Cancer Res. 2020;9:246-56
Author contact
![]() Corresponding author: Xinwei Han. fcchanxwedu.cn
Corresponding author: Xinwei Han. fcchanxwedu.cn

 Global reach, higher impact
Global reach, higher impact