10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(16):6020-6034. doi:10.7150/ijbs.74902 This issue Cite
Review
Adding fuel to the fire: The lipid droplet and its associated proteins in cancer progression
Department of Urology, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China.
#These authors have contributed equally to this work and share first authorship.
Received 2022-5-9; Accepted 2022-10-2; Published 2022-10-17
Abstract
A lipid droplet (LD) is an organelle that consists of a phospholipid monolayer and a neutral lipid core, with proteins embedded in or attached to its surface. Until recently, cancers had long been regarded as genetic disorders with the abnormal activation of oncogenes and inactivation of tumor suppressor genes before their quality of a metabolic disorder began to be recognized. The last decade has witnessed the recognition of several metabolic characteristics of cancer cells, among which one is the accumulation of lipid droplets; therefore, attention has been given to exploring the role of LDs in carcinomas. In addition, there has been a remarkable expansion in understanding the complexity of LD's function in cellular homeostasis, including but not limited to energy supply, endoplasmic reticulum (ER) stress and oxidative stress management, or lipotoxicity alleviation. Thus, lipid droplet-associated proteins, which to a great extent determine the dynamics of a lipid droplet, have attracted the interest of numerous cancer researchers and their potential as cancer diagnostic biomarkers and therapeutic targets has been affirmed by emerging evidence. In this review, we systematically summarize the critical role of LDs in cancer and then focus on four categories of lipid droplet-associated proteins having the most direct influence on LD biosynthesis (diacylglycerol acyltransferase 1 (DGAT1) and diacylglycerol acyltransferase 2 (DGAT2)), degradation (adipose triglyceride lipase (ATGL)), and two renowned protein families on the LD surface (perilipins and cell death-inducing DNA fragmentation factor alpha-like effectors (CIDEs)). In this way, we aim to highlight their important role in tumor progression and their potential in clinical applications.
Keywords: lipid droplet, cancer metabolism, perilipins, CIDEs, DGAT1, DGAT2, ATGL
Introduction
Metabolic reprogramming, an established hallmark of cancer, has been identified as a requisite for its initiation and progression in past decades [1]. The Warburg effect is the first metabolic characteristic of cancer cells that was brought forward in 1920, exhibiting their propensity for aerobic glycolysis in glucose utilization [2]. The interconnection of different metabolic pathways determines the complexity of reprogrammed metabolism, and a comprehensive alternated metabolic network involving glucose, lipids and amino acids has been uncovered by subsequent researchers [3]. Among these prominent metabolic features, deregulated lipid metabolism has attracted great research attention in recent years and is recognized as an effective tactic adopted by cancer cells to thrive in a changing microenvironment [4]. The significant role of lipids in membrane synthesis, energy supply and cellular signal transduction in cancer cells has been gradually discovered [4]. Phospholipids and glycolipids, together with sterols, constitute biological membranes, providing material to sustain the rapid proliferation of cancer cells [5]. Lipid rafts, a cholesterol- and sphingolipid-enriched dynamic microdomain on cell membranes, are reorganized in cancer cells and have been demonstrated to play a vital role in cancer metastasis [6, 7]. Triglycerides are mobilized to efficiently generate fatty acids, which can be sequentially utilized to provide energy, building blocks and antioxidants [5]. In addition, fatty acids act as precursors of small signaling molecules such as prostaglandin E2 (PGE2) to promote cancer progression [8]. This extensive participation of lipids in tumor progression reflects the dependence of cancer cells on reprogrammed lipid metabolism, thus representing it as a potential target for cancer therapy [9].
Together with the boost of de novo lipogenesis driven by overexpressed enzymes such as fatty-acid synthase (FASN) [10], enhanced lipid storage seems to be another common characteristic in multiple cancer cells [11-13], which usually refers to the accumulation of a lipid storing organelle, lipid droplets. Although first reported in 1890, LD has long been underappreciated by researchers [14]. After a century of silencing, accompanied by a surge in research interest on the relationship between obesity, lipid metabolism and different diseases, LDs finally began to come under the light. [14] Studies have revealed its dynamic structure and intricate functions beyond lipid storage [15]. Moreover, its significant role in the development of metabolic diseases began to be realized, and one of them is cancer [16, 17]. Herein, we summarized the critical role of LDs in cancer progression and focused on several significant lipid droplet proteins that determine LD biosynthesis, maturation and degradation to discuss their potential as diagnostic and therapeutic targets in clinical implications.
Lipid droplet (LD) structure and dynamics
As organelles functioning as a lipid repository, LDs are widely distributed in the cytoplasm. Different from other organelles enclosed in the phospholipid bilayer, lipid droplets consist of a phospholipid monolayer embedded with varied proteins and a hydrophobic core enriched with neutral lipid-like triacylglycerol and sterol esters [14].
Although the mechanism of LD biogenesis has not been clarified, this process is recognized to occur in the endoplasmic reticulum (ER). The enzymes responsible for the esterification of fatty acids to generate sterol esters (SEs) and triacylglycerols (TAGs) reside on ER membranes, triggering the very first step of LD synthesis and lipid accumulation [15]. Aggregating neutral lipids gradually form an oil lens between two leaflets of ER membranes, exhibiting the initial state of LD (Fig. 1).
Sketch of LD dynamics determined by LD-associated proteins. LD biosynthesis is initiated with lipid accumulation between the two ER membrane leaflets. DGAT1 and DGAT2 catalyze the last step of TAG synthesis. After budding off, LDs expand through coalescence mediated by CIDEs and local TAG synthesis catalyzed by DGAT2 on their surface. A portion of ATGL is attached to the surface and triggers the first step of TAG hydrolysis, which could be impeded by perilipins on the LD surface under basal conditions. A stress-activated cAMP-PKA pathway phosphorylates perilipins and removes the block. G0S2 is another LD protein that inhibits TAG hydrolysis. Abbreviations: LD, lipid droplet; ER, endoplasmic reticulum; DGAT, diacylglycerol acyltransferase; TAG, triacylglycerol; CIDE, cell death-inducing DNA fragmentation factor alpha (DFFA)-like effector; ATGL, adipose tissue triacylglycerol lipase; cAMP, cyclic adenosine monophosphate; PKA, protein kinase A.
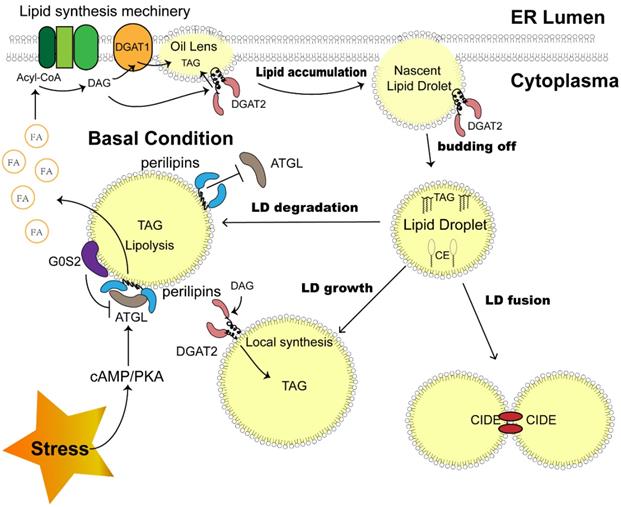
Before LD becomes an independent unit, a budding-off process is needed. The discrepancy in membrane protein distribution and phospholipid constitution between the cytosolic and luminal sides of the ER membrane results in an imbalance in surface tension, which drives LD precursors to depart from the ER into the cytosol [18]. The detailed mechanism of this process might be more complex, as other factors, such as seipin, were proven to be indispensable during LD budding off and were enriched beside the LD-ER membrane juncture [19, 20].
Investigations based on proteomics analysis revealed the presence of a variety of proteins in LDs. In LDs isolated from mammalian cells, 100-150 proteins can be reliably detected [21], including those associated with lipid synthesis and lipolysis, membrane trafficking, and protein degradation. These lipid droplet proteins determine the function and behavior of LDs after budding off (Fig. 1). The detached LDs further expand through coalescence induced by cell death-inducing DNA fragmentation factor alpha (DFFA)-like effector (CIDE) family proteins, and localized lipid synthesis takes place as the enzyme diacylglycerol acyltransferase 2 (DGAT2) translocates to the surface of LDs [22]. The dynamic LDs in the cytoplasm ensure cellular metabolic balance [15]. The degradation and sequential hydrolysis of lipids is a reliable mechanism employed by cells to fill the energy gap under metabolic stress, as the released free fatty acids (FFAs) would be efficiently utilized in mitochondria to produce ATP [5, 23]. Under this condition, lipolysis enzymes such as adipose triglyceride lipase (ATGL), hormone sensitive lipase (HSL) and monoacylglycerol lipase (MGL) will be recruited to the surface of LDs to catalyze the reaction [24], which is mediated by hormone activating signal transduction [24, 25]. In addition, perilipin family proteins also play a role in this process by coordinating the contact between lipids and lipases [26]. Recently, lipophagy was also reported as a novel autophagosome-dependent process to decompose LDs [27], suggesting the much more intricate dynamics of LDs to sustain metabolic homeostasis.
Lipid droplets in cancer progression
The biosynthesis and degradation of lipid droplets, as a pivotal part of lipid metabolism, is integral to the maintenance of normal cellular physiology and is naturally considered to be linked to disease progression, especially metabolic disorders. Previous studies have revealed its relationship with diabetes, fatty liver disease and atherosclerosis [16,17,28]. Much more recently, the metabolic characteristics of cancer have been a research focus and viewed as prospective therapeutic targets, among which abnormal lipid accumulation in cancer cells is a salient one. Moreover, investigations are beginning to shed light on the role of LDs, the lipid content depot, in the development of cancer (Fig. 2).
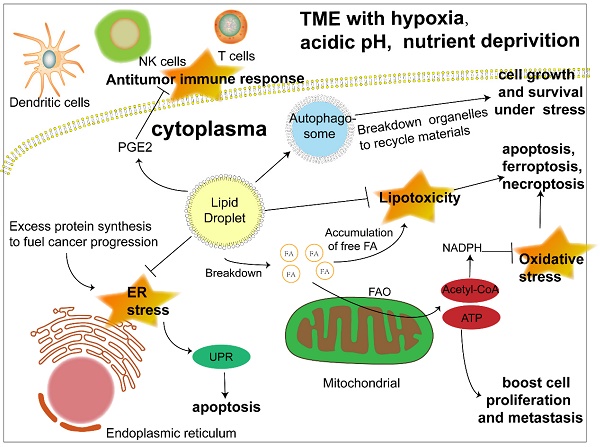
Confronted with a tumor microenvironment (TME) that is changing rapidly and sometimes inhospitable due to oxygen and glucose deprivation as well as acidosis, cancer cells depend on lipid mobilization to survive [5, 29]. The FA esterified as TAG and SE in lipid droplets can be released to generate acyl-CoA and provide energy by β-oxidation to fuel cancer cell proliferation and metastasis [30]. In addition, closer contact between LDs and mitochondria was also revealed [31, 32]. Hermes et al. reported a stronger interaction between LDs and mitochondria activated by AMPK signaling under nutrient starvation due to the translocation of both organelles from the cell center to the periphery along with a subset of microtubules, hence a more effective utilization of FAs to fill the energy gap [31]. Consistently, an earlier study proposed an intricate mechanism of channeling FAs from LDs to mitochondria under starvation, which was associated with cytoplasmic lipases, autophagy and the fusion of adjacent mitochondria [32]. However, more research on the interaction between LDs and mitochondria is still required to gain a comprehensive understanding of LD function in lipid utilization. Apart from energy, cancer cells also require membrane expansion to sustain their fast proliferation and growth, which further highlights the importance of triacylglycerol and cholesteryl esters stored in LDs as a lipid source of nascent membranes [33]. Despite cancer cell dependence on FAs as an energy source, when lipids are not urgently needed, their excess accumulation leads to lipotoxicity [34, 35]. The formation of LDs is beneficial to the survival of cancer cells by isolating lipids such as ceramides and DAG that can induce inflammation [36] and protecting polyunsaturated FAs from lipid peroxidation, which could sequentially result in DNA damage and cell death [37]. All of the above results reflect the dynamism of LDs as a flexible machinery adopted by cancer cells to cope with unstable surroundings and efficiently meet their energy demand.
Another crucial function of LDs is protecting cancer cells under ER stress and oxidative stress. The ER is an organelle responsible for the maturation of proteins owing to its capacity for protein folding [38], which, however, is not unlimited [39]. Thus, ER stress occurs when abundant proteins are synthesized in a short time, which is common in cancer cells [39]. Even though cells initiate the unfolded protein response (UPR) to cope, irreversible damage and the induction of apoptosis are still inevitable when ER stress is overwhelming. LDs are considered to participate in the management of ER stress, possibly by regulating fatty acid composition in the ER membrane [40] and removing misfolded proteins [41], although further investigations are required to elucidate the detailed mechanism. The increase of reactive oxygen species (ROS) is a characteristic of cancer cells. While they can activate protumorigenic signaling pathways, excess ROS can trigger apoptosis, necroptosis and ferroptosis [42]. Antioxidants such as NADPH are crucial to alleviate oxidative stress and sustain ROS homeostasis [43]. Acetyl-CoA, the main product of FAO, enters tricarboxylic acid cycle (TCA) and generate citrate [44], which could be transformed into isocitrate and malate to synthesize NADPH, with the catalyzation of isocitrate dehydrogenase 1 (IDH1) and malic enzyme 1 (ME1) respectively [45-47]. When the NADPH producing by pentose phosphate pathway is inhibited for a glucose deprived TME, the two forementioned pathways driven by FAO became vital to eliminate the possible detriment brought by these reduction products of oxygen in cancer cells [45, 48].
Moreover, increasing evidence has suggested an even more extensive role of LDs in cancer progression. Autophagy is a self-degradative process that maintains intercellular energy balance by breaking down damaged or senescent organelles and recycling their materials [49], an important mechanism adopted by cancer cells to cope with nutrient stress. While LDs have long been regarded as targets of autophagy, researchers have found that LDs are vital to initiate this process, which might be attributed to providing lipids to form autophagosomes and the maintenance of an appropriate phospholipid composition [50, 51]. Reprogramming metabolism is closely linked to the antitumor immune response, and LDs are involved in this connection. It has been demonstrated that LDs are the sites for the synthesis of prostaglandin E2, an established suppressor of antitumor immunity [52, 53]. In addition, LDs were reported to influence the function of immune cells such as dendritic cells and T cells [13, 54]. Although the exploration of LDs in cancer immunity is still in its infancy, it is safe to assume that LDs are a potential target to modulate antitumor immunity.
The significant role of LDs in cancer progression under stress. Lipid droplets function in multiple ways to promote cancer progression. 1) LDs release fatty acids to generate acyl-CoA and channel them to mitochondria to produce energy through FAO to boost cancer cell proliferation and metastasis. 2) Another product of FAO, acetyl-CoA, can produce NADPH, which acts as a hydrogen donor to maintain redox homeostasis and prevent cell death induced by excessive ROS accumulation. 3) LDs play an important role in ameliorating ER stress caused by abundant newly synthesized unfolded proteins in cancer cells, otherwise triggering apoptosis. 4) LDs sequester detrimental lipids such as DAG, cholesterol, ceramide and polyunsaturated FAs that tend to be oxidized into their core to prevent apoptosis, ferroptosis and necroptosis caused by lipotoxicity. 5) LD initiates autophagy by promoting the formation of autophagosomes to recycle materials from destroyed organelles in cancer cells under metabolic stress. 6) LD stores the precursor to produce signaling molecules such as PGE2, which could interfere with the antitumor immune response. Abbreviations: ATP, adenosine triphosphate; ER, endoplasmic reticulum; FA, fatty acid; FAO, fatty acid oxidation; NADPH, nicotinamide adenine dinucleotide phosphate; NK cell, natural killer cell; PGE2, prostaglandin E2; UPR, unfolded protein response.
Impacts and clinical implications of LD-associated proteins in various cancers
| Tumor type | LD associated proteins | Expression | Outcomes | Reference |
|---|---|---|---|---|
| Renal cell carcinoma | PLIN2 | Up | Promotes cell proliferation; High expression indicates good prognosis. | [12, 81, 83, 152] |
| PLIN3 | Up | Decreases sunitinib sensitivity; High expression indicates poor prognosis. | [153, 154] | |
| CIDEB | Up | High expression indicates good prognosis. | [106] | |
| Prostate cancer | PLIN3 | - | Promotes cell proliferation; Increase resistance to radiation therapy. High expression indicates poor prognosis. | [59, 155] |
| DGAT1 | Up | Promotes cell proliferation. | [156] | |
| Breast cancer | PLIN1 | Down | Reduces cell migration; High expression indicates good prognosis. | [57, 98] |
| PLIN2 | Down | Elevated in HER-2 positive subtype; High expression indicates poor prognosis. | [57] | |
| PLIN4 | Down | Elevated in doxorubicin resistant cells; High expression indicates poor prognosis in patients receive chemotherapy. | [57, 103] | |
| DGAT2 | - | Promotes cell migration, increase resistance to radiation therapy. High expression indicates poor prognosis in HER-2 patients. | [125, 157] | |
| ATGL | Up | Promotes cell proliferation and invasion; Elevated in HER-2 subtype; High expression indicates poor prognosis. | [142, 143] | |
| Hepatocellular carcinoma | PLIN5 | Up | - | [105] |
| CIDE-C | Down | Induces cell apoptosis. | [113] | |
| DGAT2 | Down | Reduces cell proliferation; High expression indicates good prognosis. | [126] | |
| ATGL | Down | Reduces cell proliferation | [135] | |
| Gastric cancer | PLIN2 | Up | Promotes cell proliferation and reduce cell apoptosis and ferroptosis. | [158] |
| DGAT2 | - | Promotes cell anoikis resistance and promote metastasis; Elevated in metastatic tumor tissue; High expression indicates poor prognosis. | [124] | |
| ATGL | - | High expression indicates good prognosis. | [139] | |
| Colorectal cancer | PLIN2 | Up | Elevated significantly even in early stage. | [92] |
| ATGL | Up | Promotes cell proliferation. | [144] | |
| Ovarian cancer | DGAT1 | - | Promote cell proliferation, tumor colonization and metastasis; Elevated in patients with venous invasion; High expression indicates poor prognosis. | [58] |
| ATGL | - | Reduces cell proliferation, migration and invasion. | [137] | |
| Cervical cancer | PLIN3 | Up | Elevated in reoccurrence; High expression indicates poor prognosis. | [94] |
| ATGL | - | Promotes cell proliferation, migration and invasion; High expression indicates poor prognosis. | [141] | |
| Glioblastoma | CIDEA | Down | Reduces cell proliferation and induce apoptosis. | [159] |
| DGAT1 | Up | Eliminates ROS and reduce cell apoptosis; High expression indicates poor prognosis. | [61] | |
| Lung cancer | PLIN2 | Up | Promotes cell proliferation; High expression indicates poor prognosis. | [89, 90] |
| DGAT1 | Up | High expression indicates good prognosis. | [160] | |
| ATGL | Down | Reduces cell proliferation, migration and invasion; Induces cell apoptosis; High expression indicates good prognosis. | [139, 161, 162] | |
| Liposarcoma | PLIN1 | Up | Promotes proliferation and migration; Reduce cell apoptosis; Elevated in liposarcoma but absent in other sarcoma subtypes. | [97, 99] |
| PLIN4 | - | Elevated in liposarcoma but absent in other sarcoma subtypes. | [97] |
Lipid droplet-associated proteins and their role in cancer
As mentioned above, emerging evidence has demonstrated the versatility of LDs in cancer progression, which promotes the thriving of cancer cells in a volatile and unfriendly environment. [4, 55] While LDs function as organelles, their biosynthesis, degradation and other behaviors essentially depend on their associated proteins [15]. Naturally, a great number of studies have focused on these proteins and revealed their differential expression in specific cancer types or their correlation with tumor stages and outcomes, suggesting that these lipid droplet proteins have diagnostic and prognostic value in clinical practice [56-58]. Furthermore, functional experiments both in vitro and in vivo further revealed their impact on cancer phenotypes such as proliferation, metastasis and chemotherapy resistance, implying their potential as therapeutic targets to interfere with homeostasis in cancer cells [12, 59-61] (Table 1). LD proteomics studies have revealed the existence of over 100 kinds of proteins on LD surfaces, some of which are enzymes involved in lipid metabolism, while others consist of histones, ribosomal proteins, and particular LD-residing proteins [21]. In this review, we focus on the proteins most directly impacting LD biosynthesis (DGAT1 and DGAT2) and degradation (ATGL) and two noted protein families enriched in the phospholipid monolayer of LDs (perilipins and CIDEs) to summarize their important role in tumor progression and outline their potential as diagnostic and therapeutic targets.
Perilipin family
Perilipins are a family of proteins detected on the surface of LDs. In mammalian cells, PLIN1-5 were identified to play an important role in sustaining lipid homeostasis, setting a balance between lipid storage and utilization [62]. All of these members share two conserved motifs in their N-terminal: a PAT domain and a repeating 11-mer helical motif, while heterogeneous structures are exhibited in their C-terminal [26]. It has been assumed that the conserved N-termini anchor these proteins to the surface of a lipid droplet, but subsequent research revealed that the C-termini were also indispensable [63, 64]. As some of the best-studied proteins reside on LDs, PLINs were found not only to be engaged in the biogenesis of lipid droplets [65, 66] but also to play a vital role in regulating both classical lipolysis [67, 68] and autophagic lipolysis [69, 70]. Their roles in traditional metabolic diseases such as fatty liver disease, type 2 disease and obesity have long been recognized. Recently, with cancer metabolism becoming a highly focused area, as proteins that determine the behaviors of LDs, the influence of PLINs in cancer progression was gradually uncovered.
PLIN2 and PLIN3
In contrast to the other three perilipin family members that appear in specific tissues, PLIN2 (also known as adipose differentiation-related protein, ADRP) and PLIN3 (known as TIP-47) are more ubiquitous [71], contributing to the formation of LDs in various cell types. The expression of PLIN2 is regarded as a marker reflecting the lipid content of cells [71]. Studies have revealed the involvement of PLIN2 in the regulation of cytosolic lipolysis by interfering with ATGL access to lipid droplets [72]. Meanwhile, unlike PLIN1 and PLIN5, whose direct binding with lipase has been identified, PLIN2 has not been found to interact with ATGL, which might account for its relatively mild effect in this classical process of lipolysis. Interestingly, PLIN2 was also shown to act as a barrier blocking another new autophagy-related approach of fat mobilization, named macroautophagy [73]. PLIN3 has been revealed to be decorating the coat of newly synthesized lipid droplets [74], and both in vivo and in vitro studies suggested that it protects lipid droplets from lipolysis, similar to PLIN2 [75, 76]. Another study reported that both PLIN2 and PLIN3 are chaperone-mediated autophagy targets whose removal could promote LD mobilization under nutrient stress by enhancing both macroautophagy and classical lipolysis [77]. Recently, Lu et al. reported that CHKα2 phosphorylates PLIN2 and PLIN3 to facilitate lipid droplet degradation and β oxidation [78].
Renal cell carcinoma is one of the most commonly diagnosed cancers worldwide, and its incidence rate has increased in recent years. Although the application of targeted drugs such as sunitinib, pazopanib and novel immune checkpoint inhibitors brought hope to patients with advanced clear cell renal cell carcinoma (ccRCC), its prognosis remains poor [79]. The accumulation of LDs has been recognized as a noticeable hallmark of ccRCC, and PLIN2 has been well studied in ccRCC to explore its biological role in cancer progression and its potential clinical application. Masahiro Yao et al. detected considerably increased expression of PLIN2 in renal cell carcinoma tissues [80]. To validate the possibility of urine PLIN2 as an early diagnosis indicator, a prospective case‒control study adopted patients with incidental radiographically discovered renal masses and presurgical presumptive diagnoses of kidney cancer, where patients who underwent other surgeries and healthy volunteers acted as controls [81]. It turned out that urine PLIN2 concentration in patients diagnosed with ccRCC or papillary cancer greatly exceeded that of the control group. Subsequent clinical research verified the specificity of urine PLIN2 to differentiate clear cell or papillary kidney cancer from other nontumorous kidney diseases [82] and other urological cancers [83].
Given the evidence from clinical studies, some studies have begun to explore the molecular mechanism of PLIN2 in renal cell carcinoma. The inactivated mutation of the Von Hippel-Lindau (VHL) gene has been identified as a key mutation in ccRCC that facilitates cancer progression [84]. The absence of its product, pVHL, leads to an abnormal accumulation of the oncogenic transcription factor HIF2α, which is strengthened by hypoxia [85]. A recent study revealed that PLIN2 expression was also upregulated by HIF-2α, subsequently promoting lipid storage [12]. The author revealed that the elevated expression of PLIN2 preserved the viability of ccRCC cells by decreasing UPR triggered by extra protein synthesis and maintaining ER homeostasis in cancer cells.
Irregular lipid accumulation is also a characteristic of prostate cancer (Pca) cells, another urological cancer with a high incidence in old males. Yue et al. identified that PTEN loss driving the accumulation of esterified cholesterol in LDs was notable in advanced prostate cancer tissue. The inhibition of cholesterol esterification substantially suppressed Pca growth and aggressiveness by downregulating the uptake of LDL [11]. Other studies revealed that the dysregulation of lipid metabolism was directly related to the development of resistance to first-line ADT treatment in prostate cancer, partly attributed to PLIN2-containing exomes released by the cells treated with ADTs in a paracrine manner [86], [87, 88]. Another study identified the role of PLIN3 in Pca, protecting cells from apoptosis induced by ER stress. Moreover, knockdown of PLIN3 was found to decrease the intratumoral synthesis of androgen and restore the ADT treatment sensitivity of enzalutamide-resistant C4-2 cells. These works collectively suggest that targeting LDs and perilipins is promising for the treatment of CRPC patients [59].
In addition to ccRCC and PCa, the involvement of PLIN2 in other cancers has also been detected. A pathological study found that PLIN2-positive lung adenocarcinoma was associated with a worse outcome reflected in overall and disease-free survival [89]. This was supported by basic research indicating that PLIN2 can promote lung adenocarcinoma cell proliferation by increasing the phosphorylation of Akt [90]. Kimberly et al. reported that PLIN2 possesses potential in breast cancer subtype differentiation, as high-grade subtypes, namely, Her2-positive and TNBC, have lower expression of this protein [91]. In addition, a better clinical outcome, reflected in the recurrence-free survival time in the PLIN2 low expression group, was presented. Another study conducting plasma proteome analysis found elevated expression of PLIN2 in colorectal cancer patient plasma, even in those with low-grade tumors, indicating that PLIN2 is a sensitive and dependable biomarker in this cancer type [92].
Studies focusing on PLIN3 have been less frequent; however, its value in cancer diagnosis and treatment is still worth exploring. Immunohistochemical analyses in a large number of specimens of multiple cancers demonstrated a positive correlation between PLIN3 staining intensity and tumor size in colorectal cancer, as well as tumor grade and stage in lung cancer [93]. In cervical cancer, PLIN3 was found to be elevated in patients with invasive tumors, lymph nodular metastasis, and recurrence after treatment [94].
PLIN1, PLIN4 & PLIN5
The distribution of these three perilipins is tissue specific. As the first identified protein in this family, PLIN1 (known as perilipin) occurs mainly in the adipocytes of white and brown adipose tissue, where the lipid droplets are relatively large. Many researchers focused on its role in lipid mobilization, and they found that the cAMP-PKA pathway activated by epinephrine and norepinephrine greatly phosphorylated PLIN1 in adipocytes under fasting or excise conditions, which sequentially recruited ATGL and HSL to separately hydrolyze TAG or DAG [95]. A delicate scaffolding complex is in charge of this efficient switch when extra energy is needed, which consists of PLIN1 and αβ hydrolase domain containing 5 (ABHD5), an activator of ATGL, on the surface of LDs in white adipocytes [96]. It was demonstrated that PLIN1 possesses prognostic value and therapeutic potential to suppress cell proliferation both in breast cancer and liposarcoma [57, 97-99]. Ronell et al. reported that PLIN1 was also upregulated in conventional ameloblastoma and ameloblastic carcinoma, two kinds of odontogenic tumors, compared with tooth germ tissue [100].
PLIN4, the largest member of this family, is mainly located in LDs in brown adipose tissue, skeletal muscle and the heart. It appears on some nascent LDs or remains unanchored [74]. Chen et al. found a decrease in TAG content in the hearts of PLIN4(-/-) mice [101]. Another recent study disclosed that it functions to decrease the size of LDs, relying on its much longer amphipathic helices compared with other perilipin members [102]. The exploration of its specific functions on lipid droplets is still in its infancy, while some studies revealed that Plin4 might have a role in cancer progression. A research group focusing on chemoresistance in triple-negative breast cancer (TNBC) demonstrated that PLIN4 was overexpressed in doxorubicin-resistant cells and could be targeted to eliminate chemoresistance [103]. Li et al. also revealed its value as a diagnostic biomarker of liposarcoma and could refer to the discrimination of its subtypes [97].
Similar to PLIN4, PLIN5 is distributed in oxidative tissue and is indispensable for maintaining lipid storage and modulating lipolysis in this tissue [26, 104]. It can bind ATGL and HSL to prevent TG hydrolysis, and similar to PLIN1, it is phosphorylated by PKA for its control to be sequentially lifted [104]. Similar to other members of the perilipin family, some evidence of its role in cancer has been uncovered. Combined with other perilipin members, it can serve as a diagnostic marker in breast cancer [57]. Researchers also detected an increase in the expression of PLIN5 in a murine HCC model and clinical specimens [105].
All the above publications suggest that PLIN1, PLIN4 and PLIN5 can be involved in cancer progression, despite their tendency to distribute in adipocytes. As paracancerous adipose tissue was identified as an essential part of the TME to influence the proliferation, migration and drug resistance of cancer cells, subsequent studies will shed light on how these perilipin members act in cancer progression.
CIDE (cell death-inducing DNA fragmentation factor alpha (DFFA)-like effector) family
CIDE family proteins, as their name implies, are characterized by their cell death-inducing DFF45-like effect domain in their N-terminus. Three members have been identified in humans: CIDE-A, which is mainly expressed in WAT and the mammary gland, CIDE-B, which is mainly present in the liver, and CIDE-C (also known as FSP27), which is mainly expressed in WAT and BAT [106]. Previous studies focused on their apoptosis-inducing effect, but the research interest in this protein family shifted as they were also found to reside on lipid droplets. These proteins were demonstrated to play an important role in promoting lipid accumulation by mediating LD fusion, since a smaller LD surface area/volume ratio reduces the accessibility of lipases to lipids [107], [108, 109]. Enriching in the LD-LD contact sites, CIDE proteins from two LDs interact with each other to form a channel to enable lipid transfer coordinated by two other lipid droplet-associated proteins, PLIN1 and RAB8 [110, 111].
The dual role played by CIDE family proteins makes it more challenging to elucidate their effect on cancer progression. Li et al. reported that CIDE-C suppressed the proliferation of cancer cells by triggering apoptosis; therefore, it could be a therapeutic target in carcinomas due to the high-frequency mutation in its chromosome region [112]. One study on hepatocellular carcinoma exhibited the downregulation of CIDE‑C in HCC tissues compared with adjacent normal tissue, as well as an obvious proapoptotic effect caused by the overexpression of CIDE-C in HCC cells, which could be explained by the interaction between CIDE‑C and lipopolysaccharide-induced tumor necrosis factor (LITAF) [113]. Another functional study revealed that this proapoptotic effect in HeLa cells could be greatly mitigated when supplementary oleic acid was added to the medium. The author inferred that CIDE-C tends to be located on LDs after extra FA supply and that subcellular distribution alteration may prevent it from activating the apoptosis process [114]. In clear cell renal cell carcinoma (ccRCC) characterized by remarkable LD accumulation, it was demonstrated that CIDE-C was upregulated in cancer as expected, but CIDE-B was downregulated. The expression of CIDE-B was also lower in high-grade ccRCC than in low-grade ccRCC, and this was associated with a better prognosis [56]. In summary, earlier studies explored the role of CIDE in cancer based on its apoptosis-inducing effect. Since its new function in LD dynamics was identified, more research was prompted to comprehensively illustrate the linkage between CIDE family members and cancer progression in the context of cancer metabolic reprogramming.
DGAT1 & DGAT2
During the process of lipid droplet formation, diacylglycerol acyltransferase (DGAT) enzymes catalyze the last step of TG synthesis from fatty acyl-CoA and diacylglycerol (DG). DGAT1 and DGAT2, although responsible for the same biochemical reaction, are encoded by genes located in different positions on chromosomes [115], which brings about a distinction in the structures of these two proteins. Their subcellular locations revealed by fluorescence imaging have shown that DGAT1 appears exclusively on ER membranes, whereas DGAT2 resides both on ER membranes and in lipid droplet phospholipid monolayers [116]. An LD target domain in DGAT2 originally embedded in the ER membrane was proven necessary for its targeting on LDs, facilitating the budding-off process to form nascent LDs after the regional accumulation of TGs [117]. It was reported that DGAT2 promotes the expansion of LDs by synthesizing TG at its surface [22]. These two enzymes also present different propensities to their substrates; the catalytic activity of DGAT1 is more concentrated on lipogenesis with exogenous FA, while DGAT2 does not discriminate the FA source [115, 118]. Reflecting this distinction in their tissue distribution, DGAT1 is ubiquitous but particularly highly expressed in intestine and adipose tissue, whereas DGAT2 is mainly expressed in hepatocytes and adipocytes [119].
Glioblastoma (GLB) is recognized as a malignant brain tumor with the hallmark of abundant LDs. Guo et al. revealed that GLB is reliant on the upregulation of DGAT1 but not DGAT2 to synthesize TG and form LDs [61]. Targeting DGAT1 in GLB leads to ROS accumulation by consequently intensifying the β-oxidation of free fatty acids. Furthermore, ROS impair the oxidative ability of mitochondria, sequentially resulting in the accumulation of acetyl-CoA, which augments oxidative stress and finally triggers apoptosis. In prostate cancer, it was demonstrated by Crawford et al. that the expression of DGAT1 increased in cancer tissues, and cell proliferation and migration could be inhibited by targeting DGAT1 [120]. Another recent study discovered that the upregulation of DGAT1 in prostate cancer is partly mediated by EPH receptor B2 (EPHB2), a tyrosine kinase ephrin receptor [121]. Studies on ovarian cancer and breast cancer also demonstrated the potential of DGAT1 as a novel target to repress cancer progression [58, 122]. The featured tumor microenvironment was considered to drive metabolic reprogramming in cancer cells. Feron et al. first reported that acidosis, a characteristic of the TME, leads to LD accumulation in cancer cells by upregulating DGAT1 and CD36, mediated by TGF-β signaling. Moreover, the authors found that LD accumulation supports cancer metastasis by abating anoikis [123].
DGAT2 deserves as much attention as DGAT1, if not more, in cancer research. A study focusing on gastric cancer revealed increased expression of DGAT2 in cancer cells cocultured with adipocytes. Silencing DGAT2 made these cells vulnerable to anoikis and efficiently inhibited cancer metastasis in vivo and in vitro [124], suggesting that it is a promising target in patients with GC. In the MCF-7 breast cancer cell line, Seco et al. found that the pharmacological inhibition of DGAT2 efficiently restrained their migration and increased their sensitivity to radiation treatment [125]. However, in some other cancers, such as hepatocellular carcinoma [126] and melanoma [127], DGAT2 was identified to play a suppressive role. Shen et al. revealed that DGAT2 is downregulated in HCC tissue, and its low expression implies a shorter survival time [126]. Overexpression of DGAT resulted in evidently inhibited proliferation in HCC cell lines, with decreased expression of cell cycle-related genes [126], suggesting that DGAT2, as a key protein in modulating lipid metabolism, may also function in other aspects.
ATGL
Adipose triglyceride lipase (ATGL) was identified as a rate-limiting enzyme in lipolysis and in charge of the very first step of TG hydrolysis by Zimmermann, R. et al. in 2004 [128]. It is highly expressed in adipose tissue but can also be detected in liver, heart and skeletal muscle. Subsequent research reported a massive amount of fat accumulation in adipocytes and muscle in mice induced by the lack of ATGL, which suggests its vital role in lipid metabolism [129]. By exploring its subcellular location in cells, researchers detected part of ATGL on the surface of LDs, as the majority of ATGL is dispersedly distributed in the cytoplasm [128]. This finding indicates that LDs provide a platform for lipid hydrolysis. Later, studies confirmed this conjecture and uncovered an intricate interaction mode between ATGL and other lipid droplet-associated proteins to modulate lipid metabolism [95, 130]. Under basal conditions, ATGL can be bound and activated by ABHD5, another protein on LDs, to moderately perform hydrolysis [131]. As mentioned previously, Perilipin family proteins impede ATGL's access to TAG in LDs by separating it from its coactivator, ABHD5. This interference ceases during fasting due to cAMP-PKA pathway-mediated phosphorylation. Liu et al. revealed that G0S2 also acts to inhibit ATGL's lipase activity [132]. Another protein, hypoxia-inducible gene 2 (HIG2), which is activated by HIF-1, was validated to inhibit ATGL and consequently promote lipid accumulation in hypoxia [133].
Even though it has been just over a decade since the mechanism of ATGL was clarified [128], growing evidence is establishing its vital role in cancer, given that it is closely related to lipid synthesis and fatty acid oxidation (FAO), two rewired pathways in cancer cells [4, 30]. As abundant lipid synthesis and LD accumulation are metabolic hallmarks of multiple cancers, ATGL, as a lipase driving the reverse reaction, was naturally revealed as a suppressor of malignancy [134, 135]. In multiple mouth cancer cell lines, ectopic overexpression of ATGL repressed cell proliferation [130, 136]. Liu et al. declared the importance of ATGL downregulation in cancer cells to survive in hypoxia, a characteristic of the TME [133]. A study in hepatocellular cancer verified that ATGL, which is significantly downregulated in HCC tissues, will switch cancer cells' preference for an energy source from glucose to FFA and inhibit cell proliferation. Moreover, the recognized tumor suppressor p53 was identified as a possible downstream target of ATGL [135]. Similar phenomena were observed for ATGL in ovarian cancer [137]. Meanwhile, its effect in lung adenocarcinoma remains somewhat elusive. Earlier, in vitro research proposed a pro-neoplasm role [138]. However, it seems that an opposite opinion prevails since Hoefler et al. revealed its reduction in tumor tissue and that knockout of ATGL in mice greatly enhanced their risk of having pulmonary neoplasm [139]. Another study adopting a 3D in vitro model found that ATGL knockout promoted the growth of spheroids and facilitated cancer cell adaptation to hypoxia [133]. Furthermore, the depletion of ATGL was validated to present a more aggressive phenotype, attributed to the consequent upregulation of SRC kinase signaling [140]. Beyond its functions in cancer cells, a new discovery shed light on ATGL's role in the crosstalk between cancer cells and the TME. Li et al. reported repressed expression of ATGL in neutrophils, which was induced by its interaction with resident mesenchymal cells, contributing to breast cancer colonization in the lung. Moreover, neutrophils sequentially transfer stored lipids to cancer cells to enable their survival and proliferation [60]. These results exhibit promising prospects to interfere with the metabolic rewiring of cancer cells by targeting ATGL.
However, in certain carcinomas, this lipase was recognized as an oncoprotein; it is reasonable to consider that many malignancies present greater reliance on FAO to fuel their progression. The increased utilization of TG in LDs through overexpression of ATGL was demonstrated to promote the migration and proliferation of HeLa cells, paralleled by activated HIF-1a-mediated mitophagy to mitigate potential damage caused by ROS accumulation [141]. Obesity has long been regarded as a risk factor in breast cancer. Obtaining free fatty acids from adipocytes in a coculture system, breast cancer cells showed an increase in proliferation and migration activity, which was dependent on ATGL inducing lipolysis both in adipocytes and cancer cells [142, 143]. Iftikhar et al. reported that ATGL might play a vital role in the development of colon cancer driven by obesity. In vitro, the increase in proliferation induced by oleic acid was eliminated through the inhibition of ATGL. Except for obvious lipid storage, some established oncogenes, such as RARA or MYC, were also found to be decreased after ATGL knockdown [144].
All these results imply that the role of ATGL in cancer is rather complex and not limited to consuming TG in LDs but also linked to other pathways. In addition to its nonenergetic functions that have begun to be clarified [145, 146], more studies are required to comprehensively address its position in cancer progression before its clinical potential can be fully realized.
Conclusions and Perspectives
Lipid droplets, as cytoplasmic organelles that were previously regarded as a pure depot of neutral lipids, are now recognized to have impacts on cells in multiple ways. Its dynamics (biosynthesis, expansion and degradation) are considered a significant indication of the metabolic status of cells in different biological contexts. Therefore, scientists employed diverse fluorescent imaging techniques to detect and observe LDs in living cells [147, 148], which enhanced the understanding of their vital role in cell metabolism and presented their connection with the development of metabolic disorders such as obesity, type 2 diabetes, cardiovascular disease, and cancer. However, unanswered questions still remain. Multiple studies have revealed the links between LDs and other cellular organelles, such as mitochondria, ER, and lysosomes, which permit material exchange and influence the trafficking of these organelles [31, 149, 150]. Nonetheless, the detailed functions and mechanism of this relationship remain elusive; thus, further studies are needed to elucidate the potential synergistic mechanism of these organelles and their impact on cellular physiology and pathological processes. In cancer research, another alluring question is how LDs mediate the crosstalk between cancer cells and their TME consisting of adipocytes, cancer-associated fibroblasts, immune cells, and so on. While some communications dependent on lipid metabolism have been deciphered [13, 124], a more comprehensive picture is awaiting construction, which will be possible by virtue of the maturation and popularization of single-cell sequencing techniques.
Great progress has been made to study lipid droplets in basic oncology research; however, their potential as clinical diagnostic and prognostic indicators has failed to be fully realized due to the unfeasibility of detection and the lack of generally applicable quantification methods. The proteins residing on LDs could be an alternative to overcome this challenge. In fact, the dynamics of LDs could be reflected by the expression of LD-associated proteins, as the behaviors and functions of LDs greatly depend on these proteins. Numerous studies have claimed the differential expression of lipid droplet-associated proteins in various neoplasms and their correlation with tumor stages and prognosis. Moreover, some studies reported that these proteins could be detected in body fluids such as serum or urine [83, 94], indicating their potential as noninvasive biomarkers in the clinic. Notwithstanding, studies involving large cohorts are required before these proteins can be practically used to guide cancer diagnosis and treatment.
Recently, targeting lipid metabolism in cancer cells has been explored as an anticancer strategy, given the pivotal role of reprogrammed lipid metabolism in tumor initiation and progression. For instance, FASN, the enzyme that functions to catalyze de novo lipogenesis, is being established as an applicable and promising therapeutic target, with its inhibitor TVB-2640 going through clinical trials in a variety of cancers, including NSCLC, CRC, HER2+ advanced breast cancer, and astrocytoma. Fatty acid uptake, namely, CD36, represents another target, and its inhibitor ABT-50 has also entered clinical trials in melanoma [151]. As another significant constituent of lipid metabolism, the storage and mobilization of intercellular lipids and their critical organelles, lipid droplets have not yet received the attention they deserve. However, cancer cell plasticity in lipid uptake, acquisition and utilization, partly owing to the mobilization of lipids stored in LDs, brings resistance to treatments targeting a single pathway. A combinational therapy with multiple targets in lipid metabolism might be conducive to disrupting lipid homeostasis in cancer cells; therefore, there is still room for LD-associated proteins in cancer treatment. To this end, as highlighted in this review, lipid droplet-associated proteins participate in cancer reprogrammed metabolism and influence tumor progression in diverse aspects, which substantiates the claims that exploring their potential as therapeutic targets is needed to expand the options for clinical cancer treatment. Looking forward to future research, with the growing understanding of LD-associated proteins and the evidence presented by large cohort studies and clinical trials expanding, we envisage that these proteins, representing one of the most intriguing metabolic characteristics of cancer cells, are to be adopted both as viable cancer biomarkers and effective oncotherapy targets.
Abbreviations
LD: lipid droplet; ER: endoplasmic reticulum; DGAT1: diacylglycerol acyltransferase 1; DGAT2: diacylglycerol acyltransferase 2; ADRP: adipose differentiation-related protein; ATGL: adipose triglyceride lipase; HSL: hormone sensitive lipase; MGL: monoacylglycerol lipase; CIDE: cell death-inducing DNA fragmentation factor alpha (DFFA)-like effector; ABHD5: αβ hydrolase domain containing 5; HIG2: hypoxia-inducible gene 2; FASN: fatty acid synthetase; FFA: free fatty acids; ROS: reactive oxygen species; ATP: adenosine triphosphate; cAMP: cyclic adenosine monophosphate; TME: tumor microenvironment; HIF: hypoxia inducible factor; PKA: protein kinase A; AMPK: AMP-activated protein kinase; TAG: triacylglycerol; DAG: diacylglycerol; CE: cholesteryl ester; ccRCC: clear cell renal cell carcinoma; Pca: prostate cancer; ADT: androgen deprivation therapy; CRC: colorectal cancer; HCC: hepatocellular carcinoma; TNBC: triple-negative breast cancer; GC: gastric cancer; NSCLC: non-small cell lung carcinoma.
Acknowledgements
Funding
This work was supported by National Natural Science Foundation of China (grant Numbers: 82173221, 81870484, 82072809; Joint construction project of Zhejiang Province and Ministry (grant number: 2020388200); Key R & D plan of Zhejiang Province (grant number: 2019C03089).
Author contributions
G.L. and L.X. performed study concept and design. W.L. and H.W. performed development of methodology and writing, review, and revision of the paper. Z.L., Q.Z., and L.D. provided acquisition and analysis. H.X., R.W. and Y.L. provided technical and material support in revision. L.R., C.Y. and Z.Z. drew the figures and the table. All authors approved the final version of the submitted manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nature reviews Cancer. 2021;21:669-80
2. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science (New York, NY). 2009;324:1029-33
3. Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell metabolism. 2016;23:27-47
4. Snaebjornsson MT, Janaki-Raman S, Schulze A. Greasing the Wheels of the Cancer Machine: The Role of Lipid Metabolism in Cancer. Cell metabolism. 2020;31:62-76
5. Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nature reviews Cancer. 2013;13:227-32
6. Greenlee JD, Subramanian T, Liu K, King MR. Rafting Down the Metastatic Cascade: The Role of Lipid Rafts in Cancer Metastasis, Cell Death, and Clinical Outcomes. Cancer research. 2021;81:5-17
7. Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science (New York, NY). 2010;327:46-50
8. Zelenay S, van der Veen AG, Böttcher JP, Snelgrove KJ, Rogers N, Acton SE. et al. Cyclooxygenase-Dependent Tumor Growth through Evasion of Immunity. Cell. 2015;162:1257-70
9. Butler LM, Perone Y, Dehairs J, Lupien LE, de Laat V, Talebi A. et al. Lipids and cancer: Emerging roles in pathogenesis, diagnosis and therapeutic intervention. Advanced drug delivery reviews. 2020;159:245-93
10. Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nature reviews Cancer. 2007;7:763-77
11. Yue S, Li J, Lee SY, Lee HJ, Shao T, Song B. et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell metabolism. 2014;19:393-406
12. Qiu B, Ackerman D, Sanchez DJ, Li B, Ochocki JD, Grazioli A. et al. HIF2α-Dependent Lipid Storage Promotes Endoplasmic Reticulum Homeostasis in Clear-Cell Renal Cell Carcinoma. Cancer discovery. 2015;5:652-67
13. Cotte AK, Aires V, Fredon M, Limagne E, Derangère V, Thibaudin M. et al. Lysophosphatidylcholine acyltransferase 2-mediated lipid droplet production supports colorectal cancer chemoresistance. Nature communications. 2018;9:322
14. Farese RV Jr, Walther TC. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139:855-60
15. Olzmann JA, Carvalho P. Dynamics and functions of lipid droplets. Nature reviews Molecular cell biology. 2019;20:137-55
16. Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nature reviews Molecular cell biology. 2008;9:367-77
17. Gluchowski NL, Becuwe M, Walther TC, Farese RV Jr. Lipid droplets and liver disease: from basic biology to clinical implications. Nature reviews Gastroenterology & hepatology. 2017;14:343-55
18. Chorlay A, Thiam AR. An Asymmetry in Monolayer Tension Regulates Lipid Droplet Budding Direction. Biophysical journal. 2018;114:631-40
19. Cartwright BR, Binns DD, Hilton CL, Han S, Gao Q, Goodman JM. Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Molecular biology of the cell. 2015;26:726-39
20. Grippa A, Buxó L, Mora G, Funaya C, Idrissi FZ, Mancuso F. et al. The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. The Journal of cell biology. 2015;211:829-44
21. Bersuker K, Peterson CWH, To M, Sahl SJ, Savikhin V, Grossman EA. et al. A Proximity Labeling Strategy Provides Insights into the Composition and Dynamics of Lipid Droplet Proteomes. Developmental cell. 2018;44:97-112.e7
22. Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A. et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Developmental cell. 2013;24:384-99
23. Heldt HW. Energy metabolism in mitochondria. Angewandte Chemie (International ed in English). 1972;11:792-8
24. Walther TC, Farese RV Jr. Lipid droplets and cellular lipid metabolism. Annual review of biochemistry. 2012;81:687-714
25. Birnbaum MJ. Lipolysis: more than just a lipase. The Journal of cell biology. 2003;161:1011-2
26. Kimmel AR, Sztalryd C. The Perilipins: Major Cytosolic Lipid Droplet-Associated Proteins and Their Roles in Cellular Lipid Storage, Mobilization, and Systemic Homeostasis. Annual review of nutrition. 2016;36:471-509
27. Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M. et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131-5
28. Yuan Y, Li P, Ye J. Lipid homeostasis and the formation of macrophage-derived foam cells in atherosclerosis. Protein & cell. 2012;3:173-81
29. Peck B, Schulze A. Lipid Metabolism at the Nexus of Diet and Tumor Microenvironment. Trends in cancer. 2019;5:693-703
30. Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr. Cellular fatty acid metabolism and cancer. Cell metabolism. 2013;18:153-61
31. Herms A, Bosch M, Reddy BJ, Schieber NL, Fajardo A, Rupérez C. et al. AMPK activation promotes lipid droplet dispersion on detyrosinated microtubules to increase mitochondrial fatty acid oxidation. Nature communications. 2015;6:7176
32. Rambold AS, Cohen S, Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Developmental cell. 2015;32:678-92
33. Skotland T, Kavaliauskiene S, Sandvig K. The role of lipid species in membranes and cancer-related changes. Cancer metastasis reviews. 2020;39:343-60
34. Williams KJ, Argus JP, Zhu Y, Wilks MQ, Marbois BN, York AG. et al. An essential requirement for the SCAP/SREBP signaling axis to protect cancer cells from lipotoxicity. Cancer research. 2013;73:2850-62
35. Fujiwara N, Nakagawa H, Enooku K, Kudo Y, Hayata Y, Nakatsuka T. et al. CPT2 downregulation adapts HCC to lipid-rich environment and promotes carcinogenesis via acylcarnitine accumulation in obesity. Gut. 2018;67:1493-504
36. Weinberg JM. Lipotoxicity. Kidney international. 2006;70:1560-6
37. Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ. et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017;171:273-85
38. Araki K, Nagata K. Protein folding and quality control in the ER. Cold Spring Harbor perspectives in biology. 2011;3:a007526
39. Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nature reviews Molecular cell biology. 2012;13:89-102
40. Volmer R, van der Ploeg K, Ron D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4628-33
41. Moldavski O, Amen T, Levin-Zaidman S, Eisenstein M, Rogachev I, Brandis A. et al. Lipid Droplets Are Essential for Efficient Clearance of Cytosolic Inclusion Bodies. Developmental cell. 2015;33:603-10
42. Moloney JN, Cotter TG. ROS signalling in the biology of cancer. Seminars in cell & developmental biology. 2018;80:50-64
43. Hayes JD, Dinkova-Kostova AT, Tew KD. Oxidative Stress in Cancer. Cancer cell. 2020;38:167-97
44. Lehninger AL. Fatty acid oxidation and the Krebs trocarboxylic acid cycle. The Journal of biological chemistry. 1945;161:413
45. Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661-5
46. Chen L, Zhang Z, Hoshino A, Zheng HD, Morley M, Arany Z. et al. NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nature metabolism. 2019;1:404-15
47. Jiang P, Du W, Mancuso A, Wellen KE, Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 2013;493:689-93
48. Ying M, You D, Zhu X, Cai L, Zeng S, Hu X. Lactate and glutamine support NADPH generation in cancer cells under glucose deprived conditions. Redox biology. 2021;46:102065
49. Kim KH, Lee MS. Autophagy-a key player in cellular and body metabolism. Nature reviews Endocrinology. 2014;10:322-37
50. Dupont N, Chauhan S, Arko-Mensah J, Castillo EF, Masedunskas A, Weigert R. et al. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Current biology: CB. 2014;24:609-20
51. Velázquez AP, Tatsuta T, Ghillebert R, Drescher I, Graef M. Lipid droplet-mediated ER homeostasis regulates autophagy and cell survival during starvation. The Journal of cell biology. 2016;212:621-31
52. Accioly MT, Pacheco P, Maya-Monteiro CM, Carrossini N, Robbs BK, Oliveira SS. et al. Lipid bodies are reservoirs of cyclooxygenase-2 and sites of prostaglandin-E2 synthesis in colon cancer cells. Cancer research. 2008;68:1732-40
53. Bonavita E, Bromley CP, Jonsson G, Pelly VS, Sahoo S, Walwyn-Brown K. et al. Antagonistic Inflammatory Phenotypes Dictate Tumor Fate and Response to Immune Checkpoint Blockade. Immunity. 2020;53:1215-29.e8
54. Veglia F, Tyurin VA, Mohammadyani D, Blasi M, Duperret EK, Donthireddy L. et al. Lipid bodies containing oxidatively truncated lipids block antigen cross-presentation by dendritic cells in cancer. Nature communications. 2017;8:2122
55. Cruz ALS, Barreto EA, Fazolini NPB, Viola JPB, Bozza PT. Lipid droplets: platforms with multiple functions in cancer hallmarks. Cell death & disease. 2020;11:105
56. Yu M, Wang H, Zhao J, Yuan Y, Wang C, Li J. et al. Expression of CIDE proteins in clear cell renal cell carcinoma and their prognostic significance. Molecular and cellular biochemistry. 2013;378:145-51
57. Zhang X, Su L, Sun K. Expression status and prognostic value of the perilipin family of genes in breast cancer. American journal of translational research. 2021;13:4450-63
58. Xia L, Wang Y, Cai S, Xu M. DGAT1 Expression Promotes Ovarian Cancer Progression and Is Associated with Poor Prognosis. Journal of immunology research. 2021;2021:6636791
59. Zhou L, Song Z, Hu J, Liu L, Hou Y, Zhang X. et al. ACSS3 represses prostate cancer progression through downregulating lipid droplet-associated protein PLIN3. Theranostics. 2021;11:841-60
60. Li P, Lu M, Shi J, Gong Z, Hua L, Li Q. et al. Lung mesenchymal cells elicit lipid storage in neutrophils that fuel breast cancer lung metastasis. Nature immunology. 2020;21:1444-55
61. Cheng X, Geng F, Pan M, Wu X, Zhong Y, Wang C. et al. Targeting DGAT1 Ameliorates Glioblastoma by Increasing Fat Catabolism and Oxidative Stress. Cell metabolism. 2020;32:229-42.e8
62. Jackson CL. Lipid droplet biogenesis. Current opinion in cell biology. 2019;59:88-96
63. McManaman JL, Zabaronick W, Schaack J, Orlicky DJ. Lipid droplet targeting domains of adipophilin. Journal of lipid research. 2003;44:668-73
64. Subramanian V, Garcia A, Sekowski A, Brasaemle DL. Hydrophobic sequences target and anchor perilipin A to lipid droplets. Journal of lipid research. 2004;45:1983-91
65. Čopič A, Antoine-Bally S, Giménez-Andrés M, La Torre Garay C, Antonny B, Manni MM. et al. A giant amphipathic helix from a perilipin that is adapted for coating lipid droplets. Nature communications. 2018;9:1332
66. Jacquier N, Mishra S, Choudhary V, Schneiter R. Expression of oleosin and perilipins in yeast promotes formation of lipid droplets from the endoplasmic reticulum. Journal of cell science. 2013;126:5198-209
67. Tansey JT, Huml AM, Vogt R, Davis KE, Jones JM, Fraser KA. et al. Functional studies on native and mutated forms of perilipins. A role in protein kinase A-mediated lipolysis of triacylglycerols. The Journal of biological chemistry. 2003;278:8401-6
68. Brasaemle DL, Rubin B, Harten IA, Gruia-Gray J, Kimmel AR, Londos C. Perilipin A increases triacylglycerol storage by decreasing the rate of triacylglycerol hydrolysis. The Journal of biological chemistry. 2000;275:38486-93
69. Cingolani F, Czaja MJ. Regulation and Functions of Autophagic Lipolysis. Trends in endocrinology and metabolism: TEM. 2016;27:696-705
70. Tasset I, Cuervo AM. Role of chaperone-mediated autophagy in metabolism. The FEBS journal. 2016;283:2403-13
71. Conte M, Franceschi C, Sandri M, Salvioli S. Perilipin 2 and Age-Related Metabolic Diseases: A New Perspective. Trends in endocrinology and metabolism: TEM. 2016;27:893-903
72. Listenberger LL, Ostermeyer-Fay AG, Goldberg EB, Brown WJ, Brown DA. Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. Journal of lipid research. 2007;48:2751-61
73. Tsai TH, Chen E, Li L, Saha P, Lee HJ, Huang LS. et al. The constitutive lipid droplet protein PLIN2 regulates autophagy in liver. Autophagy. 2017;13:1130-44
74. Wolins NE, Quaynor BK, Skinner JR, Schoenfish MJ, Tzekov A, Bickel PE. S3-12, Adipophilin, and TIP47 package lipid in adipocytes. The Journal of biological chemistry. 2005;280:19146-55
75. Bulankina AV, Deggerich A, Wenzel D, Mutenda K, Wittmann JG, Rudolph MG. et al. TIP47 functions in the biogenesis of lipid droplets. The Journal of cell biology. 2009;185:641-55
76. Carr RM, Patel RT, Rao V, Dhir R, Graham MJ, Crooke RM. et al. Reduction of TIP47 improves hepatic steatosis and glucose homeostasis in mice. American journal of physiology Regulatory, integrative and comparative physiology. 2012;302:R996-1003
77. Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nature cell biology. 2015;17:759-70
78. Liu R, Lee JH, Li J, Yu R, Tan L, Xia Y. et al. Choline kinase alpha 2 acts as a protein kinase to promote lipolysis of lipid droplets. Molecular cell. 2021;81:2722-35.e9
79. Capitanio U, Bensalah K, Bex A, Boorjian SA, Bray F, Coleman J. et al. Epidemiology of Renal Cell Carcinoma. European urology. 2019;75:74-84
80. Yao M, Tabuchi H, Nagashima Y, Baba M, Nakaigawa N, Ishiguro H. et al. Gene expression analysis of renal carcinoma: adipose differentiation-related protein as a potential diagnostic and prognostic biomarker for clear-cell renal carcinoma. The Journal of pathology. 2005;205:377-87
81. Morrissey JJ, London AN, Luo J, Kharasch ED. Urinary biomarkers for the early diagnosis of kidney cancer. Mayo Clinic proceedings. 2010;85:413-21
82. Morrissey JJ, Kharasch ED. The specificity of urinary aquaporin 1 and perilipin 2 to screen for renal cell carcinoma. The Journal of urology. 2013;189:1913-20
83. Morrissey JJ, Mobley J, Figenshau RS, Vetter J, Bhayani S, Kharasch ED. Urine aquaporin 1 and perilipin 2 differentiate renal carcinomas from other imaged renal masses and bladder and prostate cancer. Mayo Clinic proceedings. 2015;90:35-42
84. Gossage L, Eisen T, Maher ER. VHL, the story of a tumour suppressor gene. Nature reviews Cancer. 2015;15:55-64
85. Choueiri TK, Kaelin WG Jr. Targeting the HIF2-VEGF axis in renal cell carcinoma. Nature medicine. 2020;26:1519-30
86. Cai C, Chen S, Ng P, Bubley GJ, Nelson PS, Mostaghel EA. et al. Intratumoral de novo steroid synthesis activates androgen receptor in castration-resistant prostate cancer and is upregulated by treatment with CYP17A1 inhibitors. Cancer research. 2011;71:6503-13
87. Zadra G, Ribeiro CF, Chetta P, Ho Y, Cacciatore S, Gao X. et al. Inhibition of de novo lipogenesis targets androgen receptor signaling in castration-resistant prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:631-40
88. Lin LC, Gao AC, Lai CH, Hsieh JT, Lin H. Induction of neuroendocrine differentiation in castration resistant prostate cancer cells by adipocyte differentiation-related protein (ADRP) delivered by exosomes. Cancer letters. 2017;391:74-82
89. Fujimoto M, Yoshizawa A, Sumiyoshi S, Sonobe M, Menju T, Hirata M. et al. Adipophilin expression in lung adenocarcinoma is associated with apocrine-like features and poor clinical prognosis: an immunohistochemical study of 328 cases. Histopathology. 2017;70:232-41
90. Meng X, Sun R, Wang W, Zhang N, Cao S, Liu B. et al. ADFP promotes cell proliferation in lung adenocarcinoma via Akt phosphorylation. Journal of cellular and molecular medicine. 2021;25:827-39
91. Lucenay KS, Doostan I, Karakas C, Bui T, Ding Z, Mills GB. et al. Cyclin E Associates with the Lipogenic Enzyme ATP-Citrate Lyase to Enable Malignant Growth of Breast Cancer Cells. Cancer research. 2016;76:2406-18
92. Matsubara J, Honda K, Ono M, Sekine S, Tanaka Y, Kobayashi M. et al. Identification of adipophilin as a potential plasma biomarker for colorectal cancer using label-free quantitative mass spectrometry and protein microarray. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2011;20:2195-203
93. Straub BK, Herpel E, Singer S, Zimbelmann R, Breuhahn K, Macher-Goeppinger S. et al. Lipid droplet-associated PAT-proteins show frequent and differential expression in neoplastic steatogenesis. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2010;23:480-92
94. Szigeti A, Minik O, Hocsak E, Pozsgai E, Boronkai A, Farkas R. et al. Preliminary study of TIP47 as a possible new biomarker of cervical dysplasia and invasive carcinoma. Anticancer research. 2009;29:717-24
95. Lafontan M, Langin D. Lipolysis and lipid mobilization in human adipose tissue. Progress in lipid research. 2009;48:275-97
96. Egan JJ, Greenberg AS, Chang MK, Wek SA, Moos MC Jr, Londos C. Mechanism of hormone-stimulated lipolysis in adipocytes: translocation of hormone-sensitive lipase to the lipid storage droplet. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:8537-41
97. Zhang Q, Zhang P, Li B, Dang H, Jiang J, Meng L. et al. The Expression of Perilipin Family Proteins can be used as Diagnostic Markers of Liposarcoma and to Differentiate Subtypes. Journal of Cancer. 2020;11:4081-90
98. Lu C, Wang X, Zhao X, Xin Y, Liu C. Long non-coding RNA ARAP1-AS1 accelerates cell proliferation and migration in breast cancer through miR-2110/HDAC2/PLIN1 axis. Bioscience reports. 2020 40
99. Meng LX, Zheng YX, He ML, Zhou XM, Sun SY, Ding ZJ. et al. Silencing of perilipin by short hairpin RNA inhibits proliferation and induces apoptosis in liposarcoma cells. Molecular medicine reports. 2018;18:4571-6
100. Sánchez-Romero C, Carreón-Burciaga R, Gónzalez-Gónzalez R, Villarroel-Dorrego M, Molina-Frechero N, Bologna-Molina R. Perilipin 1 and adipophilin immunoexpression suggests the presence of lipid droplets in tooth germ, ameloblastoma, and ameloblastic carcinoma. Journal of oral pathology & medicine: official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 2021;50:708-15
101. Chen W, Chang B, Wu X, Li L, Sleeman M, Chan L. Inactivation of Plin4 downregulates Plin5 and reduces cardiac lipid accumulation in mice. American journal of physiology Endocrinology and metabolism. 2013;304:E770-9
102. Giménez-Andrés M, Emeršič T, Antoine-Bally S, D'Ambrosio JM, Antonny B, Derganc J. et al. Exceptional stability of a perilipin on lipid droplets depends on its polar residues, suggesting multimeric assembly. eLife. 2021 10
103. Sirois I, Aguilar-Mahecha A, Lafleur J, Fowler E, Vu V, Scriver M. et al. A Unique Morphological Phenotype in Chemoresistant Triple-Negative Breast Cancer Reveals Metabolic Reprogramming and PLIN4 Expression as a Molecular Vulnerability. Molecular cancer research: MCR. 2019;17:2492-507
104. Sztalryd C, Brasaemle DL. The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochimica et biophysica acta Molecular and cell biology of lipids. 2017;1862:1221-32
105. Asimakopoulou A, Vucur M, Luedde T, Schneiders S, Kalampoka S, Weiss TS. et al. Perilipin 5 and Lipocalin 2 Expression in Hepatocellular Carcinoma. Cancers. 2019 11
106. Chen FJ, Yin Y, Chua BT, Li P. CIDE family proteins control lipid homeostasis and the development of metabolic diseases. Traffic (Copenhagen, Denmark). 2020;21:94-105
107. Wu L, Zhou L, Chen C, Gong J, Xu L, Ye J. et al. Cidea controls lipid droplet fusion and lipid storage in brown and white adipose tissue. Science China Life sciences. 2014;57:107-16
108. Gong J, Sun Z, Wu L, Xu W, Schieber N, Xu D. et al. Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. The Journal of cell biology. 2011;195:953-63
109. Matsusue K, Kusakabe T, Noguchi T, Takiguchi S, Suzuki T, Yamano S. et al. Hepatic steatosis in leptin-deficient mice is promoted by the PPARgamma target gene Fsp27. Cell metabolism. 2008;7:302-11
110. Sun Z, Gong J, Wu H, Xu W, Wu L, Xu D. et al. Perilipin1 promotes unilocular lipid droplet formation through the activation of Fsp27 in adipocytes. Nature communications. 2013;4:1594
111. Wu L, Xu D, Zhou L, Xie B, Yu L, Yang H. et al. Rab8a-AS160-MSS4 regulatory circuit controls lipid droplet fusion and growth. Developmental cell. 2014;30:378-93
112. Liang L, Zhao M, Xu Z, Yokoyama KK, Li T. Molecular cloning and characterization of CIDE-3, a novel member of the cell-death-inducing DNA-fragmentation-factor (DFF45)-like effector family. The Biochemical journal. 2003;370:195-203
113. Min J, Zhang W, Gu Y, Hong L, Yao L, Li F. et al. CIDE-3 interacts with lipopolysaccharide-induced tumor necrosis factor, and overexpression increases apoptosis in hepatocellular carcinoma. Medical oncology (Northwood, London, England). 2011;28(Suppl 1):S219-27
114. Liu K, Zhou S, Kim JY, Tillison K, Majors D, Rearick D. et al. Functional analysis of FSP27 protein regions for lipid droplet localization, caspase-dependent apoptosis, and dimerization with CIDEA. American journal of physiology Endocrinology and metabolism. 2009;297:E1395-413
115. Bhatt-Wessel B, Jordan TW, Miller JH, Peng L. Role of DGAT enzymes in triacylglycerol metabolism. Archives of biochemistry and biophysics. 2018;655:1-11
116. Kuerschner L, Moessinger C, Thiele C. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic (Copenhagen, Denmark). 2008;9:338-52
117. McFie PJ, Banman SL, Stone SJ. Diacylglycerol acyltransferase-2 contains a c-terminal sequence that interacts with lipid droplets. Biochimica et biophysica acta Molecular and cell biology of lipids. 2018;1863:1068-81
118. Villanueva CJ, Monetti M, Shih M, Zhou P, Watkins SM, Bhanot S. et al. Specific role for acyl CoA:Diacylglycerol acyltransferase 1 (Dgat1) in hepatic steatosis due to exogenous fatty acids. Hepatology (Baltimore, Md). 2009;50:434-42
119. Liu Q, Siloto RM, Lehner R, Stone SJ, Weselake RJ. Acyl-CoA:diacylglycerol acyltransferase: molecular biology, biochemistry and biotechnology. Progress in lipid research. 2012;51:350-77
120. Nardi F, Franco OE, Fitchev P, Morales A, Vickman RE, Hayward SW. et al. DGAT1 Inhibitor Suppresses Prostate Tumor Growth and Migration by Regulating Intracellular Lipids and Non-Centrosomal MTOC Protein GM130. Scientific reports. 2019;9:3035
121. Morales A, Greenberg M, Nardi F, Gil V, Hayward SW, Crawford SE. et al. Loss of ephrin B2 receptor (EPHB2) sets lipid rheostat by regulating proteins DGAT1 and ATGL inducing lipid droplet storage in prostate cancer cells. Laboratory investigation; a journal of technical methods and pathology. 2021;101:921-34
122. Giudetti AM, De Domenico S, Ragusa A, Lunetti P, Gaballo A, Franck J. et al. A specific lipid metabolic profile is associated with the epithelial mesenchymal transition program. Biochimica et biophysica acta Molecular and cell biology of lipids. 2019;1864:344-57
123. Corbet C, Bastien E, Santiago de Jesus JP, Dierge E, Martherus R, Vander Linden C. et al. TGFβ2-induced formation of lipid droplets supports acidosis-driven EMT and the metastatic spreading of cancer cells. Nature communications. 2020;11:454
124. Li S, Wu T, Lu YX, Wang JX, Yu FH, Yang MZ. et al. Obesity promotes gastric cancer metastasis via diacylglycerol acyltransferase 2-dependent lipid droplets accumulation and redox homeostasis. Redox biology. 2020;36:101596
125. Nisticò C, Pagliari F, Chiarella E, Fernandes Guerreiro J, Marafioti MG, Aversa I. et al. Lipid Droplet Biosynthesis Impairment through DGAT2 Inhibition Sensitizes MCF7 Breast Cancer Cells to Radiation. International journal of molecular sciences. 2021 22
126. Li Y, Li T, Jin Y, Shen J. Dgat2 reduces hepatocellular carcinoma malignancy via downregulation of cell cycle-related gene expression. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;115:108950
127. Mazar J, Zhao W, Khalil AM, Lee B, Shelley J, Govindarajan SS. et al. The functional characterization of long noncoding RNA SPRY4-IT1 in human melanoma cells. Oncotarget. 2014;5:8959-69
128. Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M. et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science (New York, NY). 2004;306:1383-6
129. Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J. et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science (New York, NY). 2006;312:734-7
130. Olzmann JA, Richter CM, Kopito RR. Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1345-50
131. Lass A, Zimmermann R, Haemmerle G, Riederer M, Schoiswohl G, Schweiger M. et al. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell metabolism. 2006;3:309-19
132. Yang X, Lu X, Lombès M, Rha GB, Chi YI, Guerin TM. et al. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell metabolism. 2010;11:194-205
133. Zhang X, Saarinen AM, Hitosugi T, Wang Z, Wang L, Ho TH. et al. Inhibition of intracellular lipolysis promotes human cancer cell adaptation to hypoxia. eLife. 2017 6
134. Vegliante R, Di Leo L, Ciccarone F, Ciriolo MR. Hints on ATGL implications in cancer: beyond bioenergetic clues. Cell death & disease. 2018;9:316
135. Di Leo L, Vegliante R, Ciccarone F, Salvatori I, Scimeca M, Bonanno E. et al. Forcing ATGL expression in hepatocarcinoma cells imposes glycolytic rewiring through PPAR-α/p300-mediated acetylation of p53. Oncogene. 2019;38:1860-75
136. Xie H, Heier C, Kien B, Vesely PW, Tang Z, Sexl V. et al. Adipose triglyceride lipase activity regulates cancer cell proliferation via AMP-kinase and mTOR signaling. Biochimica et biophysica acta Molecular and cell biology of lipids. 2020;1865:158737
137. Yin L, Wang Y. Long non-coding RNA NEAT1 facilitates the growth, migration, and invasion of ovarian cancer cells via the let-7 g/MEST/ATGL axis. Cancer cell international. 2021;21:437
138. Zagani R, El-Assaad W, Gamache I, Teodoro JG. Inhibition of adipose triglyceride lipase (ATGL) by the putative tumor suppressor G0S2 or a small molecule inhibitor attenuates the growth of cancer cells. Oncotarget. 2015;6:28282-95
139. Al-Zoughbi W, Pichler M, Gorkiewicz G, Guertl-Lackner B, Haybaeck J, Jahn SW. et al. Loss of adipose triglyceride lipase is associated with human cancer and induces mouse pulmonary neoplasia. Oncotarget. 2016;7:33832-40
140. Tomin T, Fritz K, Gindlhuber J, Waldherr L, Pucher B, Thallinger GG. et al. Deletion of Adipose Triglyceride Lipase Links Triacylglycerol Accumulation to a More-Aggressive Phenotype in A549 Lung Carcinoma Cells. Journal of proteome research. 2018;17:1415-25
141. Castelli S, Ciccarone F, Tavian D, Ciriolo MR. ROS-dependent HIF1α activation under forced lipid catabolism entails glycolysis and mitophagy as mediators of higher proliferation rate in cervical cancer cells. Journal of experimental & clinical cancer research: CR. 2021;40:94
142. Balaban S, Shearer RF, Lee LS, van Geldermalsen M, Schreuder M, Shtein HC. et al. Adipocyte lipolysis links obesity to breast cancer growth: adipocyte-derived fatty acids drive breast cancer cell proliferation and migration. Cancer & metabolism. 2017;5:1
143. Wang YY, Attané C, Milhas D, Dirat B, Dauvillier S, Guerard A. et al. Mammary adipocytes stimulate breast cancer invasion through metabolic remodeling of tumor cells. JCI insight. 2017;2:e87489
144. Iftikhar R, Penrose HM, King AN, Samudre JS, Collins ME, Hartono AB. et al. Elevated ATGL in colon cancer cells and cancer stem cells promotes metabolic and tumorigenic reprogramming reinforced by obesity. Oncogenesis. 2021;10:82
145. Lettieri Barbato D, Aquilano K, Baldelli S, Cannata SM, Bernardini S, Rotilio G. et al. Proline oxidase-adipose triglyceride lipase pathway restrains adipose cell death and tissue inflammation. Cell death and differentiation. 2014;21:113-23
146. Sathyanarayan A, Mashek MT, Mashek DG. ATGL Promotes Autophagy/Lipophagy via SIRT1 to Control Hepatic Lipid Droplet Catabolism. Cell reports. 2017;19:1-9
147. Zhang C, Boppart SA. Dynamic Signatures of Lipid Droplets as New Markers to Quantify Cellular Metabolic Changes. Analytical chemistry. 2020;92:15943-52
148. Zhang F, Liu Y, Yang B, Guan P, Chai J, Wen G. et al. Tunable NIR AIE-active optical materials for lipid droplet imaging in typical model organisms and photodynamic therapy. Journal of materials chemistry B. 2021;9:2417-27
149. Salo VT, Belevich I, Li S, Karhinen L, Vihinen H, Vigouroux C. et al. Seipin regulates ER-lipid droplet contacts and cargo delivery. The EMBO journal. 2016;35:2699-716
150. Joshi AS, Nebenfuehr B, Choudhary V, Satpute-Krishnan P, Levine TP, Golden A. et al. Lipid droplet and peroxisome biogenesis occur at the same ER subdomains. Nature communications. 2018;9:2940
151. Markovic SN, Suman VJ, Rao RA, Ingle JN, Kaur JS, Erickson LA. et al. A phase II study of ABT-510 (thrombospondin-1 analog) for the treatment of metastatic melanoma. American journal of clinical oncology. 2007;30:303-9
152. Yao M, Huang Y, Shioi K, Hattori K, Murakami T, Nakaigawa N. et al. Expression of adipose differentiation-related protein: a predictor of cancer-specific survival in clear cell renal carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:152-60
153. Yao D, Xia S, Jin C, Zhao W, Lan W, Liu Z. et al. Feedback activation of GATA1/miR-885-5p/PLIN3 pathway decreases sunitinib sensitivity in clear cell renal cell carcinoma. Cell cycle (Georgetown, Tex). 2020;19:2195-206
154. Wang K, Ruan H, Song Z, Cao Q, Bao L, Liu D. et al. PLIN3 is up-regulated and correlates with poor prognosis in clear cell renal cell carcinoma. Urologic oncology. 2018;36:343.e9-e19
155. Lamprou I, Kakouratos C, Tsolou A, Pavlidis P, Xanthopoulou ET, Nanos C. et al. Lipophagy-Related Protein Perilipin-3 and Resistance of Prostate Cancer to Radiation Therapy. International journal of radiation oncology, biology, physics. 2022
156. Mitra R, Le TT, Gorjala P, Goodman OB Jr. Positive regulation of prostate cancer cell growth by lipid droplet forming and processing enzymes DGAT1 and ABHD5. BMC cancer. 2017;17:631
157. Gao C, Zhuang J, Li H, Liu C, Zhou C, Liu L. et al. Development of a risk scoring system for evaluating the prognosis of patients with Her2-positive breast cancer. Cancer cell international. 2020;20:121
158. Sun X, Yang S, Feng X, Zheng Y, Zhou J, Wang H. et al. The modification of ferroptosis and abnormal lipometabolism through overexpression and knockdown of potential prognostic biomarker perilipin2 in gastric carcinoma. Gastric cancer: official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association. 2020;23:241-59
159. Chatterjee A, Mondal P, Ghosh S, Mehta VS, Sen E. PPARγ regulated CIDEA affects pro-apoptotic responses in glioblastoma. Cell death discovery. 2015;1:15038
160. Li J, Li Q, Su Z, Sun Q, Zhao Y, Feng T. et al. Lipid metabolism gene-wide profile and survival signature of lung adenocarcinoma. Lipids in health and disease. 2020;19:222
161. Zhou Q, Sun Y. Circular RNA cMras Suppresses the Progression of Lung Adenocarcinoma Through ABHD5/ATGL Axis Using NF-κB Signaling Pathway. Cancer biotherapy & radiopharmaceuticals. 2020
162. Honeder S, Tomin T, Nebel L, Gindlhuber J, Fritz-Wallace K, Schinagl M. et al. Adipose Triglyceride Lipase Loss Promotes a Metabolic Switch in A549 Non-Small Cell Lung Cancer Cell Spheroids. Molecular & cellular proteomics: MCP. 2021;20:100095
Author contact
![]() Corresponding authors: Gonghui Li, Department of Urology, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China. Email: 3193119edu.cn; Liqun Xia, Department of Urology, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China. Email: xialiqunedu.cn.
Corresponding authors: Gonghui Li, Department of Urology, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China. Email: 3193119edu.cn; Liqun Xia, Department of Urology, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China. Email: xialiqunedu.cn.

 Global reach, higher impact
Global reach, higher impact