10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2023; 19(1):120-136. doi:10.7150/ijbs.76345 This issue Cite
Research Paper
The developmental regulator HAND1 inhibits gastric carcinogenesis through enhancing ER stress apoptosis via targeting CHOP and BAK which is augmented by cisplatin
1. Biomedical Research Center, Sir Run Run Shaw Hospital, Zhejiang University, Hangzhou, China.
2. Department of Pathology, Sir Run Run Shaw Hospital, Zhejiang University, Hangzhou, China.
3. Key Laboratory of Cancer Prevention and Intervention, Ministry of Education, Hangzhou 310016, Zhejiang, China.
4. Cancer Epigenetics Laboratory, Department of Clinical Oncology, State Key Laboratory of Translational Oncology, Sir YK Pao Center for Cancer and Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong.
Received 2022-6-20; Accepted 2022-10-2; Published 2023-1-1
Abstract
Epigenetic disruption of tumor suppressor genes, particularly aberrant CpG methylation, plays a crucial role in gastric cancer (GC) pathogenesis. Through CpG methylome and expression profiling, a developmental transcription factor - Hand-And-Neural-crest-Derivative-expressed 1 (HAND1), was identified methylated and downregulated in GC. However, its role and underlying mechanisms in GC progression are poorly understood. Here, we show that HAND1 was frequently downregulated in GC by promoter methylation, and significantly correlated with tumor progression and poor prognosis of GC patients. High expression of HAND1 in GC patients was associated with significantly higher 5-year overall survival rates. Ectopic expression of HAND1 inhibited GC cell growth and migration in vitro and in vivo. HAND1 expression increased ROS levels and cytosolic Ca2+ concentration, enhanced cisplatin-induced apoptosis through endoplasmic reticulum (ER) stress/mitochondria-mediated apoptosis. Knockdown of CHOP and BAK attenuated HAND1-induced cell apoptosis. Overexpression of CHOP increased BAK expression. HAND1 interacts with CHOP, also directly binds to CHOP and BAK promoters and positively regulates BAK transcription. Thus, the present study demonstrates that HAND1 is a tumor suppressor gene methylated in GC, induces ER stress and apoptosis via CHOP and BAK, which is augmented by cisplatin. Low HAND1 expression is an independent poor prognostic factor for GC. The tumor-specific methylation of HAND1 promoter could be a candidate biomarker for GC.
Keywords: HAND1, gastric cancer, tumor suppressor gene, methylation, apoptosis, CHOP, BAK.
Introduction
Gastric cancer (GC) is the fifth most commonly diagnosed cancer and the third leading cause of cancer death worldwide. Although its overall incidence rates in northern Europe, North America, and African regions are generally low, the rates are markedly elevated in Eastern Asia, particularly in Japan, Korea and China [1, 2]. In addition to microbial agents and environmental factors, the initiation and progression of GC are characterized by gradual accumulation of multiple genetic and epigenetic alterations. Epigenetic alterations are pervasive and multifaceted in GC, including DNA CpG methylation, histone modifications, RNA editing and noncoding RNAs [3, 4]. Aberrant CpG methylation leads to the inactivation of tumor suppressor genes (TSGs), which then fundamentally contributes to gastric carcinogenesis and development [5, 6]. Thus, identification of novel TSGs targeted by promoter methylation in GC and further exploring the related mechanisms will greatly facilitate elucidating the molecular pathogenesis of GC, as well as the development of novel effective individualized therapeutic strategies for GC patients.
We searched for novel candidate TSGs in digestive tumors through epigenomics (CpG methylome) and gene expression profiling, and discovered that a transcription factor, Hand-And-Neural-crest-Derivative-expressed 1 (HAND1), was frequently methylated and downregulated in GC. The HAND subfamily, consisting of HAND1 and HAND2, belongs to the superfamily of basic helix-loop-helix (bHLH) transcription factors. HAND factors play key roles in the regulation of cardiac, gut, and sympathetic neuronal development [7, 8]. HAND factors are highly conserved across all species and activate or suppress the transcription of multiple downstream target genes [9, 10].
HAND1, located at chromosome 5q33, is essential for trophoblast giant cell differentiation and cardiac morphogenesis as a developmental regulator [11]. Several previous studies have shown that HAND1 plays an important role in cell proliferation and carcinogenesis. HAND1 has been reported to be downregulated and methylated in several cancers, including colorectal, pancreatic, small cell lung, ovarian and thyroid cancers, as well as melanoma [12-19], although the underlying mechanism studies are scanty. HAND1 expression is negatively regulated by the high-mobility group A1 (HMGA1) protein, and restoration of HAND1 expression leads to reduced growth of thyroid cancer cells [18]. Asuthkar et al. reported that nuclear translocation of HAND1 is directly regulated by uPAR protein, which controls medulloblastoma angiogenesis; HAND1 expression attenuates epithelial-mesenchymal transition (EMT) and inhibits medulloblastoma cell invasion and metastasis via Oct-3/4/β-catenin interaction [20-22]. HAND1, which is epigenetically silenced in colon cancer, had been identified as a Polycomb target that is closely related to ES cell differentiation. Ectopic expression of HAND1 induces terminal differentiation and inhibits the growth, proliferation and xenograft tumor formation of colorectal cancer cells [16, 23]. These studies indicate that HAND1 may be a tumor suppressor involved in the development of multiple cancers. However, its functions and underlying mechanisms in GC development are poorly understood.
In this study, we discovered that HAND1 was silenced or downregulated in most GC cell lines and thus may be a TSG candidate in GC. We further investigated the inactivation of HAND1 by promoter CpG methylation and explored its functions and potential mechanisms in the initiation and progression of GC. We found that HAND1 inhibits gastric carcinogenesis through enhancing ER stress apoptosis via targeting CHOP and BAK which is augmented by cisplatin. Moreover, HAND1 promoter methylation appears to be a good prognostic epigenetic biomarker for GC patients.
Materials and methods
Cell lines, tumors, and normal control tissues
A panel of GC cell lines, AGS, MKN28, MKN45, SNU1, SNU16, Kato-III, YCC1, YCC2, YCC3, YCC6, YCC7, YCC9, YCCEL1, YCC11, YCC16, and SNU719 were studied, with YCCEL1 and SNU719 as naturally EBV+ cell lines. Cell lines were purchased from ATCC or Cell Bank of Chinese Academy of Sciences and authenticated by short tandem repeat DNA profiling analysis, or from collabrators. Cell lines were cultured in RPMI-1640 or DMEM Medium (Gibco BRL, Rockville, MD) with 10% fetal bovine serum (FBS), plus 100 U/mL penicillin and 100 mg/mL streptomycin at 37 °C with 5% CO2.
A total of 165 GC patients who underwent surgery between May 1995 and October 2009 at the Sir Run Run Shaw Hospital (Hangzhou, Zhejiang, China) were assessed by immunohistochemistry. Patients who received preoperative radiotherapy, chemotherapy or immunotherapy before surgery were excluded from the study. Ten normal gastric mucosa biopsy samples were used as normal controls. Additionally, 35 GC cases and paired normal tissues were available for MSP. This study was approved by the ethics committee of Sir Run Run Shaw Hospital, Zhejiang University.
Expression and methylation analyses of HAND1 from public databases
mRNA expression analysis of HAND1 in GC specimens was retrieved from Oncomine microarray database (www.oncomine.org). Three valid mRNA expression datasets for GC in Oncomine database were analyzed. Expression data of HAND1 were downloaded, and statistical analyses performed using GraphPad Prism version 8.0 (GraphPad Software, Inc., San Diego, CA, USA). The relevance of HAND1 methylation to mRNA expression was analyzed using online MEXPRESS (https://mexpress.be) and cBioPortal database (www.cbioportal.org).
CpG methylome analysis
We performed CpG methylome analysis by methylated DNA Immunoprecipitation (MeDIP) using NimbleGen 385K CpG Island Plus Promoter Array, as described previously [24].
RNA extraction, semi-quantitative RT-PCR and quantitative real-time RT-PCR
Total RNA was extracted using an RNA Kit (Omega Bio-tek, Norcross, GA). Semi-quantitative reverse-transcription PCR (RT-PCR) and quantitative real time RT-PCR (qRT-PCR) were performed with GoTaq polymerase (Promega, Madison, WI) and UltraSYBR Mixture (CWBio, Beijing, China) following the manufacturer's instructions, respectively. qRT-PCR was performed using the ABI QuantStudio 6 Flex (Applied Biosystems, Foster City, CA). GAPDH mRNA was amplified as internal control. The specific primers used in this study are listed in Table S4.
5-Aza-2'-deoxycytidine (5-Aza) and trichostatin A (TSA) treatment
Cells with silenced HAND1 expression were treated with 10 μM demethylation agent, 5-Aza (5-Aza; Sigma-Aldrich, St Louis, MO) for 72 h, followed by 100 nM histone deacetylase inhibitor, TSA (TSA; Sigma-Aldrich, St Louis, MO) for 24 h[25]. After the treatment, cells were harvested for DNA and RNA extraction.
Bisulfite treatment and promoter methylation analysis
Bisulfite modification of DNA, methylation-specific PCR (MSP) and bisulfate genome sequencing (BGS) were conducted as previously described [25, 26]. MSP and BGS primers are listed in Table S4.
Immunohistochemistry (IHC)
The ChemMateTM EnVisionTM Detection Kit (DAKO, Carpinteria, CA) was used for IHC. Briefly, sections were incubated with HAND1 antibody (1:250 dilution; LS-B811, LSBio, Seattle, WA, USA) overnight at 4 °C.
Immunostaining results were evaluated independently by two investigators who were blinded to the clinicopathological outcomes of patients. HAND1 protein expression was scored according to the intensity of staining (0, negative; 1, weakly positive; 2, positive; and 3, strongly positive) and percentage of positive cells (0, 0-5%; 1, 5%-25%; 2, 25%-50%; 3, 50%-75%; 4, 75%-100%). The two scores were multiplied to obtain a value ranging from 0 to 12. To examine the association of HAND1 expression levels with clinicopathological features, patients were divided into two groups: low HAND1 (0-5) or high HAND1 expression (6-12).
HAND1-expressing plasmid and cell transfection
GC cell line AGS and MKN28 were transfected with pCMV6-Entry HAND1 plasmid or empty vector (pCMV6-EntryVector) (Origene, Rockville, MD) as control, using Lipofectamine™ 3000 transfection reagent (Invitrogen, Carlsbad, CA). Stable HAND1-expressing and control vector clones were selected for further study.
Cell viability assays
Stable transfected cells were seeded in 96-well plates at a density of 5 ×103 cells/well and incubated for 24 h. Cell viability was assessed every day or after being treated with cisplatin (25 μM for AGS and MKN28; APExBio, Houston, TX) for 24 h according to the Cell Counting Kit-8 (CCK-8) protocol (Dojindo, Kumamoto, Japan). All experiments were performed at least in triplicate.
Colony formation assay
For colony formation assays, 1,000 cells were seeded in a 60-mm dish and allowed to grow for 2-3 weeks. Surviving colonies (≥50 cells/colony) were counted after crystal violet staining.
Wound healing assay
Wound healing assay was used to assess cell motility. Stably transfected cells were cultured in 6-well plates with 10 μg/mL mitomycin C (MCE, Princeton, NJ) until confluent. The cell layer was wounded using a sterile tip and washed twice with phosphate-buffered saline (PBS). Cells were incubated and photographed under a phase contrast microscope at different time points. The experiments were performed in triplicate.
Transwell cell migration assay
For the Transwell assay, 2×105 cells were resuspended in 100 μL serum-free medium and seeded in the upper chamber of a Transwell plate (Corning Inc., Corning, NY) consisting of inserts containing 8-µm pore-size PET membranes. Approximately 600 µL of medium containing 10% FBS was added to the lower chamber. After 16-24h of incubation at 37 °C, cells on the lower side of the upper chamber were fixed, stained with 0.1% crystal violet, and counted under a light microscope. The experiment was performed in triplicate.
Cell cycle and apoptosis analysis
Cell cycle distribution and percentages of apoptosis were measured using cell cycle staining Kit (MultiSciences, Hangzhou, Zhejiang, China), Annexin V-FITC Apoptosis Detection Kit I (BD, San Jose, CA), or Annexin V-APC/7-AAD apoptosis kit (MultiSciences, Hangzhou, Zhejiang, China), with flow cytometry according to manufacturer's instructions.
Protein extraction and Western blot
Cells were collected from cultured dishes and lysed in RIPA lysis buffer (Beyotime, Hangzhou, Zhejiang, China) supplemented with protease inhibitors or phosphatase inhibitors. Cytoplasmic, mitochondrial, and nuclear protein fractions were extracted using ProteoExtract Subcellular Proteome Extraction Kit (Millipore, Billerica, MA). Protein concentrations were quantified using a BCA Protein Assay Kit (Beyotime, Hangzhou, Zhejiang, China). Cell lysates (40 μg protein/line) were separated via 6%-15% SDS-PAGE and transferred to 0.45-μm thick polyvinylidene difluoride (PVDF) membranes (Millipore). The blotted membranes were blocked with 5% skim milk for 1 h at room temperature. Afterward, membranes were incubated with primary antibodies (1:1,000) overnight at 4 °C and then with HRP-labeled secondary antibody (1:2,000) for 1 h at room temperature. All the antibodies used in this study were purchased from Cell Signaling Technology (Beverly, MA). Detection was performed on a Fujifilm Las-4000 Luminescent Imaging System using ECL Kit (Pierce, Rockford, IL).
Measurement of MMP
MMP assay kit (Beyotime) with JC-1 was used to detect MMP as described by the manufacturer. Briefly, the cells were collected and stained with 0.5 mL JC-1 working solution for 20 min at 37 °C. Then, the percentage of red and green fluorescence was estimated by flowcytometry. All experiments were replicated in triplicate.
Measurement of intracellular Ca2+ levels
Intracellular Ca2+ levels were measured using Fluo-3AM (Beyotime) as previously described [27, 28]. [Ca2+]i was derived after calibration according to the following equation: [Ca2+]i (nM)=Kd (F-Fmin)/(Fmax-F). F is the base line fluorescence. Fmin is the fluorescence in the presence of EGTA. Fmax is the fluorescence detected with saturating Ca2+. Kd (400 nM) is the dissociation constant of Fluo-3AM for Ca2+ at room temperature. All experiments were replicated in triplicate.
Measurement of ROS
ROS Assay Kit (Beyotime) was used to measure ROS production levels as described by the manufacturer. Briefly, 1×106 cells were stained with 10 μM DCFH-DA for 20 min at 37 °C and analyzed by flowcytometry. All experiments were replicated in triplicate.
RNA sequencing (RNA-Seq)
Total RNA was extracted from stable HAND1-expressing and control vector cells, replicated thrice for each sample. RNA amount and purity of each sample were quantified using NanoDrop ND-1000 (NanoDrop, Wilmington, DE). Poly (A) RNA is purified from 1 μg total RNA using Dynabeads Oligo (dT)25-61005 (Thermo Fisher, Carlsbad, CA), and fragmented into small pieces using Magnesium RNA Fragmentation Module (NEB, Ipswich, MA) under elevated temperature. Cleaved RNA fragments were reverse-transcribed and the final cDNA library was constructed according to the protocol for mRNA-Seq sample preparation kit (Illumina, San Diego, CA). Subsequently, paired-end sequencing was performed on an Illumina Novaseq™ 6000 (LC-Bio, Hangzhou, Zhejiang, China) following the vendor's recommended protocol.
Sequencing reads were aligned to human reference genome (GRCh38) using HISAT2 software (Version 2.0.4) after trimming adapter sequences and removing low-quality reads using Cutadapt software (Version 1.9). FPKM were used to estimate the relative abundance of all transcripts and expression level for mRNAs. Differentially expressed mRNAs were selected with p value < 0.05 and fold change > 2, or fold change < 0.5 by R package edgeR and DESeq2, and then GO enrichment and KEGG enrichment analysis were performed.
In vivo subcutaneous tumor model
All in vivo animal experiments were approved by the animal care committee of Sir Run Run Shaw Hospital. Viable MKN28 and control cells (5 × 106 cells in 100 μL PBS) were injected subcutaneously into the right dorsal flank of 6-week-old female BALB/c nude mice (five mice per group). Tumor volume was measured every two days, and tumor weight was measured at the end of the fourth week. Tumor volume was calculated using the following formula: (Short diameter)2 × (Long diameter)/2.
RNA interference
Stable transfected cells were transfected with BAK siRNA (Ambion, Austin, TX), CHOP siRNA (Genechem, Shanghai, China) or negative control siRNA using Lipofectamine RNAiMax Transfection Reagent (Invitrogen) according to manufacturer's instructions. Approximately 72 h later, total proteins were extracted, and the efficiency of siRNA was confirmed by Western blotting.
Chromatin immunoprecipitation (ChIP)
ChIP assay was performed using Simple ChIP® Plus Enzymatic Chromatin IP Kit (Cell Signaling Technology, Danvers, MA). Sonicated nuclear fractions were incubated with anti-FLAG (Cell Signaling Technology), positive control histone H3 (Cell Signaling Technology), and negative control normal rabbit IgG (Cell Signaling Technology). Immunoprecipitated DNA was identified by PCR using specific primers for BAK or CHOP promoter. The sequences of the primers are shown in Table S4.
Luciferase reporter assay
Stable transfected cells were co-transfected with CHOP promotor-firefly luciferase reporter plasmid and pRL-TK Renilla plasmid (Genechem, Shanghai, China), or negative control and BAK-GLuc (GeneCopoeia, Rockville, MD). Luciferase activity was normalized by that of cells co-transfected with pRL-TK Renilla vector or SEAP expression vector (GeneCopoeia). Supernatants of GLuc-transfected cells or cell lysates were analyzed with a Dual-Lumi™ Luciferase Reporter Gene Assay Kit (Beyotime) or Secrete-Pair™ Dual Luminescence Assay Kit (GeneCopoeia) according to manufacturer's protocols.
Immunofluorescence
Cells grown on coverslips were fixed with 4% paraformaldehyde (Beyotime) for 10 min, permeabilized in 0.1% Triton X-100 for 4 min and blocked with 3% bovine serum albumin (Beyotime) for 20 min. Cells were subsequently incubated at 4°C overnight with HAND1 monoclonal antibody (Origene), or FLAG Tag antibody (Cell Signaling Technology) and CHOP polyclonal antibody (Proteintech, Chicago, IL), and incubated with both anti-mouse Alexa-Fluor 488 and Rhodamine phalloidin (Invitrogen) or anti-rabbit Alexa-Fluor 568 (Invitrogen) for 1 h at room temperature. Rhodamine phalloidin staining was used to visualize F-actin. Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI, Invitrogen), and fluorescence examined by Olympus BX51 microscope (Olympus, Tokyo, Japan).
Co-immunoprecipitation (Co-IP) assay
Total protein lysates were extracted from cells using RIPA lysis buffer (Beyotime) supplemented with protease inhibitors cocktail. 500 μg protein lysates, combined with 10 µg of CHOP polyclonal antibody (Proteintech), were incubated for 1-2 h at room temperature with mixing. Then, 25 µL Protein A/G magnetic beads (Thermo Fisher) were added and the mixture was incubated at 4 °C overnight with mixing. The obtained immune complexes were washed four times, boiled with 2× SDS-PAGE Sample Loading Buffer (Beyotime), and subsequently detected by Western blot.
Statistical analysis
Results were all presented as mean values ± SD. Statistical analyses were performed in SPSS version 19.0 (SPSS Inc., Chicago, IL) and GraphPad Prism software version 8.0 (GraphPad Software Inc., San Diego, CA). One-way ANOVA or two-tailed Student's t-test were used to analyze differences between groups. Chi-square tests were used to analyze the relationship between HAND1 expression and clinicopathological parameters. OS was calculated using Kaplan-Meier method with log-rank test. Univariate and multivariate Cox regression analyses were performed to evaluate prognostic factors. For all tests, p<0.05 was considered statistically significant.
Results
Expression and CpG methylome study identifies HAND1 as a methylation-silenced target in gastric cancer
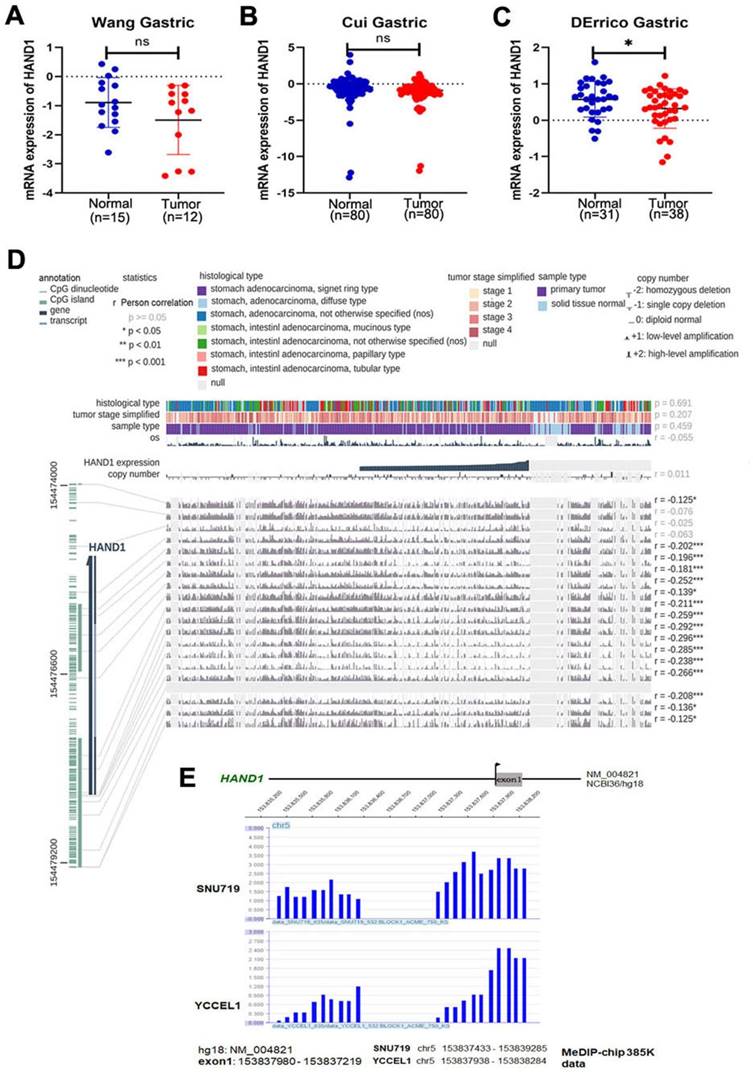
Through analyzing HAND1 mRNA expression data of GC patients in the Oncomine database, we found that HAND1 was downregulated in GC cases compared with normal gastric samples (Figure 1A-C). To further determine the mechanism of HAND1 downregulation in GC, we analyzed the correlation between HAND1 expression and CpG methylation in TCGA stomach adenocarcinoma dataset using MEXPRESS. Assessment using Pearson's correlation coefficient indicated that HAND1 expression in GC is significantly negatively correlated with its methylation levels (r up to -0.296, p<0.001, Figure 1D), indicating that promoter methylation might lead to HAND1 mRNA downregulation. Meanwhile, we performed CpG methylome analysis to identify cancer genes in GC. Methylome data showed strong signal enrichment in CpG island (CGI) of HAND1 promoter in SNU719 and YCCEL1 gastric cell lines, and defined HAND1 as a methylated target in GC (Figure 1E).
HAND1 silencing/downregulation by promoter methylation in GC cell lines and tumors
To further validate HAND1 expression and methylation status in GC tumors, we assessed HAND1 mRNA expression in GC cell lines. Results showed that HAND1 expression was silenced or downregulated in most cell lines (Figure 2A). Then, methylation-specific PCR (MSP) was conducted to analyze HAND1 promoter methylation status. HAND1 methylation was observed in 13/16 (81%) of cell lines (Figure 2A).
GC methylome study identifies HAND1 promoter methylation in gastric cancer and associated with its downregulation. (A-C) HAND1 mRNA expression is frequently downregulated in gastric tumor cases (Tumor) compared with normal gastric samples (Normal) in Oncomine database. (D) HAND1 expression is negatively correlated with promoter CpG methylation, by Pearson correlation coefficients in MEXPRESS database. (E) CpG methylome analysis by MeDIP-Chip demonstrate signal enrichment in HAND1 promoter CGI in gastric cancer. Positive signal peaks (blue) are marked. Data is presented as mean ± SD. *p<0.05; **p<0.01; ***p<0.001.
HAND1 expression and methylation status in GC cell lines and primary tumors, and its prognosis value. (A) Silencing or downregulation of HAND1 in GC cell lines due to its promoter methylation. (B) RTPCR and MSP show that HAND1 in silenced and methylated GC cell lines, but restored after treatment with demethylation agent 5-Aza and TSA (A+T). Representative results are shown. (C) MSP analysis shows that HAND1 promoter methylation or mRNA expression in silenced and methylated GC cell lines was not affected by treatment with Cisplatin for 24 h (25 μM). (D) Representative results of HAND1 promoter methylation by MSP in primary gastric tumor tissues (T) and paired adjacent normal tissues (N). (E) Representative BGS results on the methylation status of HAND1 CGI. Cloned BGS-PCR products are sequenced, and each colony shown as an individual row, representing a single allele of the CGI. Open circles represent unmethylated, and filled circles represent methylated CpG sites. (F) Representative immunohistochemical staining of HAND1 in normal gastric mucosa and GC tissues. Original magnification: 200×. (G) Kaplan-Meier survival analysis about the relationship of HAND1 expression and five-year overall survival rates in GC patients. GC patients with low HAND1 expression had poor prognosis. M: methylated; U: unmethylated. TSS: transcriptional start site.
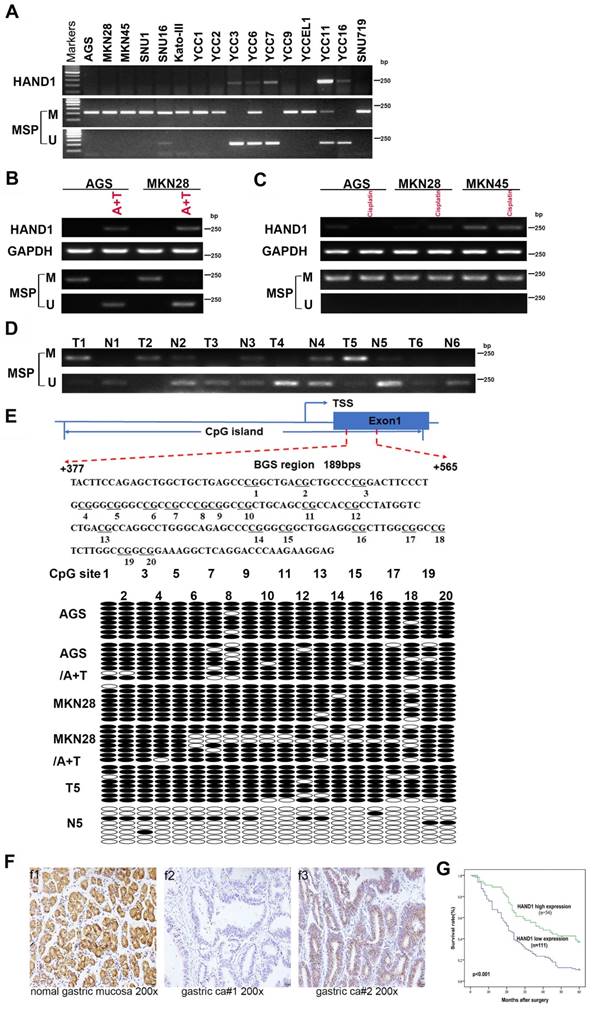
Meanwhile, HAND1 mRNA expression was restored after demethylation treatment with 5-Aza and TSA, and representative results are shown in Figure 2B. Promoter methylation and mRNA expression of HAND1 were not affected by treatment with Cisplatin (Figure 2C). Detailed methylation profiling of HAND1 CGI was further performed by bisulfite genomic sequencing (BGS) analysis of 20 CpG sites in the CGI (Figure 2E). BGS showed that HAND1 CGI was heavily methylated in GC cell lines, whereas only partial demethylation detected after 5-Aza and TSA treatment, in agreement with the MSP results (Figure 2B, 2E).
We further investigated HAND1 promoter methylation in 35 pairs of GC tissue samples by MSP. In 60% (21/35) of cases, HAND1 methylation levels were higher than paired adjacent normal controls. Similar results were obtained by BGS analysis of paired GC and adjacent normal tissues. Representative results are shown in Figure 2D and 2E. These results indicate that HAND1 expression is regulated through promoter CGI methylation.
Relationship of HAND1 expression and clinicopathological features of GC patients
HAND1 protein expression was evaluated by immunohistochemistry in 10 normal gastric mucosa biopsy specimens and 165 gastric cancer cases. HAND1 protein was mainly expressed in the cytoplasm and nuclei of tissues (Figure 2F). HAND1 protein was highly expressed in all normal gastric mucosa, but absent or downregulated in 67 % (111/165) of GC samples.
Correlation between clinicopathological parameters and HAND1 protein expression of GC patients was further analyzed (Table S1). HAND1 expression was significantly correlated with gender (p=0.019), histopathological grading (p=0.050), depth of invasion (p=0.018), lymph nodal status (p=0.017), and TNM stage (p=0.005). To ascertain the effect of HAND1 expression on the prognosis of GC patients, all GC patients were followed up for five-year overall survival (OS) after surgery. Kaplan-Meier survival analysis showed that GC patients of high HAND1 expression had higher five-year overall survival rates, compared with those with low HAND1 expression (p<0.001, Figure 2G).
Univariate and multivariate analyses with Cox regression were conducted to determine key prognostic factors of OS (Table S2 and S3). In univariate analysis, HAND1 expression (p<0.001), age (p=0.012), histopathological grading (p=0.047), depth of invasion (p<0.001), lymph node metastasis (p<0.001), distant metastasis (p<0.001), and TNM stage (p<0.001) were significantly associated with OS in GC patients. These statistically significant factors were introduced to the Cox regression model, and multivariate analyses were conducted. The results showed that HAND1 expression (p<0.001), distant metastasis (p= 0.0046), and depth of invasion (p<0.001) were independent prognostic factors for OS in GC. These results suggested that low expression of HAND1 predicted poor prognosis and was correlated with tumor progression in GC patients.
Ectopic HAND1 expression inhibits GC cell growth and migration
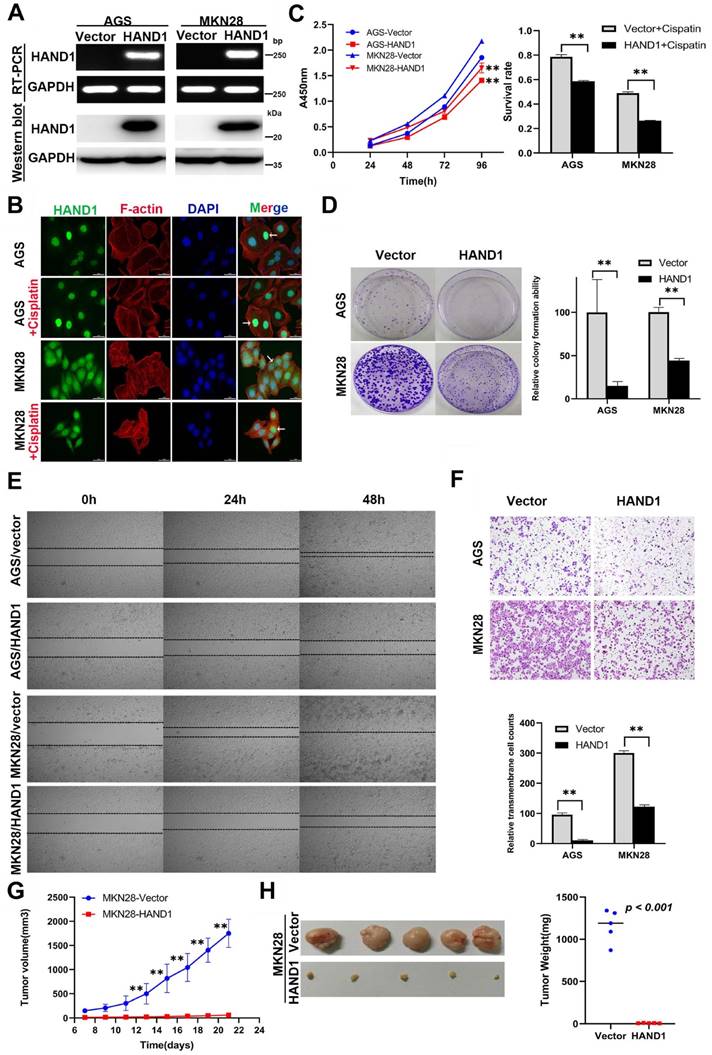
HAND1 downregulation by promoter methylation was significantly associated with malignant progression of GC, indicating its important role in GC tumorigenesis, we thus further investigated its potential biological functions in GC cells. We established two cell lines (AGS and MKN28) stably-expressing HAND1, with empty vector transfection as control. Expression levels of HAND1 mRNA and protein in these cell lines were confirmed by RT-PCR and Western blot (Figure 3A). Immunofluorescence assays showed that HAND1 protein was predominantly localized in the nucleus (Figure 3B). Cell viability assays showed that the cell survival rate of HAND1-transfected cells was significantly lower than that of control cells, especially after cisplatin treatment (p<0.05, Figure 3C). Moreover, ectopic HAND1 expression significantly inhibited the colony formation ability of transfected cells, compared with control cells (p<0.05, Figure 3D).
To assess the effects of HAND1 expression on the migration and invasion of GC cells, wound healing and transwell assays were conducted. Wound-healing assays showed that HAND1 stably-expressing cells took longer to heal the wound than that of control cells (Figure 3E). Transwell assays also showed that HAND1 inhibits GC cell migration (p<0.05, Figure 3F).
We also evaluated the effects of HAND1 on GC cell growth in vivo by injecting MKN28-HAND1 cells into nude mice. The tumor growth rate of MKN28-HAND1 cells in nude mice was significantly lower than that of control cells (p<0.05, Figure 3G). Tumor weight was significantly lower in HAND1-expressing nude mouse group, compared with control group (p<0.05, Figure 3H). These results suggested that HAND1 inhibits GC cell growth and migration in vitro and in vivo, and functions as a tumor suppressor in GC carcinogenesis.
Ectopic HAND1 expression inhibits GC cell growth and migration. (A) HAND1 mRNA and protein expression in stably transfected cells as confirmed by RT-PCR and Western blot. (B) Immunofluorescence staining is used to identify the subcellular location of HAND1 in HAND1-transfected cells before and after cisplatin treatment for 24 h (25 μM). Scale bar, 10 µm. (C) HAND1 significantly inhibits cell viability, without or with cisplatin treatment. (D) HAND1 significantly inhibits cell colony formation. (E) Representative photos of wound healing assay (Original magnification:100×). (F) Representative images of Transwell assays (Original magnification:100×). The number of migrating cells in five random fields per Transwell was counted for quantitative analysis. (G-H) HAND1 suppresses subcutaneous tumor growth in nude mice. Quantitative analyses of tumor volume and tumor weight are shown. All values are expressed as mean ± SD of three independent experiments. *p<0.05; **p<0.01.
Ectopic HAND1 expression upregulates apoptosis and intracellular reactive oxygen species levels
To investigate whether inhibition of GC cell proliferation and growth by HAND1 is related to cell apoptosis and cell cycle arrest, apoptosis and cell cycle analyses were conducted. Cell cycle analysis showed that the cell cycle distribution of HAND1-transfected cells was not significantly different from that of control cells (data not shown). We then examined spontaneous and cisplatin-induced apoptosis of HAND1-transfected cells and control cells. HAND1 expression significantly increased the spontaneous apoptosis in AGS, but not in MKN28. After cisplatin treatment, the percentage of apoptosis cells in HAND1-transfected AGS and MKN28 cells was significantly higher, compared with control cells (p<0.05, Figure 4A). To further determine the mechanism of apoptosis promotion by HAND1, we assessed the mitochondrial membrane potential (MMP), intracellular reactive oxygen species (ROS) levels, and cytosolic Ca2+ concentrations. A decrease in MMP is a near sign of cell apoptosis. Our results showed that MMP in HAND1-transfected cells with or without cisplatin treatment significantly decreased compared to control cells (p<0.05, Figure 4B). Moreover, HAND1-transfected cells had significantly higher intracellular ROS levels than control cells, regardless of cisplatin treatment (p<0.05, Figure 4C). Similarly, HAND1-transfected cells possessed higher cytosolic Ca2+ concentrations compared with control cells, especially when treated with cisplatin (p<0.05, Figure 4D).
HAND1 induces ER stress-mediated apoptosis and enhances mitochondria-mediated apoptosis
The above results predicted that HAND1 triggers ER stress-mediated apoptosis and subsequently mitochondria-mediated apoptosis. To verify that further, we carried out RNA-seq, focusing on the top 100 differentially expressed genes (DEGs) with the lowest P values for HAND1-transfected AGS cells and control cells (Figure 5A). Gene ontology (GO) and KEGG pathway enrichment analyses showed that several enriched terms were correlated with ER functions, such as “response to unfolded protein”, “response to ER stress”, “ER unfolded protein response”, and “Protein processing in ER” (Figure 5B). Consistent with this result, qRT-PCR and Western blotting analyses showed that HAND1 expression upregulated a series of ER-stress- and unfolded protein response (UPR)- related genes, including HSPA5 (also known as Bip), CHOP (also known as DDIT3), ATF6, PERK, ATF4, IRE1a, XBP-1s, and Ero1-La, especially after cisplatin treatment (Figure 5C-D). We also detected the expression of some apoptosis-related proteins and found that HAND1 expression increased the cleavage of caspase-3, caspase-7, caspase-9, caspase-12 and PARP, especially after cisplatin treatment (Figure 5E). These results suggest that HAND1 promotes ER stress-induced apoptosis and activates signaling pathways, including UPR in GC cells.
We further investigated whether HAND1 affected mitochondria-mediated apoptosis. We isolated the mitochondria and cytosol fractionation in HAND1-expressing cells, and found that the release of cytochrome c (Cyto C) and Smac, two indicators of mitochondria-mediated apoptosis, from mitochondria into cytosol, was increased in HAND1-transfected cells compared to control cells (Figure 5F). Proteins of the BCL-2 family are central regulators of mitochondria-mediated apoptosis, so some anti-apoptotic proteins and pro-apoptotic proteins were assessed. Our results showed that HAND1 expression decreased anti-apoptotic protein Bcl-2, p-Bcl-2 (Thr56), and Mcl-1 expression, and increased pro-apoptotic protein Bax, Bak, Bik, Bim, and Puma expression (Figure 5G). In addition, HAND1 expression upregulated BAK mRNA expression (Figure 5C). These findings suggested that HAND1 also promotes mitochondria-mediated apoptosis by regulating BCL-2 family proteins in GC cells.
HAND1 interacts with CHOP, targets CHOP and BAK promoter, and promotes GC cell apoptosis
The above results showed that HAND1 promotes ER stress-mediated and mitochondria-mediated apoptosis by activating UPR and regulating BCL-2 family proteins, especially upregulation of the hallmark genes CHOP and BAK. To further determine its relationship with CHOP and BAK, we silenced CHOP or BAK with specific siRNA or overexpressed CHOP, and further investigated the effects of HAND1 on GC cell apoptosis. The efficiency of CHOP and BAK knockdown was confirmed by Western blotting. HAND1 induced upregulation of CHOP and BAK protein levels. However, after knockdown of CHOP or BAK, apoptosis markers - cleaved caspase-3 and cleaved PARP markedly decreased in HAND1-transfected cells, indicating decrease of cell apoptosis (Figure 6A and S1A). Knockdown of CHOP or BAK also resulted in decreased HAND1 levels, indicating a possible feedback loop of HAND1 regulation.
Effect of HAND1 on apoptosis, mitochondrial membrane potential, ROS and cytosolic Ca2+ concentration in GC cells. (A) Representative result and quantitative analysis of Annexin V-FITC/PI staining in stably transfected GC cells, before and after cisplatin treatment. (B) HAND1 significantly decreases mitochondrial membrane potential. Representative results and quantitative analysis are shown. (C) Analysis of ROS generation in stably transfected GC cells. (D) Effect of HAND1 on the variation of cytosolic Ca2+ concentration in GC cells. All values are presented as mean ± SD of three independent experiments. *p<0.05; **p<0.01.

HAND1 induces ER stress-mediated apoptosis via the mitochondria apoptosis pathway. (A) Clustered heatmap view using the top 100 differentially expressed genes (DEGs) with the lowest P values from triplicate samples of HAND1-transfected AGS cells and control cells. The color ranged from blue to white to red indicates the level of gene expression ranged from low to high. (B) Bulb map of GO analysis and KEGG analysis of the DEGs. (C) Quantitative real-time RT-PCR (qRT-PCR) to confirm RNA-Seq results. Two up-regulated genes (HSPA5 and CHOP) from the clustered heatmap, and seven genes related to ER stress-mediated apoptosis were examined. (D) Several key UPR-related proteins were checked by Western blotting of stably transfected GC cells. (E) Cleaved caspase-3, caspase-7, caspase-9, caspase-12 and PARP are upregulated by HAND1 expression. (F) HAND1 increases the release of Cyto C and Smac from mitochondria into cytosol. (G) Several key BCL-2 family proteins checked by Western blotting in stably transfected GC cells.
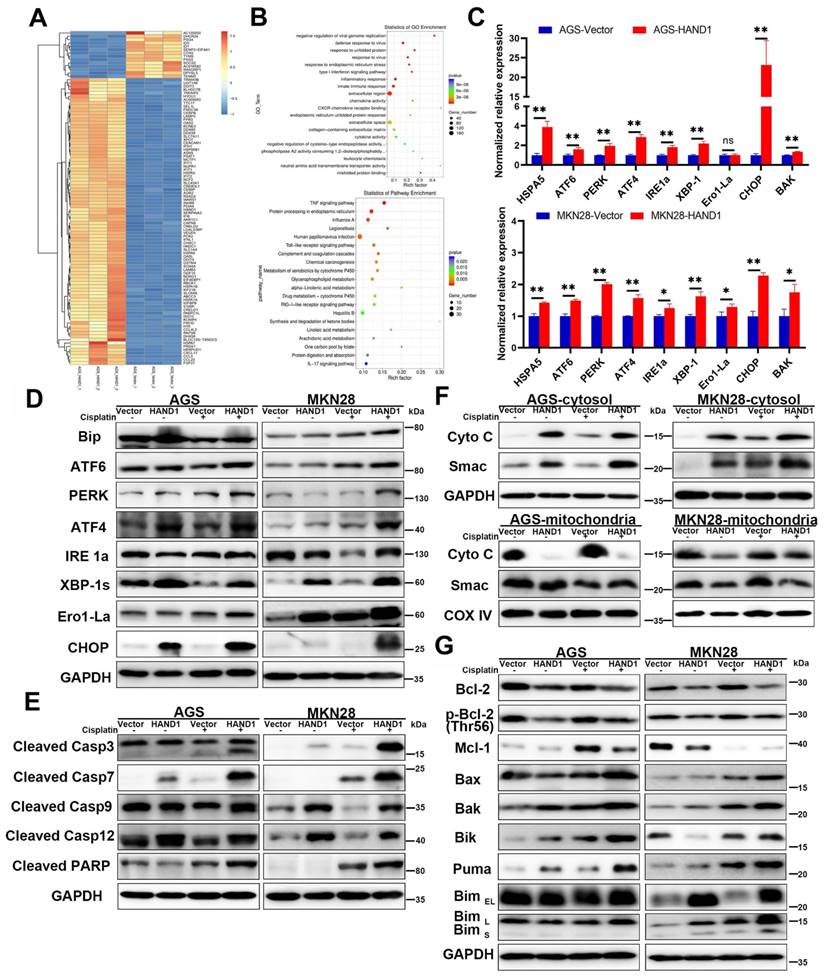
HAND1 interacts with CHOP, targets CHOP and BAK promoters and upregulates their expression and further resulting in GC cell apoptosis. (A) Knockdown of CHOP or BAK decreases Bak, HAND1, CHOP, cleaved caspase-3, and cleaved PARP in HAND1-transfected AGS cells compared with controls. (B) Representative results and quantitative analysis of apoptosis in stably transfected AGS cells with CHOP or BAK knockdown after cisplatin treatment. (C) Ectopic expression of CHOP increases the expression of BAK mRNA and protein in HAND1-transfected cells. (D) Co-IP assay examines the interaction between HAND1 and CHOP in stably transfected GC cells. (E) Immunofluorescence detects the interaction between HAND1 and CHOP in stably transfected AGS cells. Scale bar, 10 µm. (F) Binding of HAND1 to CHOP and BAK promoters as assessed by ChIP. IgG is used as a negative control, and Input as a positive control. (G) HAND1 binds to CHOP and BAK promoters and regulates their transcription in HAND1-transfected AGS cells. Data are presented as mean ± SD of three independent experiments. *p<0.05; **p<0.01.
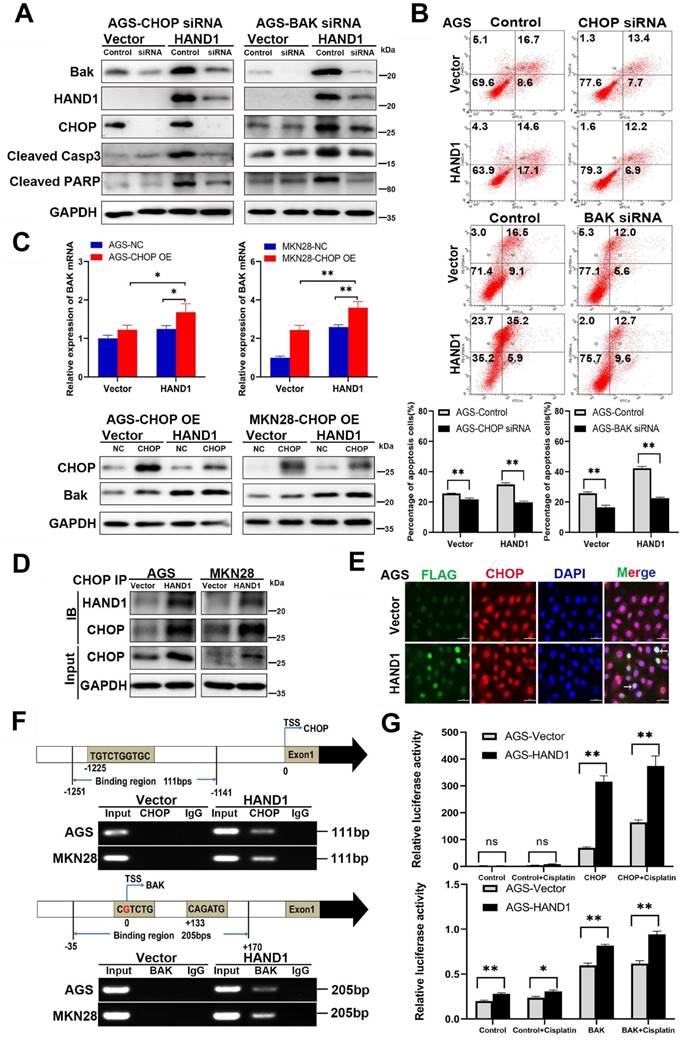
After cisplatin treatment for 24h, the percentage of apoptotic cells significantly decreased in HAND1-transfected cells with CHOP or BAK knockdown, compared to controls (p<0.05, Figure 6B and S1B). Overexpression of CHOP caused significantly upregulation of BAK mRNA and protein expression in HAND1-transfected cells (p<0.05, Figure 6C). In addition, co-IP and immunofluorescence assays revealed an interaction of HAND1 and CHOP proteins (Figure 6D-E and S1C), suggesting that HAND1 could interact with CHOP to regulate its target genes and promotes cell apoptosis via CHOP and BAK.
Previous studies showed that HAND1 binds to DNA with a consensus sequence “NNTCTG” [29, 30]. We hypothesize that HAND1 binds to CHOP or BAK promoters and regulates their transcription. We indeed found binding sites containing “NNTCTG” in CHOP or BAK promoters through analyzing JASPARCORE and TRANSFAC databases. Our further ChIP assays showed that HAND1 binds directly to CHOP promoter (nt -1251 to -1141 related to the transcription start site (TSS)) and BAK promoter (nt -35 to +170 related to TSS) (Figure 6F). To further investigate the effects of HAND1 binding to CHOP or BAK promoter, luciferase reporter assays were conducted. The result showed that their promoter luciferase activity was both markedly increased in HAND1-transfected cells, with or without cisplatin treatment, compared with control cells (p<0.05, Figure 6G and S1D), consistent with our previous finding of significant upregulation of CHOP and BAK at mRNA levels by HAND1 (Figure 5C). These results indicated that HAND1 positively regulates the transcription of CHOP and BAK, to further regulate cell apoptosis.
Discussion
The high morbidity and mortality of GC is generally caused by poor prognosis and limited treatment strategies [31]. Epigenetic inactivation of TSGs promotes GC tumorigenesis and progression, with a series of TSGs already identified in GC [32, 33]. In this study, HAND1, a transcription factor and cell differentiation regulator, is found silenced or downregulated frequently in GC due to its promoter CpG methylation. Consistent with our results, HAND1 has been reported to be silenced by methylation in other cancers including colorectal cancer [12-15, 17-19]. HAND1 methylation is closely correlated with poor prognosis in GC, especially for late-stage patients [14]. In thyroid cancer, HAND1 has been confirmed to be downregulated in differentiated and undifferentiated carcinomas, but expressed normally in benign neoplastic lesions and normal thyroid, and HAND1 restoration inhibits tumor cell growth [18]. HAND1 expression was also significantly associated with other clinicopathological features of GC patients, including depth of invasion, lymph nodal status, and TNM stage. In addition, we found that HAND1 restoration inhibited GC cell growth, proliferation and migration, suggesting that HAND1 does functions as a bona fide tumor suppressor in GC. This is consistent with previous reports that HAND1 expression suppresses uPAR-induced tumor growth and angiogenesis and inhibits tumor cell invasion and metastasis in medulloblastoma; and HAND1 acts as a tumor suppressor inhibiting colorectal cancer cell growth and xenograft tumor formation [20, 21, 23].
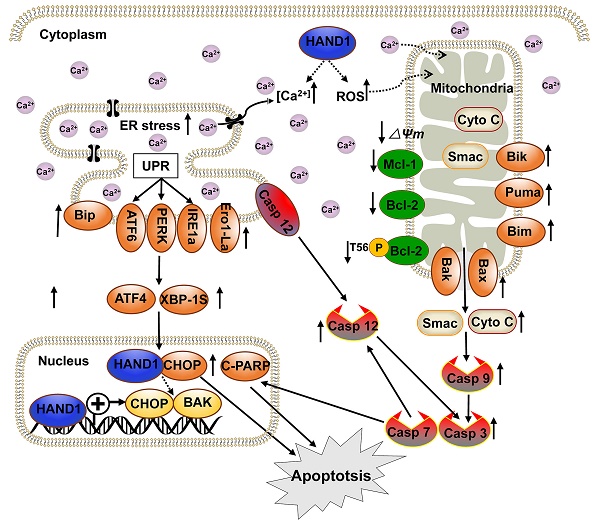
Our present study found that HAND1 induced GC cell apoptosis and ER stress. Cisplatin, one of the most commonly used chemotherapeutic agents to treat solid tumors including GC, increases ROS generation and Ca2+ release and induces cell apoptosis through ER stress and mitochondrial pathways [34-36]. Our findings showed that HAND1 enhanced cisplatin-induced apoptosis, intracellular ROS levels, and cytosolic Ca2+ concentrations. Various exogenous or endogenous factors, including ROS generation and disruption of Ca2+ homeostasis contributed to ER stress and further triggered UPR. UPR maintains ER function and reestablishes homeostasis, but if the stress occurs in excess or is sustained, then the apoptosis pathway is initiated [37, 38]. This is consistent with our finding that HAND1 increased the expression of three major ER sensors, ATF6, PERK and IRE1a, as well as other UPR-related proteins such as Bip, ATF4 and XBP-1s. In addition, HAND1 expression upregulated CHOP, Ero1, and cleaved caspase-7, caspase-12 that are involved in ER stress-indued apoptosis (Scheme 1). CHOP is activated by three UPR-related pathways and upregulates Ero1, which induces cellular ROS generation and oxidative stress in ER, thereby contributing to apoptosis [39, 40]. Our results confirmed that HAND1 elevates intracellular ROS levels in the absence of cisplatin. Caspase-12, which specifically resides on ER membrane, is activated by caspase-7 and released to cytosol in response to ER stress [41].
Dysregulated ER stress results in the transmission of ROS and Ca2+ signals from ER to mitochondria and triggers the response of BCL-2 family proteins, which then activates the mitochondrial apoptotic pathway. In this process, BH3-only proteins, which are activated by ER stress, inhibit anti-apoptotic BCL-2 family proteins and activate pro-apoptotic proteins Bax and Bak, leading to mitochondria outer membrane permeabilization (MOMP) and the release of cytochrome c and Smac [42]. Our results suggested that HAND1 upregulates BH3-only proteins Bik, Bim and PUMA and pro-apoptotic protein Bax and Bak, whereas downregulates anti-apoptotic protein Bcl-2, p-Bcl2 (Thr56) and Mcl-1. Meanwhile, HAND1 expression led to the loss of MMP and release of Cyto C and Smac from the mitochondria to cytosol that further activated caspase-9 and caspase-3. Our findings showed that HAND1 induces ER-stress-mediated apoptosis, including UPR and mitochondrial apoptosis via the caspase-dependent pathway (Scheme 1). However, the mechanisms underlying the disruption of Ca2+ homeostasis and transmission of ROS and Ca2+ signals between ER and mitochondria mediated by HAND1 in GC still remain unclear and require further investigations.
CHOP, a marker for ER stress-induced apoptosis, is well known to upregulate pro-apoptotic protein Bax/Bak and increase translocation and oligomerization of Bax/Bak in ER [43]. Bak, a key regulator of the intrinsic apoptosis pathway, is activated by cell stress and oligomerized on mitochondrial outer membrane leading to MOMP and cytochrome C release [44]. CHOP and BAK are essential for the progression of cell apoptosis. HAND1 is a transcription factor that negatively or positively regulates the transcription of its target genes by binding to their promoter region [45-47]. HAND1 targets CHOP and BAK promoters and positively regulates their transcription. Additionally, HAND1 interacts with CHOP, overexpression of CHOP increased BAK expression, and knockdown of CHOP or BAK attenuated HAND1-induced GC cell apoptosis, indicating that HAND1 induces GC cell apoptosis through CHOP and BAK. HAND1 regulates CHOP and BAK expression at the transcriptional level through interacting with CHOP, and further promotes GC tumor cells apoptosis and leads to tumor suppression.
Conclusions
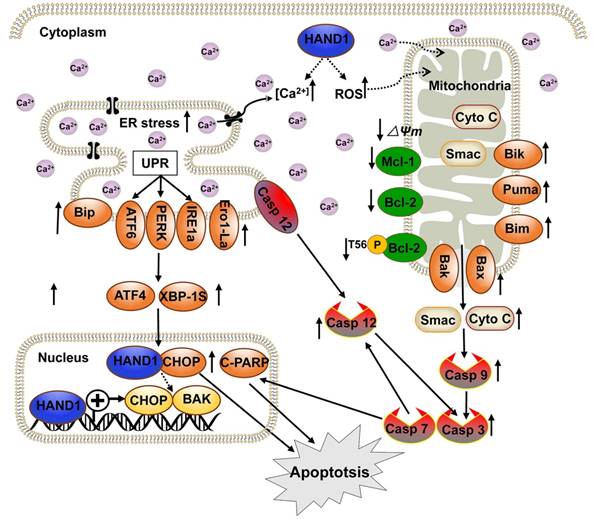
We have demonstrated that HAND1 is frequently downregulated and methylated in GC. HAND1 functions as a tumor suppressor that induces ER-stress-mediated apoptosis including UPR and mitochondrial apoptosis by targeting CHOP and BAK in GC cells. Furthermore, we confirmed that HAND1 interacts with CHOP, and positively regulates CHOP and BAK transcription in GC (Scheme 1). We also showed that HAND1 promoter methylation is a potential prognostic epigenetic biomarker for GC patients.
Diagram showing the mechanism of HAND1 tumor suppression in gastric cancer. HAND1 binds to CHOP and BAK promotors, upregulates their transcription, interacts with CHOP, and induces ER-stress mediated apoptosis including UPR and mitochondrial apoptosis via caspase-dependent pathway in gastric cancer cells. Dotted line indicates that mechanisms remain unclear. △Ψm, mitochondrial membrane potential.
Abbreviations
GC, gastric cancer; HAND1, Hand-And-Neural-crest-Derivative-expressed 1; HAND, Hand-And-Neural-crest-Derivative-expressed; ROS, reactive oxygen species; UPR, unfolded protein response; TSG, tumor suppressor gene; bHLH, basic helix-loop-helix; CGI, CpG island; MSP, methylation-specific PCR; BGS, bisulfite genomic sequencing; 5-Aza, 5-Aza-2'-deoxycytidine; TSA, trichostatin A; IHC, immunohistochemistry; OS, overall survival; MMP, mitochondrial membrane potential; RNA-Seq, RNA sequencing; Co-IP, Coimmunoprecipitation; MOMP, mitochondria outer membrane permeabilization; ER, endoplasmic reticulum; C-PARP, cleaved PARP; Cyto C, cytochrome c; siRNA, small interfering RNA; OE, overexpression; CHIP, chromatin immunoprecipitation; TSS, transcription start site.
Supplementary Material
Supplementary tables.
Acknowledgements
This study was supported by Zhejiang Provincial Natural Science Foundation (LQ20H160045); The National Natural Science Foundation of China (Grant no. 81972716) and HK RGC (GRF #14115920). We thank LetPub for its linguistic assistance during draft preparation, and Dr Sun Young Rha (Yonsei University College of Medicine) for some gastric cell lines.
Data availability
All publicly available data can be acquired from the corresponding web servers described in the Materials and methods.
Ethics approval
This study was approved by the ethics committee of Sir Run Run Shaw Hospital, Zhejiang University.
Author contributions
XH and QT conceived the study. YK, ZH, LL, CW, XC, QS, GF and JY performed experiments and collected and analyzed data. YK contributed to the interpretation of data and manuscript draft. XH and QT: assessment, revision and final approval of the manuscript. All authors have read and approved the final version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68:394-424
2. Arnold M, Abnet CC, Neale RE, Vignat J, Giovannucci EL, McGlynn KA. et al. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology. 2020;159:335-49.e15
3. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet (London, England). 2020;396:635-48
4. Padmanabhan N, Ushijima T, Tan P. How to stomach an epigenetic insult: the gastric cancer epigenome. Nature reviews Gastroenterology & hepatology. 2017;14:467-78
5. Zeng XQ, Wang J, Chen SY. Methylation modification in gastric cancer and approaches to targeted epigenetic therapy (Review). International journal of oncology. 2017;50:1921-33
6. Palii SS, Robertson KD. Epigenetic control of tumor suppression. Critical reviews in eukaryotic gene expression. 2007;17:295-316
7. Firulli AB. A HANDful of questions: the molecular biology of the heart and neural crest derivatives (HAND)-subclass of basic helix-loop-helix transcription factors. Gene. 2003;312:27-40
8. Firulli BA, George RM, Harkin J, Toolan KP, Gao H, Liu Y. et al. HAND1 loss-of-function within the embryonic myocardium reveals survivable congenital cardiac defects and adult heart failure. Cardiovascular research. 2020;116:605-18
9. Han Z, Yi P, Li X, Olson EN. Hand, an evolutionarily conserved bHLH transcription factor required for Drosophila cardiogenesis and hematopoiesis. Development (Cambridge, England). 2006;133:1175-82
10. Lu S, Nie J, Luan Q, Feng Q, Xiao Q, Chang Z. et al. Phosphorylation of the Twist1-family basic helix-loop-helix transcription factors is involved in pathological cardiac remodeling. PloS one. 2011;6:e19251
11. Riley P, Anson-Cartwright L, Cross JC. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nature genetics. 1998;18:271-5
12. Hagihara A, Miyamoto K, Furuta J, Hiraoka N, Wakazono K, Seki S. et al. Identification of 27 5' CpG islands aberrantly methylated and 13 genes silenced in human pancreatic cancers. Oncogene. 2004;23:8705-10
13. Kalari S, Jung M, Kernstine KH, Takahashi T, Pfeifer GP. The DNA methylation landscape of small cell lung cancer suggests a differentiation defect of neuroendocrine cells. Oncogene. 2013;32:3559-68
14. Shi J, Zhang G, Yao D, Liu W, Wang N, Ji M. et al. Prognostic significance of aberrant gene methylation in gastric cancer. American journal of cancer research. 2012;2:116-29
15. Yagi K, Akagi K, Hayashi H, Nagae G, Tsuji S, Isagawa T. et al. Three DNA methylation epigenotypes in human colorectal cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:21-33
16. Jin B, Yao B, Li J, Fields C, Delmas A, Liu C. et al. DNMT1 and DNMT3B modulate distinct polycomb-mediated histone modifications in colon cancer. Cancer research. 2009;69:7412-21
17. Imura M, Yamashita S, Cai LY, Furuta J, Wakabayashi M, Yasugi T. et al. Methylation and expression analysis of 15 genes and three normally-methylated genes in 13 Ovarian cancer cell lines. Cancer letters. 2006;241:213-20
18. Martinez Hoyos J, Ferraro A, Sacchetti S, Keller S, De Martino I, Borbone E. et al. HAND1 gene expression is negatively regulated by the High Mobility Group A1 proteins and is drastically reduced in human thyroid carcinomas. Oncogene. 2009;28:876-85
19. Tellez CS, Shen L, Estécio MR, Jelinek J, Gershenwald JE, Issa JP. CpG island methylation profiling in human melanoma cell lines. Melanoma research. 2009;19:146-55
20. Asuthkar S, Gogineni V, Rao J, Velpula K. Nuclear translocation of Hand-1 acts as a molecular switch to regulate vascular radiosensitivity in medulloblastoma tumors: the protein uPAR is a cytoplasmic sequestration factor for Hand-1. Molecular cancer therapeutics. 2014;13:1309-22
21. Asuthkar S, Guda MR, Martin SE, Antony R, Fernandez K, Lin J. et al. Hand1 overexpression inhibits medulloblastoma metastasis. Biochemical and biophysical research communications. 2016;477:215-21
22. Asuthkar S, Guda MR, Tsung AJ, Antony R, Velpula KK. Abstract 1153: Hand-1 overexpression inhibits medulloblastoma cell invasion and metastatic ability via Oct-3/4 / β-catenin interaction. Cancer Research. 2016;76:1153 -
23. Tan J, Yang X, Jiang X, Zhou J, Li Z, Lee P. et al. Integrative epigenome analysis identifies a Polycomb-targeted differentiation program as a tumor-suppressor event epigenetically inactivated in colorectal cancer. Cell death & disease. 2014;5:e1324
24. Li L, Zhang Y, Fan Y, Sun K, Su X, Du Z. et al. Characterization of the nasopharyngeal carcinoma methylome identifies aberrant disruption of key signaling pathways and methylated tumor suppressor genes. Epigenomics. 2015;7:155-73
25. Huang JZ, Chen M, Chen D, Gao XC, Zhu S, Huang H. et al. A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol Cell. 2017;68:171-84 e6
26. Ying J, Li H, Seng T, Langford C, Srivastava G, Tsao S. et al. Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation. Oncogene. 2006;25:1070-80
27. Yang J, Yu J, Li D, Yu S, Ke J, Wang L. et al. Store-operated calcium entry-activated autophagy protects EPC proliferation via the CAMKK2-MTOR pathway in ox-LDL exposure. Autophagy. 2017;13:82-98
28. Liu S, Fei W, Shi Q, Li Q, Kuang Y, Wang C. et al. CHAC2, downregulated in gastric and colorectal cancers, acted as a tumor suppressor inducing apoptosis and autophagy through unfolded protein response. Cell death disease. 2017;8:e3009
29. Scott IC, Anson-Cartwright L, Riley P, Reda D, Cross JC. The HAND1 basic helix-loop-helix transcription factor regulates trophoblast differentiation via multiple mechanisms. Molecular and cellular biology. 2000;20:530-41
30. Zhu H, Ren Q, Yan Z, Xu S, Luo J, Wu X. et al. Human HAND1 Inhibits the Conversion of Cholesterol to Steroids in Trophoblasts. Journal of genetics and genomics = Yi chuan xue bao. 2021;21:S1673-8527
31. Lordick F, Shitara K, Janjigian YY. New agents on the horizon in gastric cancer. Annals of oncology: official journal of the European Society for Medical Oncology. 2017;28:1767-75
32. Kang MH, Choi H, Oshima M, Cheong JH, Kim S, Lee JH. et al. Estrogen-related receptor gamma functions as a tumor suppressor in gastric cancer. Nat Commun. 2018;9:1920
33. Ma G, Liu H, Hua Q, Wang M, Du M, Lin Y. et al. KCNMA1 cooperating with PTK2 is a novel tumor suppressor in gastric cancer and is associated with disease outcome. Molecular cancer. 2017;16:46
34. Xu L, Xie Q, Qi L, Wang C, Xu N, Liu W. et al. Bcl-2 overexpression reduces cisplatin cytotoxicity by decreasing ER-mitochondrial Ca2+ signaling in SKOV3 cells. Oncology reports. 2018;39:985-92
35. Singh MP, Chauhan AK, Kang SC. Morin hydrate ameliorates cisplatin-induced ER stress, inflammation and autophagy in HEK-293 cells and mice kidney via PARP-1 regulation. International immunopharmacology. 2018;56:156-67
36. Liu H, Baliga R. Endoplasmic reticulum stress-associated caspase 12 mediates cisplatin-induced LLC-PK1 cell apoptosis. Journal of the American Society of Nephrology: JASN. 2005;16:1985-92
37. Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nature reviews Molecular cell biology. 2012;13:89-102
38. Gorman AM, Healy SJ, Jäger R, Samali A. Stress management at the ER: regulators of ER stress-induced apoptosis. Pharmacology & therapeutics. 2012;134:306-16
39. Li G, Mongillo M, Chin KT, Harding H, Ron D, Marks AR. et al. Role of ERO1-alpha-mediated stimulation of inositol 1,4,5-triphosphate receptor activity in endoplasmic reticulum stress-induced apoptosis. The Journal of cell biology. 2009;186:783-92
40. Shergalis AG, Hu S, Bankhead A 3rd, Neamati N. Role of the ERO1-PDI interaction in oxidative protein folding and disease. Pharmacology & therapeutics. 2020;210:107525
41. Rao RV, Hermel E, Castro-Obregon S, del Rio G, Ellerby LM, Ellerby HM. et al. Coupling endoplasmic reticulum stress to the cell death program. Mechanism of caspase activation. The Journal of biological chemistry. 2001;276:33869-74
42. Pihán P, Carreras-Sureda A, Hetz C. BCL-2 family: integrating stress responses at the ER to control cell demise. Cell death and differentiation. 2017;24:1478-87
43. Hu H, Tian M, Ding C, Yu S. The C/EBP Homologous Protein (CHOP) Transcription Factor Functions in Endoplasmic Reticulum Stress-Induced Apoptosis and Microbial Infection. Frontiers in immunology. 2018;9:3083
44. Cosentino K, García-Sáez AJ. Bax and Bak Pores: Are We Closing the Circle? Trends in cell biology. 2017;27:266-75
45. Cross JC, Flannery ML, Blanar MA, Steingrimsson E, Jenkins NA, Copeland NG. et al. Hxt encodes a basic helix-loop-helix transcription factor that regulates trophoblast cell development. Development (Cambridge, England). 1995;121:2513-23
46. Bounpheng MA, Morrish TA, Dodds SG, Christy BA. Negative regulation of selected bHLH proteins by eHAND. Experimental cell research. 2000;257:320-31
47. Smart N, Dubé K, Riley P. Identification of Thymosin β4 as an effector of Hand1-mediated vascular development. Nature communications. 2010;1:46
Author contact
![]() Corresponding authors: X Hu (hxt_hzedu.cn) or Q Tao (qtaoedu.hk)
Corresponding authors: X Hu (hxt_hzedu.cn) or Q Tao (qtaoedu.hk)

 Global reach, higher impact
Global reach, higher impact