Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2023; 19(2):705-720. doi:10.7150/ijbs.75466 This issue Cite
Review
Novel Insights into The Roles of N6-methyladenosine (m6A) Modification and Autophagy in Human Diseases
1. Department of Obstetrics and Gynecology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China.
2. Department of Obstetrics and Gynecology, Zhongnan Hospital of Wuhan University, Wuhan University, Wuhan 430071, China.
#Jiaxin Liang and Jingwen Sun contributed equally to this paper
Received 2022-5-25; Accepted 2022-12-9; Published 2023-1-1
Abstract
Autophagy is an evolutionarily conserved cellular degradation and recycling process. It is important for maintaining vital cellular function and metabolism. Abnormal autophagy activity can cause the development of various diseases. N6-methyladenosine (m6A) methylation is the most prevalent and abundant internal modification in eukaryotes, affecting almost all aspects of RNA metabolism. The process of m6A modification is dynamic and adjustable. Its regulation depends on the regulation of m6A methyltransferases, m6A demethylases, and m6A binding proteins. m6A methylation and autophagy are two crucial and independent cellular events. Recent studies have shown that m6A modification mediates the transcriptional and post-transcriptional regulation of autophagy-related genes, affecting autophagy regulatory networks in multiple diseases. However, the regulatory effects of m6A regulators on autophagy in human diseases are not adequately acknowledged. In the present review, we summarized the latest knowledge of m6A modification in autophagy and elucidated the molecular regulatory mechanisms underlying m6A modification in autophagy regulatory networks. Moreover, we discuss the potentiality of m6A regulators serving as promising predictive biomarkers for human disease diagnosis and targets for therapy. This review will increase our understanding of the relationship between m6A methylation and autophagy, and provide novel insights to specifically target m6A modification in autophagy-associated therapeutic strategies.
Keywords: RNA modification, N6-methyladenosine (m6A), Autophagy, Biomarkers, Therapeutic targets
Introduction
The diverse coding, structural, and biological activities of RNA are based on several alterations [1]. N6-methyladenosine (m6A) RNA methylation is the most prevalent and well-conserved post-transcriptional RNA modification in eukaryotes, which affects almost every aspect of RNA metabolisms, such as stability [2, 3], splicing [4], localization [5], translation efficiency [6], and RNA-protein interaction [7]. m6A modification plays a crucial role in many physiological and pathological processes, such as ultraviolet (UV)-induced DNA damage repair [8], circadian rhythm control [9], stem cell development [10], tumor progression [11], and drug resistance. Autophagy is a self-phagocytosis and degradation process, which maintains the steady state of the cell by decomposing non-essential cell components. The abnormal activation of autophagy is closely associated with diverse pathologies, including cancer [12-15] and numerous benign diseases [16-19]. Owing to the lack of present knowledge on the role of m6A RNA methylation in autophagy in human diseases, we aimed to summarize the effect of m6A methylation on autophagy regulatory elements in human diseases in this review and elucidated the underlying mechanisms of m6A-mediated autophagy modulating, which can be used as a potential novel technique in autophagy-associated diagnostic and therapeutic strategies.
1.1 m6A writers, erasers, and readers
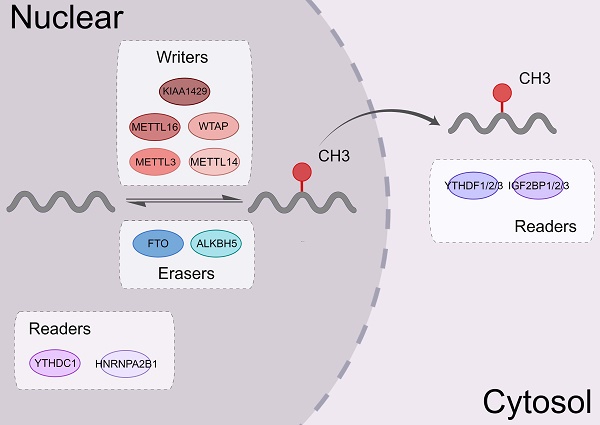
m6A modification is a dynamic and reversible process. Three complexes, namely m6A methyltransferases, m6A demethylases, and m6A binding proteins, participate in this dynamic regulation event [20] (Figure 1).
m6A methyltransferases, also known as "writers," catalyze the formation of N6-methyladenosine (m6A) by inserting a methyl substituent on the N atom of adenosine at position 6[21]. They include proteins, such as methyltransferase-like 3/14 (METTL3/14) [22, 23], Wilms tumor 1-associated protein (WTAP) [24], and KIAA1429 [25]. METTL14 binds with METTL3 to form a heterodimeric methyltransferase complex, that is extremely conserved in mammals [26]. Among them, METTL3 is the catalytically active subunit and METTL14 mainly acts on substrate recognition with its degenerated catalytic center [23]. WTAP is an important regulatory subunit of m6A methyltransferase complex. It can remarkably alter the level of m6A modification [27].
Demethylases such as fat mass and obesity-associated protein (FTO) [28] and human AlkB Homolog H5 (ALKBH5) [29], served as “erasers” to selectively remove the m6A mark via several complex mechanisms, thereby affecting certain biological processes. The amount of m6A in the RNA life cycle was found to be steady in a previous study [30], and demethylases only operate in specific situations. FTO, as the first m6A demethylase discovered, is mainly enriched in nuclear speckles but not enriched in paraspeckles [31]. The subcellular localization of FTO in cells determines its accessibility to different RNA substrates [32]. The overexpression or knockdown of FTO decreased or increased the level of m6A modifications, respectively. ALKBH5 is one of the members of the AlkB family. ALKBH5 is dysregulated in most tissues and plays a vital role in various malignancies [33-35]. The downregulation of the ALKBH5 remarkably decreases the mRNA levels in the cytoplasm, which indicated that ALKBH5 primarily affects mRNA export and RNA metabolism [36].
m6A RNA binding proteins (RBPs) called m6A “readers” participate in the specific recognition of m6A‑modified targeted RNA, thereby triggering the downstream biological events [37, 38]. YT521-B homology (YTH) domain family proteins (YTHDFs) and YTH domain-containing proteins (YTHDCs) belong to these proteins [39]. By interacting with the initiation factors, YTH domain family proteins 1 (YTHDF1) modulates the translation initiation machinery to augment the translation efficiency of target RNAs [6]. YTH domain family proteins 2 (YTHDF2) can increase the degradation of many target mRNA [2]. YTHDF2 mediates the degradation of target RNA by recruiting the carbon catabolite repression 4 (CCR4)-negative on TATA-less (NOT) complex and interacting with the SH domain of CNOT1 via its N-terminal region [40]. YTH domain family proteins 3 (YTHDF3) interacts with YTHDF1 to accelerate the translation of methylated mRNAs and also contributes to mRNA decay induced by YTHDF2. This indicates a complex cooperative mechanism between YTHDF proteins [41]. Other reader proteins that can identify the m6A motif include the heterogeneous nuclear ribonucleoproteins (hnRNP) family (hnRNPA2B1, hnRNPC, and hnRNPG) [42, 43] and insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs, including IGF2BP1/2/3) [44, 45] (Figure 2).
1.2 Characteristics, regulatory mechanisms, and biological functions of autophagy
Autophagy is a complicated process that requires the encapsulation of cytoplasmic components in the double-membrane vesicles and their transportation to lysosomes for destruction [46]. This process begins with the Unc-51-like kinase 1 (ULK1)-autophagy related gene (ATG)13-family interacting protein 200kD (FIP200) kinase complex (namely ULK complex) [47]. When the complex is exposed to environmental stress or physical and chemical damage, the ULK complex is activated cooperatively by AMP-activated protein kinase (AMPK) and ULK1. The vacuolar protein sorting (VPS)34, VPS15, ATG14, and beclin-1 proteins from the phosphatidylinositol 3-kinase (PI3K) complex, which can phosphorylate and activate the ULK complex. The formation of phagophores is mediated by the activation of the PI3K complex [48, 49]. The extension of the phagophore depends on two ubiquitin-like conjugation mechanisms. The ATG12-ATG5-ATG16 complex is initially produced by the functions of ATG7 and ATG10 [50]. Simultaneously, microtubule-associated protein light chain 3 (LC3) is cleaved to generate soluble LC3-I by ATG4. It can conjugate to the head group of the membrane lipid phosphatidylethanolamine (PE), which is mediated by the ATG7, ATG3, and ATG12-ATG5-ATG16 complex. Autophagosomes develop and transfer to lysosomes during this time. Through the synaptosomal-associated protein 29 (SNAP29) and lysosomal vesicle-associated membrane protein (VAMP) 8, the autophagosome merges with the lysosome to form an autolysosome. Many proteins and signaling pathways are involved in this process, such as ATG12, ATG5, ULK119[51], PI3K-AKT-mTORC1 signal pathway [52], and the AMPK pathway (Figure 3).
The biological functions of m6A regulators. m6A modifying enzymes include writers, erasers and readers. Writers (METTL3, METTL14, WTAP, KIAA1429, METTL16, etc.) can add a methyl group to different types of RNA. Erasers (ALKBH5 and FTO) can eliminate this modification. Readers (the YTH family, IGF2BP1, and hnRNPA2B1) could identify the m6A modification sites and regulate the downstream functional activities.
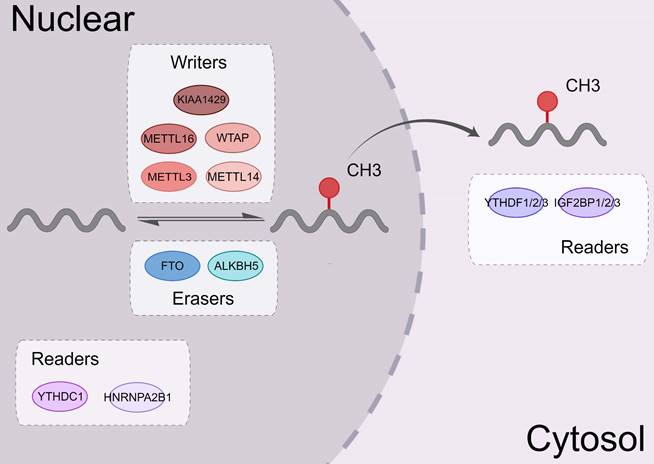
The mechanism of the YTH family proteins. In proteins containing the YTH domain, YTHDF2 promotes the decay of mRNA by recruiting the CCR4-NOT complex. YTHDF3 also helps mRNA decay. YTHDC2 and YTHDF1 can regulate the translation of mRNA. YTHDC1 can selectively splice pre-mRNA into mature transcripts and mediate the nuclear output of mature mRNA. Meanwhile, YTHDC1 can improve the stability of mRNA as well as YTHDF1.
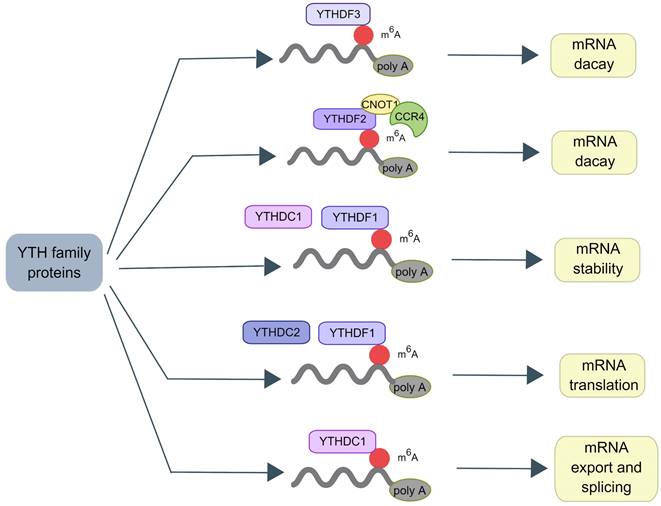
Molecular mechanisms of autophagy process. Autophagy initiation begins with the activation of the ULK1 complex under the control of AMPK and mTORC1. Induction of the ULK1 complex transfers the PI3K class III complex from the cytoplasm to the pre-autophagosomal structure, thereby promoting phagophore formation. The phagophore continues to expand, close, and form autophagosomes through the action of two ubiquitin-like conjugation systems: (1) the ATG12 system and (2) the LC3 system. Subsequently, the outer membrane of the autophagosome fuses with the lysosomal membrane to form autolysosomes where the cargo degradation occurs. UVRAG, RAB7A, and LAMP2 mediate autophagosome maturation and fusion with lysosomes. AMPK, 5' adenosine monophosphate-activated protein kinase; ULK1, Unc-51 Like Autophagy Activating Kinase 1; FIP200: Family interacting protein 200Kd; mTOR, mammalian target of rapamycin; PI3K, phosphoinositide 3-kinase; ATG, Autophagy related gene; LC3, Microtubule-associated protein 1A/1B-light chain 3; UVRAG, UV radiation resistance-associated gene protein; RAB7A, Ras-related protein Rab-7a; LAMP2, lysosomal-associated membrane protein 2.
2. Role of m6A methylation in the regulation of autophagy
2.1 Regulatory effect of m6A methyltransferases on autophagy
2.1.1 METTL3
As the key m6A methyltransferase, the expression of METTL3 is closely associated with the occurrence and development of many cancers by targeting autophagy (Figure 4). In hepatocellular carcinoma (HCC) cells, the depletion of METTL3 can promote autophagy by inducing autophagy-related gene transcription and leading to forkhead box protein O3 (FOXO3) degradation via a YTHDF1-dependent pathway [53]. Similarly, in seminoma, the overexpression of METTL3 resulted in the upregulation of the ATG5 gene with the increase of ATG5 m6A level in TCam-2 cells, thus promoting autophagy [54]. Guo et al. [55] reported that METTL3 induces autophagy in non-small cell lung cancer (NSCLC) by upregulating the expression of LC3B, ATG5, and ATG7.
The correlation between m6A epigenetic regulation and autophagy is also found in hematological tumors. A recent study showed that lncRNA-00470 decreases the phosphatase and tensin homolog (PTEN) stability by triggering METTL3-mediated m6A modification, thus inhibiting cell autophagy while promoting chemoresistance in chronic myeloid leukemia (CML) [56]. Furthermore, METTL3 also contributes to the regulation of autophagy in benign disorders. In the mouse model with temporomandibular joint osteoarthritis (TMJ OA), METTL3 inhibited the apoptosis and autophagy of chondrocytes induced by tumor necrosis factor-α (TNF-α) stimulation in vitro [57]. Chen et al. [58] reported that METTL3 overexpression can decrease the RNA stability and expression of ATG7 via m6A modification, impairing autophagy in osteoarthritis (OA) fibroblast-like synoviocytes (FLSs). A previous study showed that the upregulation of METTL3 induced by lipotoxicity promotes rubicon expression in an m6A-dependent manner, inhibiting autophagy and further suppressing the clearance of lipid droplets (LDs) via lysosomes in nonalcoholic fatty liver disease (NAFLD) [59]. However, the regulatory role of METTL3 in autophagy is still controversial. Yuan et al. reported that particulate matter 2.5 (PM2.5)-induced METTL3 upregulation can maintain oxidative the stability of stress induced growth inhibitor 1 (OSGIN1) mRNA by mediating m6A modification, thereby activating autophagy in air pollution-induced human airway epithelial cell injury [60] (Figure 4). These disparate conclusions in the field suggest that METTL3 can play more complicated roles in autophagy regulation, which will be an interesting area for research in the future.
2.1.2 METTL14
METTL14 is another key component of the methyltransferase complex. It is involved in the regulation of autophagy (Figure 4). METTL14 is upregulated in pancreatic cancer, and the downregulation of METTL14 sensitizes pancreatic cancer cells to cisplatin by activating autophagy [61]. The overexpression of METTL14 promoted autophagy by decreasing eukaryotic translation initiation factor 1 (eIF4G1) mRNA expression, thereby inhibiting the migration, invasion, and proliferation of oral squamous cell carcinoma (OSCC) cells [62]. Lu et al. reported that podocyte injury upregulated the expression of METTL14, resulting in the degradation of silencing information regulator 2 related enzymes 1 (SIRT1) mRNA via m6A modification. The downregulation of METTL14 can increase SIRT1 mRNA levels by inhibiting m6A modification of SIRT1 mRNA, resulting in autophagy activation in podocytes and consequently alleviating proteinuria and delaying the progression of podocytopathies [63].
2.1.3 WTAP
WTAP is also involved in autophagy regulation (Figure 4). In HCC, WTAP expression is upregulated. WTAP can increase the m6A modification of liver kinase B1 (LKB1) mRNA, decrease the stability of LKB1 transcripts, and downregulate its expression. As an upstream target of AMPK, the decrease of LKB1 can inhibit the phosphorylation of AMPK and damage the autophagy of liver cancer to a certain extent [64]. m6A methyltransferases for autophagy regulation are presented in Table 1.
Regulatory effects of m6A methyltransferases on autophagy in human diseases. m6A methyltransferases play an important role in nasopharyngeal carcinoma, CML, seminoma, NSCLC, HCC, PC, OSCC, H/R damage, airway epithelial cells injury, TMJ OA, Leydig cells damage, and podocytopathies, by targeting ZFAS1, PTEN, FOXO3, eIF4G1, LKB1, ULK1, TFEB, OSGIN1, Bcl2/Beclin1, CAMKK2, PPM1A, and SIRT1. The underlying mechanisms involve the activation of the PI3K/AKT pathway, AMPK and ATGs' activation. CML: chronic myeloid leukemia; NSCLC: non-small cell lung cancer; HCC: hepatocellular carcinoma; PC: pancreatic carcinoma; OSCC: oral squamous cell carcinoma; H/R: hypoxia/reoxygenation; TMJ OA: temporomandibular joint osteoarthritis.
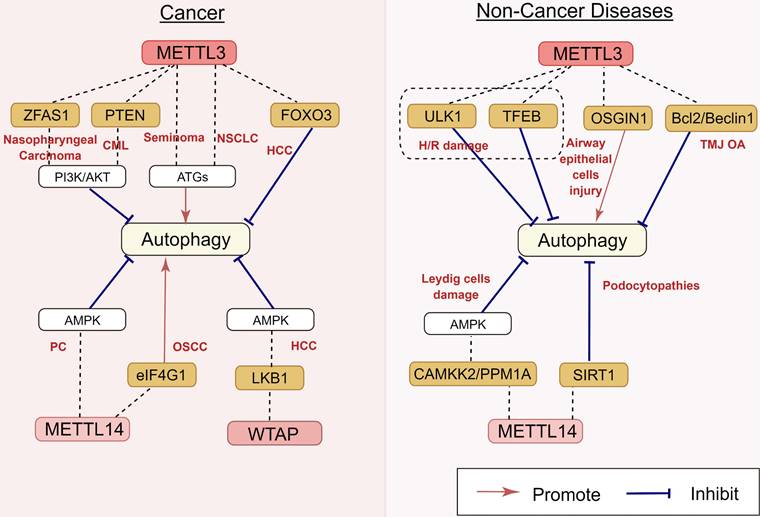
Regulatory effect of m6A methyltransferases and their molecular mechanisms
| m6A component | Related gene | Diseases | m6A level | Association between m6A and autophagy* | Autophagy regulated mechanism | References |
|---|---|---|---|---|---|---|
| METTL3 | FOXO3 | Hepatocellular carcinoma | Decrease | Negative | FOXO3 | [53] |
| METTL3 | Bcl2 | Temporomandibular joint osteoarthritis | Decrease | Negative | Bcl2 | [57] |
| METTL3 | miR-20b | Hypoxia/reoxygenation-treated endothelial cell damage | - | Negative | ULK1 | [114] |
| METTL3 | TFEB | Hypoxia/reoxygenation-treated cardiomyocyte damage | Increase | Negative | AMPK pathway | [75] |
| METTL3 | PTEN | Chronic myelocytic leukaemia | Increase | Negative | PI3K/AKT pathway | [56] |
| METTL3 | LC3B, ATG5, ATG7 | Non-small-cell lung cancer | Increase | Positive | LC3B, ATG5, ATG7 | [55] |
| METTL3 | ATG5 | Seminoma | Increase | Positive | ATG5 | [54] |
| METTL3 | OSGIN1 | Pollution-induced airway epithelial cell injury | Increase | Positive | OSGIN1 | [60] |
| METTL3 | ZFAS1 | Nasopharyngeal carcinoma | Increase | Positive | PI3K/AKT pathway | [97] |
| METTL14 | SIRT1 | Podocytopathies | Increase | Negative | SIRT1 | [63] |
| METTL14 | AMPK1/2, ERK12 | Pancreatic cancer | Increase | Negative | mTOR pathway | [61] |
| METTL14 | CAMKK2, PPM1A | Testosterone synthesis disorder | Decrease | Negative | AMPK pathway | [76] |
| METTL14 | eIF4G1 | Oral squamous cell carcinoma | Increase | Positive | - | [62] |
| WTAP | LKB1 | Hepatocellular carcinoma | Increase | Negative | AMPK pathway | [64] |
*Negative indicates that the increasing m6A level of RNA can inhibit autophagy, while positive indicates that it can promote autophagy.
2.2 Regulatory effect of m6A demethylases on autophagy
2.2.1 FTO
FTO, the first reported m6A demethylase, is found to be frequently dysregulated in its expression and functions in many human diseases [65] (Figure 5). The relationship between FTO and autophagy was gradually studied over the last few years. Jin et al. [66] demonstrated that FTO can specifically upregulate the ULK1 protein level via m6A mediated demethylation, thereby promoting the activation of autophagy. Similarly, Zhang et al. also reported that FTO promotes cisplatin resistance by facilitating autophagy by targeting ULK1 via an m6A‐dependent manner in gastric cancer cells [67]. Moreover, the downregulation of FTO suppressed the expression of ATG5 and ATG7, inhibiting autophagosome formation, thereby restaining autophagy and adipogenesis [68]. In human endometriosis, FTO overexpression can promote autophagy by m6A modification of ATG5, which could inhibit glycolysis, proliferation, and metastasis of endometriotic stromal cells (EESCs) [69]. However, previous studies have also shown that FTO can inhibit autophagy. In clear cell renal cell carcinoma (ccRCC), downregulation of FTO increases autophagic flux by targeting ATG5 and ATG7, which also impairs ccRCC growth and metastasis in vitro and vivo [70]. In oral squamous cell carcinoma, after FTO knockdown, YTHDF2 binds with eIF4G1 transcripts containing m6A, resulting in mRNA degradation and downregulating the expression of eIF4G1 protein, thereby activating autophagy and suppressing tumor growth [71]. Low-level arsenic exposure can stabilize FTO by inhibiting autophagy, whereas the increase in FTO can in turn inhibit autophagy, thus forming a positive feedback loop to maintain FTO accumulation and promote arsenic tumorigenicity [72]. This discrepancy can be explained by evidence indicating that autophagy is a highly regulated and complicated event, and the status of autophagy regulated by m6A modification depends on the different cell types and/or on the stage of disease progression. Therefore, the detailed regulatory mechanisms of FTO on autophagy should be further elucidated.
2.2.2 ALKBH5
Previous studies have shown that ALKBH5 contributes to the pathophysiology of several human diseases by increasing or inhibiting autophagy [73] (Figure 5). Furthermore, ALKBH5 can promote autophagy in many organs and tissues. Li et al. [74] reported that bone-derived mesenchymal stem cells (BMSCs) co-cultured with nucleus pulposus cells (NPCs) can increase ALKBH5 expression in the NPCs, thereby increasing autophagy by decreasing m6A modification of FIP200 mRNA and upregulating FIP200 expression. In hypoxia/reoxygenation (H/R)-induced cardiomyocytes, the expression of transcription factor EB (TFEB) was downregulated because of an increase in pre-mRNA m6A methylation. ALKBH5 can decrease the methylation of TFEB pre-mRNA, which resulted in autophagy activation [75]. In Leydig cells, human chorionic gonadotropin (HsCG) increases ALKBH5 expression by increasing ALKBH5 transcription. ALKBH5 upregulation decreases m6A levels, which alleviates m6A-mediated protein phosphatase 1A, magnesium dependent, alpha isoform (PPM1A) translation and upregulates calcium/calmodulin-dependent protein kinase kinase 2(CAMKK2) expression by attenuating m6A-mediated mRNA degradation, which subsequently contributes to autophagy activation [76].
Furthermore, ALKBH5 can also inhibit autophagy under certain circumstances. In NSCLC, ALKBH5 can maintain the transcripts of UBE2C (a ubiquitin-binding enzyme that can catalyze protein degradation in the 26s proteasome) by eliminating the m6A methylation of its pre-mRNA, thereby stabilizing UBE2C and suppressing autophagy [77]. In epithelial ovarian cancer, silencing of ALKBH5 promotes autophagy and inhibits the proliferation and invasion of SKOV3 cells by activating the PI3K-AKT-mTOR signaling pathway, whereas the overexpression of ALKBH5 exerts an opposite effect [15]. These discrepancies can be attributed to the cell-type-specific context-dependent role of autophagy, which requires further investigation. The m6A demethylases for autophagy regulation are presented in Table 2.
Regulatory effect of m6A demethylases on autophagy in human diseases. m6A demethylases play an important role in EOC, ccRCC, skin tumor, GC, OSCC, NSCLC, endometriosis, TMJ OA, Leydig cells damage, IVD, and H/R damage, by targeting circ RAB11FIP1, SIK2, eIF4G1, ULK1, UBE2C, HuR, TFEB, FIP200, CAMKK2, and PPM1A. The underlying mechanisms involve the activation of AMPK and ATGs. EOC: epithelial ovarian cancer; ccRCC: clear cell renal cell carcinoma; GC: gastric carcinoma; OSCC: oral squamous cell carcinoma; NSCLC: non-small cell lung cancer; IVD: intervertebral disc degeneration; H/R: hypoxia/reoxygenation.
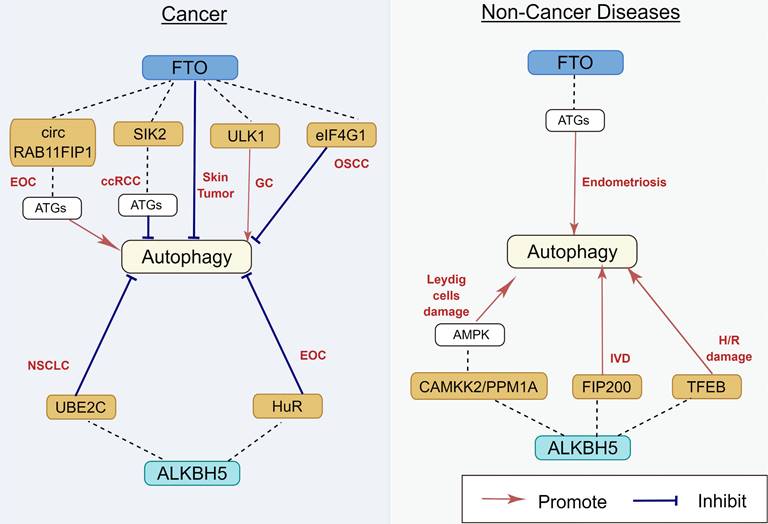
Regulatory effect of m6A demethylases and molecular mechanisms
| m6A component | Related gene | Diseases | m6A level | Association between m6A and autophagy* | Autophagy regulated mechanism | References |
|---|---|---|---|---|---|---|
| FTO | ULK1 | - | - | Positive | ULK1 | [66] |
| FTO | SIK2 | Clear cell renal cell carcinoma | Decrease | Positive | ATG5, ATG7 | [70] |
| FTO | ATG5, ATG7 | Squamous cell carcinoma | Decrease | Positive | m TOR/TFEB/AMPK | [72] |
| FTO | eIF4G1 | Oral squamous cell carcinoma | - | Positive | eIF4G1 | [71] |
| FTO | ATG5, ATG7 | Obesity | Decrease | Negative | ATG5, ATG7 | [68] |
| FTO | ULK1 | Gastric cancer | Decrease | Negative | ULK1 | [67] |
| FTO | ATG5, ATG7 | Epithelial ovarian cancer | Decrease | Negative | ATG5, ATG7 | [98] |
| FTO | ATG5 | Endometriosis | Increase | Negative | ATG5 | [69] |
| ALKBH5 | CAMKK2, PPM1A | Testosterone synthesis disorder | Increase | Negative | AMPK pathway | [76] |
| ALKBH5 | FIP200 | Intervertebral disc degeneration | - | Negative | ULK1 complex | [74] |
| ALKBH5 | TFEB | Hypoxia/reoxygenation-treated cardiomyocytes | Increase | Negative | AMPK pathway | [75] |
| ALKBH5 | UBE2C | Non-small cell lung cancer | Decrease | Positive | LC3B, ATG5, ATG7 | [77] |
| ALKBH5 | HuR, Beclin1&Bcl-2 | Epithelial ovarian cancer | Decrease | Positive | EGFR/PIK3C/AKT/mTOR pathway | [15] |
*Negative indicates that the increasing m6A level of RNA can inhibit autophagy, while positive indicates that it can promote autophagy.
Regulatory effect of m6A binding protein on autophagy in human diseases. m6A binding protein play an important role in HCC, diabetes, hair cell injury, and H/R damage, by targeting circ MDK, ELAVL1, TFEB, and SQSTM1. The underlying mechanisms involve the activation of ATGs. HCC: hepatocellular carcinoma; H/R: hypoxia/reoxygenation.
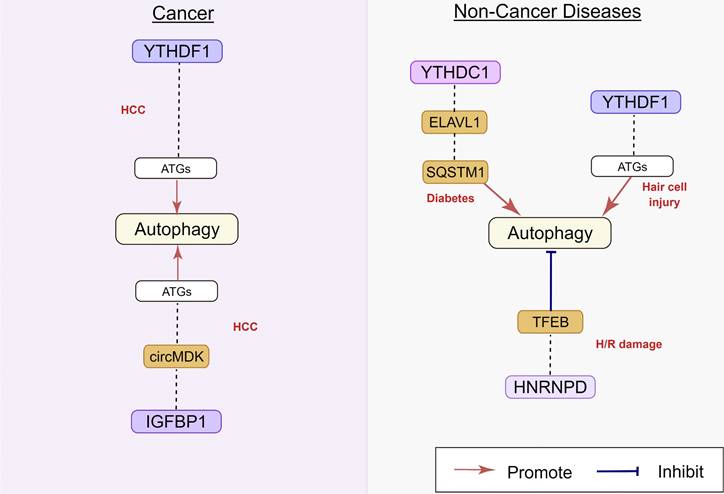
2.3 Regulatory effect of m6A binding proteins on autophagy
m6A binding proteins can recognize and bind to m6A modification sites, controlling the modified RNA's destiny. Recently studies indicated that dysregulation of m6A binding proteins might lead to misinterpretation of modified target RNAs, thus affecting autophagy (Figure 6).
2.3.1 YTHDF and YTHDC family
The YTHDF and YTHDC family proteins are the most important binding proteins for m6A modification, as they contain a YTH domain that can bind to RNA sequence motifs [78]. In human hepatocellular carcinoma (HCC), hypoxia inducible factor-1α (HIF-1α) induced YTHDF1 expression was positively associated with hypoxia-induced autophagy and autophagy-related HCC progression via promoting the translation of autophagy-related genes ATG2A and ATG14 in an m6A-dependent manner [79]. The chemotherapeutic drug cisplatin (CDP) induced YTHDF1 expression could protect sensory hair cells (HCs) against CDP-induced apoptosis by promoting the translation of autophagy-related genes ATG14, along with enhancing autophagy [80]. In addition, Hao et al. recently identified the molecular mechanism of m6A in the regulation of starvation-induced autophagy. Their results showed that YTHDF3 promotes autophagy by recognizing the modification of m6A sites near the stop codon of FOXO3 mRNA. YTHDF3 also recruits eIF3a and eIF4B to facilitate FOXO3 translation, which subsequently initiates autophagy [81]. Liang et al. [82] reported that the decrease in m6A reader YTHDC1 inhibited autophagy by accelerating sequestosome1 (SQSTM1) nuclear mRNA decay in the keratinocytes of diabetic skin, resulting in impaired migration of keratinocytes and delayed wound healing.
2.3.2 IGF2BP family
Studies have shown that the lncRNA LINRIS could inhibit the degradation of the autophagy-lysosome pathway-dependent m6A reader IGF2BP2 by binding to the K139 ubiquitination site of IGF2BP2, maintaining MYC-mediated glycolysis and colorectal cancer cell proliferation [83]. Another study showed that the lncRNA MALAT1 was a target of IGF2BP2 in NSCLC. IGF2BP2 could enhance the stabilization of MALAT1 via an m6A-dependent mechanism, activating its downstream target ATG12 and NSCLC proliferation [84].
Regulatory effect of m6A binding proteins and molecular mechanisms
| m6A component | Related gene | Diseases | m6A binding protein level | Association between m6A binding protein and autophagy* | Autophagy regulated mechanism | References |
|---|---|---|---|---|---|---|
| YTHDF1 | ATG2A, ATG14 | Hepatocellular carcinoma | Increase | Positive | ATG2A, ATG14 | [79] |
| YTHDF1 | ATG14 | Hair cell injury | Decrease | Positive | ATG14 | [80] |
| YTHDF3 | FOXO3 | - | - | Positive | ATGs | [81] |
| YTHDC1 | SQSTM1 | Diabetic skin injury | Decrease | Positive | SQSTM1 | [82] |
| IGF2BP2 | MALAT1 | Non-small cell lung cancer | Increase | Positive | ATG12 | [84] |
| hnRNPD | TFEB | Hypoxia/reoxygenation-treated cardiomyocytes | Increase | Negative | AMPK pathway | [75] |
*Negative indicates that the increasing m6A level of RNA can inhibit autophagy, while positive indicates that it can promote autophagy.
2.3.3 hnRNP family
hnRNPs represent a large subclass of RBPs that contribute to the multiple aspects of nucleic acid [85]. A previous study has shown that hnRNPD can block METTL3-mediated autophagy by downregulating TFEB mRNA expression [75]. Mechanistically, when TFEB was methylated at two m6A residues in the 3'-UTR by METTL3, hnRNPD could bind to TFEB pre-mRNA, decreasing TFEB mRNA levels. Li et al. found that hnRNPC was the decreased m6A regulator in human abdominal aortic aneurysm and a correlation analysis indicated that the level of hnRNPC was positively correlated with the infiltration degree of circulating memory T cells, macrophages, and mast cells [86].
Overall, the above findings show that diverse m6A binding proteins have been identified, which need to be thoroughly investigated in the future. As m6A binding proteins are necessary for the downstream physiological activities of m6A modification, the same m6A regulator may have opposing regulatory effects after directly binding to distinct binding proteins. To summarize, regulating the binding of m6A-modified RNA to "readers" might be a novel disease treatment strategy in the future. A list of m6A readers for autophagy regulation is provided in Table 3.
3. Role of autophagy in regulating m6A methylation
An m6A modification enzyme affects the level of autophagy, however, the relationship between m6A methylation and autophagy is not limited to this phenomenon. In parallel, autophagy activation or impairment also affects the m6A modification enzyme and participates in disease development. In melanoma cells [87], metabolic stress conditions, such as a model starvation medium (Hank's Balanced Salt Solution, HBSS), increased FTO mRNA and protein levels, accompanied by a decreased m6A level. As stress conditions can trigger autophagy, the authors reasoned that the autophagy pathway mediated the FTO increase by HBSS. Interestingly, they found that the knockdown of the autophagy key genes ATG5 or ATG7 significantly reduced HBSS-induced FTO, suggesting that autophagy-induced melanoma tumorigenesis, which was promoted by the increased FTO. Furthermore, Cui et al. [72] found that low-level arsenic could stabilize FTO by inhibiting the p62-mediated selective autophagic degradation of FTO. Their data showed that the deletion of the critical autophagy related genes ATG5 or ATG7 increased FTO stability, indicated by the increased half-life. Furthermore, increased FTO level blocked autophagy, resulting in a positive feedback loop to maintain FTO accumulation.
In summary, these findings highlight complex interactions between autophagy and m6A regulators. However, limited evidence is available on the role of autophagy in regulating m6A RNA modification. Therefore, further research is required to explore whether and how m6A regulators are tightly regulated by autophagy pathways in different pathological conditions, which will provide a broader perspective for treating diseases.
4. Potential diagnostic value of m6A modification in regulating autophagy
m6A regulators are commonly upregulated or downregulated in numerous human diseases, contributing to disease progression. Correlations between m6A regulators and clinical parameters have been confirmed in previous studies [88, 89]. Accumulating evidence has shown that targeting m6A regulators and their regulatory proteins are emerging as a novel diagnostic approach.
Previous studies indicated that YTH domain-containing proteins act as potential diagnostic biomarkers in patients with cancer [90-93]. Chen et al. [94] examined differentially expressed m6A and autophagy genes in esophageal squamous cell carcinoma (ESCC). They developed and validated a predictive model based on six characteristic autophagy genes for predicting the survival of patients with ESCC. Circulating tumor cells (CTCs) derived from tumors can truly reflect the status and progression of a tumor. A recent study showed that mass spectrometry could be used to monitor m6A levels in CTCs. Huang et al. demonstrated that compared with whole blood samples, m6A levels were significantly increased in CTCs of patients with lung cancer. This study showed that CTCs could be used as an early non-invasive diagnostic indicator of cancer [95]. Another study showed the abnormal expression of m6A regulators in endometriosis and indicated that hnRNPA2B1 and hnRNPC might be correlated with immune response, serving as useful diagnostic biomarkers for endometriosis [96]. Collectively, these findings suggest that m6A regulators may have the potential to be a specific and sensitive biomarker of human disease diagnosis.
5. Potential therapeutic value of m6A modification in regulating autophagy
5.1 Role of autophagy regulation by m6A in disease progression
m6A methylation regulates autophagy by affecting the translation or stability of several autophagy-related genes. However, the role of autophagy in the occurrence and development of human diseases cannot be generalized.
As an adaptive mechanism under stress, autophagy protects cells against pressures including hypoxia and chemotherapy, thus promoting disease progression. Peng et al. [97] found that METTL3 could increase the m6A level and stability of the lncRNA ZFAS1, subsequently activating autophagy via the PI3K/AKT pathway, thus promoting the proliferation and metastasis of nasopharyngeal carcinoma cells. In epithelial ovarian cancer, CircRNA-AB11FIP1 can increase demethylase FTO level and activate autophagy by altering the m6A level of ATG7 to promote the malignant behavior of ovarian cancer cells [98]. Wang et al. [68] reported that FTO promoted autophagy and adipogenesis by directly targeting Atg5 and Atg7 transcripts and mediating their expression in an m6A-YTHDF2-dependent manner, thus facilitating the development of therapeutic strategies for the prevention and treatment of obesity. In contrast, as a type of programmed cell death, the abnormal activation of autophagy also results in cell death in certain circumstances [15]. Recently, a study reported that downregulating ALKBH5 could increase autophagy and attenuate the proliferation and invasion potentiality of ovarian cancer cells in vitro and in vivo, whereas the ectopic expression of ALKBH5 could reverse this effect [15] (Figure 7). These findings suggest that targeting m6A to induce or inhibit autophagy might be a promising novel therapeutic strategy against several diseases. However, given the contradictory role of autophagy regulation by m6A in tumor development, the underlying mechanisms need further exploration.
5.2 Role of autophagy regulation by m6A in tumor chemotherapy resistance
Chemotherapy and targeted drug therapy are important means of tumor therapy. Accumulating evidence indicates that tumor cells can acquire resistance to death via diverse biological mechanisms, leading to anticancer drug resistance of tumors [99, 100]. Recently, multiple mechanisms by which tumor cells become resistant to anticancer drugs have been discovered, among which epigenetic change [87, 101-107]-mediated autophagy [108-110] also plays a critical role.
Lin et al. [53] discovered that METTL3-dependent sorafenib resistance exists in HCC and was mediated by promoting autophagy. The combination of autophagy inhibitors and sorafenib or treating HCC cells under hypoxia obtained showed a significant sensitivity to sorafenib, providing another novel strategy for treating drug-resistant liver cancer. In seminoma [54], METTL3 could increase autophagy by modulating ATG5, thus promoting cisplatin resistance. Therefore, METTL3 was a potential therapeutic target to reverse cisplatin resistance in seminoma. β-elements could reverse gefitinib resistance in NSCLC cells by inhibiting METTL3-mediated autophagy. Mechanistically, METTL3 can regulate autophagy by targeting ATG5, ATG7, LC3B, and SQSTM1 [55]. However, autophagy is not always conducive to chemotherapy resistance. It could also enhance sensitivity to chemotherapy. Kong et al. [61] found that the artificial regulation of METTL14 expression may significantly damage the proliferation of pancreatic cancer cells in the presence of cisplatin. METTL14 downregulation could result in autophagy activation via the mTOR signaling pathway, thus effectively improving cisplatin sensitivity in pancreatic cancer cells (Figure 7).
In summary, m6A modification regulates chemotherapy resistance by affecting various factors in different tumors, with autophagy as a downstream event. As drug resistance in cancers is becoming more common, targeted m6A modification provides a more accurate and individualized treatment option. However, the mechanism of m6A modification to regulate autophagy in tumor chemotherapy resistance is complex. This may be attributed to the heterogeneity of m6A modification enzymes in different tumors, and the “double-sword” effect of autophagy on drug resistance regulation. Thus, the mechanism underlying m6A modification in chemotherapy resistance needs further elucidation.
5.3 Role of autophagy regulation by m6A in hypoxia-reperfusion injury
Ischemia and reperfusion (I/R) injury often occurs during and after surgery, which may cause serious, even life-threatening, organ damage [111]. Previous studies have shown a significant difference between myocardial m6A levels in the ischemic myocardium and non-ischemic areas [112]. Autophagy is also involved in the occurrence and development of ischemic heart disease [113]. Song H et al. [75] found increased m6A modification in cardiomyocytes treated with H/R and I/R. METTL3 methylates the m6A residue of TFEB in the 3'-UTR, thus promoting the binding of the RBP hnRNPD to TFEB pre-mRNA and reducing TFEB mRNA levels. With TFEB as a medium, METTL3 reduced autophagy and damaged cardiomyocytes. METTL3 knockout can effectively improve the activity of cardiomyocytes treated with H/R. Similarly, under H/R conditions, propofol post-treatment prevented autophagy and cell death induced by H/R via the METTL3/miR-20b/ULK1 signaling pathway [114] (Figure 7). Overall, these studies showed that m6A regulators participating in autophagy in ischemic diseases may act as potential therapeutic targets which need further investigation.
5.4 m6A methylation mediated autophagy confers environmental damage
Nowadays, potential environmental pathogenic factors gradually enter people's line of sight. Environmental pollutants including dust, heavy metals, and plastic derivatives can damage human systems including respiration, digestion, and reproduction [115, 116]. Many studies show that autophagy regulated by m6A also plays a potential protective role in the above-mentioned processes.
As a widely used plasticizer, the toxicity of di-(2-ethylhexyl) phthalate (DEHP) has been reported [117, 118]. Zhao et al. [119] found that mono-(2-ethylhexyl) phthalate, the main metabolite of DEHP in the body, promoted m6A modification by decreasing FTO levels, leading to Leydig cell damage. Long-term low-level arsenic exposure could enhance FTO stability and decrease m6A levels by impairing selective autophagy, thus inducing the malignant transformation and tumorigenesis of keratinocytes [72]. In addition, studies showed that the air pollutant PM2.5 could activate autophagy and promote the damage of the respiratory epithelium and the development of lung cancer [60, 120]. METTL3 knockout could attenuate PM2.5-induced apoptosis and autophagy and protect the respiratory system (Figure 7). Thus, m6A methylation can regulate autophagy or protect the human body from pathogenic environmental factors considering the above-mentioned results. However, the underlying mechanisms should be further studied in depth.
Potential therapeutic value of m6A modification in regulating autophagy. m6A modification plays a key role in many aspects of disease treatment. m6Aregulators can become a potential target for therapy by affecting (1) the progression of tumor and non-tumor diseases, (2) the sensitivity of tumor cells to various chemotherapeutic drugs, (3) the response of cells to H/R damage, and (4) the damage of environmental pollutants.
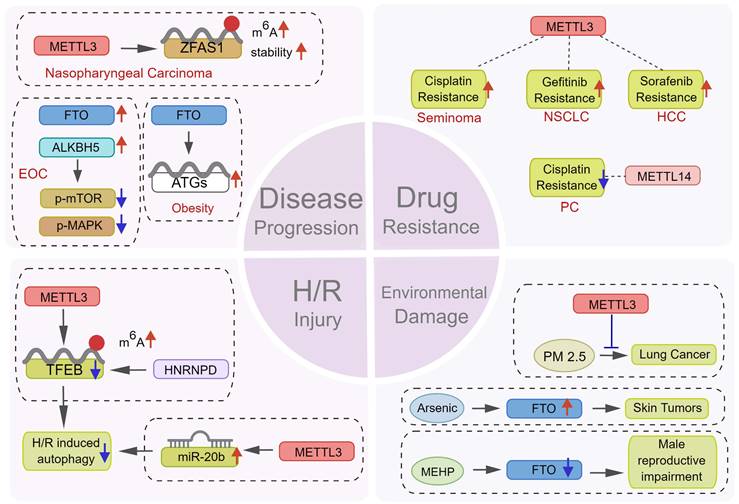
6. Conclusions and future directions
m6A modification, the most prevalent post-transcriptional epigenetic mechanism, is widely distributed in eukaryote RNAs. Many studies have indicated that alterations in m6A modification and autophagy affect the development and progression of various human diseases, and they have important implications in the diagnosis and treatment of several human diseases. This review focuses on mutual interactions between m6A regulators and autophagy and the combined effect of various m6A regulators and autophagy on different human diseases. m6A regulators could modulate autophagy processing, whereas autophagy activation or inhibition could degrade m6A regulator to change m6A levels. This evidence enhances the understanding of the pathogenesis of different types of human diseases. However, whether and how m6A regulators and m6A precisely modulate autophagy remain controversial. For instance, METTL14 inhibits autophagy in testicular tissues and pancreatic cancer, whereas METTL3 promotes autophagy in the heart, liver, and endothelial cells. Similarly, METTL3 suppresses autophagy in the lungs of patients with NSCLC. The distinction between different autophagy-mediating enzymes is more evident for m6A demethylase. FTO can induce autophagy in various tumor tissues and cell lines, including gastric cancer, ovarian cancer, and renal cell carcinoma. Two main reasons that probably contribute to this discrepancy are as follows. Firstly, m6A levels, which play diverse roles in various physiological statuses, differ in different cells and/or disease progression stages [37, 121]. Second, the function of autophagy is context-dependent and highly affected by the disease status and exposure to external stimuli [122, 123]. Hence, more research is warranted to investigate the functions and molecular regulatory mechanisms of the association between m6A regulators and autophagy.
The critical role of m6A regulators in disease initiation and progression provides new possibilities for the early diagnosis and treatment of several human diseases. m6A regulators may serve as a potential non-invasive diagnostic biomarker and be used for the diagnosis and prognosis of human diseases. Altered total m6A levels and abnormal m6A regulator expression in different human diseases have been recently reported. However, their specific roles in disease diagnosis need further exploration. Besides, antibody-based m6A detection technology has poor specificity [124]. Although various antibody-free methods have been developed to detect m6A sites, these methods have certain limitations such as low reproducibility, thus requiring further improvement [125-127]. Moreover, m6A regulators whether dysregulation regulators can be considered a potential biomarker to detect early-stage diseases, and whether the total m6A level can be a credible predictive biomarker for monitoring treatment efficacy have not been clarified [128-130]. Therefore, further research is required before applying m6A regulators to disease diagnosis.
Accumulating evidence indicates m6A modification as a new therapeutic target for disease treatment. Therefore, developing potent and specific small molecule inhibitors/activators for m6A regulatory proteins is crucial. In 2012, Chen et al. [131] discovered rhein, the first identified FTO inhibitor, which could alter the m6A levels of mRNAs inside cells, acting as a competitive inhibitor of FTO. However, rhein also biochemically inhibited demethylase ALKBH2 activity, suggesting that rhein is not an FTO-specific inhibitor. The ethyl ester form of meclofenamic acid (MA) was identified as a selective chemical inhibitor of FTO that increases m6A levels in the mRNA of HeLa cells in 2015[132]. Further studies showed that treating glioblastoma stem cells (GSCs) with the MA attenuated GSC-triggered tumorigenesis in GSC-engrafted mice [133]. Moreover, another study indicated that R-2-hydroxyglutarate produced at high levels by mutant isocitrate dehydrogenase 1/2 enzymes exhibited anticancer effects by suppressing FTO activity, thereby increasing m6A levels in acute myeloid leukemia (AML) cells [134]. Recently, Yankova et al. [135] reported that STM2457, a specific METTL3 inhibitor, could be used as a promising therapeutic drug for AML, which is expected to enter clinical trials as the first epigenetic inhibitor drug. However, whether m6A inhibitors or activators cause unpredictable side effects is unclear. Therefore, large-scale, multicenter, and collaborative clinical trials will help better elucidate the role of m6A modification in autophagy and the potential molecular mechanism of disease development, thus providing novel biomarkers and therapeutic targets for human diseases.
Taken together, with the development of Bio-Technology, the detailed regulatory mechanisms underlying the interaction networks between m6A modification and autophagy will be more extensively investigated. And more diagnostic and therapeutic targets of m6A regulators that contribute to human disease progression will be further explored.
Acknowledgements
All the figures represented in this review were created by Figdraw online software.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 82001524 and No. 81974242). Hubei Provincial Natural Science Foundation (Grant No. 2020CFB310).
Author Contributions
Hengwei Liu and Yi Liu designed this study. Jiaxin Liang drafted the paper. Jingwen Sun helped to revise the paper. Wei Zhang, Xiwen Wang, Wenqian Xiong, Ling Zhang, Ying Xu and Yuan Peng provided critical comments, suggestions of the revised paper.
Ethical Approval
All data and analyses were based on previous published studies; thus, no ethical approval and patient consent are required.
Abbreviations
m6A: N6-methyladenosine; UV: ultraviolet; METTL3: methyltransferase-like 3; METTL14: methyltransferase-like 14; WTAP: wilms tumor 1-associated protein; ALKBH5: human AlkB Homolog H5; FTO: fat mass and obesity-associated protein; RBP: RNA binding proteins; YTH: YT521-B homology; YTHDF: YTH domain family proteins; YTHDC: YTH domain-containing protein; CCR4: carbon catabolite repression 4; NOT: negative on TATA-less; HNRNP: heterogeneous nuclear ribonucleoproteins; IGF2BP: insulin-like growth factor 2 mRNA-binding proteins; ULK1: unc-51-like kinase 1; ATG: autophagy related gene; FIP200: Family interacting protein 200kD; AMPK: AMP-activated protein kinase; VPS34/15: vacuolar protein sorting 34/15; PI3K: phosphatidylinositol 3-kinase; LC3: microtubule-associated protein light chain 3; PE: phosphatidylethanolamine; SNAP29: synaptosomal-associated protein 29; VAMP: vesicle-associated membrane protein; HCC: hepatocellular carcinoma; FOXO3: forkhead box protein O3; PTEN: phosphatase and tensin homolog; CML: chronic myeloid leukemia; NSCLC: non-small cell lung cancer; TNF-α: tumor necrosis factor-α; TMJ OA: temporomandibular joint osteoarthritis; FLS: fibroblast-like synoviocytes; NAFLD: nonalcoholic fatty liver disease; LDs: lipid droplets; PM2.5: particulate matter 2.5; OSGIN1: oxidative stress induced growth inhibitor 1; eIF4G1: eukaryotic translation initiation factor 1; OSCC: oral squamous cell carcinoma; SIRT1: silencing information regulator 2 related enzyme 1silencing information regulator 2 related enzyme 1; LKB1: liver kinase B1; EESC: endometriotic stromal cells; ccRCC: clear cell renal cell carcinoma; BMSC: bone-derived mesenchymal stem cells; NPC: nucleus pulposus cells; TFEB: transcription factor EB; H/R: hypoxia/reoxygenation; HsCG: human chorionic gonadotropin; PPM1A: protein phosphatase 1A; CAMKK2: calcium/calmodulin-dependent protein kinase kinase 2; UBE2C: Ubiquitin-Conjugating Enzyme E2 C; HIF-1α: hypoxia inducible factor-1α; CDP: chemotherapeutic drug cisplatin; HCs: hair cells; SQSTM1: sequestosome1; hnRNPs: heterogeneous nuclear ribonucleoproteins; AAA: abdominal aortic aneurysm; HBSS: Hank's Balanced Salt Solution; ESCC: esophageal squamous cell carcinoma; CTC: circulating tumor cells; EOC: epithelial ovarian cancer; mTOR: mechanistic target of rapamycin; I/R: Ischemia and reperfusion; DEHP: Di-(2-ethylhexyl) phthalate; MEHP: mono-(2-ethylhexyl) phthalate; MA: meclofenamic acid; GSC: glioblastoma stem cell; R-2HG: R-2-hydroxyglutarate; IDH1/2: isocitrate dehydrogenase 1/2; AML: acute myeloid leukemia.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201-6
2. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117-20
3. Kmietczyk V, Riechert E, Kalinski L, Boileau E, Malovrh E, Malone B. et al. m(6)A-mRNA methylation regulates cardiac gene expression and cellular growth. Life science alliance. 2019 2
4. Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W. et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell research. 2014;24:1403-19
5. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular cell. 2013;49:18-29
6. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H. et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388-99
7. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560-4
8. Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W. et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543:573-6
9. Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M. et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793-806
10. Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M. et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science (New York, NY). 2015;347:1002-6
11. Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6-methyladenosine in Cancer progression. Molecular cancer. 2020;19:88
12. Yang Z, Sun Q, Guo J, Wang S, Song G, Liu W. et al. GRSF1-mediated MIR-G-1 promotes malignant behavior and nuclear autophagy by directly upregulating TMED5 and LMNB1 in cervical cancer cells. Autophagy. 2019;15:668-85
13. Li S, Song Y, Quach C, Guo H, Jang GB, Maazi H. et al. Transcriptional regulation of autophagy-lysosomal function in BRAF-driven melanoma progression and chemoresistance. Nat Commun. 2019;10:1693
14. Görgülü K, Diakopoulos KN, Ai J, Schoeps B, Kabacaoglu D, Karpathaki AF. et al. Levels of the Autophagy-Related 5 Protein Affect Progression and Metastasis of Pancreatic Tumors in Mice. Gastroenterology. 2019;156:203-17.e20
15. Zhu H, Gan X, Jiang X, Diao S, Wu H, Hu J. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J Exp Clin Cancer Res. 2019;38:163
16. Chen M, Hong MJ, Sun H, Wang L, Shi X, Gilbert BE. et al. Essential role for autophagy in the maintenance of immunological memory against influenza infection. Nat Med. 2014;20:503-10
17. Babuta M, Furi I, Bala S, Bukong TN, Lowe P, Catalano D. et al. Dysregulated Autophagy and Lysosome Function Are Linked to Exosome Production by Micro-RNA 155 in Alcoholic Liver Disease. Hepatology. 2019;70:2123-41
18. Kim J, Cheon H, Jeong YT, Quan W, Kim KH, Cho JM. et al. Amyloidogenic peptide oligomer accumulation in autophagy-deficient β cells induces diabetes. J Clin Invest. 2014;124:3311-24
19. Shirakabe A, Zhai P, Ikeda Y, Saito T, Maejima Y, Hsu CP. et al. Drp1-Dependent Mitochondrial Autophagy Plays a Protective Role Against Pressure Overload-Induced Mitochondrial Dysfunction and Heart Failure. Circulation. 2016;133:1249-63
20. Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z. et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6:74
21. Huang W, Chen TQ, Fang K, Zeng ZC, Ye H, Chen YQ. N6-methyladenosine methyltransferases: functions, regulation, and clinical potential. J Hematol Oncol. 2021;14:117
22. Śledź P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. eLife. 2016 5
23. Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Molecular cell. 2016;63:306-17
24. Ma H, Shen L, Yang H, Gong H, Du X, Li J. m6A methyltransferase Wilms' tumor 1-associated protein facilitates cell proliferation and cisplatin resistance in NK/T cell lymphoma by regulating dual-specificity phosphatases 6 expression via m6A RNA methylation. IUBMB Life. 2021;73:108-17
25. Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20:608-24
26. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2013;10:93-5
27. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell research. 2014;24:177-89
28. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nature chemical biology. 2011;7:885-7
29. Ensfelder TT, Kurz MQ, Iwan K, Geiger S, Matheisl S, Müller M. et al. ALKBH5-induced demethylation of mono- and dimethylated adenosine. Chemical communications (Cambridge, England). 2018;54:8591-3
30. Darnell RB, Ke S, Darnell JE Jr. Pre-mRNA processing includes N(6) methylation of adenosine residues that are retained in mRNA exons and the fallacy of "RNA epigenetics". RNA (New York, NY). 2018;24:262-7
31. Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M. et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell. 2020;38:79-96.e11
32. Berulava T, Ziehe M, Klein-Hitpass L, Mladenov E, Thomale J, Rüther U. et al. FTO levels affect RNA modification and the transcriptome. Eur J Hum Genet. 2013;21:317-23
33. Tang B, Yang Y, Kang M, Wang Y, Wang Y, Bi Y. et al. m(6)A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Molecular cancer. 2020;19:3
34. Zhang J, Guo S, Piao HY, Wang Y, Wu Y, Meng XY. et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75:379-89
35. Zhu Z, Qian Q, Zhao X, Ma L, Chen P. N(6)-methyladenosine ALKBH5 promotes non-small cell lung cancer progress by regulating TIMP3 stability. Gene. 2020;731:144348
36. Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z. et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017;31:591-606.e6
37. Shi H, Wei J, He C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Molecular cell. 2019;74:640-50
38. Zhao Y, Shi Y, Shen H, Xie W. m(6)A-binding proteins: the emerging crucial performers in epigenetics. J Hematol Oncol. 2020;13:35
39. Xu Y, Zhang W, Shen F, Yang X, Liu H, Dai S. et al. YTH Domain Proteins: A Family of m(6)A Readers in Cancer Progression. Frontiers in oncology. 2021;11:629560
40. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M. et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nature communications. 2016;7:12626
41. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ. et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315-28
42. Mo L, Meng L, Huang Z, Yi L, Yang N, Li G. An analysis of the role of HnRNP C dysregulation in cancers. Biomark Res. 2022;10:19
43. Wang J, Sun D, Wang M, Cheng A, Zhu Y, Mao S. et al. Multiple functions of heterogeneous nuclear ribonucleoproteins in the positive single-stranded RNA virus life cycle. Front Immunol. 2022;13:989298
44. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H. et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nature cell biology. 2018;20:285-95
45. Wang X, Ji Y, Feng P, Liu R, Li G, Zheng J. et al. The m6A Reader IGF2BP2 Regulates Macrophage Phenotypic Activation and Inflammatory Diseases by Stabilizing TSC1 and PPARγ. Adv Sci (Weinh). 2021;8:2100209
46. Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P. et al. Autophagy in major human diseases. Embo j. 2021;40:e108863
47. Shi X, Yokom AL, Wang C, Young LN, Youle RJ, Hurley JH. ULK complex organization in autophagy by a C-shaped FIP200 N-terminal domain dimer. J Cell Biol. 2020 219
48. Liang C, Feng Z, Manthari RK, Wang C, Han Y, Fu W. et al. Arsenic induces dysfunctional autophagy via dual regulation of mTOR pathway and Beclin1-Vps34/PI3K complex in MLTC-1 cells. J Hazard Mater. 2020;391:122227
49. Hill SM, Wrobel L, Ashkenazi A, Fernandez-Estevez M, Tan K, Bürli RW. et al. VCP/p97 regulates Beclin-1-dependent autophagy initiation. Nature chemical biology. 2021;17:448-55
50. Walczak M, Martens S. Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation. Autophagy. 2013;9:424-5
51. Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J. et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493-502
52. Díaz-Troya S, Pérez-Pérez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4:851-65
53. Lin Z, Niu Y, Wan A, Chen D, Liang H, Chen X. et al. RNA m(6) A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020;39:e103181
54. Chen H, Xiang Y, Yin Y, Peng J, Peng D, Li D. et al. The m6A methyltransferase METTL3 regulates autophagy and sensitivity to cisplatin by targeting ATG5 in seminoma. Transl Androl Urol. 2021;10:1711-22
55. Liu S, Li Q, Li G, Zhang Q, Zhuo L, Han X. et al. The mechanism of m(6)A methyltransferase METTL3-mediated autophagy in reversing gefitinib resistance in NSCLC cells by beta-elemene. Cell Death Dis. 2020;11:969
56. Lai X, Wei J, Gu XZ, Yao XM, Zhang DS, Li F. et al. Dysregulation of LINC00470 and METTL3 promotes chemoresistance and suppresses autophagy of chronic myelocytic leukaemia cells. J Cell Mol Med. 2021;25:4248-59
57. He Y, Wang W, Xu X, Yang B, Yu X, Wu Y. et al. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through mediating Bcl2 stability via Ythdf1-mediated m(6)A modification. Bone. 2022;154:116182
58. Chen X, Gong W, Shao X, Shi T, Zhang L, Dong J. et al. METTL3-mediated m(6)A modification of ATG7 regulates autophagy-GATA4 axis to promote cellular senescence and osteoarthritis progression. Ann Rheum Dis. 2022;81:87-99
59. Peng Z, Gong Y, Wang X, He W, Wu L, Zhang L. et al. METTL3-m(6)A-Rubicon axis inhibits autophagy in nonalcoholic fatty liver disease. Molecular therapy: the journal of the American Society of Gene Therapy. 2022;30:932-46
60. Yuan Q, Zhu H, Liu H, Wang M, Chu H, Zhang Z. METTL3 regulates PM2.5-induced cell injury by targeting OSGIN1 in human airway epithelial cells. J Hazard Mater. 2021;415:125573
61. Kong F, Liu X, Zhou Y, Hou X, He J, Li Q. et al. Downregulation of METTL14 increases apoptosis and autophagy induced by cisplatin in pancreatic cancer cells. Int J Biochem Cell Biol. 2020;122:105731
62. Wang F, Zhu Y, Cai H, Liang J, Wang W, Liao Y. et al. N6-Methyladenosine Methyltransferase METTL14-Mediated Autophagy in Malignant Development of Oral Squamous Cell Carcinoma. Front Oncol. 2021;11:738406
63. Lu Z, Liu H, Song N, Liang Y, Zhu J, Chen J. et al. METTL14 aggravates podocyte injury and glomerulopathy progression through N(6)-methyladenosine-dependent downregulating of Sirt1. Cell Death Dis. 2021;12:881
64. Li G, Deng L, Huang N, Cui Z, Wu Q, Ma J. et al. m(6)A mRNA Methylation Regulates LKB1 to Promote Autophagy of Hepatoblastoma Cells through Upregulated Phosphorylation of AMPK. Genes (Basel). 2021 12
65. Wei J, Yu X, Yang L, Liu X, Gao B, Huang B. et al. FTO mediates LINE1 m(6)A demethylation and chromatin regulation in mESCs and mouse development. Science (New York, NY). 2022;376:968-73
66. Jin S, Zhang X, Miao Y, Liang P, Zhu K, She Y. et al. m(6)A RNA modification controls autophagy through upregulating ULK1 protein abundance. Cell research. 2018;28:955-7
67. Zhang Y, Gao LX, Wang W, Zhang T, Dong FY, Ding WP. M(6) A demethylase fat mass and obesity-associated protein regulates cisplatin resistance of gastric cancer by modulating autophagy activation through ULK1. Cancer Sci. 2022;113:3085-96
68. Wang X, Wu R, Liu Y, Zhao Y, Bi Z, Yao Y. et al. m(6)A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy. 2020;16:1221-35
69. Wang H, Liang Z, Gou Y, Li Z, Cao Y, Jiao N. et al. FTO-dependent N(6)-Methyladenosine regulates the progression of endometriosis via the ATG5/PKM2 Axis. Cell Signal. 2022;98:110406
70. Xu Y, Zhou J, Li L, Yang W, Zhang Z, Zhang K. et al. FTO-mediated autophagy promotes progression of clear cell renal cell carcinoma via regulating SIK2 mRNA stability. Int J Biol Sci. 2022;18:5943-62
71. Wang F, Liao Y, Zhang M, Zhu Y, Wang W, Cai H. et al. N6-methyladenosine demethyltransferase FTO-mediated autophagy in malignant development of oral squamous cell carcinoma. Oncogene. 2021;40:3885-98
72. Cui YH, Yang S, Wei J, Shea CR, Zhong W, Wang F. et al. Autophagy of the m(6)A mRNA demethylase FTO is impaired by low-level arsenic exposure to promote tumorigenesis. Nature communications. 2021;12:2183
73. Wang J, Wang J, Gu Q, Ma Y, Yang Y, Zhu J. et al. The biological function of m6A demethylase ALKBH5 and its role in human disease. Cancer Cell Int. 2020;20:347
74. Li G, Song Y, Liao Z, Wang K, Luo R, Lu S. et al. Bone-derived mesenchymal stem cells alleviate compression-induced apoptosis of nucleus pulposus cells by N6 methyladenosine of autophagy. Cell Death Dis. 2020;11:103
75. Song H, Feng X, Zhang H, Luo Y, Huang J, Lin M. et al. METTL3 and ALKBH5 oppositely regulate m(6)A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. 2019;15:1419-37
76. Chen Y, Wang J, Xu D, Xiang Z, Ding J, Yang X. et al. m(6)A mRNA methylation regulates testosterone synthesis through modulating autophagy in Leydig cells. Autophagy. 2020:1-19
77. Guo J, Wu Y, Du J, Yang L, Chen W, Gong K. et al. Deregulation of UBE2C-mediated autophagy repression aggravates NSCLC progression. Oncogenesis. 2018;7:49
78. Shi R, Ying S, Li Y, Zhu L, Wang X, Jin H. Linking the YTH domain to cancer: the importance of YTH family proteins in epigenetics. Cell Death Dis. 2021;12:346
79. Li Q, Ni Y, Zhang L, Jiang R, Xu J, Yang H. et al. HIF-1alpha-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct Target Ther. 2021;6:76
80. Huang Y, Gao D, Wu Y, Sun L, Chen J, Chen J. et al. YTHDF1 Protects Auditory Hair Cells from Cisplatin-Induced Damage by Activating Autophagy via the Promotion of ATG14 Translation. Mol Neurobiol. 2022
81. Hao W, Dian M, Zhou Y, Zhong Q, Pang W, Li Z. et al. Autophagy induction promoted by m(6)A reader YTHDF3 through translation upregulation of FOXO3 mRNA. Nat Commun. 2022;13:5845
82. Liang D, Lin WJ, Ren M, Qiu J, Yang C, Wang X. et al. m(6)A reader YTHDC1 modulates autophagy by targeting SQSTM1 in diabetic skin. Autophagy. 2022;18:1318-37
83. Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen YX. et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Molecular cancer. 2019;18:174
84. Han L, Lei G, Chen Z, Zhang Y, Huang C, Chen W. IGF2BP2 Regulates MALAT1 by Serving as an N6-Methyladenosine Reader to Promote NSCLC Proliferation. Front Mol Biosci. 2021;8:780089
85. Liu Y, Shi SL. The roles of hnRNP A2/B1 in RNA biology and disease. Wiley interdisciplinary reviews RNA. 2021;12:e1612
86. Li T, Wang T, Jing J, Sun L. Expression Pattern and Clinical Value of Key m6A RNA Modification Regulators in Abdominal Aortic Aneurysm. J Inflamm Res. 2021;14:4245-58
87. Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y. et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10:2782
88. Qin S, Mao Y, Chen X, Xiao J, Qin Y, Zhao L. The functional roles, cross-talk and clinical implications of m6A modification and circRNA in hepatocellular carcinoma. Int J Biol Sci. 2021;17:3059-79
89. Wang H, Zhang Y, Chen L, Liu Y, Xu C, Jiang D. et al. Identification of clinical prognostic features of esophageal cancer based on m6A regulators. Front Immunol. 2022;13:950365
90. Liu S, Li G, Li Q, Zhang Q, Zhuo L, Chen X. et al. The roles and mechanisms of YTH domain-containing proteins in cancer development and progression. Am J Cancer Res. 2020;10:1068-84
91. Chen J, Sun Y, Xu X, Wang D, He J, Zhou H. et al. YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle. 2017;16:2259-71
92. Lobo J, Costa AL, Cantante M, Guimarães R, Lopes P, Antunes L. et al. m(6)A RNA modification and its writer/reader VIRMA/YTHDF3 in testicular germ cell tumors: a role in seminoma phenotype maintenance. J Transl Med. 2019;17:79
93. Fanale D, Iovanna JL, Calvo EL, Berthezene P, Belleau P, Dagorn JC. et al. Germline copy number variation in the YTHDC2 gene: does it have a role in finding a novel potential molecular target involved in pancreatic adenocarcinoma susceptibility? Expert Opin Ther Targets. 2014;18:841-50
94. Chen F, Gong E, Ma J, Lin J, Wu C, Chen S. et al. Prognostic score model based on six m6A-related autophagy genes for predicting survival in esophageal squamous cell carcinoma. J Clin Lab Anal. 2022;36:e24507
95. Huang W, Qi CB, Lv SW, Xie M, Feng YQ, Huang WH. et al. Determination of DNA and RNA Methylation in Circulating Tumor Cells by Mass Spectrometry. Anal Chem. 2016;88:1378-84
96. Jiang L, Zhang M, Wu J, Wang S, Yang X, Yi M. et al. Exploring diagnostic m6A regulators in endometriosis. Aging (Albany N Y). 2020;12:25916-38
97. Peng J, Zheng H, Liu F, Wu Q, Liu S. The m6A methyltransferase METTL3 affects autophagy and progression of nasopharyngeal carcinoma by regulating the stability of lncRNA ZFAS1. Infect Agent Cancer. 2022;17:1
98. Zhang Z, Zhu H, Hu J. CircRAB11FIP1 promoted autophagy flux of ovarian cancer through DSC1 and miR-129. Cell Death Dis. 2021;12:219
99. Boumahdi S, de Sauvage FJ. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat Rev Drug Discov. 2020;19:39-56
100. Li X, Li M, Huang M, Lin Q, Fang Q, Liu J. et al. The multi-molecular mechanisms of tumor-targeted drug resistance in precision medicine. Biomed Pharmacother. 2022;150:113064
101. Quagliano A, Gopalakrishnapillai A, Barwe SP. Understanding the Mechanisms by Which Epigenetic Modifiers Avert Therapy Resistance in Cancer. Front Oncol. 2020;10:992
102. Mudduluru G, Walther W, Kobelt D, Dahlmann M, Treese C, Assaraf YG. et al. Repositioning of drugs for intervention in tumor progression and metastasis: Old drugs for new targets. Drug Resist Updat. 2016;26:10-27
103. Jin D, Guo J, Wu Y, Du J, Yang L, Wang X. et al. m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12:135
104. Nakano M, Ondo K, Takemoto S, Fukami T, Nakajima M. Methylation of adenosine at the N(6) position post-transcriptionally regulates hepatic P450s expression. Biochem Pharmacol. 2020;171:113697
105. Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L. et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun. 2019;10:4892
106. Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R, Wang YY. et al. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog. 2018;57:590-7
107. Duan Y, Yu C, Yan M, Ouyang Y, Ni S. m6A Regulator-Mediated RNA Methylation Modification Patterns Regulate the Immune Microenvironment in Osteoarthritis. Frontiers in Genetics. 2022 13
108. Doherty J, Baehrecke EH. Life, death and autophagy. Nat Cell Biol. 2018;20:1110-7
109. Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27-42
110. Lin JF, Lin YC, Tsai TF, Chen HE, Chou KY, Hwang TI. Cisplatin induces protective autophagy through activation of BECN1 in human bladder cancer cells. Drug Des Devel Ther. 2017;11:1517-33
111. Tseng WC, Lee PY, Tsai MT, Chang FP, Chen NJ, Chien CT. et al. Hypoxic mesenchymal stem cells ameliorate acute kidney ischemia-reperfusion injury via enhancing renal tubular autophagy. Stem Cell Res Ther. 2021;12:367
112. Mathiyalagan P, Adamiak M, Mayourian J, Sassi Y, Liang Y, Agarwal N. et al. FTO-Dependent N(6)-Methyladenosine Regulates Cardiac Function During Remodeling and Repair. Circulation. 2019;139:518-32
113. Aghaei M, Motallebnezhad M, Ghorghanlu S, Jabbari A, Enayati A, Rajaei M. et al. Targeting autophagy in cardiac ischemia/reperfusion injury: A novel therapeutic strategy. J Cell Physiol. 2019;234:16768-78
114. Lu Y, Wang S, Cai S, Gu X, Wang J, Yang Y. et al. Propofol-induced MiR-20b expression initiates endogenous cellular signal changes mitigating hypoxia/re-oxygenation-induced endothelial autophagy in vitro. Cell Death Dis. 2020;11:681
115. Thessen AE, Grondin CJ, Kulkarni RD, Brander S, Truong L, Vasilevsky NA. et al. Community Approaches for Integrating Environmental Exposures into Human Models of Disease. Environ Health Perspect. 2020;128:125002
116. Qiu Z, Li G, An T. In vitro toxic synergistic effects of exogenous pollutants-trimethylamine and its metabolites on human respiratory tract cells. Sci Total Environ. 2021;783:146915
117. Erythropel HC, Maric M, Nicell JA, Leask RL, Yargeau V. Leaching of the plasticizer di(2-ethylhexyl)phthalate (DEHP) from plastic containers and the question of human exposure. Appl Microbiol Biotechnol. 2014;98:9967-81
118. Pan Y, Jing J, Dong F, Yao Q, Zhang W, Zhang H. et al. Association between phthalate metabolites and biomarkers of reproductive function in 1066 Chinese men of reproductive age. J Hazard Mater. 2015;300:729-36
119. Zhao T, Wang J, Wu Y, Han L, Chen J, Wei Y. et al. Increased m6A modification of RNA methylation related to the inhibition of demethylase FTO contributes to MEHP-induced Leydig cell injury(). Environ Pollut. 2021;268:115627
120. Lin H, Zhang X, Feng N, Wang R, Zhang W, Deng X. et al. LncRNA LCPAT1 Mediates Smoking/ Particulate Matter 2.5-Induced Cell Autophagy and Epithelial-Mesenchymal Transition in Lung Cancer Cells via RCC2. Cell Physiol Biochem. 2018;47:1244-58
121. Wang S, Lv W, Li T, Zhang S, Wang H, Li X. et al. Dynamic regulation and functions of mRNA m6A modification. Cancer cell international. 2022;22:48
122. Seo W, Silwal P, Song IC, Jo EK. The dual role of autophagy in acute myeloid leukemia. Journal of hematology & oncology. 2022;15:51
123. Dou C, Zhang Y, Zhang L, Qin C. Autophagy and autophagy-related molecules in neurodegenerative diseases. Animal Model Exp Med. 2022
124. Wang Y, Jia G. Detection methods of epitranscriptomic mark N6-methyladenosine. Essays Biochem. 2020;64:967-79
125. Meyer KD. DART-seq: an antibody-free method for global m(6)A detection. Nat Methods. 2019;16:1275-80
126. Wang Y, Xiao Y, Dong S, Yu Q, Jia G. Antibody-free enzyme-assisted chemical approach for detection of N(6)-methyladenosine. Nat Chem Biol. 2020;16:896-903
127. Yu H, Pu Q, Weng Z, Zhou X, Li J, Yang Y. et al. DNAzyme based three-way junction assay for antibody-free detection of locus-specific N(6)-methyladenosine modifications. Biosensors & bioelectronics. 2021;194:113625
128. Wang L, Zhang S, Li H, Xu Y, Wu Q, Shen J. et al. Quantification of m6A RNA methylation modulators pattern was a potential biomarker for prognosis and associated with tumor immune microenvironment of pancreatic adenocarcinoma. BMC Cancer. 2021;21:876
129. Wei C, Wang B, Peng D, Zhang X, Li Z, Luo L. et al. Pan-Cancer Analysis Shows That ALKBH5 Is a Potential Prognostic and Immunotherapeutic Biomarker for Multiple Cancer Types Including Gliomas. Frontiers in immunology. 2022;13:849592
130. Guan Q, Lin H, Miao L, Guo H, Chen Y, Zhuo Z. et al. Functions, mechanisms, and therapeutic implications of METTL14 in human cancer. Journal of hematology & oncology. 2022;15:13
131. Chen B, Ye F, Yu L, Jia G, Huang X, Zhang X. et al. Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J Am Chem Soc. 2012;134:17963-71
132. Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H. et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373-84
133. Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G. et al. m(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017;18:2622-34
134. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y. et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell. 2018;172:90-105.e23
135. Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G. et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021;593:597-601
Author contact
![]() Corresponding authors: Hengwei Liu, Department of Obstetrics and Gynecology, Zhongnan Hospital of Wuhan University, Wuhan University, Wuhan 430071, China. E-mail: hw.liuedu.cn and Yi Liu, Department of Obstetrics and Gynecology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430022, China. E-mail: Liqun94com.
Corresponding authors: Hengwei Liu, Department of Obstetrics and Gynecology, Zhongnan Hospital of Wuhan University, Wuhan University, Wuhan 430071, China. E-mail: hw.liuedu.cn and Yi Liu, Department of Obstetrics and Gynecology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430022, China. E-mail: Liqun94com.

 Global reach, higher impact
Global reach, higher impact