10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2023; 19(10):3015-3028. doi:10.7150/ijbs.83056 This issue Cite
Review
Targeting Type I Interferon Induction and Signaling: How Zika Virus Escapes from Host Innate Immunity
1. Department of Biomedical Sciences, City University of Hong Kong, Hong Kong, China.
2. City University of Hong Kong Shenzhen Research Institute, Shenzhen 518057, China.
Received 2023-1-31; Accepted 2023-5-23; Published 2023-6-4
Abstract
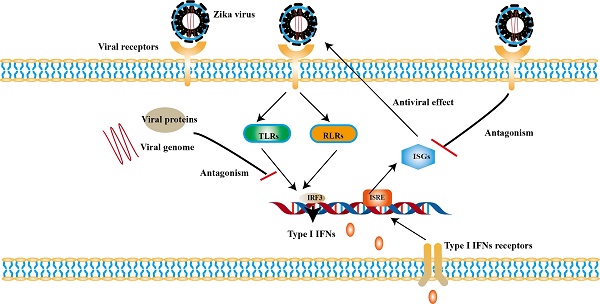
Zika virus (ZIKV) infection causes neurological disorders and draws great attention. ZIKV infection can elicit a wide range of immune response. Type I interferons (IFNs) as well as its signaling cascade play crucial role in innate immunity against ZIKV infection and in turn ZIKV can antagonize them. ZIKV genome are mainly recognized by Toll-like receptors 3 (TLR3), TLR7/8 and RIG-I-like receptor 1 (RIG-1), which induces the expression of Type I IFNs and interferon-stimulated genes (ISGs). ISGs exert antiviral activity at different stages of the ZIKV life cycle. On the other hand, ZIKV takes multiple strategies to antagonize the Type Ⅰ IFN induction and its signaling pathway to establish a pathogenic infection, especially by using the viral nonstructural (NS) proteins. Most of the NS proteins can directly interact with the factors in the pathways to escape the innate immunity. In addition, structural proteins also participate in the innate immune evasion and activation of antibody-binding of blood dendritic cell antigen 2 (BDCA2) or inflammasome also be used to enhance ZIKV replication. In this review, we summarize the recent findings about the interaction between ZIKV infection and type I IFNs pathways and suggest potential strategies for antiviral drug development.
Keywords: Zika virus, type I interferons, interferons-stimulated genes
Introduction
Innate immune response is the first line of host defense against pathogens. And Interferons (IFNs) play an important role in innate immunity, including type I, type Ⅱ and type Ⅲ IFN [1-3]. Type I IFNs, refer to a single subtype of IFN- β and multiple subtypes of IFN-α, have been found in all nucleated cells and proven to play a pivotal role in antiviral functions [4]. Generally, the production of type I IFNs is induced by cell recognition of pathogen associated molecular patterns (PAMPs) [5]. Type I IFNs bind to IFNs receptor (IFNAR), then activate receptor-associated janus kinases (JAK) and tyrosine-protein kinase (TYK), and subsequently lead to phosphorylation and activation of signal transducers and activators of transcription (STATs) [6, 7]. The phosphorylated STAT1 (P-STAT1) or the phosphorylated STAT2 (P-STAT2) and the IFN-regulatory factor 9 (IRF9) form IFN-stimulated gene factor 3 (ISGF3), and the complex enters into the nucleus to trigger expression of IFNs-stimulated genes (ISGs) [8, 9]. Some ISGs have been proven to exert diverse antiviral effects [10, 11]. Meanwhile, several studies demonstrate that type I IFNs have effects on humoral response by upregulating antibody production or suppressing B‐cell linear epitopes [12, 13].
The induction of type I IFNs expression and type I IFNs signaling are key events of antiviral innate response [14], many members of Flavivirus genus have been proven to interact with type I IFN response. Flavivirus genus consists of more than 70 RNA viruses, including Dengue virus (DENV), West Nile virus (WNV), Zika virus (ZIKV), and Japanese encephalitis virus (JEV) etc. Flavivirus genus can cause health damage to varying degrees. DENV is associated with dengue shock syndrome / dengue hemorrhagic fever and ZIKV is related with microcephaly and so on. Type I IFNs is important to resist flavivirus infection. Studies have demonstrated that IFNAR1 (Interferon Alpha and Beta Receptor Subunit 1) deficient mice were susceptible to DENV and ZIKV infection [15]. On the contrary, almost every member of the flavivirus genus can evade the type I IFN response. For example, multiple non-structural proteins (NS) of DENV have been shown to inhibit type I IFNs and the downstream signaling pathway. DENV NS4A blocks the interaction between RIG-I and MAVS [16]. DENV NS2A and NS4B suppresses TBK1 phosphorylation [17]. And WNV NS proteins are responsible for the degradation of IFNAR [18]. All of them were well summarized in previous reviews.
ZIKV, as an emerging pathogen, was first discovered in the Zika forest of Uganda in Africa in 1947 [19]. ZIKV can be sexually and vertically transmitted [20-22], and the bite by the infected female Aedes mosquitoes is the most common route of transmission [19]. Clinical symptoms of people infected with ZIKV are mostly described as asymptomatic, while 20%-25% of the infected will develop self-limiting flu-like symptoms after a duration of 4-10 days [23]. An increased risk of neurologic complications associated with ZIKV infection has been observed, such as Guillain-Barré syndrome, microcephaly and so forth [24-26]. The World Health Organization (WHO) declared ZIKV infection as a Public Health Emergency of International Concern in 2016 [27].
ZIKV has a single-stranded RNA genome with positive sense, encoding three structural and seven NS proteins [28]. The envelope (E) protein and membrane (M) protein form an icosahedral shell anchored in a lipid membrane [29]. The E protein binds to cell receptor that mediates viral attachment and membrane fusion [30]. The E protein is also an important target for immune recognition. NS proteins of ZIKV include NS1, NS2 (NS2A, NS2B), NS3, NS4 (NS4A, NS4B), NS5. Most of them are correlated with viral replication and immune response evasion [30] (Table 1).
This review focuses on the interaction between ZIKV infection and Type I IFN signaling, which would help us to identify potential strategies for antiviral drug development.
Pattern recognition receptors (PRRs) associated with ZIKV infection
The recognition of ZIKV by PRRs is fundamental in producing type I IFNs. The viral replication intermediates of double-stranded RNA (dsRNA), RNA transcripts and protein are activator for PRRs [52, 53]. It has been observed that the expression of several PRRs involved in the innate immune response in ZIKV-infected astrocytes, such as membrane-anchored Toll-like receptors (TLRs), RIG-I-like receptors (RLRs) [54]. Inhibition of PRRs signaling in testicular germ cells (TGCs) leads to the prolonged replication of ZIKV, and such phenomenon can be reversed by exogenous IFNβ [55].
Structure and functions of ZIKV proteins
| Name | Functions | Functional domains |
|---|---|---|
| E | Viral entry, membrane fusion and eliciting neutralizing antibodies [31] | Domain I, domain II and domain III [29] |
| M | Assisting E protein folding and preventing ZIKV premature fusion [31] | A loop at the N terminus (M-loop), a stem and a transmembrane region [29] |
| C | Facilitating transfer of viral genome into the host cell, recruitment viral genome and genome encapsulation [32] | Four α helices with a long pre-α1 loop [33] |
| NS1 | Viral replication, antigenic marker, immune evasion, pathogenesis and inducing a specific immune response [34, 35] | A β-hairpin domain, a wing domain and a β-ladder domain [36] |
| NS2A | Viral replication, viral assembly or secretion and immune evasion [37-39] | A membrane-traversing segment and six segments associated peripherally with the ER membrane [37] |
| NS2B/NS3 | Polyprotein processing [40] | A C-terminal fragment of NS2B and a protease domain of NS3 [41] |
| NS3 | Polyprotein processing, viral replication and immune evasion [42] | A protease domain and a helicase domain [43] |
| NS4A | Membrane binding and homo-oligomerization [44] | A N-terminal cytoplasmic region and a transmembrane segment ([45] |
| NS4B | Viral replication and immune evasion [46] | Three transmembrane helices and two helices that peripherally associate with the membrane [47] |
| NS4 | Pathogenesis, viral replication, membrane binding and homo-oligomerization [48] | A N-terminal cytosolic region and four predicted transmembrane segments (pTMSs) [49] |
| NS5 | Viral replication and immune evasion [50] | A N-terminal methyltransferase (MTase) domain and a C-terminal RNA-dependent RNA polymerase (RdRp) [51] |
Notes: C is the abbreviation of capsid
TLRs
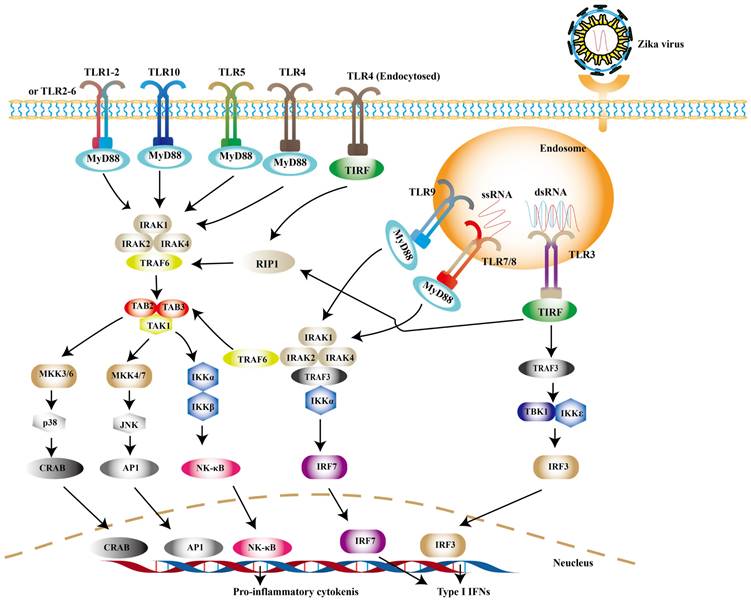
TLRs are widely distributed on the cell surface or in endosomal membranes of effector cells. TLR signaling pathways are crucial pathways in innate immune defense. Up to now, 10 human TLRs have been found in humans and the function of TLR1-9 has been confirmed. TLR-3, -7, -8 and -9, which locate in the endosome, are the key players involved in antiviral immunity [56]. After PAMPs binding to PRRs, the activation of downstream signaling depends on myeloid differentiation primary response 88 (MyD88) or toll-interleukin 1 receptor domain-containing adapter (TRIF) induces type I IFNs [57]. Among them, TRIF is required by TLR3 or TLR4, while the rest recruit MyD88 [58]. Experimental observations indicate that TLR7/8 agonist R848 is an inhibitor for blocking ZIKV replication in monocytes [59]. TLR7/8 are sensors for single-stranded RNA (ssRNA) [60]. The connection of MyD88 and TLR7/8 leads to the phosphorylation of interleukin-1 receptor-associated kinases (IRAKs), followed by the activation of TRAF6, TRAF3, IKKα and IRF7, resulting in the release of type I IFNs [61] (Fig. 1).
It is noteworthy to mention that TLR3 may exhibit opposite effects in different cell types. On one hand, TLR3 can recognize the intermediate of double-stranded RNA (dsRNA), and then recruits TRIF to phosphorylate interferon regulatory factor 3 (IRF3), nuclear factor-κB (NF-κB) successively, which leads to the production of type I IFNs ultimately [62] (Fig. 1). On the other hand, it may be related with pathogenicity of ZIKV. TLR3 is highly expressed in the early development of the brain [63]. The activation of TLR3 leads to dysregulation of neurogenesis in neural progenitor cells (NPCs) and apoptosis, the possible cause of microcephaly in newborn baby [64]. In primary human astrocytes, TLR3 contributes to ZIKV-associated neurodevelopmental disorders by releasing inflammatory factors [65]. In addition, TLR3 enhances ZIKV replication by suppressing other IFNs production and their signaling [66]. Such detrimental effect of TLR3 can also be observed in WNV, another virus of the genus Flavivirus. Studies have revealed that TLR3-dependent inflammatory response caused by WNV leads to neuronal injury [67].
RLRs
RLRs are cytoplasmic viral RNA sensors, including retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2) [68]. The short double stranded RNA with 5'- triphosphate (3P) terminal [69] and viral double-stranded RNAs (dsRNA) [70] are ideal ligands for RLRs, while the 5′ region of ZIKV's genome is such a ligand [71, 72]. In ZIKV- infected cells, RIG-1-mitochondrial antiviral signaling (MAVS) signaling pathway is a major pathway against ZIKV, especially in the central nervous system (CNS) [73]. RIG-I and MDA5 are mainly distributed in astrocytes and microglia [74]. Neural stem cells (NSCs) infected with ZIKV can activate RIG-I pathway to induce the expression of IFN-β, which limits the transmission of ZIKV [74]. And RIG-I-mediated pathway effectively protects human dermal fibroblasts and epidermal keratinocytes against ZIKV infection [73].
TLR signaling pathways. Upon ZIKV infection, TLR3 and TLR7/8 are mainly activated to induce the expression of type I IFNs. And other TLR family members can be activated during other viral infection.
RIG-I is mainly activated during ZIKV infection. The 5'ppp RNA of ZIKV activates the RIG-I with the help of Riplet and TRIM25. And the activated RIG-I translocates to mitochondria to interact with MAVS. And then the TRAF3 is phosphorylated to induce the expression of type I IFNs. Apart from this, the activation of MDA5 can be observed in other viral infections.
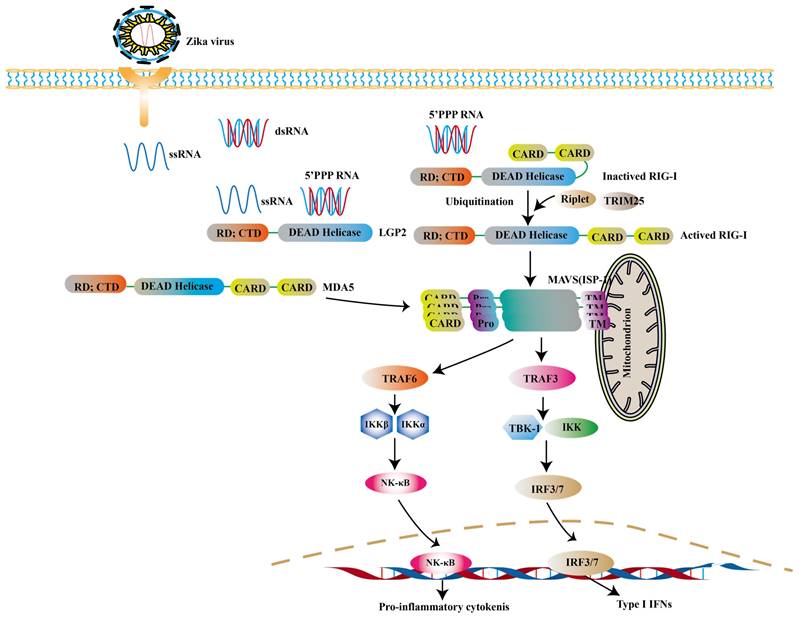
RIG-I and MDA5 are all comprised of two N-terminal caspase activation and recruitment domains (CARD), a central DExD/H box RNA helicase domain that has the ability of hydrolyzing ATP and binding or possibly unwinding RNA, and a C-terminal repressor domain (RD) embedded within the C-terminal domain (CTD) [75, 76]. After CTD binding with intracellular virus-derived, CARD is exposed, resulting in the activation of RIG-I. And then the polyubiquitination of RIG-I is triggered through two ubiquitin E3 ligases, tripartite motif-containing 25 (TRIM25) and Riplet [77]. The interaction between polyubiquitinated RIG-I and MAVS leads to further recruiting a group of molecules to activate TANK binding kinase 1 (TBK1) - IκB kinase (IKK) complex. These kinases then activate transcription factors such as IRF3, IRF7, thereby inducing the expression of genes encoding type I IFNs and the production of pro-inflammatory cytokines [78-80] (Fig. 2).
The antiviral ability of Interferon-stimulated genes (ISGs)
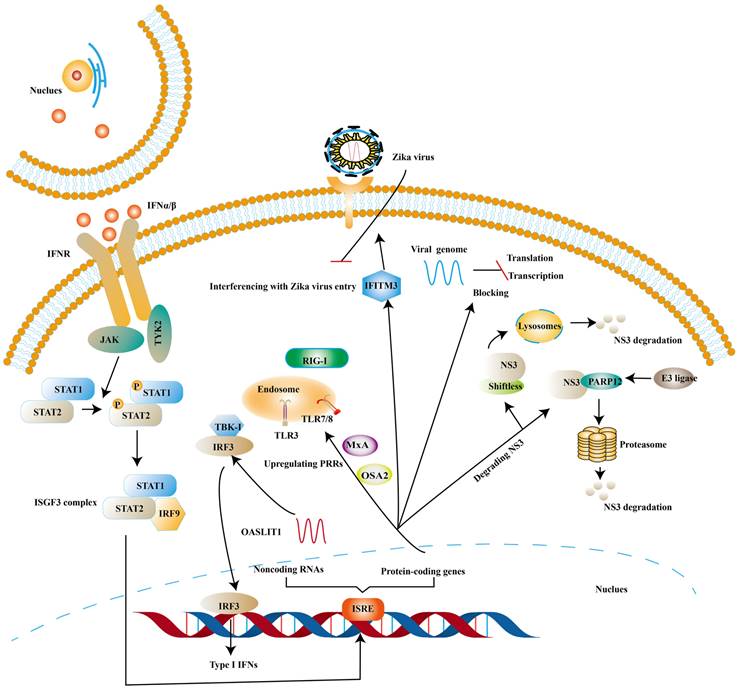
ISGs, induced by the binding of IFNs and IFNAR, have been proven to inhibit viral infection at different stages of the viral replication cycle [81, 82]. One of the canonical activation pathways to induce the transcription of ISGs is JAK-STATs pathway (Fig. 3) [83]. ISGs stimulated by type I IFNs comprise protein-coding genes and noncoding RNAs (ncRNAs). At present, protein-coding genes are more than 300 and ncRNAs can be divided into short ncRNAs and long ncRNAs based on the length [84, 85].
Some ISGs exert their antiviral effects by positive feedback on the induction of type I IFNs though ISGs are the effector molecules of type I IFNs response. OASL-IT1, a type of ncRNAs, can trigger production of IFN-β by regulating IRF3 and NF-κB positively to help epithelial cells resist ZIKV infection [85]. The same trend is mirrored in ISGs encoding antiviral protein. Myxovirus resistance protein A (MxA) is found to strengthen the expression of type I IFNs and activate JAK-STATs signaling pathway by upregulating the expression levels of PRRs [85]. In vitro experiments suggest that antigen processing type 1 (TAP1) inhibits ZIKV infection by means of phosphorylating TBK1 and IRF3 [86]. 2′, 5′-oligoadenylate synthetase (OAS) 2 can also exert its antiviral effects by means of enhancing the expression of type I IFNs [87]. The possible mechanism is OAS / RNase L pathway, in which OAS activates RNase L (a latent endoribonuclease) and the activated RNase L cleaves both host and viral RNA indiscriminately. In turn, the cleaved RNA can stimulate PRRs to reinforce the production of type I IFNs [88].
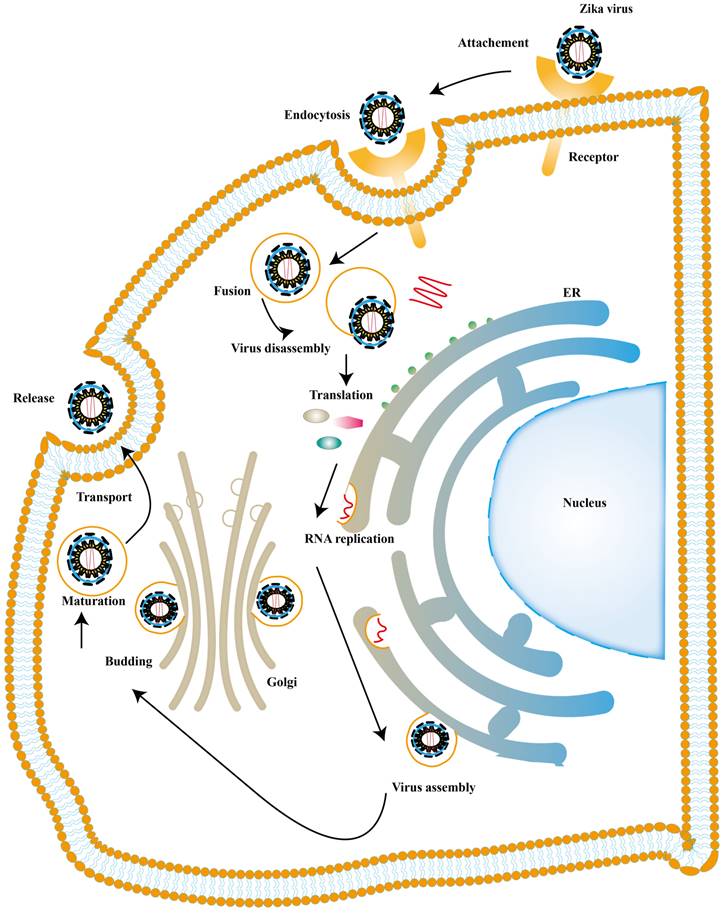
There is also a significant part of ISGs exert antiviral activity at multiple steps in the ZIKV replication cycles. The ZIKV life cycle starts with the E protein binding to cell surface receptor and ZIKV enters the cell through endocytosis. The low pH in endosome induces conformational rearrangement of E protein, which leads to the release of the genome. The viral genome is translated into a polyprotein with the help of the host translation system and it is finally cleaved into three structural and seven NS proteins. Subsequently, the viral replication takes place within vesicles. Viral RNA is packaged in the endoplasmic reticulum (ER) to an immature virion and then it is transferred into golgi vesicles to form mature virions. The mature virion can be released to the extracellular space [89] (Fig. 4). Experimental data shows that small membrane-associated interferon-inducible transmembrane proteins (IFITMs) 3 suppresses ZIKV infection by inhibiting cytosolic entry of ZIKV or its early transcription [90] and the possible mechanism is that it blocks fusion pore formation and inhibits ZIKV viral genome and proteins entry into the cytosol after ZIKV-host binding [91, 92].
ISGs elicit the antiviral state against ZIKV. The binding of Type I IFNs and IFNAR activate classical JAK/TYK2 pathway, which leads to the formation of ISGF3. And then ISGF3 translocates into the nucleus to induce the expression of ISGs. Finally, various species of ISGs exert antiviral effects at different steps of the ZIKV life cycle or strengthens the expression of Type I IFNs.
ZIKV life cycle. The ZIKV infection cycle can be divided into 7 stages, including viral attachment, membrane fusion, endocytosis, transcription and translation, genome replication, virion assembly, maturation and release.
In addition to this, ISGs can inhibit viral ZIKV replication by degrading NS3. NS3 possesses helicase and RNA triphosphatase activities, playing an essential role in virus replication [93]. Ubiquitin-proteasome system and lysosomal proteolysis are two main intracellular protein degradation pathways. Theoretically, ubiquitin system refers to the ubiquitinated protein being degraded by the proteasome. And the ubiquitination of protein is completed by a cascade of reactions, including ubiquitin-activating enzyme E1, ubiquitin-conjugating enzymes E2 and ubiquitin ligase E3 [94]. Lysosomal proteolysis is that protein is delivered into lysosomes and degraded by a series of proteases [95]. PARP12 belongs to the family of poly-adenosine 5′-diphosphate-ribose (PARPs), which consists of PARP domain, four zinc-finger (ZnF) domains and WWE domain (named after three of its conserved residues, including two conserved tryptophan (W) residues and a glutamic acid (E) residue). NS3, bound by PARP domain, is ubiquitylated by the E3 ligase and degraded by the proteasome [96]. NS1 can also be degraded by such a pathway [96]. Further analyses indicate that PARP11 and PARP12 seem to have a synergistic effect in the defense of ZIKV [97]. Viperin, a member of the radical S-adenosyl methionine (SAM) superfamily of enzymes, has been proven to degrade NS3 via proteasome-dependent manner [98] and Lys358 of NS3 is an essential amino-acid for viperin against ZIKV [99]. Shiftless (also known as C19orf66) is reported to degrade NS3 by lysosomal proteolysis. Shiftless is a conserved ISG in mammals, can bind NS3 protease domain and then NS3-shiftless is localized in lysosomes to promote the degradation of NS3 [100]. However, according to Natasha et al. shiftless can bind the ZIKV RNA to inhibit viral replication [101].
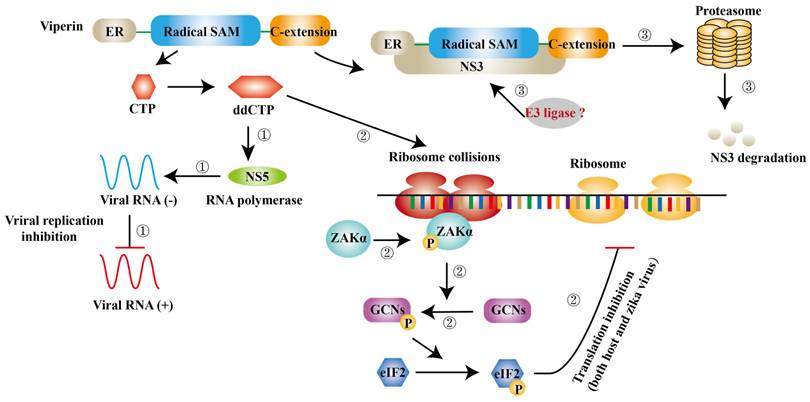
ISGs have also been reported to inhibit viral transcription and translation. Apart from degrading NS3, viperin is found to block the minus-strand RNA or plus-strand RNA synthesis to limit viral protein expression [59]. Jack et al. further suggest that viperin restricts the translation of ZIKV genome via triggering ribosome collisions pathway, and it even restricts the translation of other genomes in cells [102]. ddhCTP is the enzymatic product of viperin, which can activate the GCN2, an eIF2α kinase and the activated eIF2α blocks translation initiation to restrict protein expression immediately after [103, 104]. ISGs can also impair viral RNA to prevent viral infection. ISG20 is a 3′-5′ exonuclease and can degrade ZIKV RNA to block viral replication in cytrophoblast cells of first‐trimester placenta [105] (Fig. 5).
Type I IFNs and type I IFNs- mediated ISGs empower host antiviral ability, however, they may involve in the pathogenesis of ZIKV. Type I IFNs induced by ZIKV interferes with the development of placental labyrinthine zone in mice, finally resulting in fetal (mouse) death and the exact mechanism still needs to be explored [106].
Antagonism of innate immunity by ZIKV
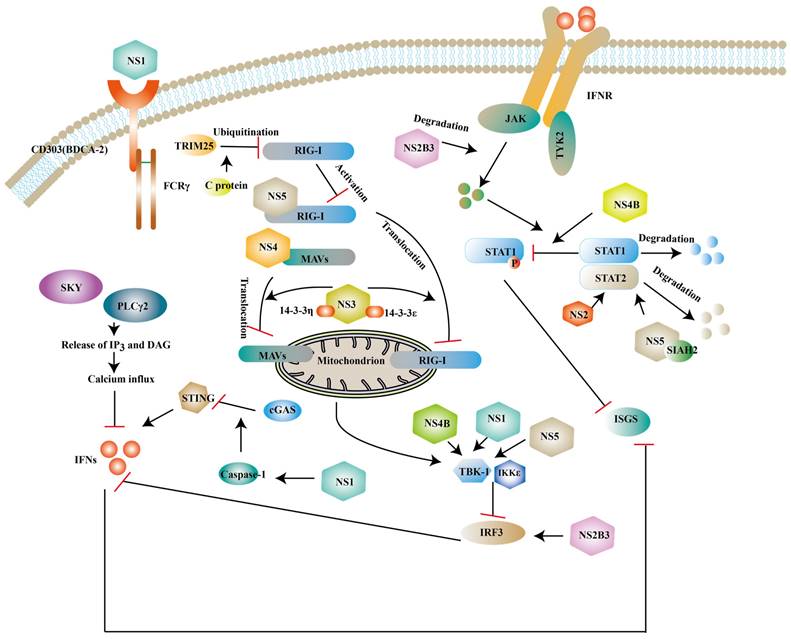
Although innate immunity is an important immune response against ZIKV infection, studies have shown that ZIKV can escape innate immunity by different ways. Human dendritic cells have limited immunogenicity after ZIKV infection, partly due to the viral antagonism of type I IFN response. For example, IFN- β upregulates the expression of major histocompatibility complex class I (MHC-I) and inhibits the killing effect of natural killer cells against ZIKV [107] and NS proteins of ZIKV inhibit the expression of type I IFNs or its downstream molecules to antagonize the innate immunity of host [108, 109] (Fig. 6).
Viperin inhibits ZIKV infection via different mechanisms. ① ddCTP can be a chain terminator for RNA polymerase and then inhibit viral replication. ② ddCTP is able to trigger ribosome collisions and activates ZAKα and induces the phosphorylation of GCNs and eIF2 successively. As a result, translation of both host cells and ZIKV is inhibited. ③ The complex of NS3 and viperin initiates NS3 degradation via a ubiquitin proteasome pathway.
Structural and NS proteins of ZIKV antagonize the host innate immune response. NS2B3, NS3, NS4B, NS5 can block the expression of type I IFNs and ISGs via inhibit phosphorylation of some factors. NS2B3 also can degrade JAK. NS1 can interact with TBK1 or enhance active caspase-1 stability to inhibit the expression of type I IFNs. While NS5 and NS4 can also prevent RIG-I/ MAVs translocation.
NS1
NS1 is a glycoprotein and it can be secreted into the extracellular space in the form of a hexameric lipoprotein particle (sNS1) [110]. BDCA2 (also known as CD303) is a C-type lectin and the activation of it leads to a reduction of type I IFNs, which presumably be related to calcium mobilization and PLCγ2 phosphorylation [111, 112]. One study has revealed that NS1 can activate BDCA2 to limit type I IFNs production based on its N-glycosylation sites [113]. In the CNS, NS1 can upregulate the miR-146a expression and followed a decrease of TRAF6 in human microglial cells, which leads to the reduction of type Ⅰ IFNs [114]. NS1 also interacts with TBK1 to inhibit the expression of IFNs [115]. Specifically, the NS1 A188V mutation leads to less phosphorylation of TBK1 [116]. In addition to interfering with the typical type I IFNs activated pathway, NS1 is also reported to weaken the expression of type I IFNs via activating inflammasome. Caspase-1 is a protease and it mediates the cleavage of cyclic GMP-AMP synthase (cGAS) [117]. cGAS is a DNA sensor located in the cytosolic and cells without it are more vulnerable to some flavivirus according to John et.al [118]. ZIKV can enhance active caspase-1 stability via activating the nucleotide-binding domain and leucine-rich repeat protein-3 (NLRP3) inflammasome or lowering the caspase-1 degradation, which leads to the cleavage of cGAS [119].
NS2/NS3
As revealed by recent studies, NS2A impairs the activation of the NF-κB promoter and the exact mechanism needs further exploration [120]. NS2A consists of 226 amino acids and NS2A51-100 localized in the ER has been proven to mediate the degradation of STAT1 and STAT2, but it is still unclear that such a degradation occurs in proteasomes or lysosomes [121]. Structural study has shown that NS3 protease domain is folded as chymotrypsin-like and it is enwrapped by NS2B polypeptide [122]. NS3 can serve as a protease with the help of NS2B, which is responsible for polyprotein processing and maturation of structural/NS proteins [40]. Mediator of IRF3 activation (MITA, also known as STING or ERIS) is a scaffold protein located in the ER and it can recruit TBK1 or IRF3 to MAVS to transmit signals to downstream molecules [123]. The interaction of NS2B3 with the MITA leads to the degradation of MITA via the ubiquitin proteasome pathway [124]. And the overexpression of NS2B3 can degrade JAK protein levels in a proteasome-dependent manner to impair downstream signaling pathway [115]. Apart from the above, NS3 has been reported to interrupt the translocation of RIG-I and MDA5 from the cytosol to the mitochondria via interacting with 14-3-3ε protein and 14-3-3η protein and such a translocation is required for the activation of TRAF3 [125].
NS4
NS4A is divided into a water-soluble N-terminal cytoplasmic domain and three predicted transmembrane (pTMs) segments. It has been demonstrated that NS4A binds to N-terminal CARD of MAVS to inhibit the interaction of RIG-I and MAVS and which part of NS4 mediates this binding needs further investigation [126]. NS4A also plays an antagonistic role in the production of NF-κB [120]. NS4B can reduce TBK1 activation. One possible mechanism is that NS4B promotes Cholesterol metabolic enzyme 7-dehydrocholesterol reductase (DHCR7) expression. DHCR7 is the inhibitor of TBK1 and IRF3 activation [127]. In vitro studies have shown that NS4B can strongly inhibit the phosphorylation of STAT1 without affecting the total expression of STAT1 [46]. Some studies have showed that NS4B and NS1 protein have synergistic effects to inhibit type I IFN response. IFNβ degrades NS2B-NS3 by autophagic degradation. Wu et al suggested that NS1 or NS4B impaired NS2B3 degradation to attenuate type I IFN response and the detailed mechanisms needs to be explored [115].
NS5
NS5 is composed of an N-terminal MTase and a C-terminal RdRp domain, plays an important role in viral replication and suppresses the RIG-I/MAVs pathway at different levels. The interaction of polyubiquitinated RIG-I and MAVS is necessary for activating downstream signaling pathways. NS5 is proven to impair the polyubiquitination of RIG-I [128]. Some studies suggest that NS5 suppresses the activation of TBK1 [129] or interacts with IKKε to decrease IRF3 phosphorylation [130]. Recently, it has been shown that NS5 interacts with IRF3 localized in the nucleus to inhibit the transcription of type I IFNs during ZIKV infection [131]. Another important target of NS5 is STAT2, which consists of an N-terminal domain (ND), a coiled coil domain (CCD), a DNA binding domain, a linker domain (LD), a SH2 domain, and a transcriptional activation domain (TAD) [132]. It has been demonstrated that the expression of NS5 degrades CCD of STAT2 via the proteasome pathway [132, 133], which may be related to the MTase domain of NS5 [134]. A computer simulation indicates that seven in absentia homolog (SIAH) 2, an E3 ubiquitin ligase that mediates ubiquitination and proteasomal degradation, can be recruited by NS5 to degrade STAT2, which needs to be experimentally validated [135]. However, Jun Shu et. al suggest that NS5 only slightly degrades STAT2 and it is the restriction of ZIKV on host de novo protein synthesis that accelerates the degradation of STAT2 [136]. Certain members of the genus Flavivirus are also proven to repress host protein synthesis [137]. And it is unclear whether the suppression of host protein synthesis of other Flavivirus accelerates STAT2 degradation. Additionally, a study demonstrated that small-ubiquitin-like modifier (SUMO) NS5 of ZIKV can form discrete punctate nuclear bodies (NBs) with STAT2 and thereby remove promyelocytic leukemia (PML) protein from NBs. STAT2/NS5 NBs repress ISG transcription [138].
Structural proteins
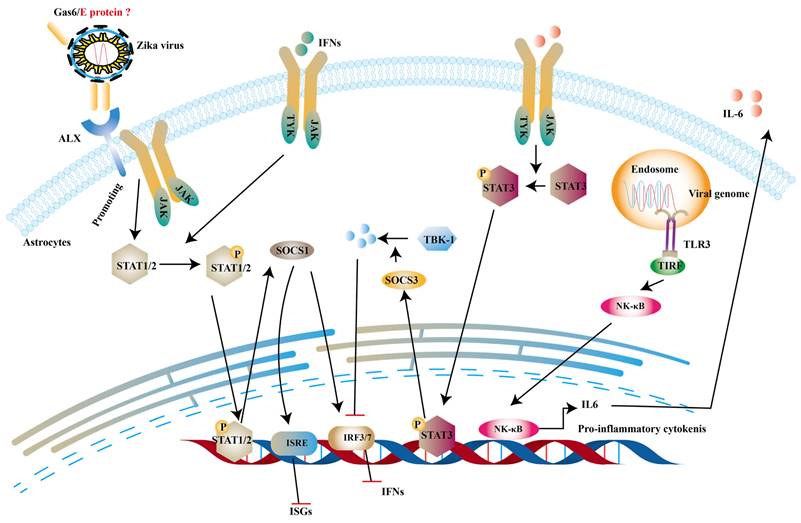
Structural proteins are necessary components of ZIKV virions along with the nucleic acid. And they are also involved in innate immune evasion. Tripartite motif protein 25 (TRIM25) is a type of ubiquitin ligase E3 and mediates polyubiquitination of RIG-I, thereby activating downstream signal transduction [139]. One study reported that capsid protein of ZIKV can bind to TRIM25 to prevent ubiquitination RIG-I [140]. TAM receptor tyrosine kinases include Tyro3, Axl, Mer and Axl are important co-factors in ZIKV entry into human fetal endothelial cells [141]. It is stated that growth arrest specific gene 6 (Gas6), a ligand of Axl, firstly binds to phosphatidylserine on the E protein of ZIKV and subsequently helps ZIKV to bind to TAM [142]. Chen et al. indicates that the activation of TAM leads to the production of SOCS1, which inhibits type I IFNs and ISGs [143]. SOCS1 belongs to SOCS proteins and negatively regulate JAK-STATs pathway to restrict the expression of ISGs [144]. In addition to the this, interleukin-6 (IL-6) promotes production of SOCS3. And then SOCS3 suppresses the levels of type I IFNs and ISGs through degrading TBK1 [145] and decreasing STAT1 phosphorylation [146] (Fig. 7).
ZIKV inhibits type 1 IFNs pathway via IL-6 and ALX receptor. The dsRNA of ZIKV can activate TLR3 to induce the expression of IL-6 and in turn IL-6 phosphorylates STAT3 to trigger the production of SOCS3 to degrade TKB-1. On the other hand, the complex of Gas6 and E protein can promote the phosphorylation of STATs, which induce the expression of SOCS1. And it can inhibit the production of IFNs and ISGs.
Subgenomic flavivirus RNAs and the genomic RNA of ZIKV
Subgenomic flavivirus RNAs (sfRNAs) are products of incomplete degradation of viral genome. They have been proven to be involved in antagonizing type I IFNs responses [147, 148]. Two species of sfRNAs are produced during ZIKA infection, as a consequence of stalling of host 5′- 3′ exoribonucleases in the 3′ untranslated region (UTR) of ZIKV genome [149, 150]. It has been reported that ZIKV sfRNAs inhibited type 1 IFN response, as evidence by that ZIKV-derived sfRNA suppressed type I response at the cellular level [150, 151]. Some evidence suggests that ZIKV sfRNAs and ZIKV NS5 act in cooperation to inhibit STAT1 phosphorylation [152].
The genomic RNA (gRNA) can be chemical modificated during flavivirus infection and RNA methylation modification is the most common. ZIKV RNA methylation occurs at different sites. For example, the ribose 2′-oxygen (2′-O), the position N-6 of adenosine, and the nitrogen on position 7 (N-7) of guanosine [153]. Some studies indicate that 2′-O-methylation of the cap structure can escape the recognization of PRRs [154, 155]. Experimental evidence shows that 2′-O methylation of WNV can evade the ISG, tetra-tricopeptide repeats (IFIT) response [156] and DENV deficient 2′-O methylation mutant was more sensitive to IFN [157].
Conclusions and future perspectives
Currently, there are no licensed vaccines or approved drugs to prevent or treat ZIKV infection. Drug targets can be either host proteins or viral proteins. Targeting host factors may trigger immune antiviral responses or disrupt the viral life cycle, while targeting viral protein could directly damage or suppress the virus life cycle [158].
Type I IFNs plays an important role in defending against viral infection and type 1 IFNs system has been used as a host-targeting antiviral method. However, the interaction between ZIKV and type 1 IFNs system is complicated. Better understanding their interactions would help us to identify potential molecular targets for treating ZIKV infection. As mentioned in this article, ZIKV has different mechanisms to evade the innate immunity, especially the inhibition of the type I IFN response. At present, many research studies focus on ZIKV proteins directly act on the key factors involved in type I IFNs induction pathway and type I IFNs signaling pathway. A NS protein can interfere with multiple factors involved in the typical type I IFNs system and there are cascade reactions between those factors, such as TBK1 and IRF3. The inhibitory mechanism of NS proteins on these interrelated factors needs further exploration. NS proteins are usually composed of several domains and some of the inhibitory effects of NS proteins on type I IFNs are not localized to specific domains or sites. This allows us to conduct detailed studies about this aspect in the future. And some proteins induced by ZIKV can also indirectly inhibit the expression of type I IFNs and ISGs, such as SOCS proteins and this field needs further investigation. A recent study showed that ZIKV infection stimulates Pim1 kinase expression, which serves as a negative regulator of type I IFN signaling. The Pim1 inhibitors potently inhibited ZIKV reproduction [159], suggest a strategy for developing anti-ZIKV drugs although the underlying mechanism is to be further investigated.
Type I IFNs therapy has been considered an effective antiviral therapy, for example, IFN-β treatment can repress the transcription of ZIKV in primary human vaginal and cervical epithelial cells [160]. Apart from this, the combination of type 1 IFNs and drugs has evident limitations in viral replication. Sofosbuvir [161], ribavirin [162] and bromocriptine [163] combined with IFN-α/β have also been proved to protect against ZIKV infection in vitro. In addition to that, antiviral proteins encoded by ISGs may applied in protecting against ZIKV infection and some drugs with positive feedback on the type I IFNs induction pathway and type I IFNs signaling pathway are considered. After all, there are some disadvantages in using type I IFNs as drug therapy, such as short half-life in vivo, high cost, unexpected side effects and high dosage [164]. In addition to this, some researchers think it is an effective vaccine strategy to modify the NS proteins sites. ZIKV NS4BC100S mutant has been reported to induce higher type 1 IFNs and it enhances CD4+ and CD8+ T-cells responses in immunized mice [165]. Whether the same effect is achieved by modifying the NS proteins sites involved in innate immune evasion should be investigated in the future study.
Acknowledgements
Funding
The work was partially supported by grants from The Science Technology and Innovation Committee of Shenzhen Municipality [JCYJ20180507181627057]; RGC General Research Fund of Hong Kong Special Administrative Region [11104020] and Strategic funds from The City University of Hong Kong to M. He.
Author contributions
Huan Hu built the structure and wrote the first draft. Yaxiu Feng contributed to revise and prepare the manuscript. Mingliang He supervised the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Stark GR, Darnell JE Jr. The JAK-STAT pathway at twenty. Immunity. 2012;36:503-14
2. Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80:4501-9
3. Darnell JE Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415-21
4. Paz S, Vilasco M, Werden SJ, Arguello M, Joseph-Pillai D, Zhao T. et al. A functional C-terminal TRAF3-binding site in MAVS participates in positive and negative regulation of the IFN antiviral response. Cell Res. 2011;21:895-910
5. Dai P, Cao H, Merghoub T, Avogadri F, Wang W, Parikh T. et al. Myxoma virus induces type I interferon production in murine plasmacytoid dendritic cells via a TLR9/MyD88-, IRF5/IRF7-, and IFNAR-dependent pathway. J Virol. 2011;85:10814-25
6. Stancato LF, David M, Carter-Su C, Larner AC, Pratt WB. Preassociation of STAT1 with STAT2 and STAT3 in separate signalling complexes prior to cytokine stimulation. J Biol Chem. 1996;271:4134-7
7. Tang X, Gao JS, Guan YJ, McLane KE, Yuan ZL, Ramratnam B. et al. Acetylation-dependent signal transduction for type I interferon receptor. Cell. 2007;131:93-105
8. Yuliantie E, Dai X, Yang D, Crack PJ, Wang MW. High-throughput screening for small molecule inhibitors of the type-I interferon signaling pathway. Acta Pharm Sin B. 2018;8:889-99
9. de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM. et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912-20
10. Yang E, Nguyen LP, Wisherop CA, Kan RL, Li MMH. The Role of ZAP and TRIM25 RNA Binding in Restricting Viral Translation. Front Cell Infect Microbiol. 2022;12:886929
11. Hao X, Li Y, Chen H, Chen B, Liu R, Wu Y. et al. Canine Circovirus Suppresses the Type I Interferon Response and Protein Expression but Promotes CPV-2 Replication. Int J Mol Sci. 2022;23(12):6382
12. Lee CY, Carissimo G, Chen Z, Lum FM, Abu Bakar F, Rajarethinam R. et al. Type I interferon shapes the quantity and quality of the anti-Zika virus antibody response. Clin Transl Immunology. 2020;9:e1126
13. Fallet B, Narr K, Ertuna YI, Remy M, Sommerstein R, Cornille K. et al. Interferon-driven deletion of antiviral B cells at the onset of chronic infection. Sci Immunol. 2016 21;1(4):eaah6817
14. Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373-81
15. Züst R, Toh YX, Valdés I, Cerny D, Heinrich J, Hermida L. et al. Type I interferon signals in macrophages and dendritic cells control dengue virus infection: implications for a new mouse model to test dengue vaccines. Journal of virology. 2014;88:7276-85
16. He Z, Zhu X, Wen W, Yuan J, Hu Y, Chen J. et al. Dengue Virus Subverts Host Innate Immunity by Targeting Adaptor Protein MAVS. Journal of virology. 2016;90:7219-30
17. Dalrymple NA, Cimica V, Mackow ER. Dengue Virus NS Proteins Inhibit RIG-I/MAVS Signaling by Blocking TBK1/IRF3 Phosphorylation: Dengue Virus Serotype 1 NS4A Is a Unique Interferon-Regulating Virulence Determinant. MBio. 2015;6:e00553-15
18. Evans JD, Crown RA, Sohn JA, Seeger C. West Nile virus infection induces depletion of IFNAR1 protein levels. Viral immunology. 2011;24:253-63
19. Weaver SC, Costa F, Garcia-Blanco MA, Ko AI, Ribeiro GS, Saade G. et al. Zika virus: History, emergence, biology, and prospects for control. Antiviral Res. 2016;130:69-80
20. Musso D, Roche C, Robin E, Nhan T, Teissier A, Cao-Lormeau VM. Potential sexual transmission of Zika virus. Emerg Infect Dis. 2015;21:359-61
21. Venturi G, Zammarchi L, Fortuna C, Remoli ME, Benedetti E, Fiorentini C. et al. An autochthonous case of Zika due to possible sexual transmission, Florence, Italy, 2014. Euro Surveill. 2016;21:30148
22. Mesci P, Macia A, Moore SM, Shiryaev SA, Pinto A, Huang CT. et al. Blocking Zika virus vertical transmission. Sci Rep. 2018;8:1218
23. Duffy MR, Chen TH, Hancock WT, Powers AM, Kool JL, Lanciotti RS. et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med. 2009;360:2536-43
24. Şahiner F. [Global spread of Zika virus epidemic: current knowledges and uncertainties]. Mikrobiyol Bul. 2016;50:333-51
25. Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. Zika Virus and Birth Defects-Reviewing the Evidence for Causality. N Engl J Med. 2016;374:1981-7
26. Wang JN, Ling F. Zika Virus Infection and Microcephaly: Evidence for a Causal Link. Int J Environ Res Public Health. 2016 20;13(10):1031
27. Lessler J, Chaisson LH, Kucirka LM, Bi Q, Grantz K, Salje H. et al. Assessing the global threat from Zika virus. Science. 2016;353:aaf8160
28. Miner JJ, Diamond MS. Zika Virus Pathogenesis and Tissue Tropism. Cell Host Microbe. 2017;21:134-42
29. Sirohi D, Chen Z, Sun L, Klose T, Pierson TC, Rossmann MG. et al. The 3.8 Å resolution cryo-EM structure of Zika virus. Science. 2016;352:467-70
30. Shi Y, Gao GF. Structural Biology of the Zika Virus. Trends Biochem Sci. 2017;42:443-56
31. Shi Y, Gao GF. Structural Biology of the Zika Virus. Trends in Biochemical Sciences. 2017;42:443-56
32. Ambroggio EE, Costa Navarro GS, Pérez Socas LB, Bagatolli LA, Gamarnik AV. Dengue and Zika virus capsid proteins bind to membranes and self-assemble into liquid droplets with nucleic acids. J Biol Chem. 2021;297:101059
33. Shang Z, Song H, Shi Y, Qi J, Gao GF. Crystal Structure of the Capsid Protein from Zika Virus. J Mol Biol. 2018;430:948-62
34. Young PR, Hilditch Pa Fau - Bletchly C, Bletchly C Fau - Halloran W, Halloran W. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. 2000; 38(3): 1053-7.
35. Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, Bianchi S. et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. 2016; 353(6301): 823-6.
36. Brown WC, Akey DL, Konwerski JR, Tarrasch JT, Skiniotis G, Kuhn RJ. et al. Extended surface for membrane association in Zika virus NS1 structure. Nat Struct Mol Biol. 2016;23:865-7
37. Zhang X, Xie X, Zou J, Xia H, Shan C, Chen X. et al. Genetic and biochemical characterizations of Zika virus NS2A protein. 2019; 8(1):585-602.
38. He J, Yang LA-O, Chang P, Yang S, Lin S, Tang QA-O. et al. Zika virus NS2A protein induces the degradation of KPNA2 (karyopherin subunit alpha 2) via chaperone-mediated autophagy. 2020; 16(12): 2238-2251.
39. Zhang X, Xie XA-OX, Xia H, Zou J, Huang L, Popov VL. et al. Zika Virus NS2A-Mediated Virion Assembly. mBio. 2019;10(5):e02375-19
40. Voss S, Nitsche C. Inhibitors of the Zika virus protease NS2B-NS3. Bioorg Med Chem Lett. 2020;30(5):126965
41. Phoo WW, Li Y, Zhang Z, Lee MY, Loh YR, Tan YB. et al. Structure of the NS2B-NS3 protease from Zika virus after self-cleavage. Nat Commun. 2016Nov15;7:13410
42. Xu S, Ci Y, Wang L, Yang Y, Zhang L, Xu C. et al. Zika virus NS3 is a canonical RNA helicase stimulated by NS5 RNA polymerase. Nucleic Acids Res. 2019;47:8693-707
43. Luo D, Xu T, Hunke C, Grüber G, Vasudevan SG, Lescar J. Crystal structure of the NS3 protease-helicase from dengue virus. J Virol. 2008;82:173-83
44. To J, Torres J. Trimerization of the N-Terminal Tail of Zika Virus NS4A Protein: A Potential In vitro Antiviral Screening Assay. Membranes (Basel). 2021;11(5):335
45. Klaitong P, Smith DR. Roles of Non-Structural Protein 4A in Flavivirus Infection. Viruses. 2021 15;13(10):2077
46. Fanunza E, Grandi N, Quartu M, Carletti F, Ermellino L, Milia J. et al. INMI1 Zika Virus NS4B Antagonizes the Interferon Signaling by Suppressing STAT1 Phosphorylation. Viruses. 2021 6;13(12):2448
47. Kundharapu S, Chowdary TA-O. Dengue Virus NS4b N-Terminus Disordered Region Interacts with NS3 Helicase C-Terminal Subdomain to Enhance Helicase Activity. Viruses. 2022 3;14(8):1712
48. Surya W, Liu Y, Torres J. The cytoplasmic N-terminal tail of Zika virus NS4A protein forms oligomers in the absence of detergent or lipids. Sci Rep. 2023;13(1):7360
49. Stern O, Hung YF, Valdau O, Yaffe Y, Harris E, Hoffmann S. et al. An N-terminal amphipathic helix in dengue virus nonstructural protein 4A mediates oligomerization and is essential for replication. J Virol. 2013;87:4080-5
50. Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M. et al. Zika Virus Targets Human STAT2 to Inhibit Type I Interferon Signaling. Cell Host Microbe. 2016;19(6):882-90
51. Coloma J, Jain R, Rajashankar KR, García-Sastre A, Aggarwal AK. Structures of NS5 Methyltransferase from Zika Virus. Cell Rep. 2016;16:3097-102
52. Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732-8
53. Jensen S, Thomsen AR. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. Journal of virology. 2012;86:2900-10
54. Hamel R, Ferraris P, Wichit S, Diop F, Talignani L, Pompon J. et al. African and Asian Zika virus strains differentially induce early antiviral responses in primary human astrocytes. Infect Genet Evol. 2017;49:134-7
55. Kuassivi ON, Abiven H, Satie AP, Cartron M, Mahé D, Aubry F. et al. Human Testicular Germ Cells, a Reservoir for Zika Virus, Lack Antiviral Response Upon Zika or Poly(I:C) Exposure. Front Immunol. 2022;13:909341
56. Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335-76
57. Kim AY, Shim HJ, Kim SY, Heo S, Youn HS. Differential regulation of MyD88- and TRIF-dependent signaling pathways of Toll-like receptors by cardamonin. Int Immunopharmacol. 2018;64:1-9
58. Yamashita M, Chattopadhyay S, Fensterl V, Saikia P, Wetzel JL, Sen GC. Epidermal growth factor receptor is essential for Toll-like receptor 3 signaling. Sci Signal. 2012;5(233):ra50
59. Vanwalscappel B, Tada T, Landau NR. Toll-like receptor agonist R848 blocks Zika virus replication by inducing the antiviral protein viperin. Virology. 2018;522:199-208
60. Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529-31
61. van der Sluis RM, Cham LB, Gris-Oliver A, Gammelgaard KR, Pedersen JG, Idorn M. et al. TLR2 and TLR7 mediate distinct immunopathological and antiviral plasmacytoid dendritic cell responses to SARS-CoV-2 infection. EMBO J. 2022;41(10):e109622
62. Matsumoto M, Oshiumi H, Seya T. Antiviral responses induced by the TLR3 pathway. Rev Med Virol. 2011;21:67-77
63. Lathia JD, Okun E, Tang SC, Griffioen K, Cheng A, Mughal MR. et al. Toll-like receptor 3 is a negative regulator of embryonic neural progenitor cell proliferation. J Neurosci. 2008;28:13978-84
64. Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, Eroshkin AM. et al. Zika Virus Depletes Neural Progenitors in Human Cerebral Organoids through Activation of the Innate Immune Receptor TLR3. Cell Stem Cell. 2016;19:258-65
65. Ojha CR, Rodriguez M, Karuppan MKM, Lapierre J, Kashanchi F, El-Hage N. Toll-like receptor 3 regulates Zika virus infection and associated host inflammatory response in primary human astrocytes. PLoS One. 2019;14(2):e0208543
66. Plociennikowska A, Frankish J, Moraes T, Del Prete D, Kahnt F, Acuna C. et al. TLR3 activation by Zika virus stimulates inflammatory cytokine production which dampens the antiviral response induced by RIG-I-like receptors. J Virol. 2021;95(10):e01050-20
67. Wang T, Town T, Alexopoulou L, Anderson JF, Fikrig E, Flavell RA. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat Med. 2004;10(12):1366-73
68. Yoneyama M, Fujita T. Function of RIG-I-like receptors in antiviral innate immunity. J Biol Chem. 2007;282(21):15315-8
69. Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L. et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82(1):335-45
70. Uchikawa E, Lethier M, Malet H, Brunel J, Gerlier D, Cusack S. Structural Analysis of dsRNA Binding to Anti-viral Pattern Recognition Receptors LGP2 and MDA5. Mol Cell. 2016;62(4):586-602
71. Chazal M, Beauclair G, Gracias S, Najburg V, Simon-Lorière E, Tangy F. et al. RIG-I Recognizes the 5' Region of Dengue and Zika Virus Genomes. Cell Rep. 2018;24(2):320-8
72. Lin JY, Kuo RL, Huang HI. Activation of type I interferon antiviral response in human neural stem cells. Stem Cell Res Ther. 2019;10(1):387
73. Kim JA, Seong RK, Son SW, Shin OS. Insights into ZIKV-Mediated Innate Immune Responses in Human Dermal Fibroblasts and Epidermal Keratinocytes. J Invest Dermatol. 2019;139(2):391-9
74. de Rivero Vaccari JP, Minkiewicz J, Wang X, De Rivero Vaccari JC, German R, Marcillo AE. et al. Astrogliosis involves activation of retinoic acid-inducible gene-like signaling in the innate immune response after spinal cord injury. Glia. 2012;60(3):414-21
75. Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC. et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A. 2007;104(2):582-7
76. Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K. et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175(5):2851-8
77. Oshiumi H, Miyashita M, Matsumoto M, Seya T. A distinct role of Riplet-mediated K63-Linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog. 2013;9(8):e1003533
78. Kell AM, Gale M Jr. RIG-I in RNA virus recognition. Virology. 2015;479-480:110-21
79. Said EA, Tremblay N, Al-Balushi MS, Al-Jabri AA, Lamarre D. Viruses Seen by Our Cells: The Role of Viral RNA Sensors. J Immunol Res. 2018;2018:9480497
80. Zevini A, Olagnier D, Hiscott J. Crosstalk between Cytoplasmic RIG-I and STING Sensing Pathways. Trends Immunol. 2017;38(3):194-205
81. Rohaim MA, Al-Natour MQ, Abdelsabour MA, El Naggar RF, Madbouly YM, Ahmed KA. et al. Transgenic Chicks Expressing Interferon-Inducible Transmembrane Protein 1 (IFITM1) Restrict Highly Pathogenic H5N1 Influenza Viruses. Int J Mol Sci. 2021;22(16):8456
82. Subramanian G, Popli S, Chakravarty S, Taylor RT, Chakravarti R, Chattopadhyay S. The interferon-inducible protein TDRD7 inhibits AMP-activated protein kinase and thereby restricts autophagy-independent virus replication. J Biol Chem. 2020;295(20):6811-22
83. Binelli M, Subramaniam P, Diaz T, Johnson GA, Hansen TR, Badinga L. et al. Bovine interferon-tau stimulates the Janus kinase-signal transducer and activator of transcription pathway in bovine endometrial epithelial cells. Biol Reprod. 2001;64(2):654-65
84. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nature Reviews Genetics. 2014;15(1):7-21
85. Wang Y, Huo Z, Lin Q, Lin Y, Zhang P. Positive Feedback Loop of Long Noncoding RNA OASL-IT1 and Innate Immune Response Restricts the Replication of Zika Virus in Epithelial A549 Cells. Journal of Innate Immunity. 2021;13(3):1-15
86. Zhao J, Li R, Li Y, Chen J, Feng F, Sun C. Broadly Antiviral Activities of TAP1 through Activating the TBK1-IRF3-Mediated Type I Interferon Production. Int J Mol Sci. 2021;22(9):4668
87. Liao X, Xie H, Li S, Ye H, Li S, Ren K. et al. 2', 5'-Oligoadenylate Synthetase 2 (OAS2) Inhibits Zika Virus Replication through Activation of Type Ι IFN Signaling Pathway. Viruses. 2020;12(4):418
88. Chakrabarti A, Jha BK, Silverman RH. New insights into the role of RNase L in innate immunity. J Interferon Cytokine Res. 2011;31(1):49-57
89. Anwar MN, Akhtar R, Abid M, Khan SA, Rehman ZU, Tayyub M. et al. The interactions of flaviviruses with cellular receptors: Implications for virus entry. Virology. 2022;568:77-85
90. Savidis G, Perreira JM, Portmann JM, Meraner P, Guo Z, Green S. et al. The IFITMs Inhibit Zika Virus Replication. Cell Rep. 2016;15(11):2323-30
91. Perreira JM, Chin CR, Feeley EM, Brass AL. IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol. 2013;425(24):4937-55
92. Lin TY, Chin CR, Everitt AR, Clare S, Perreira JM, Savidis G. et al. Amphotericin B increases influenza A virus infection by preventing IFITM3-mediated restriction. Cell Rep. 2013;5(4):895-908
93. Cao X, Liu K, Yan S, Li S, Li Y, Jin T. et al. Mechanical regulation of the helicase activity of Zika virus NS3. Biophys J. 2022;121(24):4900-4908
94. Galant C, Marchandise J, Stoenoiu MS, Ducreux J, De Groof A, Pirenne S. et al. Overexpression of ubiquitin-specific peptidase 15 in systemic sclerosis fibroblasts increases response to transforming growth factor β. Rheumatology (Oxford). 2019;58(4):708-18
95. Li C, Wang X, Li X, Qiu K, Jiao F, Liu Y. et al. Proteasome Inhibition Activates Autophagy-Lysosome Pathway Associated With TFEB Dephosphorylation and Nuclear Translocation. Front Cell Dev Biol. 2019;7:170
96. Li L, Zhao H, Liu P, Li C, Quanquin N, Ji X. et al. PARP12 suppresses Zika virus infection through PARP-dependent degradation of NS1 and NS3 viral proteins. Sci Signal. 2018;11(535):eaas9332
97. Li L, Shi Y, Li S, Liu J, Zu S, Xu X. et al. ADP-ribosyltransferase PARP11 suppresses Zika virus in synergy with PARP12. Cell Biosci. 2021;11(1):116
98. Panayiotou C, Lindqvist R, Kurhade C, Vonderstein K, Pasto J, Edlund K. et al. Viperin Restricts Zika Virus and Tick-Borne Encephalitis Virus Replication by Targeting NS3 for Proteasomal Degradation. J Virol. 2018;92(7):e02054-17
99. Vanwalscappel B, Gadea G, Desprès P. A Viperin Mutant Bearing the K358R Substitution Lost its Anti-ZIKA Virus Activity. Int J Mol Sci. 2019;20(7):1574
100. Wu Y, Yang X, Yao Z, Dong X, Zhang D, Hu Y. et al. C19orf66 interrupts Zika virus replication by inducing lysosomal degradation of viral NS3. PLoS Negl Trop Dis. 2020;14(3):e0008083
101. Hanners NW, Mar KB, Boys IN, Eitson JL, De La Cruz-Rivera PC, Richardson RB. et al. Shiftless inhibits flavivirus replication in vitro and is neuroprotective in a mouse model of Zika virus pathogenesis. Proc Natl Acad Sci U S A. 2021;118(49):e2111266118
102. Hsu JC, Laurent-Rolle M, Pawlak JB, Xia H, Kunte A, Hee JS. et al. Viperin triggers ribosome collision-dependent translation inhibition to restrict viral replication. Mol Cell. 2022;82(9):1631-42.e6
103. Wu CC, Peterson A, Zinshteyn B, Regot S, Green R. Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate. Cell. 2020;182(2):404-16.e14
104. Yan LL, Zaher HS. Ribosome quality control antagonizes the activation of the integrated stress response on colliding ribosomes. Mol Cell. 2021;81(3):614-28.e4
105. Ding J, Aldo P, Roberts CM, Stabach P, Liu H, You Y. et al. Placenta-derived interferon-stimulated gene 20 controls ZIKA virus infection. EMBO Rep. 2021;22(10):e52450
106. Yockey LA-O, Jurado KA, Arora N, Millet AA-O, Rakib TA-O, Milano KM. et al. Type I interferons instigate fetal demise after Zika virus infection. Sci Immunol. 2018;3(19):eaao1680
107. Glasner A, Oiknine-Djian E, Weisblum Y, Diab M, Panet A, Wolf DG. et al. Zika Virus Escapes NK Cell Detection by Upregulating Major Histocompatibility Complex Class I Molecules. J Virol. 2017;91(22):e00785-17
108. Ye J, Zhu B, Fu ZF, Chen H, Cao S. Immune evasion strategies of flaviviruses. Vaccine. 2013;31(3):461-71
109. Bowen JR, Quicke KM, Maddur MS, O'Neal JT, McDonald CE, Fedorova NB. et al. Zika Virus Antagonizes Type I Interferon Responses during Infection of Human Dendritic Cells. PLoS Pathog. 2017;13(2):e1006164
110. Song H, Qi J, Haywood J, Shi Y, Gao GF. Zika virus NS1 structure reveals diversity of electrostatic surfaces among flaviviruses. Nat Struct Mol Biol. 2016;23(5):456-8
111. Röck J, Schneider E, Grün JR, Grützkau A, Küppers R, Schmitz J. et al. CD303 (BDCA-2) signals in plasmacytoid dendritic cells via a BCR-like signalosome involving Syk, Slp65 and PLCgamma2. Eur J Immunol. 2007;37(12):3564-75
112. Dzionek A, Sohma Y, Nagafune J, Cella M, Colonna M, Facchetti F. et al. BDCA-2, a novel plasmacytoid dendritic cell-specific type II C-type lectin, mediates antigen capture and is a potent inhibitor of interferon alpha/beta induction. J Exp Med. 2001;194(12):1823-34
113. Bos S, Poirier-Beaudouin B, Seffer V, Manich M, Mardi C, Desprès P. et al. Zika Virus Inhibits IFN-α Response by Human Plasmacytoid Dendritic Cells and Induces NS1-Dependent Triggering of CD303 (BDCA-2) Signaling. Front Immunol. 2020;11:582061
114. Shukla A, Rastogi M, Singh SK. Zika virus NS1 suppresses the innate immune responses via miR-146a in human microglial cells. Int J Biol Macromol. 2021;193(Pt B):2290-6
115. Wu Y, Liu Q, Zhou J, Xie W, Chen C, Wang Z. et al. Zika virus evades interferon-mediated antiviral response through the co-operation of multiple nonstructural proteins in vitro. Cell Discov. 2017;3:17006
116. Xia H, Luo H, Shan C, Muruato AE, Nunes BTD, Medeiros DBA. et al. An evolutionary NS1 mutation enhances Zika virus evasion of host interferon induction. Nat Commun. 2018;9(1):414
117. Shrivastava G, León-Juárez M, García-Cordero J, Meza-Sánchez DE, Cedillo-Barrón L. Inflammasomes and its importance in viral infections. Immunol Res. 2016;64(56):1101-17
118. Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL. et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505(7485):691-5
119. Zheng Y, Liu Q, Wu Y, Ma L, Zhang Z, Liu T. et al. Zika virus elicits inflammation to evade antiviral response by cleaving cGAS via NS1-caspase-1 axis. Embo j. 2018;37(18):e99347
120. Lee JY, Nguyen TTN, Myoung J. Zika Virus-Encoded NS2A and NS4A Strongly Downregulate NF-κB Promoter Activity. J Microbiol Biotechnol. 2020;30(11):1651-8
121. Fanunza E, Carletti F, Quartu M, Grandi N, Ermellino L, Milia J. et al. Zika virus NS2A inhibits interferon signaling by degradation of STAT1 and STAT2. Virulence. 2021;12(1):1580-96
122. Lei J, Hansen G, Nitsche C, Klein CD, Zhang L, Hilgenfeld R. Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor. Science. 2016;353(6298):503-5
123. Liu S, Cai X, Wu J, Cong Q, Chen X, Li T. et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347(6227):aaa2630
124. Li W, Li N, Dai S, Hou G, Guo K, Chen X. et al. Zika virus circumvents host innate immunity by targeting the adaptor proteins MAVS and MITA. Faseb j. 2019;33(9):9929-44
125. Riedl W, Acharya D, Lee JH, Liu G, Serman T, Chiang C. et al. Zika Virus NS3 Mimics a Cellular 14-3-3-Binding Motif to Antagonize RIG-I- and MDA5-Mediated Innate Immunity. Cell Host Microbe. 2019;26(4):493-503.e6
126. Ma J, Ketkar H, Geng T, Lo E, Wang L, Xi J. et al. Zika Virus Non-structural Protein 4A Blocks the RLR-MAVS Signaling. Front Microbiol. 2018;9:1350
127. Chen W, Li Y, Yu X, Wang Z, Wang W, Rao M. et al. Zika virus non-structural protein 4B interacts with DHCR7 to facilitate viral infection. Virol Sin. 2023;38(1):23-33
128. Li A, Wang W, Wang Y, Chen K, Xiao F, Hu D. et al. NS5 Conservative Site Is Required for Zika Virus to Restrict the RIG-I Signaling. Front Immunol. 2020;11:51
129. Lin S, Yang S, He J, Guest JD, Ma Z, Yang L. et al. Zika virus NS5 protein antagonizes type I interferon production via blocking TBK1 activation. Virology. 2019;527:180-7
130. Lundberg R, Melén K, Westenius V, Jiang M, Österlund P, Khan H. et al. Zika Virus Non-Structural Protein NS5 Inhibits the RIG-I Pathway and Interferon Lambda 1 Promoter Activation by Targeting IKK Epsilon. Viruses. 2019;11(11):1024
131. Zhao Z, Tao M, Han W, Fan Z, Imran M, Cao S. et al. Nuclear localization of Zika virus NS5 contributes to suppression of type I interferon production and response. J Gen Virol. 2021;102(3):001376
132. Parisien JP, Lenoir JJ, Alvarado G, Horvath CM. The Human STAT2 Coiled-Coil Domain Contains a Degron for Zika Virus Interferon Evasion. J Virol. 2022;96(1):e0130121
133. Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M. et al. Zika Virus Targets Human STAT2 to Inhibit Type I Interferon Signaling. Cell Host Microbe. 2016;19(6):882-90
134. Kumar A, Hou S, Airo AM, Limonta D, Mancinelli V, Branton W. et al. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep. 2016;17(12):1766-75
135. Dar HA, Zaheer T, Paracha RZ, Ali A. Structural analysis and insight into Zika virus NS5 mediated interferon inhibition. Infect Genet Evol. 2017;51:143-52
136. Shu J, Ma X, Zhang Y, Zou J, Yuan Z, Yi Z. NS5-independent Ablation of STAT2 by Zika virus to antagonize interferon signalling. Emerg Microbes Infect. 2021;10(1):1609-25
137. Roth H, Magg V, Uch F, Mutz P, Klein P, Haneke K. et al. Flavivirus Infection Uncouples Translation Suppression from Cellular Stress Responses. MBio. 2017;8(2):PMID 28074025
138. Conde JN, Schutt WR, Mladinich M, Sohn SY, Hearing P, Mackow ER. NS5 Sumoylation Directs Nuclear Responses That Permit Zika Virus To Persistently Infect Human Brain Microvascular Endothelial Cells. J Virol. 2020;94(19):e01086-20
139. Sánchez-Aparicio MT, Ayllón J, Leo-Macias A, Wolff T, García-Sastre A. Subcellular Localizations of RIG-I, TRIM25, and MAVS Complexes. J Virol. 2017;91(2):e01155-16
140. Airo AM, Felix-Lopez A, Mancinelli V, Evseev D, Lopez-Orozco J, Shire K. et al. Flavivirus Capsid Proteins Inhibit the Interferon Response. Viruses. 2022;14(5):968
141. Richard AS, Shim BS, Kwon YC, Zhang R, Otsuka Y, Schmitt K. et al. AXL-dependent infection of human fetal endothelial cells distinguishes Zika virus from other pathogenic flaviviruses. Acad Sci U S A. 2017;114(8):2024-2029
142. Richard AS, Shim BS, Kwon YC, Zhang R, Otsuka Y, Schmitt K. et al. AXL-dependent infection of human fetal endothelial cells distinguishes Zika virus from other pathogenic flaviviruses. Proc Natl Acad Sci U S A. 2017;114(8):2024-9
143. Chen J, Yang YF, Yang Y, Zou P, Chen J, He Y. et al. AXL promotes Zika virus infection in astrocytes by antagonizing type I interferon signalling. Nat Microbiol. 2018;3(3):302-309
144. Yoshimura A, Suzuki M Fau - Sakaguchi R, Sakaguchi R Fau - Hanada T, Hanada T Fau - Yasukawa H, Yasukawa H. SOCS, Inflammation, and Autoimmunity. Front Immunol. 2012 12;3:20
145. Liu D, Sheng C, Gao S, Yao C, Li J, Jiang W. et al. SOCS3 Drives Proteasomal Degradation of TBK1 and Negatively Regulates Antiviral Innate Immunity. Mol Cell Biol. 2015;35(14):2400-13
146. Plociennikowska A, Frankish J, Moraes T, Del Prete D, Kahnt F, Acuna C. et al. TLR3 activation by Zika virus stimulates inflammatory cytokine production which dampens the antiviral response induced by RIG-I-like receptors. J Virol. 2021Apr26;95(10):e01050-20
147. Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, Ong EZ. et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015;350(6257):217-21
148. Moon SL, Dodd BJ, Brackney DE, Wilusz CJ, Ebel GD, Wilusz J. Flavivirus sfRNA suppresses antiviral RNA interference in cultured cells and mosquitoes and directly interacts with the RNAi machinery. Virology. 2015;485:322-9
149. Akiyama BM, Laurence HM, Massey AR, Costantino DA, Xie X, Yang Y. et al. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science. 2016;354(6316):1148-52
150. Donald CL, Brennan B, Cumberworth SL, Rezelj VV, Clark JJ, Cordeiro MT. et al. Full Genome Sequence and sfRNA Interferon Antagonist Activity of Zika Virus from Recife, Brazil. PLoS Negl Trop Dis. 2016;10(10):e0005048
151. Pallarés HM, Costa Navarro GS, Villordo SM, Merwaiss F, de Borba L, Gonzalez Lopez Ledesma MM. et al. Zika Virus Subgenomic Flavivirus RNA Generation Requires Cooperativity between Duplicated RNA Structures That Are Essential for Productive Infection in Human Cells. Journal of virology. 2020;94(18):e00343-20
152. Slonchak A, Wang X, Aguado J, Sng JDJ, Chaggar H, Freney ME. et al. Zika virus noncoding RNA cooperates with the viral protein NS5 to inhibit STAT1 phosphorylation and facilitate viral pathogenesis. Sci Adv. 2022;8(48):eadd8095
153. Dong H, Fink K, Züst R, Lim SP, Qin CF, Shi PY. Flavivirus RNA methylation. J Gen Virol. 2014;95(Pt 4):763-778
154. Devarkar SC, Wang C, Miller MT, Ramanathan A, Jiang F, Khan AG. et al. Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc Natl Acad Sci U S A. 2016;113(3):596-601
155. Schuberth-Wagner C, Ludwig J, Bruder AK, Herzner AM, Zillinger T, Goldeck M. et al. A Conserved Histidine in the RNA Sensor RIG-I Controls Immune Tolerance to N1-2'O-Methylated Self RNA. Immunity. 2015;43(1):41-51
156. Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J. et al. 2'-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468(7322):452-6
157. Li SH, Dong H, Li XF, Xie X, Zhao H, Deng YQ. et al. Rational design of a flavivirus vaccine by abolishing viral RNA 2'-O methylation. Journal of virology. 2013;87(10):5812-9
158. Lou Z, Sun Y, Rao Z. Current progress in antiviral strategies. Trends Pharmacol Sci. 2014;35(2):86-102
159. Zhou F, Wan Q, Chen Y, Chen S, He ML. PIM1 kinase facilitates Zika virus replication by suppressing host cells' natural immunity. Signal Transduct Target Ther. 2021;6(1):207
160. Caine EA, Scheaffer SM, Arora N, Zaitsev K, Artyomov MN, Coyne CB. et al. Interferon lambda protects the female reproductive tract against Zika virus infection. Nat Commun. 2019;10(1):280
161. Snyder B, Goebel S, Koide F, Ptak R, Kalkeri R. Synergistic antiviral activity of Sofosbuvir and type-I interferons (α and β) against Zika virus. J Med Virol. 2018;90(1):8-12
162. Kamiyama N, Soma R, Hidano S, Watanabe K, Umekita H, Fukuda C. et al. Ribavirin inhibits Zika virus (ZIKV) replication in vitro and suppresses viremia in ZIKV-infected STAT1-deficient mice. Antiviral Res. 2017;146:1-11
163. Chan JF, Chik KK, Yuan S, Yip CC, Zhu Z, Tee KM. et al. Novel antiviral activity and mechanism of bromocriptine as a Zika virus NS2B-NS3 protease inhibitor. Antiviral Res. 2017;141:29-37
164. Pryke KM, Abraham J, Sali TM, Gall BJ, Archer I, Liu A. et al. A Novel Agonist of the TRIF Pathway Induces a Cellular State Refractory to Replication of Zika, Chikungunya, and Dengue Viruses. mBio. 2017;8(3):e00452-17
165. Li G, Adam A, Luo H, Shan C, Cao Z, Fontes-Garfias CR. et al. An attenuated Zika virus NS4B protein mutant is a potent inducer of antiviral immune responses. NPJ Vaccines. 2019;4:48
Author contact
![]() Corresponding author: Dr. Ming-Liang He, mlhe7788com.
Corresponding author: Dr. Ming-Liang He, mlhe7788com.

 Global reach, higher impact
Global reach, higher impact