Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2023; 19(15):4898-4914. doi:10.7150/ijbs.84970 This issue Cite
Research Paper
Inonotus obliquus upregulates muscle regeneration and augments function through muscle oxidative metabolism
1. Department of Molecular Cell Biology, Sungkyunkwan University School of Medicine, Suwon 16419, Republic of Korea.
2. Research Institute of Aging Related Disease, AniMusCure Inc., Suwon 16419, Republic of Korea.
3. School of Pharmacy, Sungkyunkwan University, Suwon 16419, Republic of Korea.
4. Department of Chemistry, Vanderbilt University, Nashville, TN 37235, United States.
5. Department of Biological Sciences, Vanderbilt University, Nashville, TN 37235, United States.
6. Drug Information Research Institute, Muscle Physiome Research Center, College of Pharmacy, Sookmyung Women's University, Seoul 04310, Republic of Korea.
#These authors contributed equally to this work.
Received 2023-4-5; Accepted 2023-9-1; Published 2023-9-11
Abstract
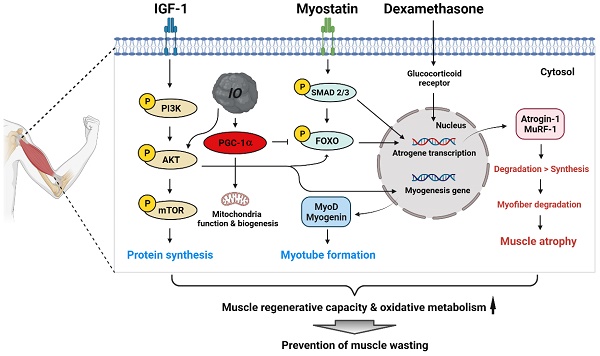
Skeletal muscle wasting related to aging or pathological conditions is critically associated with the increased incidence and prevalence of secondary diseases including cardiovascular diseases, metabolic syndromes, and chronic inflammations. Much effort is made to develop agents to enhance muscle metabolism and function. Inonotus obliquus (I. obliquus; IO) is a mushroom popularly called chaga and has been widely employed as a folk medicine for inflammation, cardiovascular diseases, diabetes, and cancer in Eastern Europe and Asia. However, its effect on muscle health has not been explored. Here, we aimed to investigate the beneficial effect of IO extract in muscle regeneration and metabolism. The treatment of IO in C2C12 myoblasts led to increased myogenic differentiation and alleviation of dexamethasone-induced myotube atrophy. Network pharmacological analysis using the identified specific chemical constituents of IO extracts predicted protein kinase B (AKT)-dependent mechanisms to promote myogenesis and muscle regeneration. Consistently, IO treatment resulted in the activation of AKT, which suppressed muscle-specific ubiquitin E3 ligases induced by dexamethasone. IO treatment in mice improved the regeneration of cardiotoxin-injured muscles accompanied by elevated proliferation and differentiation of muscle stem cells. Furthermore, it elevated the mitochondrial content and muscle oxidative metabolism accompanied by the induction of peroxisome proliferator-activated receptor γ coactivator α (PGC-1α). Our current data suggest that IO is a promising natural agent in enhancing muscle regenerative capacity and oxidative metabolism thereby preventing muscle wasting.
Keywords: Inonotus obliquus, PGC-1α, Muscle regeneration, Muscle atrophy, Muscle oxidative metabolism
Introduction
Inonotus obliquus (I. obliquus) is a mushroom belonging to the family Hymenochaetaceae (Basidiomycota) popularly called chaga in Russian folk medicine [1]. Chaga mushroom is a parasitic fungus that grows on birch in the forests of Northern European countries [2]. The mushroom is widely consumed as tea, syrup, bath agents, or concentrate [1,3] and has been widely employed as a folk medicine for inflammation, cardiovascular diseases, diabetes, and cancer in Eastern Europe and Asia [1,4,5]. The main bioactive compounds of I. obliquus are polysaccharides, polyphenols, melanin, and triterpenes [6]. By using whole I. obliquus extract or its components, studies have shown antitumor, anti-inflammatory, immunomodulatory, antioxidant, hypoglycemic, and hypolipidemic activity of I. obliquus. However, its potential effect on myogenesis and muscle regeneration has not been investigated so far.
Decline in skeletal muscle mass and function is a serious public health issue without effective cure. It not only severely compromises body movement and daily activities, but also increases the incidence and prevalence of secondary diseases leading to frailty and reduced healthy life span [7]. The age-associated decline in muscle mass and strength, which is referred to as sarcopenia, is attributed to reduced regenerative capacity of the muscle, declined mitochondrial function, and dysregulated immune responses [8,9]. Among diverse strategies, so far exercise seems to be most effective measure to improve muscle function in sarcopenic patients so far [10,11]. Thus, much efforts have been made to elucidate the underlying molecular mechanisms of the exercise effect on muscle mass and function. Peroxisome proliferator-activated receptor γ coactivator-1 alpha (PGC-1α) is a key regulator exerting exercise effect by eliciting gene expression related to mitochondrial biogenesis, fatty acid oxidation, angiogenesis, suppression of muscle atrophy and muscle regenerative capacity [12]. Mice lacking PGC-1α display impaired oxidative muscle metabolism and exercise capacity [13,14]. Thus, elevation of PGC-1α seems to be an attractive strategy to intervene muscle atrophy and weakness related to aging. Indeed, several studies have reported the beneficial effect of PGC-1α increase in aging related metabolic diseases and muscle wasting [15,16]. Two PGC-1α inducers, AICAR (AMPK agonist) and metformin (anti-diabetes) have shown to replicate the effect of exercise on muscle function [17-19]. Protein kinase B (AKT) signaling is another promyogenic signaling pathway critical for cell survival and differentiation of myoblasts triggered by the external signals like insulin-like growth factor [20-23]. AKT can activate the action of myogenic regulatory factors like MyoD and MEF2 thereby inducing muscle specific gene expression contributing to myogenesis and muscle regeneration [22,24]. AKT signaling also plays a key role in anabolic responses associated with increased protein synthesis and resulting in muscle growth [23,25].
The wide range of biological activity of I. obliquus in cells to modulate oxidative stress, immune response, and metabolic properties make the natural resource an attractive candidate to promote muscle health. Therefore, we investigated the effect of I. obliquus extract on myogenesis and myotube atrophy models of C2C12 myoblasts and muscle regeneration model of mice. We show that I. obliquus treatment improves myoblast differentiation and prevents myotube atrophy induced by dexamethasone (DEX). In addition, I. obliquus enhances muscle stem cell proliferation and regeneration post muscle injury. Further analysis revealed an increase in mitochondrial content and oxidative muscle metabolism in I. obliquus-treated muscles. Finally, we present the activation of AKT and PGC-1α expression as the potential underlying mechanism of I. obliquus's biological activities in muscle.
Materials and methods
Chemicals and Reagents
Wild-type C57BL/6 male mice were purchased from (Orient-Bio, Seongnam, Korea). Fetal bovine serum (FBS), horse serum (HS) and Dulbecco modified Eagle's medium (DMEM) were purchased from Thermo Scientific (Waltham, MA). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), DEX and all other chemicals were from Sigma-Aldrich (St. Louis, MO). Antibodies were purchased as following: Myosin heavy chain (MHC, Developmental Studies Hybridoma Bank (DSHB), Iowa, IA), MyoD (Novus biologicals, Littleton, CO), Myogenin, Myoglobin, total-OxPHOS (Abcam, Cambridge, MA), total AKT, phospho-AKT, total p38, phospho-p38, β-actin (Cell Signaling Technology, Beverly, MA), MuRF1, Atrogin-1, HSP90 (Santa Cruz Biotechnology, Santa Cruz, CA), PGC-1α (Calbiochem, San Diego, CA) and β-tubulin (Zymed, South San Francisco, CA).
Plant material
The fruiting bodies of chaga mushroom (I. obliquus) were purchased at Kyungdong herbal market, Seoul, Korea, in July 2019, and were identified by one of the authors (K. H. Kim). A voucher specimen (SKKU CG-2019-07) has been deposited in the herbarium of the School of Pharmacy, Sungkyunkwan University, Suwon, Korea.
Extraction procedure of IO1 and IO2
Dried fruiting bodies of I. obliquus (500 g) were dried, chopped, and the chopped material was extracted using distilled water (3 × 500 mL × 5 h) under reflux, and then filtered. The filtrate was evaporated under a vacuum to obtain an aqueous extract (20.2 g) of I. obliquus (IO1). Dried fruiting bodies of I. obliquus (500 g) were partially chopped and extracted with 70% EtOH (3 × 500 mL) for 2 days at room temperature and filtered. The filtrate was concentrated under vacuum pressure, generating a crude EtOH extract (22.5 g) of I. obliquus (IO2). The IO1 and IO2 extracts were prepared as stock solutions at 1.0 μg/mL in dimethyl sulfoxide (DMSO, Sigma-Aldrich). All stock solutions were aliquoted and stored at -80 °C until use.
UPLC Conditions
The samples were analyzed on an Agilent 1290 Infinity II UPLC coupled to a G6545B Q-TOF MS system with dual ESI source (Agilent Technologies, USA). All samples were separated on an Agilent ZORBAX RRHD Eclipse Plus C18 column (50 × 2.1 mm, 1.8 µm) using 0.1% formic acid-deionized water (A) and acetonitrile (B). The optimized gradient elution program was as follows: 0-10 min, 10-100% B; 10-12 min, 100% B; 12-15 min, 10% B. The temperature was set at 20 °C, and the injection volume was 1 µL. The spectral acquisition rate and time were set at 1 spectra/s and 1000 ms/spectrum each. The flow rate was 0.3 mL/min. The wavelength was set at 210 nm. The concentration of samples (distilled water extract and EtOH extract) was prepared as 1000 ppm and injected 3 times in cases of positive ion-mode and negative ion-mode respectively.
ESI Q-TOF MS Analysis
The Agilent Q-TOF G6545B mass spectrometer (Agilent Technologies) was operated in positive-ion and negative-ion mode. The parameters of the ESI source were optimized as follows: gas temperature 320 °C, gas flow 8 L/min, nebulizer pressure 35 psi, sheath gas temperature 350 °C, sheath gas flow 11 L/min, capillary voltage 3500 V, nozzle voltage 1000 V, and fragmentor voltage 100 V. Internal references (Purine and HP-0921) were adopted to modify the measured masses in real time. The reference masses in positive ion mode were at m/z 121.0508 and 922.0097. The reference masses in negative ion mode were at m/z 119.0363 and 1033.9881. The full scan range of the mass spectrometer was m/z 100-1700 for MS.
Data processing
Data processing process was conducted in cases of positive-ion mode and negative-ion mode respectively. The obtained UPLC Q-TOF MS raw data were further processed by Agilent MassHunter Profinder software (version 10.0, Agilent, America). The batch recursive feature extraction (small molecules/peptide) algorithm was applied to extract compounds from the total ion chromatograms (TICs) according to their molecular features including m/z, retention time and ion intensities, and the main parameters of MFE were optimized. Also, this algorithm used to bind and align compounds within the batch by retention time and mass tolerance; the tolerance windows of retention time and accurate mass were 0.3 min and 20 ppm, respectively. The restrict retention time range and m/z range were set at 0.5-13.0 min and 100 to 1700 m/z respectively. Low-abundance ions can be hard to identify if the precursor ion intensity is low, generally below 1000 counts for an Agilent Q-TOF. To produce a matrix containing fewer biased and redundant data, the thresholds of peak filters was set at 1000 counts. Missing peaks were filtered according to their frequency, and metabolites that appeared in 100% of samples in at least one group were retained. All the extracted compounds were output to create a .pfa (Profinder Archive) file, which can be imported into Mass Profiler Professional (MPP) software (version 15.1, Agilent) for further data analysis. Normalization (percentile shift), defining the sample sets, baselining (median of all samples), filtering by frequency, and significance analysis (T-test; p-value cut-off: 0.05; fold change cut-off: 2.0) were applied to process the data. The generated data were then processed for principal component analysis (PCA) by MPP software (version 15.1, Agilent). The successfully obtained specific metabolites (Table S1) for IO1 were identified by their accurate mass-to-charge ratio (m/z) values and the MS/MS fragmentation ion for each of the corresponding accurate m/z values aided by CFM-ID 4.0, a software tool for MS/MS spectral prediction and MS-based compound identification at http://cfmid.wishartlab.com as well as literature survey of I. obliquus compounds reported and comparison to authentic standards.
Network pharmacology analysis
Predicted protein targets of metabolites were identified using the STITCH [26], SwissTargetPrediction [27], and ChEMBL [28] databases. We included a target in the network if it was a human protein and had a greater than 90% probability in SwissTarget or was predicted to be active at 90% confidence in ChEMBL or if a connection was present between the molecule and the target in the STITCH network. Under these criteria, there were no hits from SwissTarget or STITCH, but there were hits from ChEMBL. A network of these compounds and targets was built using STITCH and a network of the protein-protein interactions was built using STRING [29]. In the STRING network, we observed that the following gene ontologies relevant to muscular function were enriched: skeletal muscle tissue growth, neuromuscular synaptic transmission, skeletal muscle contraction, muscle contraction mainly due to the presence of acetylcholine receptor subunits in the network. Important target nodes were identified by comparing the degree, betweenness centrality, and closeness centrality to the median values. Betweenness and closeness centrality were calculated using the Networkx python package [30].
Animal experimental design
The wild-type C57Bl/6 male mice were obtained from Orient-Bio (Seongnam, Korea) and maintained until sacrificed. All mice were maintained at 23 ℃ with a 12:12 h light-dark cycle and free access to food and water. The mice were orally administrated a daily dose of 4 mg/kg IO for 4 weeks (4-month-old mice, young) and control mice were administrated the same amount of vehicle drinking water. For the muscle atrophy experiment, the 4-month-old mice were orally administrated either vehicle or IO for 1 week prior to injecting cardiotoxin (CTX) and were administrated until be sacrificed. Then, they were victimized on Day 3 and Day 21 after injection. All animals were sacrificed after fasting for 16 h with ad libitum to water, and their muscles were harvested 4 h after the last administration of IO. All animal experiments were approved by the Institutional Animal Care and Research Advisory Committee at Sungkyunkwan University school of Medicine (SUSM) and complied with the regulations of the institutional ethics committee.
Cell culture and cell viability assay
C2C12 myoblasts were cultured as previously described [31]. They were grown in DMEM (Dulbecco's Modified Eagle Medium high glucose; Thermo Scientific, Waltham, MA) containing 15% FBS (growth medium, GM), 10 units/mL penicillin and 10 µg/mL streptomycin (Welgene, Daegu, Korea) at 37 ℃, 5% CO2. To induce differentiation of C2C12 myoblasts, cells at near confluence were changed growth medium into DMEM containing 2% HS (differentiation medium, DM) and myotube formation was observed at 2 or 3 days after differentiation. For the DEX-induced atrophy study, C2C12 cells were induced to differentiate in differentiation medium for 3 days and treated with 100 µM DEX and indicated concentration of IO, followed by incubation in differentiation medium for additional 1 day [32].
Cell viability assay was quantified using MTT colorimetric assay. In briefly, C2C12 cells were seeded in a 96-well plate (5 × 104 cells/well) overnight and treated with the indicated concentration of IO for 24 h. MTT solution was added to the each well, and the cells were incubated for 4 h at 37 ℃. The optical density was measured at 540 nm.
Western blotting and immunostaining
Western blot analysis was performed as previously described [33]. Briefly, cells were lysed in cell extraction buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) containing complete protease inhibitor cocktail (Roche Diagnostics, Basel, Switzerland), followed by SDS-PAGE and incubation with primary and secondary antibodies.
Immunostaining for MHC expression was carried out as previously described [33]. Briefly, the differentiated cultures were then immunostained for MHC antibodies and Alexa 568- or 488-conjugated secondary antibodies (Molecular Probes, Eugene, OR). Images were captured and processed with a Nikon ECLIPSE TE-2000U microscope and NIS-Elements F software (Nikon, Tokyo, Japan). To analyze the efficiency of myotube formation, the MHC-positive myotubes containing two to nine, or ten or more nuclei, were quantified at least three times and measured using IMAGE J software (version 1.53e; National Institutes of Health, Bethesda, MD). MHC-positive myotubes with 10 and more nuclei were measured transverse diameter in six fields. Quantification of myotube diameter was performed with IMAGE J software. Average myotube diameter is presented as means determination of six fields ± 1 standard deviation (SD).
RNA isolation and quantitative RT-PCR
Total RNA extraction and quantitative RT-PCR analysis was performed as previously described [31]. Tissues were homogenized by FastPrepR-24 (MP Biomedicals, Santa Ana, CA) and extracted using the easy-spin Total RNA Extract kit (iNtRON, Seongnam, Korea). Gene expression fold change was normalized against the expression of 18S ribosomal RNA. The sequences of the primers used in this study are provided in Table 1.
Cryosections, staining analysis, and fiber size measurement
Muscle tissue was embedded in Tissue-Tek OCT Compound (Sakura Finetek, Nagano, Japan), and 7mm thick serial sections for staining were cut using a cryomicrotome. To analyze the NADH dehydrogenase activity, we dried the sectioned tissues for 10 min in room temperature and incubated in 0.9 mM NADH and 1.5 mM nitro blue tetrazolium (NBT; Sigma-Aldrich) in 3.5 mM phosphate buffer (pH 7.4) for 30 min. To analyze the succinate dehydrogenase (SDH) activity, we incubated the sections for 1 h in 50 µM sodium succinate and 0.3 mM nitro blue tetrazolium in 114 mM phosphate buffer containing K-EGTA (Sigma-Aldrich). Myh immunostaining of muscle tissue sections was performed in the sequence of fixation, permeation, and incubation with primary antibodies against MyhIIa and MyhⅡb (DSHB) and laminin (Abcam). Images were captured and proceed with a Nikon ECLIPSE TE-2000U using NIS-Elements F software. Myofiber area was measured with ImageJ software. For muscle histology, the cryosections were stained with Mayer's hematoxylin and eosin (BBC Biomedical, McKinney, TX). The images were captured using a Nikon ECLIPS TE-2000U.
The primers used in this study
| Primer | Sequence | |
|---|---|---|
| eMHC | Forward | 5'-CTGGAGTTTGAGCTGGAAGG-3' |
| Reverse | 5'-CAGCCTGCCTCTTGTAGGAC-3' | |
| Myogenin | Forward | 5'-ATCTCCGCTACAGAGGCGGG-3' |
| Reverse | 5'-TAGGGTCAGCCGCGAGCAAA-3' | |
| Atrogin-1 | Forward | 5'-CAACATTAACATGTGGGTGTAT-3' |
| Reverse | 5'-GTCACTCAGCCTCTGCATG-3' | |
| MuRF1 | Forward | 5'-GAGAACCTGGAGAAGCAGCT-3' |
| Reverse | 5'-CCGCGGTTGGTCCAGTAG-3' | |
| MyhI | Forward | 5'-ACAAGCTGCAGCTGAAGGTG-3' |
| Reverse | 5'-TCATTCAGGCCCTTGGCAC-3' | |
| MyhIIa | Forward | 5'-CCAGCTGCACCTTCTCGTTTGCCAG-3' |
| Reverse | 5'-CATGGGGAAGATCTGGTCTTCTT-3' | |
| MyhIIb | Forward | 5'-CCTGGAACAGACAGAGAGGAGCAGGAGAG-3' |
| Reverse | 5'-GTGAGTTCCTTCACTCTGCGCTCGTGC-3' | |
| MyhIIX | Forward | 5'-TGCAACAGTTCTTCAACCAC-3' |
| Reverse | 5'-GCCAGGTCCATCCCAAAGT-3' | |
| Myh3 | Forward | 5'-CTTCACCTCTAGCCGGATGGT-3' |
| Reverse | 5'-AATTGTCAGGAGCCACGAAAAT-3' | |
| Pgc-1α | Forward | 5'-ATGTGTCGCCTTCTTGCTCT-3' |
| Reverse | 5'-CGGTGTCTGTAGTGGCTTGA-3' | |
| Mtco1 | Forward | 5'-CTACTATTCGGAGCCTGAGC-3' |
| Reverse | 5'-GCATGGGCAGTTACGATAAC-3' | |
| Mcad | Forward | 5'-GGTTTGGCTTTTGGACAATG-3' |
| Reverse | 5'-TGACGTGTCCAATCTACCACA-3' | |
| Sdhb | Forward | 5'-CAGAGTCGGCCTGCAGTTTC-3' |
| Reverse | 5'-GGTCCCATCGGTAAATGGCA-3' | |
| Cox7a1 | Forward | 5'-GTCTCCCAGGCTCTGGTCCG-3' |
| Reverse | 5'-CTGTACAGGACGTTGTCCATTC-3' | |
| Cox4 | Forward | 5'-CTATGTGTATGGCCCCATCC-3' |
| Reverse | 5'-AGCGGGCTCTCACTTCTTC-3' | |
| Ucp2 | Forward | 5'-ACTGTCGAAGCCTACAAGAC-3' |
| Reverse | 5'-CACCAGCTCAGTACAGTTGA-3' | |
| Tnfα | Forward | 5'-AGCCCCCAGTCTGTATCCTT-3' |
| Reverse | 5'-CTCCCTTTGCAGAACTCAGG-3' | |
| Il6 | Forward | 5'-GGTGACAACCACGGCCTTCCC-3' |
| Reverse | 5'-AAGCCTCCGACTTGTGAAGTGGT-3' | |
| Il10 | Forward | 5'-GCCAAGCCTTATCGGAAATG-3' |
| Reverse | 5'-CACCCAGGGAATTCAAATGC-3' | |
| Il1RA | Forward | 5'-TTCTTGTTGCCTCTGCCACTCG-3' |
| Reverse | 5'-GATTGGTCTGGACTGTGGAAGTG-3' | |
| Ccl5 | Forward | 5'-TGCCCACGTCAAGGAGTATTT-3' |
| Reverse | 5'-TTCTCTGGGTTGGCACACACT-3' | |
| Ccl22 | Forward | 5'-AAGACAGTATCTGCTGCCAGG-3' |
| Reverse | 5'-GATCGGCACAGATATCTCGG-3' | |
| Cxcl1 | Forward | 5'-TGAGCTGCGCTGTCAGTGCC-3' |
| Reverse | 5'-AGAAGCCAGCGTTCACCAGA-3' | |
| Pax7 | Forward | 5'-GAGTTCGATTAGCCGAGTGC-3' |
| Reverse | 5'-CGGGTTCTGATTCCACATCT-3' | |
| MyoD | Forward | 5'-GATGGCATGATGGATTACAGCGGC-3' |
| Reverse | 5'-GTGGAGATGCGCTCCACTATGCTG-3' | |
| Ki67 (mouse) | Forward | 5'-CTGCCTCAGATGGCTCAAAGA-3' |
| Reverse | 5'-GAAGACTTCGGTTCCCTGTAAC-3' | |
| Ki67 (human) | Forward | 5'-TCCCGCCTGTTTTCTTTCTGAC-3' |
| Reverse | 5'-CTCTCCAAGGATGATGATGCTTTAC-3' | |
| Myf5 | Forward | 5'-CTCAGGAATGCCATCCGCTACATTGAGA-3' |
| Reverse | 5'-ATCCAAGCTGGATAAGGAGCTTTTATCCG-3' | |
| 18S rRNA (mouse) | Forward | 5'-AGGGGAGAGCGGGTAAGAGA-3' |
| Reverse | 5'-GGACAGGACTAGGCGGAACA-3' | |
| 18S rRNA (human) | Forward | 5'-GTAACCCGTTGAACCCCATT-3' |
| Reverse | 5'-CCATCCAATCGGTAGTAGCG-3' |
PGC-1α Luciferase Assay
PGC-1α luciferase assay was performed as previously described [34]. C2C12 cells were transfected with an expression plasmid for luciferase responsive to the 2 kb promoter region of PGC-1α (Addgene plasmid #8887, Addgene, Cambridge, MA, USA), PGC-1α promoter luciferase delta CRE site (Addgene plasmid #8888, Addgene) and PGC-1α promoter luciferase delta MEF site (Addgene plasmid #8889, Addgene) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After 24 h of transfection, the cells were incubated in differentiation media with IO1. The cells were lysed with Reporter Lysis Buffer (Promega), and luciferase assays were performed using a Luciferase Assay System kit and a luminometer (Berthold Technology, Bad Wildbad, Germany). The Transfection efficiency was normalized based on the co-transfected b-galactosidase enzyme activity measured using an assay kit (Promega). Experiments were performed in triplicates and repeated at least three times independently.
Statistical analysis
Values are expressed as mean ± SD or ± standard error of mean (SEM), as indicated in the figure legends. Differences were considered statistically significant at or under values of P < 0.05.
Results
Effects of Inonotus obliquus extracts on myogenic differentiation of C2C12 myoblasts
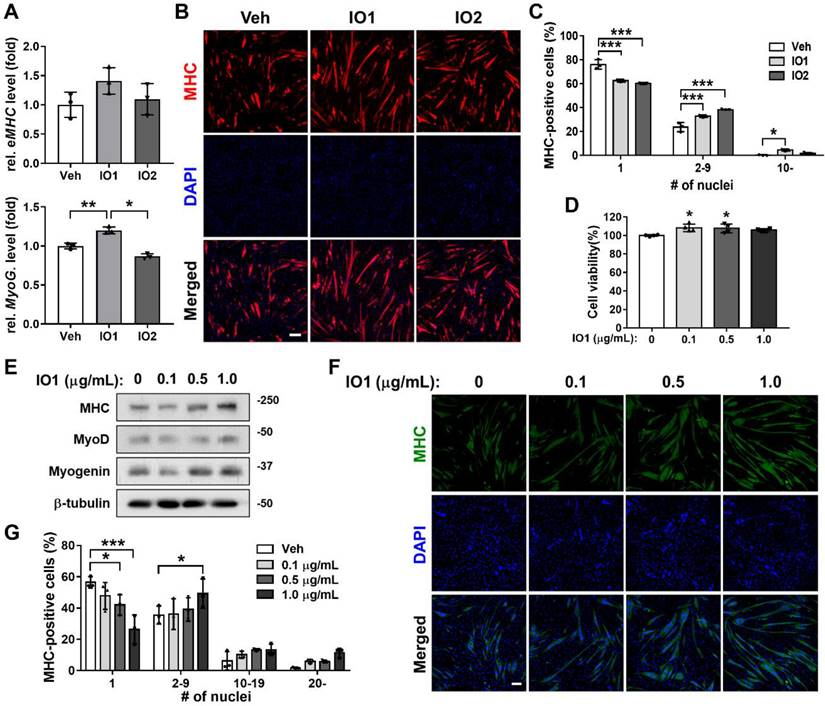
To investigate the effect of Inonotus obliquus extract (IO) on myogenic differentiation, we prepared two types of IO: IO1 (aqueous extract) and IO2 (ethanol extract). C2C12 myoblasts were induced to differentiate for 1.5 days in the presence of vehicle, IO1, or IO2 in DM and subjected to the assessment of myoblast differentiation by quantitative real-time polymerase chain reaction (qRT-PCR). IO1 treatment substantially elevated the expression of muscle-specific genes, embryonic myosin heavy chain (eMHC) and Myogenin, compared to vehicle or IO2 treatment (Figure 1A) and in a dose-dependent manner (Figure S1), while IO2 did not show much effect on the expression of muscle-specific genes. To further examine the promyogenic effects of IOs, myotube formation was assessed by MHC immunostaining. Both IO1 and IO2 treatment in C2C12 myoblasts promoted the formation of larger MHC-positive multinucleated myotubes compared to vehicle-treated cells (Figure 1B and C). In particular, IO1 treatment effectively elevated the proportion of larger MHC-positive myotubes containing ten or more nuclei. However, the effects of IO2 were lesser than IO1. Therefore, we decided to place more emphasis on the investigation of IO1 (hereafter designated as IO). Up to a concentration of 10 μg/mL, IO did not show any cytotoxicity, as assessed by the MTT assay (Figure 1D). However, significant cytotoxicity was observed with the treatment of IO starting at a concentration of 50 μg/mL (Figure S2). IO-treated C2C12 cells displayed incremental increase in the expression of MHC, MyoD, and Myogenin in a dose-dependent manner (Figure 1E), which was also confirmed by immunostaining of MHC (Figure 1F). For the quantification of myotube formation, the nuclei MHC-positive myotubes were counted. Treatment with IO significantly increased the proportion of larger myotubes with 20 or more nuclei, while mononucleated myocytes decreased in a concentration-dependent manner (Figure 1G). These data suggest that IO enhances C2C12 myoblast differentiation without overt cytotoxicity.
Effect of IO on myogenic differentiation of C2C12 myoblast cells. (A) Comparison of IO1 and IO2 in the degree of myoblast differentiation. Expression of eMHC and Myogenin in C2C12 cells treated with vehicle, IO1 (1.0 µg/mL), or IO2 (1.0 µg/mL) for 1 day in DM were analyzed by qRT-PCR. The data were normalized using 18s RNA, n = 3. eMHC and Myogenin expression levels were further normalized to the expression level of vehicle (DMSO). (B) Immunostaining for MHC expression in C2C12 cells treated with DM for 1 day and treated with vehicle, IO1 (1.0 µg/mL), or IO2 (1.0 µg/mL) for 1 day in DM. Scale bar, 50 µM. (C) Quantification of myotube formation from data shown in panel B, n = 3. (D) Cell viability of C2C12 cells in the presence of 0, 0.1, 0.5, and 1.0 µg/mL IO for 1 day, n = 4. (E) Immunoblot analysis of C2C12 cells treated with indicated concentration of IO and differentiated in DM for 2 days. Cell lysates were subjected to antibodies against MHC, MyoD, Myogenin, and β-tubulin. (F) Immunostaining of C2C12 cells treated with indicated concentration of IO for MHC expression (green). Scale bar, 50 µM. (G) Quantification of myotube formation from data shown in panel F, n = 3. Data are expressed as the means ± SD. Asterisks indicate significant difference from the control. *P < 0.01, **P < 0.05, and ***P < 0.001.
Characterization of the chemical constituents in the active IO and network pharmacological analysis
To investigate the potential underlying mechanism of the effects of I. obliquus in muscle, the chemical constituents of I. obliquus extracts were analyzed by ultra-high-performance liquid chromatography (UPLC) coupled with quadrupole time-of-flight mass spectrometry (Q-TOF MS). Since IO1 showed greater enhancement of C2C12 myoblast differentiation than IO2, we presumed that the specific metabolites in the aqueous extract of I. obliquus (IO1) are the potential bioactive compounds. The obtained typical total ion chromatograms (TICs) for the two types of IO were similar (Figure S3) with most of the major peaks appearing in TICs of both IO1 and IO2. However, the principal component analysis (PCA) of the UPLC-Q-TOF MS data showed that the IO1 and IO2 samples were clearly separated (Figure 2A), indicating the different metabolite profiles for IO1 and IO2. In the positive-ion mode, there were 201 chemical constituents in IO1 and 328 chemical constituents in IO2. In the negative-ion mode, there were 69 chemical constituents in IO1 and 138 chemical constituents in IO2. To verify the specific metabolites in IO1, pairwise analysis of IO1 versus IO2 was performed using a Venn diagram (Figure 2B). The results indicated that 27 and 3 specific metabolites of IO1 were found in the positive-ion mode and negative-ion mode, respectively. The specific metabolites obtained were checked manually, since they contained redundant signals caused by different isotopes and in-source fragmentation, and the redundant signals were manually removed. Finally, the highly reproducible and non-redundant metabolite signals were taken as 6 specific chemical constituents in IO1 (Figure 2C).
Analysis of the chemical constituents in IO1 and network pharmacological analysis. (A) Principal component analysis (PCA) of IO1 and IO2 samples based on the metabolomics analysis in negative-ion mode. (B) Venn diagrams of pairwise analyses (IO1 versus IO2). (C) The identified specific chemical constituents in IO1. (D) Pharmacological network of IO1. Pharmacological network was built using SwissTargetPrediction, ChEMBL, STRING, and STITCH. The specific chemical constituents identified from IO1 are shown in red, predicted targets in blue, proteins of interest in green, and proteins added to the network by STRING or STITCH in yellow. Edges hypothesized to be important in the mechanism of action based on node importance and literature search are highlighted in red.
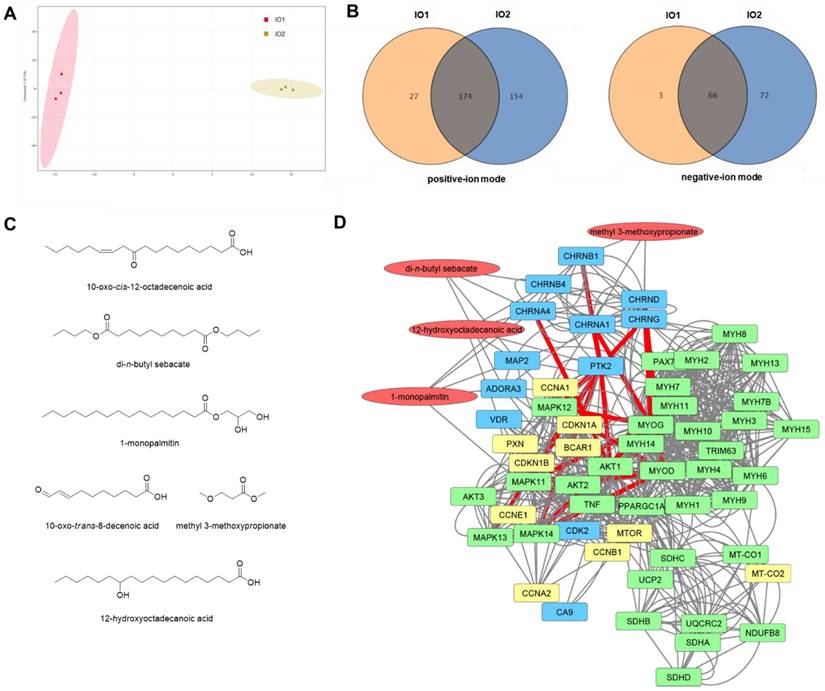
Next, network pharmacology analysis was performed to explore the potential mechanisms of I. obliquus on myoblast differentiation and muscle regeneration using the identified specific chemical constituents of IO1. We used STITCH [26], SwissTargetPrediction [27], and ChEMBL [28] to identify possible protein targets of IO1. The ChEMBL database was the only tool that identified candidate target proteins with high confidence. We then constructed a network of the target proteins identified by ChEMBL and proteins of interest associated with skeletal muscle tissue growth, contraction, and neuromuscular synaptic transmission using STRING [29] and STITCH [26]. In this network, the neuronal acetylcholine receptor was a predicted target of di-n-butyl sebacate, 1-monopalmitin, and 12-hydroxyoctadecanoic acid and the acetylcholine receptor was a predicted target of methyl 3-methoxypropionate (Figure 2D). The acetylcholine receptors were in turn linked to many of the proteins of interest including protein kinase B (AKT1), myoblast differentiation protein (MYOD), myogenin (MYOG), myosin heavy chain 2, 3, 8 (MYH2, MYH3, MYH8), E3 ubiquitin-protein ligase (TRIM63), and peroxisome proliferator-activated receptor g coactivator (Pgc-1α). In addition, 12-hydroxyoctadecanoic acid was predicted to interact with focal adhesion kinase 1 (PTK2), which was identified to be a major node that links between the acetylcholine receptors and downstream kinases such as AKT and MAP in the network. Accordingly, the network pharmacological analysis suggests that the most likely mechanism of IO1 activity is through activation of acetylcholine receptors and PTK2 leading to downstream activation of the AKT and MAPK pathways to promote myoblast differentiation. To verify the prediction obtained from the network pharmacology analysis, the promyogenic effects of four constituents (di-n-butyl sebacate, 1-monopalmitin, 12-hydroxyoctadecanoic acid, and methyl 3-methoxypropionate) were examined in C2C12 myoblasts (Figure S4). Consistent with the network pharmacology analysis, these compounds exhibited promyogenic effects, as evidenced by the expression of Ki67 (a proliferation marker), Myf5, and MyoD (markers for muscle progenitors), as well as MyoG and Myh3 (differentiation markers) (Figure S4).
Preventive effects of IO (IO1) on DEX-induced myotube atrophy through AKT activation
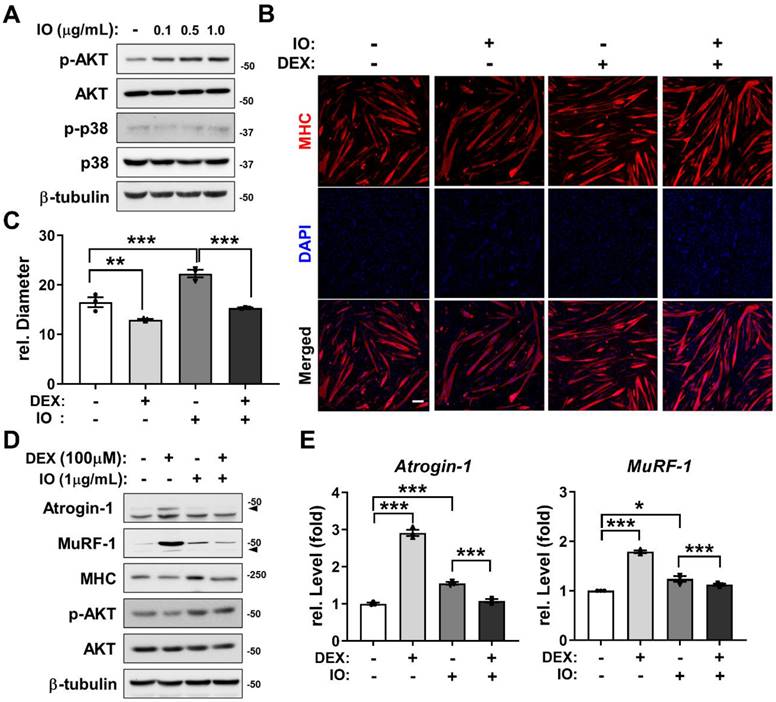
AKT functions as a promyogenic kinase in myoblast differentiation and as an essential regulator of muscle protein synthesis and hypertrophy [33, 35]. To examine whether the promyogenic effect of IO (IO1) is through AKT activation as predicted from the network pharmacological analysis, C2C12 myoblasts were induced to differentiate in DM for 1 day, treated with the indicated concentrations of IO, and the activation status of AKT and p38 was assessed by immunoblotting. The treatment of IO dramatically increased p-AKT levels in a dose-dependent manner without changes in the levels of p-p38 and total form of AKT and p38 (Figure 3A). AKT activation attenuates the induction of muscle-specific ubiquitin ligases triggered by synthetic glucocorticoid DEX thereby preventing muscle atrophy [36]. To examine whether IO can protect DEX-induced C2C12 myotube atrophy, C2C12 cells were treated with DEX alongside with vehicle DMSO or IO and subjected to MHC immunostaining to assess the thickness of the myotubes. The DEX-elicited myotube atrophy was suppressed by IO treatment, as evident by the larger multinucleated myotubes in IO-treated cultures (Figure 3B and C). In addition, the IO treatment abrogated the DEX-induced elevation of protein levels of muscle-specific E3 ligases, Atrogin-1 and MuRF-1 (Figure 3D). The rescue effect of IO treatment on DEX-induced atrophic myotubes was also reflected by the restored MHC expression close to the level of vehicle control. Furthermore, DEX-treated myotubes exhibited decreased level of p-AKT while IO treatment in DEX-treated myotubes abrogated this decrease. Consistent with the immunoblotting analysis, qRT-PCR analysis showed that DEX treatment greatly elevates the expression of muscle-specific E3 ubiquitin ligases, Atrogin-1 and MuRF-1, while the treatment with IO in DEX-treated myotubes significantly diminishes the mRNA level of Atrogin-1 and MuRF-1 (Figure 3E). Taken together, these results suggest that IO rescues DEX-induced myotube atrophy through inhibition of muscle-specific ubiquitin ligases mediated by Akt activation.
Effect of IO on skeletal muscle regeneration in CTX-injury mouse model
To further examine the role of IO in muscle regeneration, acute injury was induced by cardiotoxin (CTX) injection in the tibialis anterior (TA) muscle of four-month-old wild type male mice. Starting from a week before CTX injection, mice were orally administered with vehicle or 4 mg/kg IO daily and dissected on day 21 after CTX injection (Figure 4A). IO treatment did not incur changes in body weight, food intake, and blood glucose level compared with the vehicle treatment (Figure 4B-D). The hindlimb muscles of the IO-treated mice appeared darker than those of control mice, which could be readily detected in gastrocnemius (GAS) muscle (Figure 4E). Among four hindlimb muscles, TA muscle displayed the most significant increase in mass per body weight with approximately 12% increase in the IO-fed mice compared to the vehicle-fed mice (Figure 4F). Sections of the TA muscle also revealed an increase in the cross-sectional areas (CSA) of myofibers by IO treatment (Figure 4G-I). In addition, treatment with IO increased the number of myonuclei per CSA in TA muscle fibers compared to the treatment with vehicle (Figure 4J). Next, we examined the effects of IO on the activation of AKT, a key anabolic mediator. IO-treated muscles had significantly enhanced p-AKT expression levels compared with the vehicle muscles (Figure 4K). Taken together, these data suggest that IO treatment improves muscle regeneration in CTX-injured muscles.
To investigate the effect of IO on myofiber types, CTX-injected TA muscle sections were analyzed by immunostaining with antibodies against MyhIIa and MyhIIb. The IO treatment significantly increased the number and CSA of MyhIIb-positive myofibers and increased the number and CSA of MyhIIa-positive myofibers without statistical significance compared with the vehicle treatment (Figure 4L-N). Furthermore, qRT-PCR analysis of TA muscles from vehicle- and IO-fed mice revealed that IO treatment significantly increases the expression of both oxidative myofiber markers, MyhI and MyhIIa, and glycolytic myofiber markers, MyhIIb and MyhIIx (Figure 4O). These data indicate that IO treatment enhances the regeneration of both oxidative and glycolytic myofiber types.
Effect of IO on destruction of DEX-induced muscle atrophy model through AKT activation. (A) C2C12 cells were differentiated in DM for 1 day and treated with indicated concentration of IO. Cell lysates were subjected to immunoblotting with antibodies against p-AKT, AKT, p-p38, p38, and β-tubulin. (B) Immunostaining for MHC expression in C2C12 cells treated with normal DM for 3 days and treated with vehicle, IO (1.0 µg/mL), DEX (100 µM), or both IO and DEX for 1 day in DM. Scale bar, 50 µm. (C) Quantification of MHC-positive myotube diameter per field, n = 3. (D) C2C12 cells were differentiated in DM for 3 days and treated with vehicle, IO (1.0 µg/mL), DEX (100 µM), or both IO and DEX for 1 day in DM. Cell lysates were subjected to immunoblotting with antibodies against Atrogin-1, MuRF-1, MHC, p-AKT, AKT, and β-tubulin. (E) Expression of Atrogin-1 and MuRF-1 in C2C12 cells treated with normal DM for 3 days and treated with vehicle, IO (1.0 µg/mL), DEX (100 µM), or both IO and DEX for 1 day in DM. The data were normalized using 18s RNA. Atrogin-1 and MuRF-1 expression levels were further normalized to the expression level of vehicle (DMSO), n = 3. Data are expressed as the means ± SD. To determine statistical significance, an unpaired two-tailed Student's t-test was used. *P < 0.05, **P < 0.01 and ***P < 0.001.
Effect of IO on skeletal muscle regeneration in in vivo mice model at day 21 post-CTX-injection. (A) Experimental set-up. (B-D) Body weights (B), changes in food intake (C), and blood glucose level (D) of Vehicle- and IO-treated mice, n = 5. (E) Photographs of isolated hindlimb muscles from 4-month-old mice ingested with vehicle or 4 mg/kg IO for 4 weeks. (F) Weights of four muscle types from vehicle or 4 mg/kg IO ingested mice for 4 weeks, n = 4. (G) Representative H&E and Laminin staining of TA muscle sections from vehicle- and IO-ingested mice. Scale bar, 100 µm. (H) Distribution of the myofibers based on their cross-sectional area (CSA) from panel G, n = 5. (I) Quantification of the average myofiber CSA from panel G, n = 5. (J) Quantification of myonuclei per CSA from panel G, n = 5. (K) Western blot analysis for the expression of p-AKT and AKT in the TA muscles from 4-month-old mice ingested with Vehicle or 4 mg/kg IO for 4 weeks, n = 5. (L) Immunostaining of MyhIIa (green), MyhIIb (green), and laminin (red) in the TA muscles of vehicle- or 4 mg/kg IO-ingested 4-month-old mice for 4 weeks. Scale bar, 100 µm. (M) Distribution of MyhIIa- and MyhIIb- positive myofibers based on their CSA and average myofiber CSA from panel K. Quantification of MyhIIa- and MyhIIb-positive myofibers, n = 5. (N) Quantification of fiber-type content in TA muscle, n = 5. (O) Expression of slow and fast muscle-associated genes in TA muscles. Myh gene expressions by qRT-PCR. MyhI is represented as a slow muscle type, whereas MyhIIa, MyhIIb, and MyhIIX are fast muscle types, n = 5 (Veh), n = 4 (IO). Data are expressed as the means ± SD. Asterisks indicate significant difference from the control. *P < 0.05 and **P < 0.01.
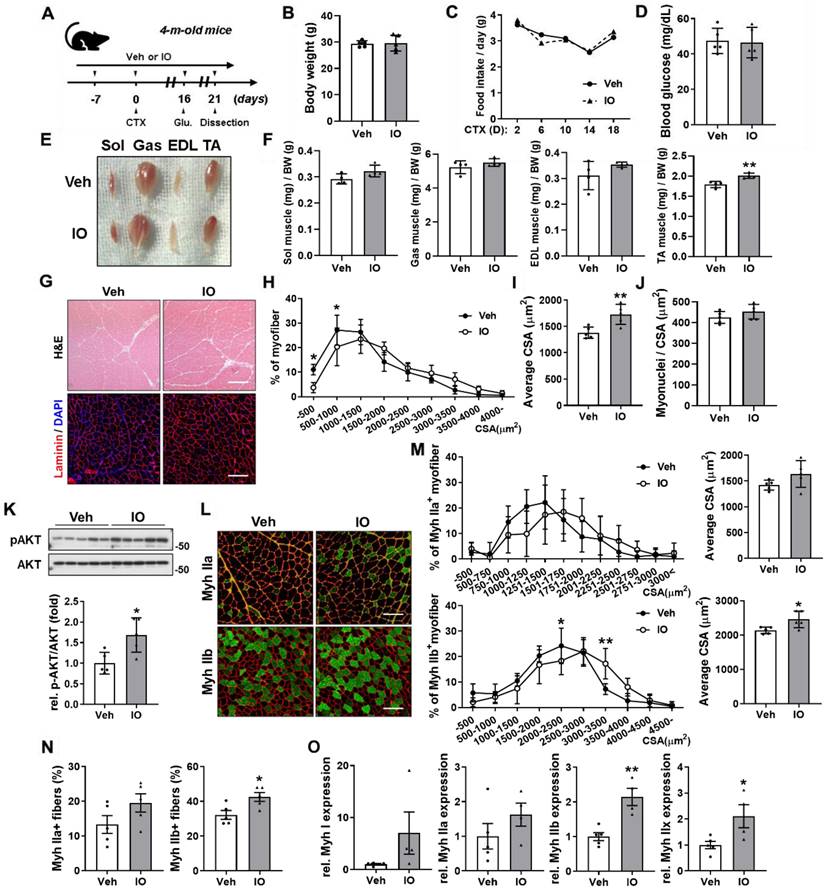
Effect of IO on muscle stem cell proliferation in in vivo mice model at 3 days post-CTX-injection. (A) The experimental scheme. (B) The change of body weight for total 10 days of the experiment, n = 3. (C) The relative TA muscle weight normalized to body weight, n = 3. (D) Immunostaining of BrdU incorporation and quantification of BrdU-positive cells in Vehicle- or IO treated TA muscles post 3 days of CTX injury, n = 3. (E) Immunstaining for Ki67 and quantification of Ki67-positive cells in TA muscles, n = 3. (F & G) Quantitative RT-PCR analysis of cell cycle markers (F) and early myogenic regulatory factors (G) in TA muscles, n = 3. The data were normalized to 18s RNA levels and were further normalized to the expression level of vehicle. (H) Immunostaining of BrdU incorporation in Vehicle- or IO-treated aged human myoblasts (66-year-old). (I) Quantification of BrdU-positive cells and cell number per field from data shown in panel H, n = 9. (J) Quantitative RT-PCR analysis of Ki67 in Vehicle- or IO-treated aged human myoblasts (66-year-old), n = 3. Data are expressed as the means ± SD. Asterisks indicate significant difference from the control. *P < 0.05, **P < 0.01 and ***P < 0.001.
Effect of IO on muscle stem cell proliferation in regenerating muscles at day 3 post-CTX-injury
To investigate the effect of IO at the early stage of muscle regeneration, four-month-old wild type male mice were orally administered with either vehicle or 4 mg/kg IO daily starting from 7 days prior to CTX injection in TA muscles and the muscles were harvested post 3 days of injury (Figure 5A). The IO treatment did not induce changes in body weight and relative TA muscle weight compared with the vehicle treatment (Figure 5B and C). To monitor the proliferation in the regenerating muscles, cryosections of TA muscles were subjected to immunostaining for BrdU incorporation and Ki67. IO treatment elevated the number of BrdU and Ki67-positive cells compared to the vehicle treatment (Figure 5D and E). Furthermore, IO treatment significantly increased the expression levels of genes expressed during cell proliferation (Ccnb1, Ccnb2, Ccne1, Ccnf, Aurkb, Mcm6, and p21) and markers for activated proliferating muscle stem cells (Pax7 and MyoD), suggesting that the improved muscle regeneration by IO treatment is attributable to the increased proliferation of muscle stem cells (Figure 5F and G). Aged human myoblasts treated with IO also exhibited increased proliferation compared with the vehicle-treated cells, further verifying the effect of IO on the muscle stem cell proliferation (Figure 5H and I). In parallel with Figure 5H & I, IO treatment significantly increased the Ki67 expression (Figure 5J). These results suggest that IO enhances the activation and proliferation of muscle stem cells in response to muscle injury.
Effect of IO on the oxidative muscle metabolism
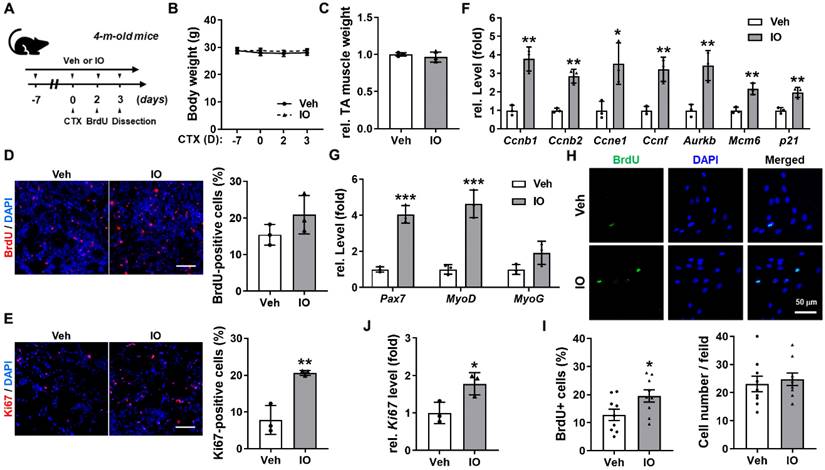
Since IO-treated mice exhibited darker hindlimb muscles and substantially increased expression of MyhI and MyhIIa compared with the vehicle-treated mice (Figure 4E and N), we presumed that IO may also have an effect in promoting oxidative muscle metabolism. TA muscles of 4-month-old mice treated with either vehicle or 4 mg/kg of IO for 4 weeks were subjected to histochemical staining for the activity of mitochondrial enzyme succinate dehydrogenase (SDH). IO treatment elevated the proportion of myofibers with strong (dark) and intermediate staining for SDH compared to the vehicle treatment (Figure 6A and B). In addition, IO significantly enhanced the expression of total OxPHOS (Atp5a, Uqcr2, Mtco1, NdufB8, and Sdhd) and mitochondrial genes (Mtco1, Mcad, Sdhd, Cox7a1, Cox4, and Ucp2) (Figure 6C-E). The relative mitochondrial DNA content was also significantly increased by IO treatment, indicating that IO enhances oxidative muscle metabolism (Figure 6F). To confirm these effects of IO, C2C12 myoblasts were induced to differentiate with the indicated concentrations of IO in DM and subjected to immunoblotting analysis. In agreement with the results from the mice model, IO treatment significantly increased the expression of total OxPHOS and mitochondrial genes in a dose dependent manner (Figure 6G and H). Finally, IO-treated C2C12 cells exhibited increased mitochondrial membrane potential as evidenced by the increased JC-1 polymer/monomer ratio compared with the vehicle-treated cells (Figure 6I and J). These results suggest that IO enhances the oxidative muscle metabolism through upregulation of mitochondrial gene expression.
Effect of IO on the expression of PGC-1α in young mice skeletal muscle and C2C12 cells
PGC-1α is a transcriptional coactivator that activates the expression of genes involved in mitochondrial biogenesis, stimulation of fatty acid oxidation, and resistance to muscle atrophy [12]. Thus, we investigated whether IO regulates the oxidative muscle metabolism through regulation of PGC-1α expression. The mRNA and protein levels of PGC-1α and myoglobin were elevated in the IO-treated TA muscles compared with the vehicle-treated muscles (Figure 7A and B). In agreement with the data of TA muscles, IO treatment increased the level of PGC-1α mRNA as well as protein up to approximately 3.0-fold in C2C12 myoblasts (Figure 7C and D). To further define the mechanism of IO in the activation of PGC-1α expression, the reporter activity of full-length or mutant PGC-1α promoters (with a deletion of either MEF2 or CRE motif) was measured in C2C12 cells. The full-length promoter-driven luciferase activity was significantly upregulated by IO1 in a dose-dependent manner, with 1.0 µg/mL IO treatment resulting in higher luciferase activity than the treatment with 0.5 mM (1.313 mg/mL) AICAR, which is a known activator of PGC-1α through AMPK activation (Figure 7E). The deletion of MEF2 or CRE motif abrogated the reporter activity upregulation by IO to a greater degree than and it did to the reporter activity upregulation by AICAR (Figure 7F), suggesting for the requirement of both elements to mediate the effect of IO on PGC-1α expression. Taken together, these data suggest that IO upregulates the expression of PGC-1α to promote oxidative muscle metabolism.
Discussion
I. obliquus has been reported to have various health promoting properties which include antitumor, antioxidant, antiviral, anti-inflammatory, and immunomodulatory activity without prominent side effects (reviewed in [37,38]). However, most studies were performed in the context of cancer cells with a few exceptions of normal cell lines from digestive system such as pancreatic and hepatic cell lines. There have been limited investigation to our knowledge on its effect on myogenesis and regeneration of skeletal muscle. In this study, we investigated the effect of IO treatment on muscle health and the potential mechanism underlying the effect. IO treatment promoted C2C12 myoblast differentiation and alleviated DEX-induced myotube atrophy. IO treatment also improved muscle regeneration of CTX-injury mouse accompanied by an increase in muscle stem cell proliferation and oxidative muscle metabolism. Network pharmacological analysis of IO predicted that the promyogenic function of IO is mediated by the activation of AKT pathway, which was confirmed in C2C12 myoblasts. Taken together, the data here present a positive effect of IO on myogenesis and muscle regeneration in both in vivo and in vitro model of muscle atrophy through AKT-dependent mechanisms.
Investigation on the bioactive ingredients of IO and the molecular mediators of its biological effect is yet minimal and still ongoing. By comparing the metabolite profiles of IO1 and IO2, we identified 6 chemical constituents of IO as the potential bioactive compounds responsible for the myogenic effect of IO (Figure 2C; Table S1). Based on the network pharmacological analysis using the identified 6 chemical constituents of IO, we propose two major mediators of IO in the skeletal muscle: AKT and PGC-1α. Previous studies have shown the activation of PI3K/AKT signaling pathway by treatment of IO or its component in other contexts [39,40]. In both in vivo and in vitro muscle models, we also observed an increase in phosphorylated AKT in response to IO. We present here the AKT activation as one of the potential mechanisms underlying the rescue effect of IO on muscle atrophy. Currently, how IO regulates the crosstalk between AKT/muscle hypertrophy and PGC-1α/mitochondria pathways is unclear. It is plausible that AKT activation might induce AMPK/PGC-1α signaling as feedback, leading to mitochondrial biogenesis.
Effect of IO on the oxidative muscle metabolism. (A) Histochemical staining for SDH enzymatic activities in Vehicle- or IO-treated TA muscle. Scale bar, 100 µm. (B) The staining intensities of SDH are quantified as three different grades (dark, intermediate, and pale) and plotted as a percentile, n = 3. (C) Relative mitochondrial DNA content in TA muscles from 4-month-old mice ingested with Vehicle or 4 mg/kg IO for 4 weeks, n = 5. (D) Western blot analysis for the expression of total-OxPHOS in TA muscles from 4-month-old mice ingested with Vehicle or 4 mg/kg IO for 4 weeks. (E) Quantification of the relative levels of total-OxPHOS proteins from panel D, n = 5. (F) The expression of genes involved in the regulation of mitochondrial function examined by qRT-PCR analysis. The data were normalized using 18s RNA and were further normalized to the expression level of vehicle, n = 5 (Veh), 4 (IO). (G) Western blot analysis for the expression of total-OxPHOS in C2C12 cells treated with indicated concentration of IO. (H) Quantification of the relative levels of total-OxPHOS proteins from panel G, n = 3. (I) JC-1 fluorescence in C2C12 cells treated with either Vehicle or IO1. (J) Quantification of the relative levels of JC-1 from panel I. Data are expressed as the means ± SD. To determine statistical significance, an unpaired two-tailed Student's t-test was used. *P < 0.05, **P < 0.01 and ***P < 0.001.
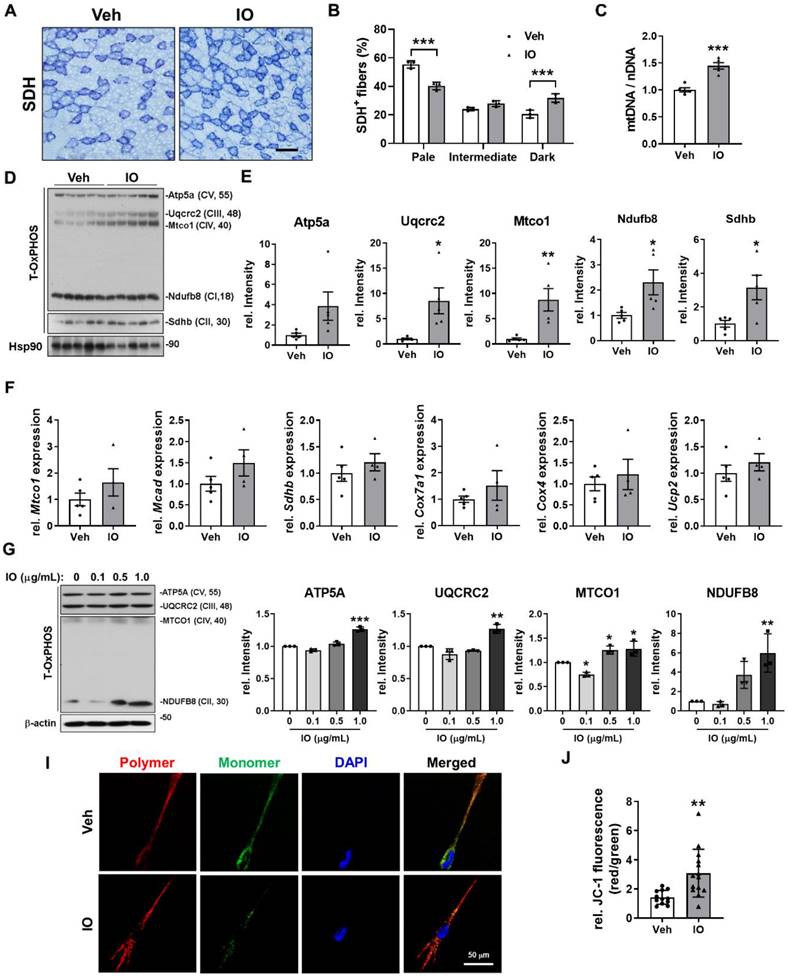
Effect of IO on the expression of PGC-1α in young mice skeletal muscle and C2C12 myoblasts. (A) Relative expression of PGC-1α in TA muscles from 4-month-old mice ingested with Vehicle or 4 mg/kg IO for 4 weeks, n = 5 (Veh), 4 (IO). (B) Western blot analysis and quantification of the relative levels of PGC-1α and myoglobin proteins in TA muscles from 4-month-old mice ingested with Vehicle or 4 mg/kg IO for 4 weeks, n = 5. (C) Western blot analysis for the expression of PGC-1α in C2C12 cells treated with indicated concentration of IO. And (D) quantification of the relative levels of PGC-1α protein, n = 3. (E) The relative PGC-1α luciferase activity in C2C12 cells treated with vehicle (DMSO) or indicated concentration of IO or AICAR as a positive control, n = 3. (F) The relative PGC-1α luciferase activity in C2C12 cells treated with vehicle (DMSO), IO (1.0 µg/mL), or AICAR (0.5 mM), n = 4. Data are expressed as the means ± SD. To determine statistical significance, an unpaired two-tailed Student's t-test was used. Asterisk indicates statistical significance. *P < 0.05, **P < 0.01 and ***P < 0.001 (IO vs. Vehicle).
A similar mechanism has been observed in resistance exercise-mediated enhancement in muscle mass and strength [41]. A recent study has also reported that muscle Akt is required for the maintenance of oxidative muscle metabolism and protein synthesis to enhance muscle mass and strength [42]. Akt signaling also prevents immobilization-induced muscle wasting [43]. Conversely, the inhibition of Akt signaling in muscle causes a shift towards non-oxidative muscle and mitochondrial dysfunction, with the concomitant loss of exercise performance [42,43]. AKT-deficient skeletal muscles display a substantial reduction in Pgc1α/β expression, resulting in the loss of mitochondrial function [42]. Thus, it is likely that IO might regulate both AKT/muscle hypertrophy and PGC1α/mitochondria via AKT signaling. Further studies are needed to define the exact molecular mechanism.
PGC-1α is a key regulator of muscle metabolism via modulating mitochondria biosynthesis [44]. Of the 6 potential bioactive compounds of IO, 10-oxo-cis-12-octadecenoic acid and 12-hydroxyoctadecanoic acid were shown to strongly activate peroxisome proliferator-activated receptors (PPARs), which in turn regulate the activity and expression of PGC-1α [45,46]. The expression of PGC-1α was indeed significantly increased by IO treatment in both mice and C2C12 myoblast (Figure 7A-D). Moreover, increased oxidative muscle metabolism by IO treatment was indicated by the increase in SDH enzymatic activity, expression of total OxPHOS and mitochondrial genes, and relative mitochondrial DNA content in TA muscles of IO-treated mice (Figure 6A-F). Since mitochondrial function is important during regeneration for energy production, homeostasis of reactive oxygen species, cross talk with immune cells, and modulation of stem cell fate, the enhanced mitochondrial function by IO through PGC-1α may also contribute to the improved muscle regeneration.
Taken together, our study demonstrates a promyogenic effect of IO in the context of muscle regeneration following injury through modulation of AKT signaling pathway and oxidative muscle metabolism. As imbalance in protein homeostasis and mitochondrial dysfunction are the key characteristics of aged and diseased muscle, IO is a promising drug candidate to promote muscle health.
Conclusions
IO treatment resulted in the activation of AKT, which inhibits the muscle-specific ubiquitin E3 ligase induced by dexamethasone. IO treatment showed improved regeneration of muscle with increased proliferation and differentiation of muscle stem cells and increased mitochondrial content and muscle oxidative metabolism with induction of PGC-1α. These results suggest that IO is considered a promising drug candidate for promoting muscle health with promyogenic effects that overcome the imbalance of protein homeostasis and mitochondrial dysfunction seen in aging through modulation of the AKT signaling pathway and oxidative muscle metabolism.
Abbreviations
AICAR: aminoimidazole carboxamide ribonucleotide; AKT: protein kinase B; Atp5a: ATP synthase subunit alpha; Atrogin-1: F-box only protein 32; Ccl22: c-c motif chemokine ligand 22; Ccl5: c-c motif chemokine ligand 5; Cox4: cytochrome c oxidase subunit 4; Cox7a1: cytochrome c oxidase subunit 7a1; CSA: cross section area; Cxcl1: c-x-c motif chemokine ligand 1; DEX: dexamethasone; DMSO: dimethyl sulfoxide; EDL: extensor digitorum longus; GAS: gastrocnemius; H&E: hematoxylin and eosin; Hsp90: heat shock protein 90; Il10: interleukin-10; Il1Ra: interleukin-1 receptor antagonist; Il6: interleukin-6; Mcad: medium-chain acyl-coenzyme a dehydrogenase; MHC: myosin heavy chain; Mtco1: mitochondrially encoded cytochrome c Oxidase 1; MuRF-1: E3 ubiquitin-protein ligase TRIM63; Myh: myosin heavy chain; MyoD: Myoblast determination protein 1; MyoG: myogenin; NdufB8: NADH dehydrogenase ubiquinone 1 beta subcomplex subunit 8; p38MAPK: p38 mitogen-activated protein kinases; Pax7: paired box 7; Pgc-1α: peroxisome proliferator-activated receptor γ coactivator α; SDH: succinate dehydrogenase; Sdhb: succinate dehydrogenase complex iron sulfur subunit B; SOL: soleus; TA: tibialis anterior; Tnfα: tumor necrosis factor-alpha; Upc2: mitochondrial uncoupling protein 2.
Supplementary Material
Supplementary figures and table.
Acknowledgements
This work was supported by National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIT) (grant numbers 2019R1A5A2027340, 2020R1A2C1007555 and 2021R1A2C2007937). This work was supported by Industry cooperation grant funded by Sungkyunkwan University (grant number S-2021-1372-000). This work was also supported by the Korea Environment Industry & Technology Institute (KEITI) through a project to improve the development of multi-ministerial national biological research resources, funded by the Republic of Korea Ministry of Environment (MOE) (grant number 2021003420003).
Ethics approval and consent to participate
The present study was reviewed and approved by the Sungkyunkwan University Animal Ethics Committee. All experimental procedures were performed in accordance with the Korea Food and Drug Administration (KFDA) Guidelines for the Care and Use of Laboratory Animals, and animal handling followed the dictates of the national animal Welfare Law of Korea. All experimental procedures were performed in accordance with guidelines of the Committee for the Purpose of Control and Supervision of Experiments on Animals of Sungkyunkwan University.
Author contributions
The authors' responsibilities were as follows—GUB, KHK, and JSK designed the project; CLY, SJL performed most of the experiments and analyzed data; CLY, SJL, JL, TAV, and HYL performed the molecular biology and cell biology experiments; SYJ and AA performed extraction and LC/MS analysis; ASW performed network pharmacology analysis; CLY, SJL, GUB, KHK, and JSK wrote the manuscript; All authors contributed and approved the final version of the manuscript.
Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Duru KC, Kovaleva EG, Danilova IG, van der Bijl P. The pharmacological potential and possible molecular mechanisms of action of Inonotus obliquus from preclinical studies. Phytother Res. 2019;33(8):1966-80
2. Lee MW, Hur H, Chang KC, Lee TS, Ka KH, Jankovsky L. Introduction to Distribution and Ecology of Sterile Conks of Inonotus obliquus. Mycobiology. 2008;36(4):199-202
3. Kim YO, Park HW, Kim JH, Lee JY, Moon SH, Shin CS. Anti-cancer effect and structural characterization of endo-polysaccharide from cultivated mycelia of Inonotus obliquus. Life Sci. 2006;79(1):72-80
4. Zhong XH, Ren K, Lu SJ, Yang SY, Sun DZ. Progress of research on Inonotus obliquus. Chin J Integr Med. 2009;15(2):156-60
5. Balandaykin ME, Zmitrovich IV. Review on Chaga Medicinal Mushroom, Inonotus obliquus (Higher Basidiomycetes): Realm of Medicinal Applications and Approaches on Estimating its Resource Potential. Int J Med Mushrooms. 2015;17(2):95-104
6. Lee IK, Yun BS. Styrylpyrone-class compounds from medicinal fungi Phellinus and Inonotus spp, and their medicinal importance. J Antibiot (Tokyo). 2011;64(5):349-59
7. Verschuren O, Smorenburg ARP, Luiking Y, Bell K, Barber L, Peterson MD. Determinants of muscle preservation in individuals with cerebral palsy across the lifespan: a narrative review of the literature. J Cachexia Sarcopenia Muscle. 2018;9(3):453-64
8. Argiles JM, Busquets S, Stemmler B, Lopez-Soriano FJ. Cachexia and sarcopenia: mechanisms and potential targets for intervention. Curr Opin Pharmacol. 2015;22:100-6
9. Dasarathy S, Merli M. Sarcopenia from mechanism to diagnosis and treatment in liver disease. J Hepatol. 2016;65(6):1232-44
10. Cruz-Jentoft AJ, Sayer AA. Sarcopenia. Lancet. 2019;393(10191):2636-46
11. Dennison EM, Sayer AA, Cooper C. Epidemiology of sarcopenia and insight into possible therapeutic targets. Nat Rev Rheumatol. 2017;13(6):340-7
12. Arany Z. PGC-1 coactivators and skeletal muscle adaptations in health and disease. Curr Opin Genet Dev. 2008;18(5):426-34
13. Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S. et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3(4):e101
14. Arany Z, Foo SY, Ma Y, Ruas JL, Bommi-Reddy A, Girnun G. et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature. 2008;451(7181):1008-12
15. Gan Z, Fu T, Kelly DP, Vega RB. Skeletal muscle mitochondrial remodeling in exercise and diseases. Cell Res. 2018;28(10):969-80
16. Handschin C, Spiegelman BM. The role of exercise and PGC1alpha in inflammation and chronic disease. Nature. 2008;454(7203):463-9
17. Aschenbach WG, Hirshman MF, Fujii N, Sakamoto K, Howlett KF, Goodyear LJ. Effect of AICAR treatment on glycogen metabolism in skeletal muscle. Diabetes. 2002;51(3):567-73
18. Suwa M, Nakano H, Radak Z, Kumagai S. Endurance exercise increases the SIRT1 and peroxisome proliferator-activated receptor gamma coactivator-1alpha protein expressions in rat skeletal muscle. Metabolism. 2008;57(7):986-98
19. Narkar VA, Downes M, Yu RT, Embler E, Wang YX, Banayo E. et al. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134(3):405-15
20. Hribal ML, Nakae J, Kitamura T, Shutter JR, Accili D. Regulation of insulin-like growth factor-dependent myoblast differentiation by Foxo forkhead transcription factors. J Cell Biol. 2003;162(4):535-41
21. Lawlor MA, Rotwein P. Insulin-like growth factor-mediated muscle cell survival: central roles for Akt and cyclin-dependent kinase inhibitor p21. Mol Cell Biol. 2000;20(23):8983-95
22. Serra C, Palacios D, Mozzetta C, Forcales SV, Morantte I, Ripani M. et al. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol Cell. 2007;28(2):200-13
23. Glass DJ. Signaling pathways perturbing muscle mass. Curr Opin Clin Nutr Metab Care. 2010;13(3):225-9
24. Zanou N, Gailly P. Skeletal muscle hypertrophy and regeneration: interplay between the myogenic regulatory factors (MRFs) and insulin-like growth factors (IGFs) pathways. Cell Mol Life Sci. 2013;70(21):4117-30
25. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274-93
26. Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44(D1):D380-4
27. Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357-W64
28. Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D. et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017;45(D1):D945-D54
29. Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605-D12
30. Hagberg AA SD, Swart PJ, editor Exploring network structure, dynamics, and function using NetworkX. Proceedings of the 7th Python in Science Conference; 2008; Pasadena, CA USA
31. Jeong HJ, Lee HJ, Vuong TA, Choi KS, Choi D, Koo SH. et al. Prmt7 Deficiency Causes Reduced Skeletal Muscle Oxidative Metabolism and Age-Related Obesity. Diabetes. 2016;65(7):1868-82
32. Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO. et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14(3):395-403
33. Bae GU, Lee JR, Kim BG, Han JW, Leem YE, Lee HJ. et al. Cdo interacts with APPL1 and activates Akt in myoblast differentiation. Mol Biol Cell. 2010;21(14):2399-411
34. Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci U S A. 2003;100(12):7111-6
35. Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA. et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704-8
36. Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168(6):960-76
37. Lu Y, Jia Y, Xue Z, Li N, Liu J, Chen H. Recent Developments in Inonotus obliquus (Chaga mushroom) Polysaccharides: Isolation, Structural Characteristics, Biological Activities and Application. Polymers (Basel). 2021 13(9)
38. Szychowski KA, Skora B, Pomianek T, Gminski J. Inonotus obliquus - from folk medicine to clinical use. J Tradit Complement Med. 2021;11(4):293-302
39. Zhang Z, Liang X, Tong L, Lv Y, Yi H, Gong P. et al. Effect of Inonotus obliquus (Fr.) Pilat extract on the regulation of glycolipid metabolism via PI3K/Akt and AMPK/ACC pathways in mice. J Ethnopharmacol. 2021;273:113963
40. Zhao Y, Zheng W. Deciphering the antitumoral potential of the bioactive metabolites from medicinal mushroom Inonotus obliquus. J Ethnopharmacol. 2021;265:113321
41. Schoenfeld B.J. The Mechanisms of Muscle Hypertrophy and Their Application to Resistance Training. J. Strength Cond. Res. 2010 24, 2857-2872
42. Natasha Jaiswal, Matthew Gavin, Emanuele Loro, Jaimarie Sostre-Colón, Paul A. Roberson, Kahealani Uehara, Nicole Rivera-Fuentes, Michael Neinast, Zoltan Arany, Scot R. Kimball, Tejvir S. Khurana, Paul M. Titchenell. AKT controls protein synthesis and oxidative metabolism via combined mTORC1 and FOXO1 signalling to govern muscle physiology. J Cachexia Sarcopenia Muscle. 2022;13(1):495-514
43. Das S, Morvan F, Jourde B, Meier V, Kahle P, Brebbia P. et al. ATP citrate lyase improves mitochondrial function in skeletal muscle. Cell Metab. 2015;21:868-876
44. Liang H, Ward WF. PGC-1alpha: a key regulator of energy metabolism. Adv Physiol Educ. 2006;30(4):145-51
45. Goto T, Kim YI, Furuzono T, Takahashi N, Yamakuni K, Yang HE. et al. 10-oxo-12(Z)-octadecenoic acid, a linoleic acid metabolite produced by gut lactic acid bacteria, potently activates PPARgamma and stimulates adipogenesis. Biochem Biophys Res Commun. 2015;459(4):597-603
46. Umeno A, Sakashita M, Sugino S, Murotomi K, Okuzawa T, Morita N. et al. Comprehensive analysis of PPARgamma agonist activities of stereo-, regio-, and enantio-isomers of hydroxyoctadecadienoic acids. Biosci Rep. 2020 40(4)
Author contact
![]() Corresponding authors: Jong-Sun Kang, Ph.D., Department of Molecular Cell Biology, Single Cell Network Research Center, Sungkyunkwan University School of Medicine, Suwon 16419, Republic of Korea; Tel.: +82-31-299-6135, Fax.: +82-31-299-6157, E-mail: kangj01edu. Ki Hyun Kim, Ph.D., School of Pharmacy, Sungkyunkwan University, Suwon 16419, Republic of Korea; Tel: +82-31-290-7700; Fax: +82-31-290-7730; E-mail: khkim83edu. Gyu-Un Bae, Ph.D., Research Institute of Aging Related Disease, AniMusCure Inc., Suwon 16419, Republic of Korea; Drug Information Research Institute, Muscle Physiome Research Center, College of Pharmacy, Sookmyung Women's University, Seoul 04310, Republic of Korea; Tel.: +82-2-2077-7629, Fax.: +82-2-2077-7629, E-mail: gbaeac.kr.
Corresponding authors: Jong-Sun Kang, Ph.D., Department of Molecular Cell Biology, Single Cell Network Research Center, Sungkyunkwan University School of Medicine, Suwon 16419, Republic of Korea; Tel.: +82-31-299-6135, Fax.: +82-31-299-6157, E-mail: kangj01edu. Ki Hyun Kim, Ph.D., School of Pharmacy, Sungkyunkwan University, Suwon 16419, Republic of Korea; Tel: +82-31-290-7700; Fax: +82-31-290-7730; E-mail: khkim83edu. Gyu-Un Bae, Ph.D., Research Institute of Aging Related Disease, AniMusCure Inc., Suwon 16419, Republic of Korea; Drug Information Research Institute, Muscle Physiome Research Center, College of Pharmacy, Sookmyung Women's University, Seoul 04310, Republic of Korea; Tel.: +82-2-2077-7629, Fax.: +82-2-2077-7629, E-mail: gbaeac.kr.

 Global reach, higher impact
Global reach, higher impact