Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2024; 20(10):4029-4043. doi:10.7150/ijbs.94156 This issue Cite
Review
Helicobacter pylori Outer Membrane Vesicles: Biogenesis, Composition, and Biological Functions
1. Center of Infectious Diseases, West China Hospital, Sichuan University, Chengdu, China.
2. Laboratory of Infectious and Liver Diseases, Institute of Infectious Diseases, West China Hospital of Sichuan University, Chengdu 610041, China.
3. QuTEM AB, Gävlegatan 22, 11330 Stockholm, Sweden.
4. Helicobacter Research Laboratory, The Marshall Centre for Infectious Disease Research and Training, University of Western Australia, Nedlands WA 6009, Australia.
Received 2024-1-11; Accepted 2024-7-6; Published 2024-7-15
Abstract
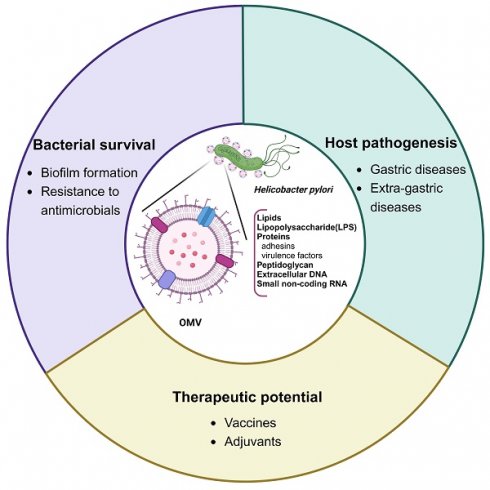
Helicobacter pylori has been recognized not only as a causative agent of a spectrum of gastroduodenal diseases including chronic gastritis, peptic ulcer, mucosa-associated lymphoid tissue lymphoma, and gastric cancer, but also as the culprit in several extra-gastric diseases. However, the association of H. pylori infection with extra-gastric diseases remains elusive, prompting a reevaluation of the role of H. pylori-derived outer membrane vesicles (OMVs). Like other gram-negative bacteria, H. pylori constitutively sheds biologically active OMVs for long-distance delivery of bacterial virulence factors in a concentrated and protected form, averting the need of direct bacterial contact with distant host cells to induce extra-gastric diseases associated with this gastric pathogen. Additionally, H. pylori-derived OMVs contribute to bacterial survival and chronic gastric pathogenesis. Moreover, the immunogenic activity, non-replicable nature, and anti-bacterial adhesion effect of H. pylori OMVs make them a desirable vaccine candidate against infection. The immunogenic potency and safety concerns of the OMV contents are challenges in the development of H. pylori OMV-based vaccines. In this review, we discuss recent advances regarding H. pylori OMVs, focusing on new insights into their biogenesis mechanisms and biological functions.
Keywords: Helicobacter pylori, outer membrane vesicles, biogenesis, composition, biological functions
Introduction
Helicobacter pylori, a gram-negative and spiral-shaped bacterium, colonizes the human gastric mucosa of about half of the world's population [1]. Helicobacter pylori is usually acquired in childhood and can persist lifelong within the human stomach unless treated with antibiotics. Upon infection with H. pylori, the host responds by initiating inflammation of the gastric mucosa, causing gastritis, but failing to eradicate the pathogen. H. pylori-induced gastritis is usually asymptomatic, but prolonged infection is a major risk factor for the development of peptic ulcer, mucosa-associated lymphoid tissue (MALT) lymphoma, and gastric cancer [2, 3]. Therefore, H. pylori is listed as a group Ⅰ carcinogen by the World Health Organization (WHO) [4].
In addition to gastric diseases, emerging evidence indicates that H. pylori infection is associated with various extra-gastric diseases, such as cardiovascular diseases [5], diabetes [6], liver diseases [7], and Alzheimer's disease [8]. However, the mechanisms underlying the remote effects of H. pylori infection remain largely unknown. As an extracellular bacterium with poor invasiveness, most H. pylori reside in the gastric mucus layer, with only a small proportion attached to the surface of epithelial cells [9, 10]. Since there is no evidence that H. pylori can enter the blood circulation [11], H. pylori-derived virulence factors, especially the outer membrane vesicles (OMVs), may play an important role in the development of the reported extra-gastric diseases. Shedding spherical and membranous OMVs, ranging between 20-300 nm in diameter, is a common feature of virtually all gram-negative bacteria [12]. Emerging evidence indicates that OMVs from the host microbiota, particularly the commensal bacteria in the gastrointestinal tract, can enter blood circulation [13]. Compared with soluble secretion pathways, OMVs provide a relatively superior secretion option for pathogens to deliver virulence factors in a highly concentrated form, whilst offering additional protection from host proteases during travelling to distant organs and cells [14]. Helicobacter pylori constitutively sheds OMVs both in vivo and in vitro [15-17]. A recent study showed that the transcriptomic changes induced by H. pylori OMVs in MKN74 gastric adenocarcinoma cell line were largely similar to those triggered by the parental bacteria [18], indicating that OMVs may largely contribute to or amplify the pathogenic effects induced by the bacterium itself. Moreover, the detection of H. pylori-derived OMVs in the sera of mice infected with H. pylori provided further evidence for the potential involvement of H. pylori OMVs in extra-gastric diseases [19].
Apart from the role as a delivery vehicle for virulence factors, OMVs exhibit a large variety of other biological roles, such as horizontal gene transfer, nutrient acquisition, bacterial biofilm formation, and host immune response modulation [20, 21]. OMVs contain abundant bacterial antigens, including lipids, proteins, carbohydrates, and nucleic acids. By eliciting and modulating immune responses, as well as preventing pathogen localization and proliferation, OMVs may play an anti-infection role in attenuating and even preventing pathogen-associated diseases [22]. This is supported by the good safety and efficacy profiles of OMV-based vaccines against Neisseria meningitidis [22-24]. However, the potential use of H. pylori OMVs as vaccines against H. pylori infection is just at the early stage.
In this review, we discuss recent advances regarding H. pylori OMVs biogenesis and composition with a special focus on their pathological roles in gastric and extra-gastric diseases, and their potential use as vaccines.
OMV biogenesis
Models for OMV biogenesis in other gram-negative bacteria
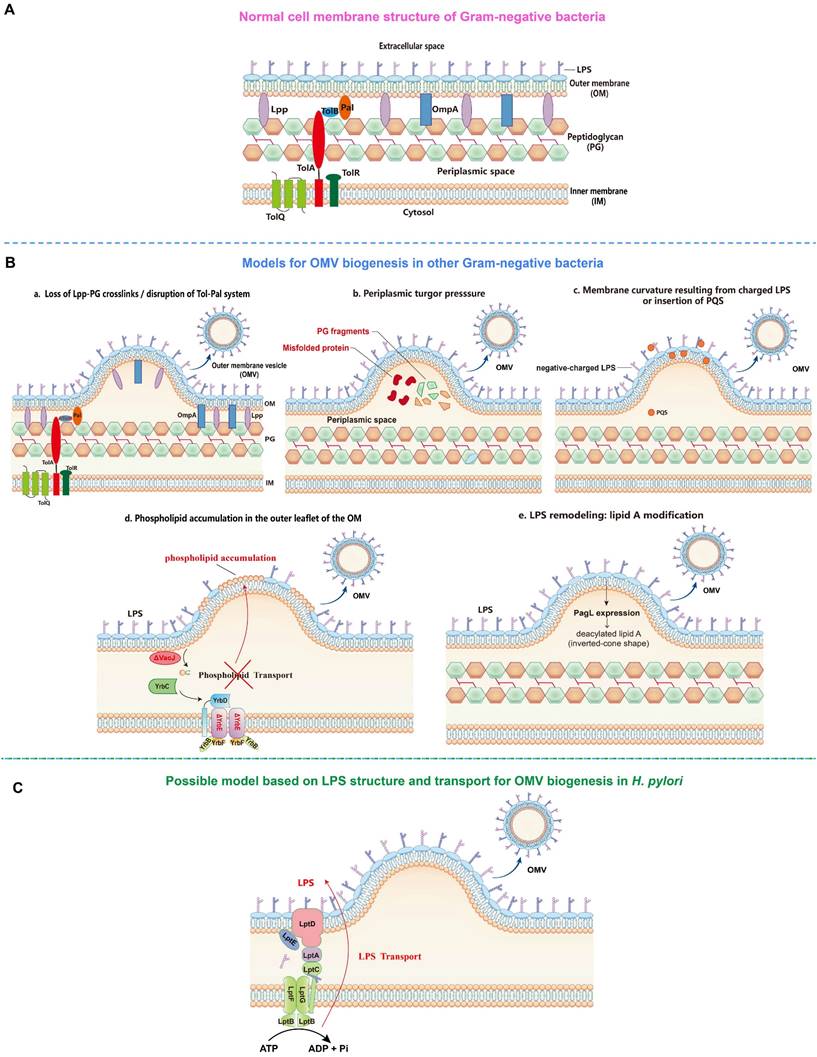
The first model proposes that the formation of OMVs is due to the loss of covalent or non-covalent crosslinks between the outer membrane (OM) and the underlying peptidoglycan layer (Figure 1). For instance, the lack or reduction of covalent crosslinks of lipoprotein (Lpp) or outer membrane protein A (OmpA) with the peptidoglycan layer allows a faster growth rate of the OM relative to the peptidoglycan layer, which then leads to the detachment and release of protruded OM as OMVs in several gram-negative bacteria including Escherichia coli, Acinetobacter baumannii, and Salmonella spp. [25-28]. In addition, the disruption of one or more components of the Tol-Pal system (a protein complex comprising the inner membrane proteins TolQ-TolR-TolA, the periplasmic protein TolB, and the outer membrane protein Pal), which is involved in non-covalent interaction with the peptidoglycan, has also been shown to significantly increase OMV production in E. coli, Salmonella choleraesuis, Shigella flexneri, and Bordetella pertussis [29-33].
The second model proposes that turgor pressure, caused by the accumulation of peptidoglycan fragments or misfolded proteins in the periplasmic space, induces the bulging and pinching-off of the bacterial OM (Figure 1). This model is supported by the detection of low molecular weight muramic acid, a precursor of peptidoglycan, in OMVs isolated from Porphyromonas gingivalis [34], and the deletion of peptidoglycan amidase in the same bacterial species that has resulted in the increase of OMV production, possibly due to the disruption of the peptidoglycan recycling process and thus the buildup of turgor pressure following the periplasmic accumulation of peptidoglycan fragments [35]. Furthermore, McBroom et al. demonstrated that the extent of vesiculation is correlated with the level of protein accumulation in the cell envelope [36]. Another study also reported that the inactivation of DegQ periplasmic serine protease has led to abnormal accumulation of proteins in the periplasmic space, causing increased OMV production in Shewanella oneidensi [37].
The third model involves the induction of membrane curvature, which is contributed mainly by lipopolysaccharide (LPS) and Pseudomonas quinolone signal (PQS) (Figure 1). Bacterial LPS, typically composed of three domains including lipid A, core oligosaccharide, and the outermost O-antigen polysaccharide, is a major constituent of the outer leaflet of the OM of gram-negative bacteria [38]. The LPS of most gram-negative bacteria is negatively charged due to the presence of anionic phosphate groups in the lipid A and inner core. The negatively charged LPS molecules are crosslinked by divalent cations, such as Ca2+ and Mg2+, to stabilize the OM [39]. Thus, the imbalance of the ionic interaction may lead to the strong electrostatic repulsion between neighbouring LPS molecules, inducing membrane curvature and subsequent OMV formation. This is supported by the finding that the treatment of cells with divalent cation chelator EDTA enhanced the release of OMVs, whereas the addition of exogenous Mg2+ suppressed OMV formation [40]. It has also been found that OMVs from Pseudomonas aeruginosa consist primarily of the negatively charged B-band LPS, but not the neutral A-band LPS present in the OM [41, 42]. Similarly, in P. gingivalis, only the negatively charged LPS has been found in OMVs [43], suggesting the involvement of charge-to-charge repulsion by LPS in OMV formation. Interestingly, PQS, a membrane curvature-inducing and LPS-binding signaling molecule, is not only packaged into OMVs, but also required for OMV production in P. aeruginosa [44, 45]. It has been shown that PQS stimulates OMV biogenesis through its interaction with LPS. By sequestering divalent cations, PQS enhances anionic repulsions between LPS molecules, causing a bilayer couple effect that induces membrane curvature and the budding of OMVs [46, 47]. Of note, the PQS-based model is species-specific as PQS is only produced by P. aeruginosa [22].
The fourth model involves the VacJ/Yrb ABC (ATP-binding cassette) transport system, a phospholipid (PL) transporter, which functions to prevent PL accumulation in the outer leaflet of the OM by transporting PLs retrogradely from the OM to the inner membrane (IM), thereby maintaining the lipid asymmetry in the OM [48] (Figure 1). It has been reported that the deletion or downregulated expression of VacJ or YrbE resulted in increased OMV production in several distantly related bacteria, including Haemophilus influenzae, Vibrio cholerae, and E. coli [48]. Lipidomic analyses indicated that phosphatidylethanolamine (PE) was the most dominant PL species in OM and OMVs preparations, and the total PE content of OMVs derived from the transporter mutants was two-fold higher than that of the wild-type OMVs. The accumulation of PLs in the outer leaflet of the OM induced by deletion or reduced expression of the VacJ/Yrb ABC transport system induces an outward bulging of the OM, which pinches off to form OMVs [48].
The fifth model involves LPS lipid A deacylation-mediated OM remodeling (Figure 1). It has been reported that the expression of the “latent” lipid A deacylase PagL in intracellular Salmonella typhimurium results in hypervesiculation without inducing an envelope stress response [49]. Mass spectrometry analysis further revealed considerable differences in the patterns of lipid A between OM and OMVs, with deacylated lipid A accumulating exclusively in OMVs. In contrast, the S. typhimurium ΔpagL strain displayed a significant reduction in intracellular OMV production compared to the wild-type strain. It was proposed that intracellular S. typhimurium PagL is activated to deacylate lipid A, leading to a decrease in hydrophobic cross-section area. The deacylated lipid A adopts an inverted-cone shaped structure, leading to membrane curvature and OMV formation [49].
Possible OMV biogenesis model in H. pylori
Although the mechanistic models for OMV biogenesis are increasingly being unraveled, whether these models are applicable to the OMV biogenesis in H. pylori remains to be further investigated. Interestingly, a study demonstrated that knocking out hp0044 and hp1275 genes responsible for GDP-fucose biosynthesis have led to a truncated LPS (truncating from the conserved O-antigen Trio fucose residue) and altered protein sorting into OMVs [50]. In addition, a recent study showed that inactivation of HP0860 responsible for ADP-heptose biosynthesis also caused truncation of the LPS core and altered protein sorting into OMVs [51]. These results suggest that H. pylori LPS may play an important role in OMV cargo sorting and biogenesis. Considering that OMV production requires a significant energy cost, whereas ATP or other energy sources directly at the site of budding are not available [52], we hypothesize that the ATP used to export a continuous stream of newly synthesized LPS from the IM to the outer leaflet of the OM through the transenvelope Lpt machinery comprising seven LPS transport proteins (LptA-G) is likely to be source of energy for OMV biogenesis in H. pylori [53] (Figure 1).
Composition of H. pylori OMVs
Lipids and LPS
The total lipid content of H. pylori consists of about 70% phospholipids, up to 25% of unique cholesteryl glucosides (CGs), and the rest being neutral lipids [54]. In another study, it was shown that the lipid profile of H. pylori OMVs is highly similar to that of the OM, with PE being the most abundant phospholipid detected in H. pylori OMVs and then followed by cardiolipin. Additional phospholipids found in H. pylori OMVs include lyso-PE, phosphatidylglycerol (PG), lyso-PG, phosphatidylcholine (PC) and lyso-PC [55].
OMV biogenesis models (By Figdraw). A. Cell membrane structure of gram-negative bacteria consists of inner membrane (IM), periplasmic space containing peptidoglycan (PG) and outer membrane (OM). B. OMV biogenesis models in other gram-negative bacteria: a. Loss of covalent crosslinks between the OM and PG or disruption of non-covalent crosslinks through Tol-Pal system; b. Accumulation of misfolded proteins and PG fragments in the periplasmic space causes turgor pressure; c. Electrostatic repulsion between negative-charged lipopolysaccharide (LPS) molecules or insertion of Pseudomonas quinolone signal (PQS) into the OM induce membrane curvature; d. Phospholipid accumulation in the outer leaflet of the OM resulted from the disruption within VacJ/Yrb ABC transport system; e. LPS remodeling mediated by PagL expression-induced deacylation of lipid A. C. Hypothetical OMV biogenesis model based on LPS structure and transport in Helicobacter pylori.
Most gram-negative bacterial OMVs characterized to date are enriched with LPS that is structurally identical to the parental bacteria [9, 56, 57]. The H. pylori LPS contains lipid A, core oligosaccharide, and the O-antigen. The O-antigen portion contains Lewis antigen composed of a Gal-GlcNAc backbone chain, which is divided into two types based on the linkage. Namely, type 1 composed of Gal-β-(1,3)-GlcNAc forms Lewis a (Lea) and Lewis b (Leb), and type 2 composed of Gal-β-(1,4)-GlcNAc forms Lewis x (Lex) and Lewis y (Ley) [58]. It was reported that the abundance of LPS in H. pylori OMVs is influenced by the iron availability in the parental bacteria growth conditions, since less and shorter LPS was detected in OMVs from iron-limiting bacteria than that from iron-replete bacteria. More specifically, structural analysis and serological detection revealed that iron-limiting bacterial whole cells and OMVs exhibited reduced Ley expression [59].
Proteins
A mass spectrometry (MS)-based proteomic analysis of OMVs isolated from H. pylori strain J99 identified 162 distinct proteins, of which 30.86% (50), 21.6% (35), 7.4% (12) and 4.9% (8) were known or predicted OM, cytoplasmic, periplasmic and IM proteins, respectively, and proteins with no confirmed or predicted subcellular localization accounting for 29.6% [60]. In another study, more than 300 different H. pylori proteins were identified in the OMV proteome of strain CCUG17875. Interestingly, OM proteins constituted only 16% of the OMV proteome [55]. More recently, Wei et al. identified 436 proteins in the reference strain NCTC11637 OMVs and 372 proteins in the clinical strain Hp-400 OMVs [61]. The discrepancies exhibited by these studies lead to the conclusion that H. pylori OMVs show great heterogeneity in their protein composition, which may be determined by strain genotypes, different bacterial growth conditions, growth stages, OMV preparation, and the sensitivity of MS methods used for proteomic analysis [62]. Of note, a recent study revealed that the OMV size can determine its protein cargo content, as smaller OMVs contained significantly fewer and less diverse proteins compared to larger ones, and most H. pylori adhesins were packaged within larger OMVs [63].
Among the protein components of H. pylori, CagA and VacA cytotoxins are the most studied virulence factors [64]. VacA is a pore-forming toxin that induces vacuolation in gastric epithelial cells [65, 66]. Although it is an autotransporter protein, VacA has also been identified in H. pylori OMVs. By electron microscopy and immunocytochemistry, VacA was confirmed to be released by H. pylori in both free-soluble and OMV-associated forms, both of which were observed to be internalized by MKN28 gastric adenocarcinoma cell line in vitro, and were detected in the gastric mucosa isolated from H. pylori-infected patients [67]. Quantitative analysis revealed that OMV-associated VacA accounted for about 25% of total VacA, with the remaining 75% being free-soluble [68]. CagA cytotoxin, also known for its role as an oncoprotein, can induce a “hummingbird” phenotype in gastric epithelial cells [69]. Typically, CagA is injected into host cells by the bacteria through the type IV secretion system (T4SS) [70]. Intriguingly, CagA was found to be associated with the surface of H. pylori OMVs, suggesting that OMVs might provide an alternative mechanism for H. pylori to transport CagA directly into host cells independently of the T4SS [55, 71]. Furthermore, additional proteins involved in H. pylori colonization and virulence, such as SabA, BabA, AlpA, AlpB, OipA, and HopZ adhesins, and several other enzymes including urease, catalase and HtrA serine protease were identified in the OMVs [55, 60, 61, 71-73].
Peptidoglycan and nucleic acids
Peptidoglycan component can be found in H. pylori OMV as demonstrated by Kaparakis et al. that OMVs isolated from H. pylori strain 251 were shown to contain approximately 0.3-0.5 ng of the muramic acid moiety of peptidoglycan, per μg of OMV protein [74]. Besides, genetic materials including extracellular DNA (eDNA) and small non-coding RNA (sncRNA) have also been detected in OMVs from multiple gram-negative bacteria [56, 75]. In H. pylori, eDNA implicated in communication between bacteria was detected on the surface of OMVs from H. pylori strain NCTC 11639 [76]. Moreover, Zhang et al. identified the presence of regulatory sncRNAs (sR-2509025 and sR-989262) in OMVs isolated from H. pylori strain J99 by RNA sequencing [77].
Biological functions of H. pylori OMVs
H. pylori OMVs favor bacterial biofilm formation and resistance to antimicrobials
Biofilm formation represents a survival strategy by which bacteria are protected from the attack of both host immune system and antimicrobials, thus contributing to bacterial persistence [78, 79]. Notably, the ability of H. pylori to form biofilms on abiotic surfaces in vitro as well as on human gastric mucosa in vivo has been well characterized [78, 80]. The favorable role of OMVs in H. pylori biofilm formation was confirmed in in vitro studies. Yonezawa et al. demonstrated that biofilm formation of H. pylori was dependent on the cell-cell aggregation mediated by OMVs [81]. In addition, the presence of OMVs was detected in the biofilm extracellular polymeric substance matrix of TK1402 clinical isolate with strong biofilm-forming ability, and the addition of TK1402-derived OMVs to H. pylori cultures resulted in remarkably enhanced biofilm formation [81]. By further analyzing the protein profiles of OMVs from different H. pylori strains with strong or weak biofilm-forming ability, an uncharacterized 22-KDa protein and the outer membrane protein AlpB were identified as crucial factors contributing significantly to the biofilm formation in the strain TK1402 [82, 83]. In addition to protein molecules, eDNA, mainly associated with the surface of OMVs, appears to play a pivotal role during the development of H. pylori biofilms [76]. Helicobacter pylori produces OMVs in both biofilm (bOMVs) and planktonic (pOMVs) phenotypes. In H. pylori strain NCTC11639, bOMVs were found to have a broader size range, more negative charges, increased OMV aggregation, as well as a four-fold increase in eDNA as compared with pOMVs, suggesting that H. pylori bOMVs could prevent the degradation of eDNA. The eDNA may then act as a bridge to facilitate OMV-OMV and cell-cell aggregation, ultimately promoting biofilm formation [76]. More recently, the α-class carbonic anhaydrase (α-CA), a periplasmic enzyme essential for the acid acclimation of H. pylori within human stomach, was also detected in bOMVs and pOMVs of four H. pylori strains, with a higher level seen in pOMVs than in bOMVs. Moreover, α-CA was confirmed to induce the release of eDNA, thereby stabilizing the biofilm formation [84]. Correspondingly, carvacrol and thymol, inhibitors of α-CA, could prevent H. pylori OMV production and biofilm development via the inhibition of eDNA release [85]. Taken together, H. pylori-derived OMVs are both “components” and “boosters” of bacterial biofilm, which represents a survival strategy exploited by the microorganism to protect itself against the attack of both host immune system and antimicrobials [86].
Apart from facilitating biofilm formation, OMVs confer bacterial resistance to antimicrobials in two possible ways: 1) OMV-mediated transfer of antibiotic resistance genes between bacteria; 2) OMVs acting as decoys to bind to or absorb antimicrobials [14, 56]. For instance, in E. coli, the use of a hypervesiculating mutant or the addition of OMVs enhanced immediate resistance to the antimicrobial peptides polymyxin B and colistin [87]. Similarly, OMVs from Pseudomonas syringae reduced the levels of colistin and melittin in solution by sequestering these compounds [88]. In H. pylori, it was found that the addition of purified OMVs from strain 60190 facilitated H. pylori survival upon exposure to antimicrobial peptide LL-37 as well as clarithromycin [89]. Given that clarithromycin is a hydrophobic antibiotic that enters bacterial cells via lipid-mediated passive diffusion [31, 32], and it is also a macrolide antibiotic that can bind directly to lipid membranes [33], H. pylori OMVs of lipid bilayer may act as a decoy sequestering clarithromycin that would otherwise diffuse into bacterial cells [89].
Therefore, the OMVs produced by H. pylori play a crucial role in its survival by promoting H. pylori biofilm formation and/or acting as a decoy to sequester host-derived antimicrobials or antibiotics (Figure 2).
H. pylori OMVs contribute to the pathogenesis of gastric and extra-gastric diseases
Uptake of H. pylori OMVs by host cells
Uptake of H. pylori OMVs by host cells is the initial step required for delivering bacterial virulence factors into host cells and exerting pathological effects. By utilizing an in vivo imaging system, it was found that orally administrated H. pylori OMVs remained in the stomach of mice for at least 24 h and could enter gastric epithelial cells [17]. In vitro assays also observed that H. pylori OMVs adhered to and were internalized by primary human antral epithelial cells, as well as by AGS gastric adenocarcinoma cell line [90, 91]. Fluorescent confocal microscopy revealed that internalized vesicles colocalized with markers for early endosomes and lysosomes, suggesting their entry through the endocytic pathway [91, 92]. Additionally, UreA was observed to localize to the cytoplasm and nucleus of AGS cells incubated with OMVs from H. pylori strain 26695, reinforcing the entry of H. pylori OMVs into host cells and subsequent release of virulence factors [93]. Cellular uptake of H. pylori OMVs has been reported to occur through both clathrin-dependent and clathrin-independent pathways [92], with clathrin-mediated endocytosis identified as the primary mechanism [94]. For instance, treatment of AGS cells with chlorpromazine, a known inhibitor of clathrin-mediated endocytosis, reduced the uptake of H. pylori OMVs [92, 95]. Disruption of lipid rafts in AGS cells using Fumonisin B1, an inhibitor of sphingomyelin, or methyl-β-cyclodextrin (MβCD), a cholesterol-depleting agent, also reduced OMV uptake, suggesting the involvement of lipid raft-mediated endocytosis [55, 74]. Additionally, caveolin- and dynamin-mediated endocytosis, macropinocytosis, and phagocytosis are also identified as important mechanisms mediating the entry of H. pylori OMVs into host cells [63, 91]. Notably, the size of H. pylori OMVs was reported to determine the mechanisms of their uptake by host cells. Vesicles smaller than 100 nm are typically taken up through endocytosis, whereas the vesicles larger than 200 nm are usually taken up through macropinocytosis and phagocytosis [96]. Moreover, Parker et al. reported that the presence of VacA in OMVs significantly enhanced vesicle association with host cells. They found that the internalization of VacA+ OMVs was less inhibited by chlorpromazine compared to VacA- OMVs, suggesting that VacA allows vesicles to use more than one pathway of internalization [95].
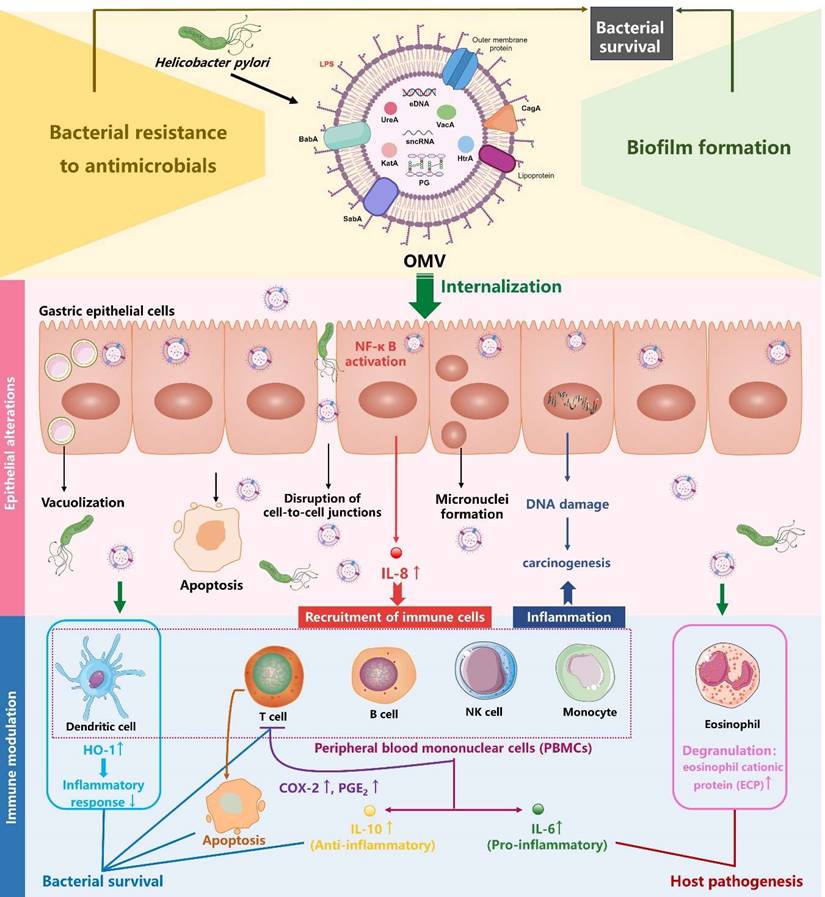
Role of H. pylori OMVs in bacterial survival and host gastric pathogenesis (created with BioRender.com). On the one hand, H. pylori OMVs promote biofilm formation and enhance bacterial resistance to antimicrobials, both of which synergistically admit H. pylori to survive and aggravate the chronic infection. On the other hand, H. pylori OMVs containing multiple virulence factors can be internalized by gastric epithelial cells and induced a plethora of pathological effects: elevation of proinflammatory factor IL-8, apoptosis, disruption of gastric epithelial cell-to-cell junctions, vacuolization, micronuclei formation. These alterations in gastric epithelial cells contribute to the development of gastric diseases. Additionally, H. pylori OMVs can modulate immune cells within the lamina propria, either suppressing the immune response to promote bacterial survival or stimulating the inflammatory response, thus contributing to host pathogenesis.
Pathogenic effects of H. pylori OMVs on gastric epithelial cells
It has been well established that H. pylori OMVs stimulate IL-8 production by gastric epithelial cells in a dose-dependent manner, both in vivo and in vitro [17, 61, 97]. IL-8 serves as a pro-inflammatory cytokine and chemokine, triggering the recruitment of immune cells such as macrophages, neutrophils, and T lymphocytes to the infected tissues, thereby inducing an intense mucosal inflammatory response [98]. Kaparakis et al. demonstrated that the OMV-mediated delivery of peptidoglycan, rather than LPS, induced cytosolic NOD1-dependent NF-κB activation and subsequent IL-8 production in AGS cells. Notably, microinjection of peptidoglycan into AGS cells failed to initiate a NOD1-dependent inflammatory response, highlighting the crucial role of H. pylori OMVs in the delivery of bacterial peptidoglycan to induce host cell inflammation [74]. Interestingly, it was also demonstrated in another study that OMVs were involved in the delivery of LPS into host cell cytosol, triggering caspase-11-dependent pyroptotic cell death and IL-1 responses [57]. Future study is needed to investigate whether H. pylori LPS can rely on OMVs to gain access into cytosol for caspase-11 activation.
H. pylori OMVs have also been reported to induce apoptosis in AGS cells through the activation of caspase-8 and caspase-3, independently of the mitochondrial pathway, as indicated by the absence of cytochrome c release [99]. Moreover, OMVs can enhance the carcinogenic potential of H. pylori as increased micronuclei formation (a cellular event associated with carcinogenesis) was observed in AGS cells when treated with OMVs from H. pylori strain 60190 [100]. Additionally, a transcriptomic study demonstrated that OMVs from H. pylori strain 26695 induced transcriptomic remodeling of MKN74 cells, characterized by downregulation of cell cycle, DNA replication, and DNA repair [18]. This remodeling may lead to accumulated DNA mutations and genome instability, predisposing cells to carcinogenesis. Consistently, a proteomic study reported that GES-1 human normal gastric epithelial cell line treated with OMVs from H. pylori strains NCTC11637 and Hp400 exhibited proteomic changes related to cancer signaling pathways, including increased expression of the pro-carcinogenic proteins VTN and complement C3 [61]. Notably, OMV-induced transcriptomic and proteomic alterations in MKN74 and GES-1 cells largely coincided with H. pylori-induced alterations [18, 61], implying that H. pylori induces pathogenicity in gastric epithelial cells in part by producing OMVs. Collectively, H. pylori OMVs contribute to the development of gastric diseases by promoting inflammation, apoptosis, and tumorigenesis in gastric epithelial cells (Figure 2).
Immunomodulatory effects of H. pylori OMVs on immune cells within gastric lamina propria
The ability of H. pylori OMVs to transmigrate across gastric epithelial monolayers allows these vesicles to directly deliver antigens and virulence factors to immune cells infiltrating the lamina propria, which is triggered by H. pylori infection [70]. For instance, an in vitro study showed that eosinophils treated with H. pylori OMVs or co-cultured with OMV-exposed gastric epithelial cells were activated and underwent degranulation, releasing cytotoxic granule proteins such as eosinophil cationic protein (ECP) which exacerbate local inflammatory responses and tissue damage [101]. Additionally, Winter et al. reported that OMVs from H. pylori stains SS1 and 60190 strongly triggered the production of both pro-inflammatory cytokine IL-6 and anti-inflammatory cytokine IL-10 by human peripheral blood mononuclear cells (PBMCs) in a dose-dependent manner, suggesting a delicate role for H. pylori OMVs in the modulation of host cell innate immune response [102]. Another study revealed that exposure of both human PBMC-derived dendritic cells (DCs) and murine bone marrow-derived DCs to H. pylori OMVs upregulated heme oxygenase-1 (HO-1) expression via the activation of Akt-Nrf2 and mTOR-NF-κB signaling pathways, potentially inhibiting the maturation of DCs to help regulate inflammatory responses during H. pylori infection [103]. In terms of T cells, H. pylori OMVs induced apoptosis in both Jurkat T cell line and native CD4+ T cells, suggesting a role for H. pylori OMVs in the repression of T cell immunity [102]. Consistently, a more recent study revealed that H. pylori OMVs indirectly inhibited T cell responses by inducing cyclooxygenase-2 expression in monocytes, thereby increasing levels of prostaglandin-E2 and IL-10 [104]. Taken together, these findings suggest that H. pylori OMVs are associated with gastric pathologies and the bacterium's persistence in the gastric mucosa through exerting immunomodulatory effects on immune cells within the lamina propria (Figure 2).
Extra-gastric pathogenic effects of H. pylori OMVs
Owing to their nanometric structure with the ability to serve as delivery vehicles for their cargo virulence factors, H. pylori-derived OMVs have increasingly been recognized as a mechanistic link between H. pylori infection and several extra-gastric diseases, such as liver fibrosis, Alzheimer's disease, and atherosclerosis [105-108].
In liver fibrosis, an in vitro study demonstrated that OMVs isolated from clinical strains of H. pylori can trigger the activation of hepatic stellate cells (HSCs) and enhance the expression of hepatic fibrosis markers (α-SMA, E-cadherin, vimentin, snail, and β-catenin) [106]. Additionally, exposure of HSCs to H. pylori OMVs was found to upregulate autophagy inhibitory makers (PI3K, AKT, and MTOR) while downregulating autophagy core proteins (BECN1, ATG16 and LC3B), suggesting that H. pylori OMV-induced autophagy inhibition in HSCs may contribute to the development of H. pylori-mediated liver fibrosis [109]. Furthermore, another study revealed that hepatocytes treated with H. pylori OMVs induced changes to the hepatocyte-derived exosomes, which promoted HSC activation and the progression of liver fibrosis [110] (Figure 3).
In Alzheimer's disease, an in vivo study demonstrated that OMVs isolated from H. pylori strain 26695 can cross the blood-brain barrier to reach the brain, where they were taken up by astrocytes and induced astrocyte reactivation followed by microglial activation via activation of the C3a-C3aR signaling pathway. Reactive phagocytic microglia then actively participated in the elimination of synapses and the phagocytosis of neurons, leading to synaptic deficits and excessive neuronal loss, exacerbating amyloid-β pathology and ultimately, resulting in cognitive decline [107]. Interestingly, another study indicated that H. pylori OMVs induced astrocyte reactivation, characterized by the production of the neurotoxic factor IFN-γ through activation of the NF-κB signaling pathway, thereby causing neuronal dysfunction including neurite retraction and increased neuronal death [111]. Furthermore, an in vitro study demonstrated that the STAT3 signaling pathway was involved in the detrimental effects of H. pylori OMVs on the brain, thus contributing to the development of Alzheimer's disease [112] (Figure 3).
Role of H. pylori OMVs in extra-gastric pathogenesis related to H. pylori infection (created with BioRender.com). A. Helicobacter pylori OMVs induce the activation of hepatic stellate cells (HSCs) and autophagy inhibition, or indirectly induce altered hepatocyte exosomes and then trigger HSC activation, promoting liver fibrosis. B. Helicobacter pylori OMVs induce astrocyte reactivity and then cause neuronal damage by activating C3/C3aR, NF-κB or STAT3 signaling pathways, thereby promoting Alzheimer's disease. C. Helicobacter pylori OMVs either directly impair endothelial function by activating the ROS/ NF-κB signaling, or indirectly stimulate endothelial cells to release CagA-containing exosomes to promote foam cell formation, ultimately accelerating atherosclerotic plaque formation.
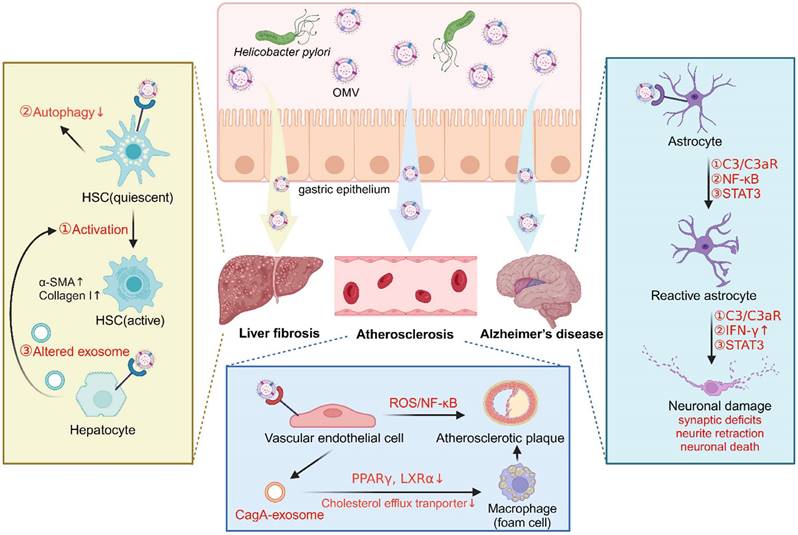
In atherosclerosis, a study reported that OMVs from H. pylori strain 26695 accelerated atherosclerotic plaque formation in ApoE -/- mice, and triggered apoptotic cell death of human umbilical vein endothelial cells (HUVECs) via activation of the ROS/NF-κB signaling pathway [108]. Notably, LPS and CagA were identified as two important contributors in these processes, as demonstrated by significantly ameliorated endothelial cell dysfunction upon treatment with LPS-depleting or CagA-negative H. pylori OMVs [108]. Interestingly, another study revealed that OMVs from H. pylori stain PMSS1 stimulated gastric epithelial cells to release CagA-containing exosomes into blood circulation, promoting macrophage-derived foam cell formation and augmenting atherosclerotic plaque growth and instability. Exosomal CagA was reported to downregulate the expression of transcription factors PPARγ and LXRα, leading to the dysfunction of the cholesterol efflux transporter. Consequently, the excessive accumulation of cholesterol in the macrophages induced foam cell formation, thus contributing to the progression of atherosclerosis [113] (Figure 3).
Collectively, H. pylori-derived OMVs represent nanocarriers of H. pylori virulence factors to both local or distant organs, playing a crucial role in the pathogenesis of H. pylori infection-related gastric and extra-gastric diseases.
Exploiting H. pylori OMVs as vaccines
Potential of H. pylori OMVs as vaccine candidates
OMVs share great similarity with their parental bacteria in immunomodulatory components such as lipids, lipoproteins, enzymes, peptidoglycan, and genetic materials (DNA, RNA), by which they can elicit strong humoral and cellular immune responses against pathogens [14, 114]. This highly immunogenic potential of bacteria-derived OMVs presents them as promising vaccine candidates against pathogen infection. Currently available OMV vaccines including VA-MENGOC-BC® (1987), MenBVac® (1991), and MeNZB™ (2004) were successfully developed to control meningococcal serogroup B outbreaks in Cuba, Norway and New Zealand, respectively [115-117]. Notably, the same OMV used to formulate MeNZB™ was also added to the recently developed MenB vaccine (Bexsero®) as an adjuvant in combination with three recombinant proteins (NHBA, NadA, fHBP) to help protect against meningococcal serogroup B infection [118, 119].
Despite more than 30 years of effort in the development of H. pylori vaccine, no effective vaccine against H. pylori has been developed yet. Lessons learned from the past failures will help in the identification of vaccine candidates with new approaches. The use of H. pylori OMVs as potential vaccine candidates may prove to be a promising strategy, as some preliminary cellular and animal studies have provided a basis for their potential protective effects. For instance, an in vitro study reported that OMVs from H. pylori strain SS1 were able to induce a T helper 2 (Th2) immune response, as demonstrated by high levels of IL-10 and IL-4 in macrophage RAW 264.7 cells. Importantly, compared with the total antigens isolated from H. pylori SS1, OMVs showed comparable immunogenic activity and better biocompatibility, hemocompatibility, and safety due to their lack of toxicity [120]. An in vivo study by Das et al. demonstrated that immunization of mice with OMVs from H. pylori strain A61C(1) elicited higher level of adaptive immune responses compared to the non-immunized group, which significantly lowered bacterial loads upon challenge with H. pylori strain SS1 post immunization [121]. In terms of immune responses, another study reported that IFN-γ and IL-17, representative cytokines for Th1 and Th17-mediated immune responses, respectively, were both significantly induced in the spleens of mice immunized with OMVs form H. pylori strain HP99 [17]. Regarding immunization routes, it was observed that intraperitoneal immunization of H. pylori OMVs stimulated even higher levels of immunoglobulin (IgG) compared to oral immunization [17]. Additionally, Liu et al. reported that oral immunization of mice with OMVs from H. pylori strain 7.13 induced stronger humoral and mucosal immune responses without inducing mucosal inflammation, compared with H. pylori whole cell immunization. This study revealed that H. pylori OMVs predominantly induced a Th2-biased immune response, dramatically lowering bacterial loads in a mouse model challenged with H. pylori strain SS1 [122]. Interestingly, when using OMVs from H. pylori strain 7.13 as adjuvants during the immunization of mice with 7.13 outer membrane proteins (OMPs) or whole cell vaccine (WCV), more humoral and mucosal immunity was elicited and gastric colonization of H. pylori in mice were significantly reduced when compared to the administrations of OMPs or WCV, alone or coupled with standard adjuvant cholera toxin (CT) [123].
In addition to its potential role as a vaccine, H. pylori OMVs may also help combat H. pylori infection by preventing bacterial adherence to host cells. OMVs mimic their parental bacteria and encapsulate various adhesins, enabling them to inhibit the adhesion of parental bacteria to host cells by competitively binding to the same target sites [124]. For instance, H. pylori OMV-coated nanoparticles have been reported to adhere to and block H. pylori adhesion to AGS cells in a dose-dependent manner. Furthermore, the adherence of these OMV-coated nanoparticles facilitated the detachment of adherent H. pylori from mouse stomach tissues [125]. The anti-bacterial adhesion efficacy of OMVs can be further enhanced through genetic engineering of bacteria to modulate adhesin expression on the OMVs and by selecting optimal nanoparticle cores with the desired characteristics [126].
Challenges in developing H. pylori OMVs as vaccines
Current vaccine development using H. pylori OMVs as targets faces many challenges including the optimization of OMVs isolation conditions including growth conditions and the selection of a well-characterized strain with an intricate balance between OMV content immunogenicity and toxicity [127]. For instance, bacterial LPS is a major content of OMVs in most gram-negative bacteria. It is a very potent activator of immune cells such as monocytes and macrophages, triggering NF-κB and IRF3-mediated pro-inflammatory responses via recognition of the lipid A moiety of LPS by the TLR4/MD2 receptor. This activation is important in initiating and directing the adaptive immune response, which is a highly important vaccine property for effective clearance of bacterial infections [128]. However, overly strong reactogenicity triggered by LPS may lead to septic shock. Thus, OMVs are usually prepared with detergent extraction to reduce endotoxin levels, but with some limitations including potential toxicity from residual detergent particles and the unintentional removal of essential LPS-bound OMPs required for immunogenic activity [129]. Hence, the use of mutants with defects in lipid A biosynthesis seems to provide a balanced response, generating sufficient adjuvant activity while preventing unwanted effects limiting the vaccine efficacy [24]. For instance, N. meningitidis ΔlpxL mutant was reported to produce LPS with penta- instead of hexa-acylated lipid A, retaining adjuvant activity but with reduced toxicity [130]. Unlike other gram-negative bacteria expressing bi-phosphorylated and hexa-acylated LPS, H. pylori constitutively modifies its LPS into an under-acylated and dephosphorylated lipid A, effectively reducing its reactogenicity [131, 132]. In vivo studies using mice and rabbits have shown that H. pylori LPS exhibits 500 to 1000 times lower levels of cytotoxicity, mitogenicity, and pyrogenicity as compared to that of other gram-negative pathogens, such as E. coli and S. typhimurium [133]. The constitutively modified LPS structure in H. pylori may alleviate the need for detergent extraction or genetic modification of lipid A. However, genetic engineering of H. pylori to remove LPS Lewis antigen is required in the development of H. pylori OMV vaccines. This is due to the fact that the O-antigen of H. pylori LPS usually contains Lewis antigens that resemble human Lewis molecules and blood group antigens, resulting in the potential development of autoimmunity if present [51, 134, 135].
In addition to LPS, other well-known virulence factors present in H. pylori OMVs including CagA oncoprotein, VacA vacuolating cytotoxin, and HtrA serine protease (also a known gastric cancer risk factor) also pose a big safety concern for vaccine development [71, 100, 136]. One possible option would be to create a genetically-modified strain lacking CagA, VacA, and HtrA virulence factors. However, a drawback of this option may be the reduction or loss of immunogenicity attributed to these virulence factors, resulting in suboptimal immune response. This warrant further investigations. An alternative approach is to select strains containing non-toxigenic virulence factor genotypes or to genetically modify the corresponding genes to generate detoxified or non-virulent subtypes while retaining their immunogenicity. For instance, strains with the VacA subtype s1/m1 exhibit high cytotoxic activity, while those with the subtype s2/m2 show no cytotoxicity [137]. This variability necessitates careful selection or genetic modification of non-toxic VacA variants, such as s2/m2, for safe vaccine formulations. Similar approaches can be applied to other virulence factors including CagA and HtrA.
Summary
In light of the data above, H. pylori OMVs play an important role in bacterial colonization, survival and pathogenesis. Due to their nanosized structure, H. pylori OMVs can be internalized and translocated across the gastric mucosal epithelial layer to reach the underlying blood circulation system and then migrate to distant cells and organs, causing a plethora of cellular responses and that may explain the association of H. pylori infection with extra-gastric diseases. Moreover, the immunogenic, non-replicable, and anti-bacterial adhesion properties of H. pylori OMVs make them a desirable vaccine candidate against infection. However, there are many challenges in the development of H. pylori OMVs-based vaccines including safety concerns and how to achieve an intricate balance between OMV content immunogenicity and reactogenicity/toxicity. Helicobacter pylori OMVs are a double-edged sword. While they may contribute to disease pathogenesis by causing inflammatory tissue damage, they may also hold therapeutic potential in the form of vaccine to help protect against H. pylori-related diseases. Future experimental studies are needed for a comprehensive analysis of H. pylori OMVs biogenesis and biological functions.
Abbreviations
MALT: mucosa-associated lymphoid tissue; WHO: world Health Organization; OMVs: outer membrane vesicles; OM: outer membrane; Lpp: lipoprotein; OmpA: outer membrane protein A; LPS: lipopolysaccharide; PQS: pseudomonas quinolone signal; PL: phospholipids; IM: inner membrane; PE: phosphatidylethanolamine; CGs: cholesteryl glucosides; PG: phosphatidylglycerol; PC: phosphatidylcholine; Lea: Lewis a; MS: mass spectrometry; T4SS: type IV secretion system; eDNA: extracellular DNA; sncRNA: small non-coding RNA; bOMVs: biofilm OMVs; pOMVs: planktonic OMVs; α-CA: α-class carbonic anhydrase; PBMCs: peripheral blood mononuclear cells; DCs: dendritic cells; HO-1: heme oxygenase-1; HSCs: hepatic stellate cells; Th2: T helper 2; OMPs: outer membrane proteins; WCV: whole cell vaccine.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant 82072248), the International Cooperation Excellence Initiative Grant, West China Hospital, Sichuan University (grant 139220012). The authors acknowledge the use of Figdraw (created Figure 1) and Biorender (created Graphic abstract, Figure 2, and Figure 3).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mezmale L, Coelho LG, Bordin D, Leja M. Review: Epidemiology of Helicobacter pylori. Helicobacter. 2020;25(Suppl 1):e12734
2. Ansari S, Yamaoka Y. Helicobacter pylori Virulence Factors Exploiting Gastric Colonization and its Pathogenicity. Toxins (Basel). 2019;11:677
3. Topi S, Santacroce L, Bottalico L, Ballini A, Inchingolo AD, Dipalma G. et al. Gastric Cancer in History: A Perspective Interdisciplinary Study. Cancers (Basel). 2020;12:264
4. Navashenaq JG, Shabgah AG, Banach M, Jamialahmadi T, Penson PE, Johnston TP. et al. The interaction of Helicobacter pylori with cancer immunomodulatory stromal cells: New insight into gastric cancer pathogenesis. Semin Cancer Biol. 2021;86:951-9
5. Jamkhande PG, Gattani SG, Farhat SA. Helicobacter pylori and cardiovascular complications: a mechanism based review on role of Helicobacter pylori in cardiovascular diseases. Integr Med Res. 2016;5:244-9
6. Polyzos SA, Papaefthymiou A, Doulberis M, Mavridoglou G, Kountouras J. Helicobacter pylori infection and diabetes mellitus. Diabetes Metab Syndr. 2021;15:845-6
7. Mohammadi M, Attar A, Mohammadbeigi M, Peymani A, Bolori S, Fardsanei F. The possible role of Helicobacter pylori in liver diseases. Arch Microbiol. 2023;205:281
8. Doulberis M, Kotronis G, Gialamprinou D, Polyzos SA, Papaefthymiou A, Katsinelos P. et al. Alzheimer's disease and gastrointestinal microbiota; impact of Helicobacter pylori infection involvement. Int J Neurosci. 2021;131:289-301
9. Parker H, Keenan JI. Composition and function of Helicobacter pylori outer membrane vesicles. Microbes Infect. 2012;14:9-16
10. Oh JD, Karam SM, Gordon JI. Intracellular Helicobacter pylori in gastric epithelial progenitors. Proc Natl Acad Sci U S A. 2005;102:5186-91
11. Xia X, Zhang L, Chi J, Li H, Liu X, Hu T. et al. Helicobacter pylori Infection Impairs Endothelial Function Through an Exosome-Mediated Mechanism. J Am Heart Assoc. 2020;9:e014120
12. Aytar Celik P, Derkus B, Erdogan K, Barut D, Blaise Manga E, Yildirim Y. et al. Bacterial membrane vesicle functions, laboratory methods, and applications. Biotechnol Adv. 2022;54:107869
13. Stentz R, Carvalho AL, Jones EJ, Carding SR. Fantastic voyage: the journey of intestinal microbiota-derived microvesicles through the body. Biochem Soc Trans. 2018;46:1021-7
14. Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. 2015;13:605-19
15. Snider CA, Voss BJ, McDonald WH, Cover TL. Growth phase-dependent composition of the Helicobacter pylori exoproteome. J Proteomics. 2016;130:94-107
16. Melo J, Pinto V, Fernandes T, Malheiro AR, Osorio H, Figueiredo C. et al. Isolation Method and Characterization of Outer Membranes Vesicles of Helicobacter pylori Grown in a Chemically Defined Medium. Front Microbiol. 2021;12:654193
17. Choi HI, Choi JP, Seo J, Kim BJ, Rho M, Han JK. et al. Helicobacter pylori-derived extracellular vesicles increased in the gastric juices of gastric adenocarcinoma patients and induced inflammation mainly via specific targeting of gastric epithelial cells. Exp Mol Med. 2017;49:e330
18. Melo J, Cavadas B, Pereira L, Figueiredo C, Leite M. Transcriptomic remodeling of gastric cells by Helicobacter pylori outer membrane vesicles. Helicobacter. 2023;29:e13031
19. Park AM, Tsunoda I. Helicobacter pylori infection in the stomach induces neuroinflammation: the potential roles of bacterial outer membrane vesicles in an animal model of Alzheimer's disease. Inflamm Regen. 2022;42:39
20. Domingues S, Nielsen KM. Membrane vesicles and horizontal gene transfer in prokaryotes. Curr Opin Microbiol. 2017;38:16-21
21. Tiku V, Tan MW. Host immunity and cellular responses to bacterial outer membrane vesicles. Trends Immunol. 2021;42:1024-36
22. Ahmed AAQ, Besio R, Xiao L, Forlino A. Outer Membrane Vesicles (OMVs) as Biomedical Tools and Their Relevance as Immune-Modulating Agents against H. pylori Infections: Current Status and Future Prospects. Int J Mol Sci. 2023;24:8542
23. Semchenko EA, Seib KL. Outer membrane vesicle vaccines for Neisseria gonorrhoeae. Nat Rev Urol. 2022;19:5-6
24. Tan K, Li R, Huang X, Liu Q. Outer Membrane Vesicles: Current Status and Future Direction of These Novel Vaccine Adjuvants. Front Microbiol. 2018;9:783
25. Furuyama N, Sircili MP. Outer Membrane Vesicles (OMVs) Produced by Gram-Negative Bacteria: Structure, Functions, Biogenesis, and Vaccine Application. Biomed Res Int. 2021;2021:1490732
26. Schwechheimer C, Rodriguez DL, Kuehn MJ. NlpI-mediated modulation of outer membrane vesicle production through peptidoglycan dynamics in Escherichia coli. Microbiologyopen. 2015;4:375-89
27. Moon DC, Choi CH, Lee JH, Choi CW, Kim HY, Park JS. et al. Acinetobacter baumannii outer membrane protein A modulates the biogenesis of outer membrane vesicles. J Microbiol. 2012;50:155-60
28. Sonntag I, Schwarz H, Hirota Y, Henning U. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J Bacteriol. 1978;136:280-5
29. Bernadac A, Gavioli M, Lazzaroni JC, Raina S, Lloubes R. Escherichia coli tol-pal mutants form outer membrane vesicles. J Bacteriol. 1998;180:4872-8
30. Li Q, Li Z, Fei X, Tian Y, Zhou G, Hu Y. et al. The role of TolA, TolB, and TolR in cell morphology, OMVs production, and virulence of Salmonella Choleraesuis. AMB Express. 2022;12:5
31. Reimer SL, Beniac DR, Hiebert SL, Booth TF, Chong PM, Westmacott GR. et al. Comparative Analysis of Outer Membrane Vesicle Isolation Methods With an Escherichia coli tolA Mutant Reveals a Hypervesiculating Phenotype With Outer-Inner Membrane Vesicle Content. Front Microbiol. 2021;12:628801
32. Pastor Y, Camacho AI, Zuniga-Ripa A, Merchan A, Rosas P, Irache JM. et al. Towards a subunit vaccine from a Shigella flexneri DeltatolR mutant. Vaccine. 2018;36:7509-19
33. de Jonge EF, van Boxtel R, Balhuizen MD, Haagsman HP, Tommassen J. Pal depletion results in hypervesiculation and affects cell morphology and outer-membrane lipid asymmetry in bordetellae. Res Microbiol. 2022;173:103937
34. Zhou L, Srisatjaluk R, Justus DE, Doyle RJ. On the origin of membrane vesicles in gram-negative bacteria. FEMS Microbiol Lett. 1998;163:223-8
35. Hayashi J, Hamada N, Kuramitsu HK. The autolysin of Porphyromonas gingivalis is involved in outer membrane vesicle release. FEMS Microbiol Lett. 2002;216:217-22
36. McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 2007;63:545-58
37. Ojima Y, Mohanadas T, Kitamura K, Nunogami S, Yajima R, Taya M. Deletion of degQ gene enhances outer membrane vesicle production of Shewanella oneidensis cells. Arch Microbiol. 2017;199:415-23
38. Li H, Yang T, Liao T, Debowski AW, Nilsson HO, Fulurija A. et al. The redefinition of Helicobacter pylori lipopolysaccharide O-antigen and core-oligosaccharide domains. PLoS Pathog. 2017;13:e1006280
39. Lam NH, Ha BY. Surface-lattice model describes electrostatic interactions of ions and polycations with bacterial lipopolysaccharides: ion valence and polycation's excluded area. Langmuir. 2014;30:13631-40
40. Lugtenberg B, Van Alphen L. Molecular architecture and functioning of the outer membrane of Escherichia coli and other gram-negative bacteria. Biochim Biophys Acta. 1983;737:51-115
41. Kadurugamuwa JL, Beveridge TJ. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol. 1995;177:3998-4008
42. Sabra W, Lunsdorf H, Zeng AP. Alterations in the formation of lipopolysaccharide and membrane vesicles on the surface of Pseudomonas aeruginosa PAO1 under oxygen stress conditions. Microbiology (Reading). 2003;149:2789-95
43. Haurat MF, Aduse-Opoku J, Rangarajan M, Dorobantu L, Gray MR, Curtis MA. et al. Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem. 2011;286:1269-76
44. Florez C, Raab JE, Cooke AC, Schertzer JW. Membrane Distribution of the Pseudomonas Quinolone Signal Modulates Outer Membrane Vesicle Production in Pseudomonas aeruginosa. mBio. 2017;8:e01034-17
45. Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437:422-5
46. Mashburn-Warren L, Howe J, Garidel P, Richter W, Steiniger F, Roessle M. et al. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol Microbiol. 2008;69:491-502
47. Schertzer JW, Whiteley M. A bilayer-couple model of bacterial outer membrane vesicle biogenesis. mBio. 2012;3:e00297-11
48. Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, Eichmann TO. et al. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat Commun. 2016;7:10515
49. Elhenawy W, Bording-Jorgensen M, Valguarnera E, Haurat MF, Wine E, Feldman MF. LPS Remodeling Triggers Formation of Outer Membrane Vesicles in Salmonella. mBio. 2016;7:e00940-16
50. Liu AN, Teng KW, Chew Y, Wang PC, Nguyen TTH, Kao MC. The Effects of HP0044 and HP1275 Knockout Mutations on the Structure and Function of Lipopolysaccharide in Helicobacter pylori Strain 26695. Biomedicines. 2022;10:145
51. Chiu SF, Teng KW, Wang PC, Chung HY, Wang CJ, Cheng HC. et al. Helicobacter pylori GmhB enzyme involved in ADP-heptose biosynthesis pathway is essential for lipopolysaccharide biosynthesis and bacterial virulence. Virulence. 2021;12:1610-28
52. Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163-84
53. Simpson BW, Trent MS. Pushing the envelope: LPS modifications and their consequences. Nat Rev Microbiol. 2019;17:403-16
54. Hirai Y, Haque M, Yoshida T, Yokota K, Yasuda T, Oguma K. Unique cholesteryl glucosides in Helicobacter pylori: composition and structural analysis. J Bacteriol. 1995;177:5327-33
55. Olofsson A, Vallstrom A, Petzold K, Tegtmeyer N, Schleucher J, Carlsson S. et al. Biochemical and functional characterization of Helicobacter pylori vesicles. Mol Microbiol. 2010;77:1539-55
56. Yu YJ, Wang XH, Fan GC. Versatile effects of bacterium-released membrane vesicles on mammalian cells and infectious/inflammatory diseases. Acta Pharmacol Sin. 2018;39:514-33
57. Vanaja SK, Russo AJ, Behl B, Banerjee I, Yankova M, Deshmukh SD. et al. Bacterial Outer Membrane Vesicles Mediate Cytosolic Localization of LPS and Caspase-11 Activation. Cell. 2016;165:1106-19
58. Moran AP. Relevance of fucosylation and Lewis antigen expression in the bacterial gastroduodenal pathogen Helicobacter pylori. Carbohydr Res. 2008;343:1952-65
59. Keenan JI, Davis KA, Beaugie CR, McGovern JJ, Moran AP. Alterations in Helicobacter pylori outer membrane and outer membrane vesicle-associated lipopolysaccharides under iron-limiting growth conditions. Innate Immun. 2008;14:279-90
60. Mullaney E, Brown PA, Smith SM, Botting CH, Yamaoka YY, Terres AM. et al. Proteomic and functional characterization of the outer membrane vesicles from the gastric pathogen Helicobacter pylori. Proteomics Clin Appl. 2009;3:785-96
61. Wei S, Li X, Wang J, Wang Y, Zhang C, Dai S. et al. Outer Membrane Vesicles Secreted by Helicobacter pylori Transmitting Gastric Pathogenic Virulence Factors. ACS Omega. 2022;7:240-58
62. Bitto NJ, Zavan L, Johnston EL, Stinear TP, Hill AF, Kaparakis-Liaskos M. Considerations for the Analysis of Bacterial Membrane Vesicles: Methods of Vesicle Production and Quantification Can Influence Biological and Experimental Outcomes. Microbiol Spectr. 2021;9:e0127321
63. Turner L, Bitto NJ, Steer DL, Lo C, D'Costa K, Ramm G. et al. Helicobacter pylori Outer Membrane Vesicle Size Determines Their Mechanisms of Host Cell Entry and Protein Content. Front Immunol. 2018;9:1466
64. Chauhan N, Tay ACY, Marshall BJ, Jain U. Helicobacter pylori VacA, a distinct toxin exerts diverse functionalities in numerous cells: An overview. Helicobacter. 2019;24:e12544
65. Genisset C, Puhar A, Calore F, de Bernard M, Dell'Antone P, Montecucco C. The concerted action of the Helicobacter pylori cytotoxin VacA and of the v-ATPase proton pump induces swelling of isolated endosomes. Cell Microbiol. 2007;9:1481-90
66. McClain MS, Beckett AC, Cover TL. Helicobacter pylori Vacuolating Toxin and Gastric Cancer. Toxins (Basel). 2017;9:316
67. Fiocca R, Necchi V, Sommi P, Ricci V, Telford J, Cover TL. et al. Release of Helicobacter pylori vacuolating cytotoxin by both a specific secretion pathway and budding of outer membrane vesicles. Uptake of released toxin and vesicles by gastric epithelium. J Pathol. 1999;188:220-6
68. Ricci V, Chiozzi V, Necchi V, Oldani A, Romano M, Solcia E. et al. Free-soluble and outer membrane vesicle-associated VacA from Helicobacter pylori: Two forms of release, a different activity. Biochemical and Biophysical Research Communications. 2005;337:173-8
69. Sukri A, Hanafiah A, Kosai NR, Mohammed Taher M, Mohamed R. New insight on the role of Helicobacter pylori cagA in the expression of cell surface antigens with important biological functions in gastric carcinogenesis. Helicobacter. 2022;27:e12913
70. Tegtmeyer N, Wessler S, Necchi V, Rohde M, Harrer A, Rau TT. et al. Helicobacter pylori Employs a Unique Basolateral Type IV Secretion Mechanism for CagA Delivery. Cell Host Microbe. 2017;22:552-60 e5
71. Chew Y, Chung HY, Lin PY, Wu DC, Huang SK, Kao MC. Outer Membrane Vesicle Production by Helicobacter pylori Represents an Approach for the Delivery of Virulence Factors CagA, VacA and UreA into Human Gastric Adenocarcinoma (AGS) Cells. Int J Mol Sci. 2021;22:3942
72. Lekmeechai S, Su YC, Brant M, Alvarado-Kristensson M, Vallstrom A, Obi I. et al. Helicobacter pylori Outer Membrane Vesicles Protect the Pathogen From Reactive Oxygen Species of the Respiratory Burst. Front Microbiol. 2018;9:1837
73. Loharch S, Berlicki L. Rational Development of Bacterial Ureases Inhibitors. Chem Rec. 2022;22:e202200026
74. Kaparakis M, Turnbull L, Carneiro L, Firth S, Coleman HA, Parkington HC. et al. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell Microbiol. 2010;12:372-85
75. Koeppen K, Hampton TH, Jarek M, Scharfe M, Gerber SA, Mielcarz DW. et al. A Novel Mechanism of Host-Pathogen Interaction through sRNA in Bacterial Outer Membrane Vesicles. PLoS Pathog. 2016;12:e1005672
76. Grande R, Di Marcantonio MC, Robuffo I, Pompilio A, Celia C, Di Marzio L. et al. Helicobacter pylori ATCC 43629/NCTC 11639 Outer Membrane Vesicles (OMVs) from Biofilm and Planktonic Phase Associated with Extracellular DNA (eDNA). Front Microbiol. 2015;6:1369
77. Zhang H, Zhang Y, Song Z, Li R, Ruan H, Liu Q. et al. sncRNAs packaged by Helicobacter pylori outer membrane vesicles attenuate IL-8 secretion in human cells. Int J Med Microbiol. 2020;310:151356
78. Pinho AS, Seabra CL, Nunes C, Reis S, MC LM, Parreira P. Helicobacter pylori biofilms are disrupted by nanostructured lipid carriers: A path to eradication? J Control Release. 2022;348:489-98
79. Jeong GJ, Khan F, Tabassum N, Cho KJ, Kim YM. Bacterial extracellular vesicles: Modulation of biofilm and virulence properties. Acta Biomater. 2024;178:13-23
80. Zou Y, Chen X, Sun Y, Li P, Xu M, Fang P. et al. Antibiotics-free nanoparticles eradicate Helicobacter pylori biofilms and intracellular bacteria. J Control Release. 2022;348:370-85
81. Yonezawa H, Osaki T, Kurata S, Fukuda M, Kawakami H, Ochiai K. et al. Outer membrane vesicles of Helicobacter pylori TK1402 are involved in biofilm formation. BMC Microbiol. 2009;9:197
82. Yonezawa H, Osaki T, Fukutomi T, Hanawa T, Kurata S, Zaman C. et al. Diversification of the AlpB Outer Membrane Protein of Helicobacter pylori Affects Biofilm Formation and Cellular Adhesion. J Bacteriol. 2017;199:e00729-16
83. Yonezawa H, Osaki T, Woo T, Kurata S, Zaman C, Hojo F. et al. Analysis of outer membrane vesicle protein involved in biofilm formation of Helicobacter pylori. Anaerobe. 2011;17:388-90
84. Ronci M, Del Prete S, Puca V, Carradori S, Carginale V, Muraro R. et al. Identification and characterization of the alpha-CA in the outer membrane vesicles produced by Helicobacter pylori. J Enzyme Inhib Med Chem. 2019;34:189-95
85. Grande R, Carradori S, Puca V, Vitale I, Angeli A, Nocentini A. et al. Selective Inhibition of Helicobacter pylori Carbonic Anhydrases by Carvacrol and Thymol Could Impair Biofilm Production and the Release of Outer Membrane Vesicles. Int J Mol Sci. 2021;22:11583
86. Krzyzek P, Grande R, Migdal P, Paluch E, Gosciniak G. Biofilm Formation as a Complex Result of Virulence and Adaptive Responses of Helicobacter pylori. Pathogens. 2020;9:1062
87. Manning AJ, Kuehn MJ. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 2011;11:258
88. Kulkarni HM, Swamy Ch V, Jagannadham MV. Molecular characterization and functional analysis of outer membrane vesicles from the antarctic bacterium Pseudomonas syringae suggest a possible response to environmental conditions. J Proteome Res. 2014;13:1345-58
89. Murray BO, Dawson RA, Alsharaf LM, Anne Winter J. Protective effects of Helicobacter pylori membrane vesicles against stress and antimicrobial agents. Microbiology (Reading). 2020;166:751-8
90. Heczko U, Smith VC, Mark Meloche R, Buchan AM, Finlay BB. Characteristics of Helicobacter pylori attachment to human primary antral epithelial cells. Microbes Infect. 2000;2:1669-76
91. Khan A, Sardar A, Tarafdar PK, Mallick AI. Heterogeneity and Compositional Diversities of Campylobacter jejuni Outer Membrane Vesicles (OMVs) Drive Multiple Cellular Uptake Processes. ACS Infect Dis. 2023;9:2325-39
92. Olofsson A, Nygard Skalman L, Obi I, Lundmark R, Arnqvist A. Uptake of Helicobacter pylori vesicles is facilitated by clathrin-dependent and clathrin-independent endocytic pathways. mBio. 2014;5:e00979-14
93. Lee JH, Jun SH, Kim JM, Baik SC, Lee JC. Morphological changes in human gastric epithelial cells induced by nuclear targeting of Helicobacter pylori urease subunit A. J Microbiol. 2015;53:406-14
94. Kaksonen M, Roux A. Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2018;19:313-26
95. Parker H, Chitcholtan K, Hampton MB, Keenan JI. Uptake of Helicobacter pylori outer membrane vesicles by gastric epithelial cells. Infect Immun. 2010;78:5054-61
96. Rennick JJ, Johnston APR, Parton RG. Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nature Nanotechnology. 2021;16:266-76
97. Ismail S, Hampton MB, Keenan JI. Helicobacter pylori outer membrane vesicles modulate proliferation and interleukin-8 production by gastric epithelial cells. Infect Immun. 2003;71:5670-5
98. Ikuse T, Blanchard TG, Czinn SJ. Inflammation, Immunity, and Vaccine Development for the Gastric Pathogen Helicobacter pylori. Curr Top Microbiol Immunol. 2019;421:1-19
99. Ayala G, Torres L, Espinosa M, Fierros-Zarate G, Maldonado V, Melendez-Zajgla J. External membrane vesicles from Helicobacter pylori induce apoptosis in gastric epithelial cells. FEMS Microbiol Lett. 2006;260:178-85
100. Chitcholtan K, Hampton MB, Keenan JI. Outer membrane vesicles enhance the carcinogenic potential of Helicobacter pylori. Carcinogenesis. 2008;29:2400-5
101. Ko SH, Jeon JI, Kim YJ, Yoon HJ, Kim H, Kim N. et al. Helicobacter pylori outer membrane vesicle proteins induce human eosinophil degranulation via a beta2 Integrin CD11/CD18- and ICAM-1-dependent mechanism. Mediators Inflamm. 2015;2015:301716
102. Winter J, Letley D, Rhead J, Atherton J, Robinson K. Helicobacter pylori membrane vesicles stimulate innate pro- and anti-inflammatory responses and induce apoptosis in Jurkat T cells. Infect Immun. 2014;82:1372-81
103. Ko SH, Rho DJ, Jeon JI, Kim YJ, Woo HA, Kim N. et al. Crude Preparations of Helicobacter pylori Outer Membrane Vesicles Induce Upregulation of Heme Oxygenase-1 via Activating Akt-Nrf2 and mTOR-IkappaB Kinase-NF-kappaB Pathways in Dendritic Cells. Infect Immun. 2016;84:2162-74
104. Hock BD, McKenzie JL, Keenan JI. Helicobacter pylori outer membrane vesicles inhibit human T cell responses via induction of monocyte COX-2 expression. Pathog Dis. 2017 75
105. Wang C, Li W, Shao L, Zhou A, Zhao M, Li P. et al. Both extracellular vesicles from helicobacter pylori-infected cells and helicobacter pylori outer membrane vesicles are involved in gastric/extragastric diseases. Eur J Med Res. 2023;28:484
106. Bolori S, Shegefti S, Baghaei K, Yadegar A, Moon KM, Foster LJ. et al. The Effects of Helicobacter pylori-Derived Outer Membrane Vesicles on Hepatic Stellate Cell Activation and Liver Fibrosis In vitro. Biomed Res Int. 2023;2023:4848643
107. Xie J, Cools L, Van Imschoot G, Van Wonterghem E, Pauwels MJ, Vlaeminck I. et al. Helicobacter pylori-derived outer membrane vesicles contribute to Alzheimer's disease pathogenesis via C3-C3aR signalling. J Extracell Vesicles. 2023;12:e12306
108. Wang N, Zhou F, Chen C, Luo H, Guo J, Wang W. et al. Role of Outer Membrane Vesicles From Helicobacter pylori in Atherosclerosis. Front Cell Dev Biol. 2021;9:673993
109. Shegefti S, Bolori S, Nabavi-Rad A, Dabiri H, Yadegar A, Baghaei K. Helicobacter pylori-derived outer membrane vesicles suppress liver autophagy: A novel mechanism for H. pylori-mediated hepatic disorder. Microb Pathog. 2023;183:106319
110. Zahmatkesh ME, Jahanbakhsh M, Hoseini N, Shegefti S, Peymani A, Dabin H. et al. Effects of Exosomes Derived From Helicobacter pylori Outer Membrane Vesicle-Infected Hepatocytes on Hepatic Stellate Cell Activation and Liver Fibrosis Induction. Front Cell Infect Microbiol. 2022;12:857570
111. Palacios E, Lobos-Gonzalez L, Guerrero S, Kogan MJ, Shao B, Heinecke JW. et al. Helicobacter pylori outer membrane vesicles induce astrocyte reactivity through nuclear factor-kappaappa B activation and cause neuronal damage in vivo in a murine model. J Neuroinflammation. 2023;20:66
112. Kandpal M, Baral B, Varshney N, Jain AK, Chatterji D, Meena AK. et al. Gut-Brain Axis Interplay via STAT3 pathway: Implications of Helicobacter pylori derived secretome on inflammation and Alzheimer's disease. Virulence. 2024;15:2303853
113. Yang S, Xia YP, Luo XY, Chen SL, Li BW, Ye ZM. et al. Exosomal CagA derived from Helicobacter pylori-infected gastric epithelial cells induces macrophage foam cell formation and promotes atherosclerosis. J Mol Cell Cardiol. 2019;135:40-51
114. Xie J, Li Q, Haesebrouck F, Van Hoecke L, Vandenbroucke RE. The tremendous biomedical potential of bacterial extracellular vesicles. Trends Biotechnol. 2022;40:1173-1194
115. Bjune G, Hoiby EA, Gronnesby JK, Arnesen O, Fredriksen JH, Halstensen A. et al. Effect of outer membrane vesicle vaccine against group B meningococcal disease in Norway. Lancet. 1991;338:1093-6
116. Sierra GV, Campa HC, Varcacel NM, Garcia IL, Izquierdo PL, Sotolongo PF. et al. Vaccine against group B Neisseria meningitidis: protection trial and mass vaccination results in Cuba. NIPH Ann. 1991;14:195-207 discussion 8-10
117. Loring BJ, Turner N, Petousis-Harris H. MeNZB vaccine and epidemic control: when do you stop vaccinating? Vaccine. 2008;26:5899-904
118. Fredriksen JH, Rosenqvist E, Wedege E, Bryn K, Bjune G, Froholm LO. et al. Production, characterization and control of MenB-vaccine "Folkehelsa": an outer membrane vesicle vaccine against group B meningococcal disease. NIPH Ann. 1991;14:67-79 discussion -80
119. Vernikos G, Medini D. Bexsero(R) chronicle. Pathog Glob Health. 2014;108:305-16
120. Ahmed AAQ, Qi F, Zheng R, Xiao L, Abdalla AME, Mao L. et al. The impact of ExHp-CD (outer membrane vesicles) released from Helicobacter pylori SS1 on macrophage RAW 264.7 cells and their immunogenic potential. Life Sci. 2021;279:119644
121. Das S, Halder P, Banerjee S, Mukhopadhyay AK, Dutta S, Koley H. Establishment of an intragastric surgical model using C57BL/6 mice to study the vaccine efficacy of OMV-based immunogens against Helicobacter pylori. Biol Open. 2024: bio.060282.
122. Liu Q, Li X, Zhang Y, Song Z, Li R, Ruan H. et al. Orally-administered outer-membrane vesicles from Helicobacter pylori reduce H. pylori infection via Th2-biased immune responses in mice. Pathog Dis. 2019;77:ftz050
123. Song Z, Li B, Zhang Y, Li R, Ruan H, Wu J. et al. Outer Membrane Vesicles of Helicobacter pylori 7.13 as Adjuvants Promote Protective Efficacy Against Helicobacter pylori Infection. Front Microbiol. 2020;11:1340
124. Zhang Y, Chen Y, Lo C, Zhuang J, Angsantikul P, Zhang Q. et al. Inhibition of Pathogen Adhesion by Bacterial Outer Membrane-Coated Nanoparticles. Angew Chem Int Ed Engl. 2019;58:11404-8
125. Berne C, Ellison CK, Ducret A, Brun YV. Bacterial adhesion at the single-cell level. Nat Rev Microbiol. 2018;16:616-27
126. Gasperini G, Biagini M, Arato V, Gianfaldoni C, Vadi A, Norais N. et al. Outer Membrane Vesicles (OMV)-based and Proteomics-driven Antigen Selection Identifies Novel Factors Contributing to Bordetella pertussis Adhesion to Epithelial Cells. Mol Cell Proteomics. 2018;17:205-15
127. Johnston EL, Guy-Von Stieglitz S, Zavan L, Cross J, Greening DW, Hill AF. et al. The effect of altered pH growth conditions on the production, composition, and proteomes of Helicobacter pylori outer membrane vesicles. Proteomics. 2023;24:e2300269
128. Maeshima N, Fernandez RC. Recognition of lipid A variants by the TLR4-MD-2 receptor complex. Front Cell Infect Microbiol. 2013;3:3
129. Mamat U, Wilke K, Bramhill D, Schromm AB, Lindner B, Kohl TA. et al. Detoxifying Escherichia coli for endotoxin-free production of recombinant proteins. Microb Cell Fact. 2015;14:57
130. van der Ley P, Steeghs L, Hamstra HJ, ten Hove J, Zomer B, van Alphen L. Modification of lipid A biosynthesis in Neisseria meningitidis lpxL mutants: influence on lipopolysaccharide structure, toxicity, and adjuvant activity. Infect Immun. 2001;69:5981-90
131. Cullen TW, Giles DK, Wolf LN, Ecobichon C, Boneca IG, Trent MS. Helicobacter pylori versus the host: remodeling of the bacterial outer membrane is required for survival in the gastric mucosa. PLoS Pathog. 2011;7:e1002454
132. Matsuura M. Structural Modifications of Bacterial Lipopolysaccharide that Facilitate Gram-Negative Bacteria Evasion of Host Innate Immunity. Front Immunol. 2013;4:109
133. Muotiala A, Helander IM, Pyhala L, Kosunen TU, Moran AP. Low biological activity of Helicobacter pylori lipopolysaccharide. Infect Immun. 1992;60:1714-6
134. Sheu BS, Odenbreit S, Hung KH, Liu CP, Sheu SM, Yang HB. et al. Interaction between host gastric Sialyl-Lewis X and H. pylori SabA enhances H. pylori density in patients lacking gastric Lewis B antigen. Am J Gastroenterol. 2006;101:36-44
135. Sijmons D, Guy AJ, Walduck AK, Ramsland PA. Helicobacter pylori and the Role of Lipopolysaccharide Variation in Innate Immune Evasion. Front Immunol. 2022;13:868225
136. Sharafutdinov I, Tegtmeyer N, Linz B, Rohde M, Vieth M, Tay AC. et al. A single-nucleotide polymorphism in Helicobacter pylori promotes gastric cancer development. Cell Host Microbe. 2023;31:1345-58 e6
137. Palframan SL, Kwok T, Gabriel K. Vacuolating cytotoxin A (VacA), a key toxin for Helicobacter pylori pathogenesis. Front Cell Infect Microbiol. 2012;2:92
Author contact
![]() Corresponding authors: Hong Tang, htang6198com; Hong Li, E-mail addresses: lihongcn.
Corresponding authors: Hong Tang, htang6198com; Hong Li, E-mail addresses: lihongcn.

 Global reach, higher impact
Global reach, higher impact