10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2025; 21(8):3351-3378. doi:10.7150/ijbs.110447 This issue Cite
Review
HSF1 Activation Mechanisms, Disease Roles, and Small Molecule Therapeutics
1. Ministry of Education Key Laboratory of Molecular and Cellular Biology; Hebei Research Center of the Basic Discipline of Cell Biology, Hebei Collaborative Innovation Center for Eco-Environment, Hebei Province Key Laboratory of Animal Physiology, Biochemistry and Molecular Biology, College of Life Sciences, Hebei Normal University, Shijiazhuang, 050024, China.
2. Key Laboratory of Systems Biomedicine (Ministry of Education), Shanghai Center for Systems Biomedicine, Shanghai Jiao Tong University, Shanghai, 200240, China.
Received 2025-1-14; Accepted 2025-4-14; Published 2025-4-28
Abstract
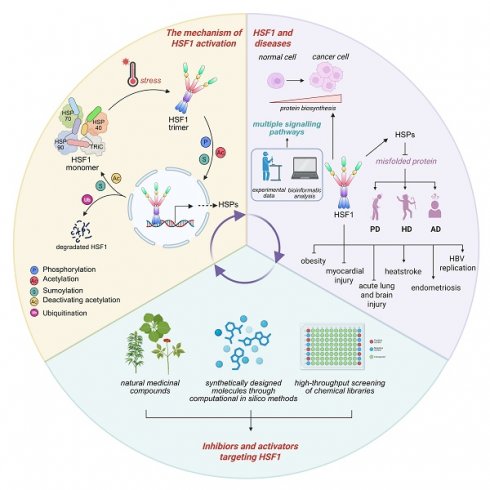
The heat shock factor 1 (HSF1) is a master transcription regulator that orchestrates the expression of heat shock proteins (HSPs) in response to various cellular stresses. Dysfunction of HSF1 contributes to the pathogenesis of a spectrum of acute and chronic diseases, including cancer. Consequently, the modulation of HSF1 activity through the development of small molecules emerges as a promising therapeutic strategy for disease treatment. The activation of HSF1 is a multifaceted process, governed by a complex interplay of regulatory mechanisms, including post-translational modifications, protein-protein interactions, and a balance between its activation and inactivation. Recently, a plethora of compounds, ranging from synthetic to naturally derived, that either inhibit or activate HSF1 was identified, holding considerable potential for the treatment of numerous human diseases. In this comprehensive review, we elucidate the sophisticated mechanisms underlying activation of human HSF1, introduce its role in the etiology of diseases, and provide a comprehensive summary of the inhibitors and activators of HSF1 that have been discovered to date. This review not only offers novel insights for the development of small molecule therapeutics targeting HSF1 but also charts new territories in the design of innovative interventions for the amelioration of disease.
Keywords: HSF1, inhibitors, activators, cancer, therapeutic strategy
1. Introduction
Heat shock response (HSR) is an evolutionarily highly conserved protective process that regulates the expression of molecular chaperones in response to cellular stresses, such as temperature changes, oxidative stresses, lack of glucose, or the accumulation of misfolded proteins [1]. The human heat shock factor (HSF) family consists of six identified members: HSF1, HSF2, HSF4, HSF5, HSFX and HSFY [2]. Among them, HSF1 is the most ubiquitously expressed HSF, renowned for its swift and robust ability to induce transcription of HSPs [2]. These HSPs are essential for cellular recovery from stress-induced damage and are systematically classified into several major families according to their molecular weights, including large HSPs (HSP110 family), HSP90 family (HSPC), HSP70 family (HSPA), chaperonin/HSP60 family (HSPD), HSP40 co-chaperones (DNAJ), and small HSPs (HSPB) [3, 4] . There is an increasing acknowledgment of the therapeutic potential in manipulating HSF1 activity for the development of novel treatments for various diseases. For instance, the activation of HSF1 has demonstrated to exert cytoprotective effects on cardiomyocytes [5, 6], whereas the inhibition of HSF1 could increase the chemosensitivity [7]. To realize these therapeutic benefits, the development of inhibitors and activators that specifically target HSF1 is imperative.
In the following sections, we provide an overview of the activation process of human HSF1, discuss its specific roles in human diseases, and categorize the known activators and inhibitors of HSF1. Our aim is to lay the groundwork for the advancement of innovative therapeutic strategies that target HSF1, offering new avenues for clinical intervention.
2. The regulatory mechanism of HSF1 activation and inactivation
2.1 Glimpse into the process of HSF1 activation and inactivation
HSF1 is a multi-domain protein composed of distinct domains that collectively enable its regulatory role. It has an N-terminal DNA-binding domain capable of recognizing and binding to the specific sequences known as heat shock element (HSE) on target gene promoters [8]. Additionally, HSF1 contains two heptad repeat regions (HR-A and HR-B), along with a third heptad repeat region (HR-C), which are responsible for its oligomerization [8-11]. A regulatory domain is also present, serving as a platform for various post-translational modifications (PTMs) that modulate HSF1 activity [12]. Moreover, HSF1 has a C-terminal transactivation domain (TAD) that could recruit the transcriptional cofactors TAF9 and CBP/p300 and initiate the transcription of downstream target genes (Fig. 1A) [13, 14].
Under normal conditions, HSF1 is a cytoplasmic and monomeric molecule that lacks DNA binding activity. Its inactivity is preserved by a cohort of cytosolic HSPs, such as HSP70, HSP90, and chaperonin TCP-1 ring complex (TRiC) (Fig. 1A) [15, 16]. Simultaneously, the interactions between HR-A, HR-B and HR-C domains also contribute to the stabilization of HSF1 in its monomeric form [11]. Interestingly, even in the absence of stress, the inactive monomeric HSF1 is capable of shuttling between the nucleus and cytoplasm, highlighting the dynamic nature of HSF1 [17].
The activation mechanism of HSF1. (A) Upper panel: An architectural blueprint of the HSF1 protein. It consists of a DNA-binding domain (DBD), a heptad repeat region (HR) that exhibits leucine-zipper-like characteristics, a regulatory domain (RD), and a trans-activation domain (TAD). Lower panel: Under stress, the quiescent HSF1, which is normally sequestered within a complex with HSPs and the chaperonin TRiC, is released. This liberation triggers a conformational change, leading to the formation of active HSF1 trimers that migrate to the nucleus. (B) The HSF1 trimers subsequently bind to the HSE sequences, thereby initiating the transcription of HSPs. Throughout this transcriptional cascade, the activity of HSF1 is finely tuned by an array of PTMs such as phosphorylation, dephosphorylation, acetylation, and sumoylation, along with interactions from various proteins that can either enhance or repress its activity. (C) The activity of active HSF1 is gradually attenuated through inhibitory acetylation and sumoylation. Additionally, the binding of HSP70 to HSF1 acts as a crucial feedback loop that inhibits HSF1 activity and leads to the cessation of HSP transcription. (D) Following attenuation, HSF1 is released from the DNA and undergoes deactivation. It can either revert to monomeric form, associate with inhibitory protein complexes, or be targeted for ubiquitination and degradation. Figure was created in BioRender.
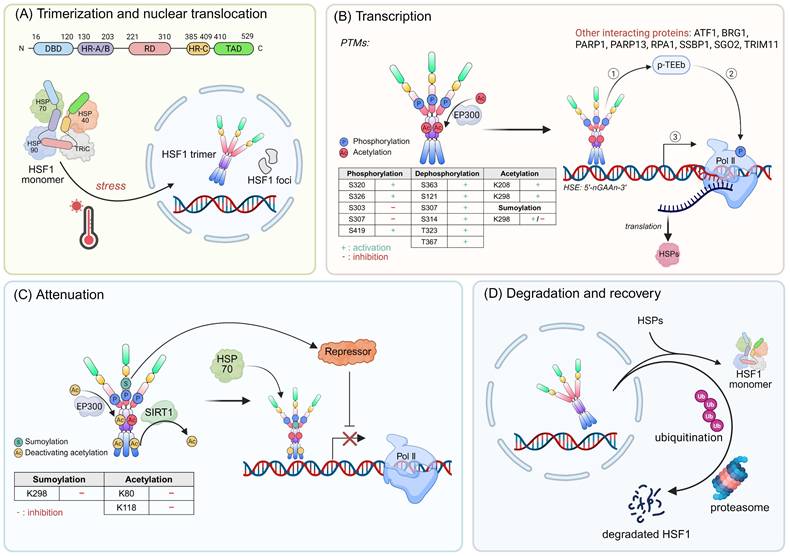
The HSF1 activation during stress responses is a multi-step process that can be delineated into four consecutive phases: (I) Trimerization and Nuclear Translocation: The HR-A and HR-B regions form a highly elongated structure through intermolecular hydrophobic interactions [18]. Subsequently, HSF1 undergoes numerous PTMs and interacts with some important regulatory factors, facilitating its conversion to an active trimeric state and nuclear translocation (Fig. 1A) [19]. (II) Transcription: Upon translocating into the nucleus, HSF1 acquires DNA-binding activity and binds to the HSEs within promoter regions, thereby inducing the expression of chaperones [8, 20, 21]. In yeast genome, transcriptional induction of HSP genes is accompanied by dynamic changes in their 3D structure and spatial organization. For example, Hsf1 can accumulate in distinct nuclear foci (also known as nuclear stress bodies). The formation of these foci is correlated with an enhanced transactivation potential of Hsf1 (Fig. 1B) [22]. In another case within the yeast genome, Hsf1, in conjunction with Mediator and RNA polymerase II (Pol II), forms dynamic transcriptional condensates. This process is associated with the 3D reorganization of the genome, which fosters cellular fitness and ensures robust transcriptional activity under stress conditions [23]. However, only modest changes in distal regulatory elements on chromatin are observed in human cells following heat shock [24]. These results suggests that the global chromatin structure necessary for HSR is preestablished in human enabling cells to respond rapidly to stress. (III) Attenuation: The interaction of HSP70 with HSF1, along with the sumoylation events, contribute to the reduced activity of HSF1 (Fig. 1C) [25-27]. (IV) Degradation and Recovery: HSF1 is released from DNA and transformed into monomer with the assistance of the HSP70 system, and this is followed by acetylation and ubiquitination [28]. Ultimately, HSF1 is degraded via the ubiquitin-proteasome system (UPS), thus completing the cycle and allowing for cellular recovery (Fig. 1D) [28].
2.2 The regulators in HSF1 activation
2.2.1 The PTMs of HSF1
2.2.1.1 Phosphorylation of HSF1
The PTMs of HSF1 include phosphorylation, sumoylation, acetylation and ubiquitination. Notably, phosphorylation events are predominant among these PTMs [29]. The PhosphoSitePlus database provides a comprehensive of phosphorylation of HSF1, identifying a total of 59 residues that undergo such modification. Phosphorylation of HSF1 exerts either stimulatory or inhibitory effects on its activation (Fig. 1B) [30, 31]. For example, phosphorylation at serine 230 (Ser230) and Ser326 promotes HSF1 activation and are critical for the enhanced interaction between HSF1 and general transcription factors, such as CDK9 and TFIIB [32]. In contrast, the constitutive phosphorylation of Ser303 and Ser307 attenuates HSF1 transcriptional activity [33]. Moreover, dephosphorylation events can also trigger HSF1 activation. The immediate-early response gene IER5, along with B55 regulatory subunits, induces the dephosphorylation of HSF1 at Ser363, thereby enhancing the expression of HSF1 target genes [34]. Additionally, IER5 participates in a ternary complex with phosphatase PP2A and HSF1, dephosphorylating HSF1 at multiple sites, including Ser121, Ser307, Ser314, threonine 323 (Thr323) and Thr367, resulting in the formation of a new hypo-phosphorylated active state of HSF1 [35]. HSF1 phosphorylation is also related to the active chromatin status and tumorigenesis. A recent study reveals that phosphorylation of HSF1 at Ser419 recruits the TRRAP-TIP60 acetyltransferase complex at the HSP70 promoter during HSR. Subsequently, TRIM33 cooperates with TRIM24 to facilitate mono-ubiquitination of histone H2B at lysine 120 (K120), which stabilizes the HSF1-containing transcription complex in the HSP70 promoter [36].
2.2.1.2 Sumoylation of HSF1
HSF1 undergoes rapid and transient SUMOylation following heat stress [37-39]. This dynamic PTM occurs at K298 within a phosphorylation-dependent SUMOylation motif (PDSM), representing the first identified example of this regulatory mechanism [38, 40]. SUMOylation efficiency is enhanced by HSF1 trimerization and phosphorylation at Ser303/Ser307, indicating this modification occurs in the transcriptionally active state [25]. Non-SUMOylatable mutants (K298R and S303A) of HSF1 exhibit increased transcriptional activity, demonstrating SUMOylation serves as a negative regulatory switch [40]. Mechanistically, SUMOylation reduces transactivation potential without affecting DNA binding capacity and HSC70-mediated dissociation from chromatin [25]. The PDSM-mediated coupling of phosphorylation and SUMOylation represents a precisely timed attenuation mechanism for HSR. However, another study suggested sumoylation of HSF1 at K298 can enhance its nuclear translocation and stability and HSPs expression, thereby promoting the mitochondrial unfolded protein response (UPRmt) activation [41]. This process resulted in increased cell proliferation, migration, invasion and reduced apoptosis, ultimately promoting tumor growth in glioblastoma (GBM) xenograft models [41]. Nevertheless, a critical limitation of this study is that the K298R mutation concurrently abolishes both SUMOylation and acetylation modifications, making it difficult to definitively attribute the observed effects solely to SUMOylation. Given these conflicting findings, the biological functions of HSF1 SUMOylation need further investigation using more precise genetic tools.
2.2.1.3 Acetylation of HSF1
The acetylation of HSF1 exerts a highly intricate and comprehensive regulatory influence on HSF1 function. The acetylation occurs across a broad spectrum of domains within the HSF1 protein, thereby modulating its activity in a multifaceted manner [9]. For instance, under stress conditions, the acetyltransferase EP300 modulates HSF1 stability by acetylating K208 and K298, thereby inhibiting HSF1 degradation and sustaining its activity (Fig. 1B) [28, 42]. Conversely, EP300 can also impair the interaction between HSF1 and DNA phosphate backbone by acetylating K80 of HSF1, which attenuated the HSR (Fig. 1C) [28]. Similarly, the interplay between HSF1 and BRCA1-associated protein-1 (BAP1) sustains the acetylation status of HSF1 at K80, thereby augmenting the interaction between HSF1 and HSP70 [43]. This process facilitates the dissociation of HSF1 from chromatin, ultimately suppressing its transcriptional activity [43]. However, the NAD-dependent protein deacetylase sirtuin 1 (SIRT1), which deacetylates HSF1 at K80, can counteract this effect by promoting the retention of HSF1 in a DNA-binding state, thereby activating HSPs following heat stress (Fig. 1C) [44]. Additionally, acetylation of K118 hinders the binding of HSF1 to chromatin, restricting its DNA binding capacity and initiating the process of attenuation (Fig. 1C) [27, 28].
2.2.1.4 Ubiquitination of HSF1
HSF1 protein is degraded by the UPS during the attenuation phase of HSR (Fig. 1D) [28]. The ubiquitin ligase FBXW7 directly binds to HSF1 and mediates its ubiquitination and proteasomal degradation [45]. Moreover, the E3 ubiquitin ligase NEDD4 (neuronally expressed downregulated protein 4) promotes HSF1 ubiquitination in neuroblastoma cells overexpressing the A53T mutant variant of α-synuclein [46].
2.2.2 Interactome of HSF1
2.2.2.1 Various types of HSP
HSP70 consistently suppresses HSF1 activity by binding to it and maintaining it in an inactive state (Fig. 1A) [47, 48]. Mechanistically, the co-chaperone DNAJB1 aids HSP70 in binding to a region proximal to the HR-B domain of HSF1. This binding induces a conformational flexibility by generating a locally crowded conformation, resulting in a low-entropy state. This process is marked by repeating cycles of entropic pulling, which lead to the dissociation of HSF1 trimers into monomers and the forcibly unzip the triple leucine zipper present within HSF1 trimers [49]. Jumonji domain-containing protein 6 (JMJD6) also participates in the regulation of HSF1-HSP70 complex. Specifically, HSF1 binds and promotes the expression of JMJD6, which in turn reduces mono-methylation of HSP70 at arginine 469 (R469), thereby disrupting HSP70-HSF1 repressive complex formation [50]. Additionally, the HSP70-HSP40 complex facilitates the interaction between HSP70 and client protein by guiding substrates to the peptide-binding site of HSP70, thereby inducing a conformational change [51, 52]. Notably, a recent study suggests that HSP70 also represses Hsf1 by binding to its CE2 domain, a sequence of 14 amino acids, which represses HSF1 cluster formation in yeast [23].
In unstressed cells, HSP90 maintains HSF1 in an inactive state by forming a stable HSP90-HSF1 complex [53, 54]. The assembly of HSF1-HSP90 complex suppresses the trimerization of HSF1 (Fig. 1A) [53]. HSP90 inhibitors, such as radicicol, celastrol, geldanamycin, and the analogs of geldanamycin (17-AAG and 17-DMAG), target the N-terminal ATP-binding site of HSP90 [55-59]. This interaction prevents the conformational change of HSP90 to its “closed” dimeric state and competitively disrupts the interaction between HSF1 and HSP90, thus contributing to the transcriptional activation of HSF1 [60]. Moreover, the HSP90-associated co-chaperone p23 and histone acetyltransferase KAT2A/GCN5 can remove HSF1 and other transcription factors from DNA, although the exact mechanisms remain to be elucidated [27].
TRiC, also referred as CCT (chaperonin containing TCP-1), is composed of multiple subunits. Similar to HSP70 and HSP90, TRiC/CCT interacts with HSF1 and maintains it in an inactive state (Fig. 1A) [16]. Specifically, TRiC/CCT is capable of folding substrates within the central cavity of its barrel-like structure, a feature that underpins its chaperone activity [61].
2.2.2.2 Interactions among HSF family members
HSF2 is involved in the developmental processes and spermatogenesis. HSF2 is also implicated in modulating cellular responses to specific proteotoxic stressors, such as proteasome inhibition and febrile-range thermal stress [62, 63]. Despite its capacity to bind to consensus HSEs, HSF2 appears to play a relatively limited role in directly promoting the HSR [64]. Notably, HSF1 and HSF2 exhibit co-expression across tissues and can form heterotrimers, which functionally impacts gene regulation [64, 65]. Recent studies have revealed a physical interaction between HSF2 and HSF1, mediated by their coiled-coil domains [66]. This interaction results in similar chromatin occupancy profiles and the coordinated regulation of a shared set of genes. The gene repertoire not only includes HSPs but also encompasses noncanonical transcriptional targets that are crucial for supporting malignant processes [67]. Furthermore, a novel interrelationship between HSF1 and HSF2 has been proposed based on findings that HSF1 transcriptionally regulates HSF2 levels following proteasome inhibition [68]. This discovery highlights the intricate regulatory network between these two HSFs and their combined roles in cellular stress responses and gene regulation.
HSF1 and HSF4 exhibit distinct and sometimes opposing roles in regulating gene expression and maintaining cellular homeostasis [64, 69]. HSF4, particularly its splice variant HSF4b, has been shown to inhibit HSF1 transcriptional activity. By binding to N-terminal hydrophobic region of HSF1, HSF4b disrupts HSF1's intramolecular interactions, promoting its cytoplasmic retention and degradation, thereby attenuating HSF1-driven HSP expression [70]. While HSF1 primarily activates HSP genes during stress, HSF4 can suppress this activation by competing for HSE binding or directly inhibiting HSF1's transcriptional function [71, 72]. Therefore, HSF1 and HSF4 collaborate or compete in modulating gene expressions, highlighting their context-dependent roles [72, 73]. The HSF1-HSF4 relationship exemplifies a sophisticated regulatory mechanism that balances stress adaptation with developmental and homeostatic processes.
2.2.2.3 Transcription factors and other interacting proteins
HSP70 is a classic gene transcribed by Pol II. In the absence of stress, Pol II interacted with DRB sensitivity-inducing factor (DSIF) and negative elongation factor (NELF) to maintain the pausing state [74, 75]. However, when subjected to stress, HSF1 recruits transcription factor p-TEFb, which phosphorylates Pol II, DSIF and NELF, thereby removing NELF from Pol II, allowing transcription to proceed [76-78]. Thus, the recruitment of p-TEFb contributes to the transcriptional activation of HSF1 (Fig. 1B). In addition, the poly (ADP)-ribose polymerase 1 (PARP1) is liberated from chromatin during heat shock, which is followed by the loss of nucleosomes from the HSP70 coding regions, thereby activating transcriptional process [79-81]. Moreover, PARP1 and PARP13 interact with HSF1 to form a ternary complex in response DNA damage, which promotes PARP1 activation and auto-PARylation, and then facilitates the redistribution to genomic lesions [82].
HSF1 recruits diverse transcription factors to modulate gene expression (Fig. 1B). For example, HSF1 recruits mitochondrial single-stranded DNA binding protein 1 (SSBP1) to the promoters of mitochondrial chaperones, subsequently recruiting the chromatin-remodeling factor BRG1. This collaboration is crucial for sustaining mitochondrial membrane potential during proteotoxic stress, thus initiating the UPRmt [83-87]. The physical interaction between replication protein A (RPA) and HSF1 is also important. The deletion of RPA1 impairs HSF1's binding to the HSP70 promoter in non-stress conditions, and delays the rapid activation of HSF1 in response to heat shock [88]. The nuclear translocation of HSF1 is increased by tripartite motif containing 11 (TRIM11), which interacts with and stabilizes HSF1 [89]. The pericentromeric adaptor protein shugoshin 2 (SGO2), acting as a coactivator, supports cell survival by promoting the recruitment of Pol II to the HSP70 promoter [90]. The phosphorylation of activating transcription factor 1 (ATF1) is essential for the assembly of HSF1 transcription complex during heat shock [91]. The ATF1-BRG1 complex facilitates the formation of an active chromatin state and elevates HSP70 expression, and the ATF1-p300/CBP complex accelerates the decline of HSF1 DNA-binding activity during recovery from acute stress, potentially through the acetylation of HSF1 [91].
3. HSF1 is an ideal therapeutic target for human diseases
3.1 HSF1 and cancer
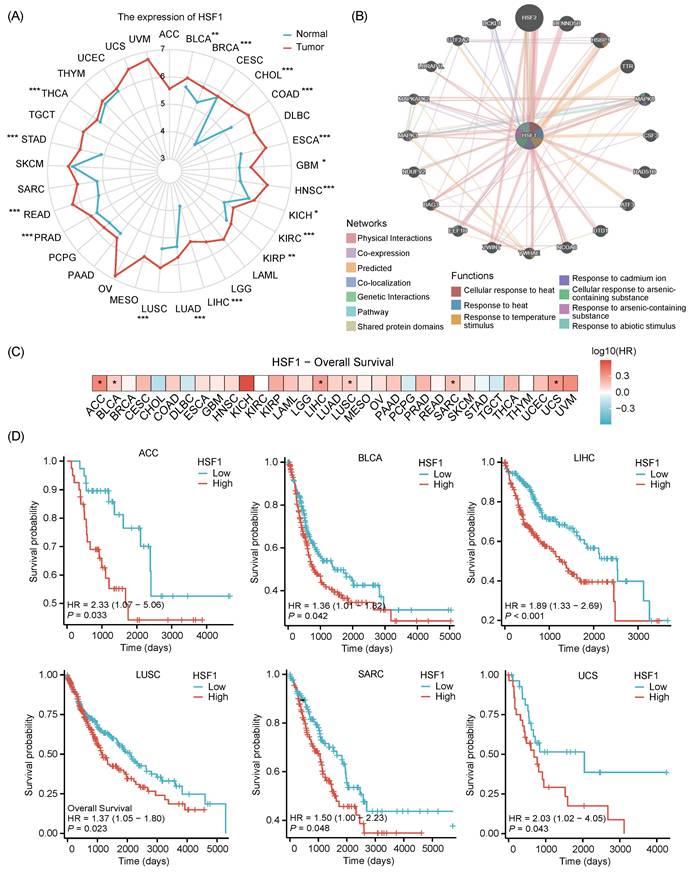
The role of HSF1 in cancer extends far beyond its classical function in stress response. By promoting cancer cell survival, metabolic reprogramming, epithelial-mesenchymal transition (EMT) process and immune evasion, HSF1 emerges as a multifaceted driver of tumorigenesis [92-94]. Therefore, targeting HSF1 represents a promising therapeutic strategy. Consistent with previous studies, HSF1 is significantly upregulated in a diverse range of 17 cancer types based on comprehensive analysis of the TCGA database (Fig. 2A) [92, 94]. This upregulation is particularly intriguing as HSF1 interacts with a multitude of proteins that are strategically positioned upstream of the tumor growth signaling pathways (Fig. 2B). These interactions suggest a complex regulatory network involving HSF1, which is consistent with its pivotal role in driving the proliferation and growth of cancer cells. The Kaplan-Meier overall survival (OS) curves reveal that patients with low HSF1 expression trend to have longer OS (Fig. 2C, 2D) [95, 96]. These findings are consistent with previous pan-cancer analysis, which has implicated a correlation between the elevated HSF1 expression and poor outcomes in patients with different types of cancer [97]. These insights reinforce the established connection between HSF1 and tumorigenesis, highlighting its potential as a significant factor in cancer development and progression.
The expression and prognostic value of HSF1 in pan-cancer. (A) Radar chart represents the expression of HSF1 in various cancer types. Notably, HSF1 is highly expressed in BLCA, BRCA, CHOL, COAD, ESCA, GBM, HNSC, KICH, KIRC, KIRP, LIHC, LUAD, LUSC, PRAD, READ, STAD and THCA. (B) The HSF1 interactome was generated through GeneMania database analysis, integrating experimental evidence and bioinformatic predictions to produce an unbiased protein-protein interaction (PPI) network. This network offers valuable insights into the interactive partners of HSF1. (C) The prognostic potential of HSF1 in pan-cancer. (D) Kaplan-Meier overall survival (OS) curves are presented to compare the survival rates between patients with high and low HSF1 expression. The elevated HSF1 expression is associated with a poorer prognosis in patients with ACC, BLCA, LIHC, LUSC, SARC, and UCS.
Cancer cells are tasked with the challenge of increased protein biosynthesis and the buildup of mutated or misfolded proteins [98]. HSF1 is capable of enhancing the expression of protein-folding chaperones and an array of pro-survival factors, including DNA replication and repair factors, metabolic enzymes and cell cycle regulators, which increases the malignancy of cancer and shortens the lifespan of cancer patients (Fig. 3A) [21, 94, 99-102]. Consistently, previous studies have demonstrated that cancer cells exhibit a tendency to be addicted to HSF1 [103, 104].
Accumulating evidence indicates that HSF1 promotes tumor growth across multiple cancer types (Fig. 3A). For instance, in breast cancer, HSF1 stimulates the growth of the stromal cells within the tumor microenvironment (TME), which in turn influences tumor progression [95]. DYRK2, a dual specificity tyrosine-regulated kinase, can phosphorylate HSF1, thereby increasing nuclear HSF1 stability and transcriptional potency in breast cancer cells (Fig. 3A) [105]. In multiple myeloma, the demethylase fat mass and obesity-associated protein (FTO) exhibits tumor-promoting and pro-metastatic functions by targeting HSF1/HSPs pathway in a m6A reader protein YTHDF2-dependent manner (Fig. 3A) [106]. In hepatocellular carcinoma (HCC), HSF1 upregulates PD-L1 expression through inducing apolipoprotein J (APOJ) expression and activating the STAT3 signaling pathway to neutralize the cytotoxic effects of CD8+ T cell against cancer cells (Fig. 3A) [107]. In acute myeloid leukemia (AML) cells, HSF1 deletion downregulates succinate dehydrogenase C (SDHC) expression and suppresses the mitochondrial oxidative phosphorylation (OXPHOS), thereby inhibiting the growth of tumor cells (Fig. 3A) [108]. Additionally, HSF1 also activates the forkhead box protein O3a (FOXO3a)-dependent transcription of ΔNp63α (p63 isoform in the p53 family) to promote the growth of head and neck squamous cell carcinoma (HNSCC) (Fig. 3A) [109]. Moreover, epidermal growth factor receptor (EGFR)-HSF1 axis facilitates the initiation of pancreatic cancer (Fig. 3A) [110].
Several microRNAs (miRNAs) regulate the development of human cancers by influencing HSF1 activity. In HCC, miR-644a targets 3'-untranslated region (3'-UTR) of HSF1, leading to a decrease in HSF1 expression, inhibition of HCC cell proliferation and the initiation of apoptosis (Fig. 3A) [111]. Similarly, microRNA-455-3p curbs osteosarcoma progression through suppressing HSF1 expression [112]. HSF1 also upregulates the acetyltransferase EP300-mediated lncRNA-LINC00857, which facilitates SLC1A5-mediated glutamine transport, ultimately accelerating the growth of colorectal cancer (CRC) cells (Fig. 3A) [113].
3.2 HSF1 and neurodegenerative diseases
HSPs are instrumental in preventing the aggregation of misfolded proteins and assisting in the correct folding of newly synthesized polypeptides [114]. Therefore, dysfunctional HSF1-mediated downregulation of HSPs expression has been implicated in initiating or exacerbating neurodegenerative diseases, which are characterized by misfolded protein aggregation [19, 115, 116]. Targeting HSF1 to increase the expression of HSPs represents a promising therapeutic strategy that may effectively halt or reverse the pathological progression of neurodegenerative disorders while minimizing potential side effects [101, 115]. Accumulating evidences suggest that HSF1 mitigates α-synuclein toxicity [46], inhibits the polyglutamine aggregation [117], and prevents Tau-related pathologies [118], thereby delaying the progression of Parkinson's disease (PD), Huntington's disease (HD) and Alzheimer's disease (AD), respectively (Fig. 2B).
PD is characterized by the accumulation of pathological α-synuclein aggregates and microglial overactivation (Fig. 3B) [119, 120]. HSF1 has emerged as a critical regulator of neuroprotection in PD, primarily through its induction of HSP70, a chaperone protein that facilitates the clearance of α-synuclein aggregates and modulates microglial activation [121]. Furthermore, paeoniflorin, a bioactive monoterpene glycoside, can activate the HSF1-NRF1 (nuclear respiratory factor 1) pathway to mitigate oxidative stress and neuroinflammation in PD [122]. These findings highlight HSF1 activation and its downstream effectors as promising therapeutic targets for slowing or preventing PD-associated neurodegeneration.
HD is caused by a CAG repeat expansion in the HTT gene, resulting in an elongated polyglutamine (polyQ) tract in the huntingtin protein. This mutation leads to protein misfolding, aggregation, and subsequent neurodegeneration (Fig. 3B) [123]. An active form of HSF1 significantly extends the lifespan of HD model mice by suppressing polyQ inclusion formation [117]. HSF1 and NFAT (nuclear factor of activated T cells) synergistically promote the expression of the scaffold protein PDZK3 and αB-crystallin, which together contribute to the degradation of polyQ proteins [124]. Beyond its role in protein quality control, HSF1 also upregulates brain-derived neurotrophic factor (BDNF) under acute stress, highlighting its neuroprotective function and importance in hippocampal plasticity [125]. However, HD pathogenesis involves mechanisms that impair HSF1 activity. Mutant huntingtin upregulates protein kinase CK2α and the E3 ubiquitin ligase FBXW7, both of which promote HSF1 degradation. This loss of HSF1 function exacerbates protein aggregation and striatal neurodegeneration [126]. These findings collectively suggest HSF1 activation as a promising therapeutic strategy to counteract HD progression.
HSF1 plays a pivotal role in the progression of numerous diseases, highlighting its complex and multifaceted involvement in cellular processes. (A) In the context of cancer, HSF1 promotes the initiation and progression of the disease by enhancing protein biosynthesis. This increased biosynthetic activity is particularly advantageous for cancer cells, as it supports their rapid growth and proliferation. By upregulating the transcription of genes encoding ribosomal proteins and other factors involved in protein synthesis, HSF1 contributes to the creation of a favorable microenvironment for tumor development. (B) Misfolded protein aggregates lead to the occurrence of neurodegenerative diseases, and the expression of HSPs mediated by HSF1 can exert protective effects by mitigating cellular stress and promoting cell survival, which prevent the progress of this diseases. (C) Beyond cancer and neurodegenerative diseases, HSF1 has also been implicated in a wide range of other human diseases. For instance, it has been shown to play a role in obesity, where it may contribute to metabolic dysregulation and the development of insulin resistance. In myocardial injury, HSF1 activation is linked to the protection of cardiomyocytes from stress-induced damage, suggesting a potential therapeutic target for heart disease. Similarly, in acute lung and brain injury, HSF1-mediated HSPs expression may provide cytoprotective effects, although the precise mechanisms involved remain under investigation. Additionally, HSF1 regulates the expression of genes involved in endometriosis, suggesting a potential role in the disease's pathogenesis. HSF1 is a key player in the progression of multiple diseases, with its effects ranging from promotional to protective depending on the specific context. Figure was created in BioRender.
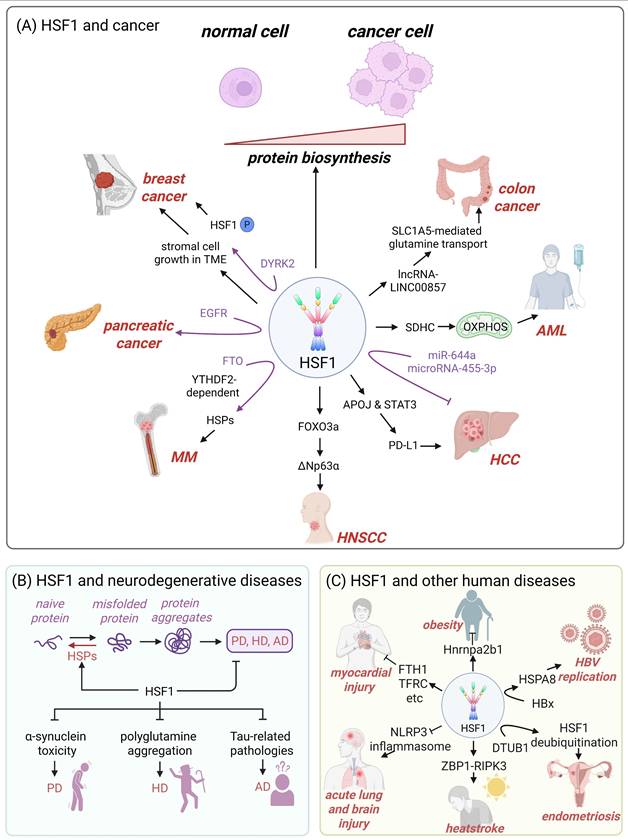
AD is neuropathologically defined by extracellular amyloid-β (Aβ) plaques and intracellular tau neurofibrillary tangles (Fig. 3B) [127]. In N2a cells stably expressing the pro-aggregation mutant TauRD ΔK280, HSF1 upregulation mitigates tau-induced proteotoxicity through downregulating C/EBP homologous protein (CHOP), a key mediator of ER stress, and enhancing BiP/GRP78 expression to promote proper protein folding [118]. Moreover, HSF1 directly counteracts amyloidogenic processes by neutralizing soluble amyloid oligomers through physical interaction [128]. HSF1 also protects HSP60 from amyloid oligomer-induced destabilization, thereby preserving mitochondrial proteostasis and preventing mitophagy/apoptosis cascades [128]. However, AD progression involves mechanisms that compromise HSF1 function. The non-coding RNA Alu inhibits HSF1-mediated transcription by blocking RNA polymerase II binding at HSF1 target promoters, thereby exacerbating protein misfolding [129]. These evidences strongly support the development of HSF1-targeted therapies that could simultaneously mitigate multiple aspects of AD pathogenesis.
3.3 HSF1 and other human diseases
Beyond its role in oncology and neurodegenerative disorders, HSF1 is a key player in many other human diseases (Fig. 3C). For obesity treatment, HSF1-mediated upregulation of heterogeneous nuclear ribonucleoprotein A2/B1 (Hnrnpa2b1) results in white fat beige, therefore inducing weight loss by increasing energy expenditure (Fig. 3C) [130]. Additionally, overexpression of HSF1 ameliorates palmitic acid-induced myocardial injury by regulating the expression of iron metabolism-related genes, such as FTH1, TFRC, SLC40A1 and GPX4 (Fig. 3C) [5]. Notably, HSF1 is also identified as a critical protective factor against various acute and chronic diseases. Recent research has demonstrated that HSF1 protects cells from sepsis-induced acute lung or brain injury by inhibiting the activation of NLRP3 inflammasome (Fig. 3C) [131, 132]. Moreover, HSF1 can foster the development of heatstroke by activating the Z-DNA binding protein 1 (ZBP1), which initiates receptor-interacting protein kinase 3 (RIPK3)-dependent cell death (Fig. 3C) [133]. In the case of endometriosis, the ubiquitin thioesterase OTUB1 interacts with HSF1, enhancing its protein stability through deubiquitination, and thus contributing to the disease's development (Fig. 3C) [134]. Moreover, the hepatitis B X protein (HBx) can upregulate the expression of HSPA8 in an HSF1-dependent manner, which in turn enhances hepatitis B virus (HBV) replication (Fig. 3C) [135].
4. HSF1 inhibitors and activators
A vast repertoire of small molecules targeting HSF1, serving as inhibitors or activators, has been identified. These molecules encompass derivatives sourced from natural medicinal compounds, synthetically designed molecules through computational in silico methods, and candidates pinpointed by high-throughput screening of expansive synthetic chemical libraries [136]. Notably, the majority of these modulators indirectly regulate HSF1 activity. The intricate mechanisms by which certain molecules act at multiple points within the HSF1 activation cascade have been clearly elucidated, while the precise effects of others remain incompletely understood. Based on the distinct stages at which these molecules operate, we have summarized the inhibitors and activators in Table 1 and Table 2, respectively.
4.1 Inhibitors of HSF1
4.1.1 Inhibit HSF1 at the pre-transcriptional level
4.1.1.1 Targeting HSF1-interacting proteins
Doxorubicin (DOX), a widely used anthracycline chemotherapeutic agent originally isolated from Streptomyces peucetius [137], exerts its anti-tumor effects through DNA intercalation and Topoisomerase II inhibition. Cardiotoxicity is a major limitation of DOX, primarily due to mitochondrial dysfunction and ROS overload [138]. DOX impedes the binding of HSF1 to the carboxyl-terminus of HSP70 interacting protein (CHIP), a co-chaperone and ubiquitin ligase, leading to the destabilization of HSF1 [139]. Disruption of this interaction by DOX leads to the removal of active HSF1 from the nucleus and triggers its degradation via the proteasome pathway (Table 1) [139]. However, the inactivation of HSF1 leads to the loss of its inhibitory effect on insulin-like growth factor receptor II (IGF-IIR), which ultimately accelerates the onset of apoptosis-induced cardiotoxicity [139]. These findings not only clarify the molecular mechanisms underlying DOX-induced cardiotoxicity but also reveal that the antitumor efficacy of DOX is partially mediated through suppression of the HSF1 pathway.
4.1.1.2 Inhibitors influencing PTMs of HSF1
Phosphorylation at Ser326 modulates HSF1 activation, and a variety of inhibitors have been designed to target this residue to suppress HSF1 activity. For example, 2,4-bis (4-hydroxybenzyl) phenol, a compound derived from the rhizomes of Gastrodia elata, significantly dephosphorylates HSF1 at Ser326, leading to the downregulation of HSP27 and HSP70 expression and the induction of apoptosis in lung cancer cells (Table 1) [140]. Bisamide 26 (CCT251236) has inhibitory effects on HSF1-mediated transcriptional activity, leading to a reduction in mRNA expression levels of HSP70 and HSP27. This phenomenon may be associated with its binding affinity to pirin, a redox-sensitive transcription factor regulator. However, the precise implications of this interaction in regulating the HSF1 signaling pathway require further investigation (Table 1) [141]. Bisamide 26 exhibits significant antitumor activity by suppressing cancer cell proliferation through the inhibition of the HSF1 pathway [142]. It's worth noting that NXP800 (CCT361814), a derivative of Bisamide 26, reduces HSF1 phosphorylation at Ser326, resulting in a concentration-dependent decrease in the expression of HSP27 and HSP70 in myeloma cell lines [143]. Importantly, NXP800 has advanced to Phase 1 clinical trials, highlighting its potential as a promising therapeutic agent for refractory ovarian cancer and other malignancies (Table 1) [144].
Summary of HSF1 inhibitors
| Compounds | Chemical structure | Source | Mechanism of action |
|---|---|---|---|
| Targeting DNA binding ability of HSF1 | |||
| Doxorubicin (DOX) | 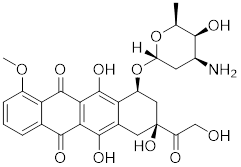 | Streptomyces peucetius | Impedes the binding of HSF1 to CHIP, leading to the destabilization and degradation of HSF1 [139]. |
| 2,4-bis(4-hydroxybenzyl) phenol | 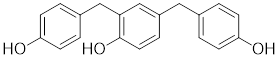 | Gastrodiaelata | Dephosphorylates HSF1 at Ser326 and decreased the expression of HSP27 and HSP70 [140]. |
| NXP800 (CCT361814) | 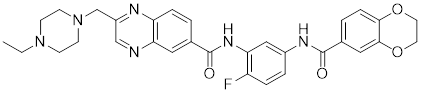 | Derivatives of bisamide (CCT251236) | Suppresses HSF1 phosphorylation at Ser326 and decreased the expression of HSP27 and HSP70 [143]. |
| PW3405 |  | A compound identified by phenotypic screen | Suppresses the phosphorylation of HSF1 at Ser326 [145]. |
| CB14649 | 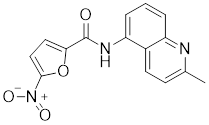 | A compound identified by phenotypic screen | Suppresses the phosphorylation of HSF1 at Ser326 [145]. |
| CB01587 | 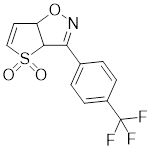 | A compound identified by phenotypic screen | Suppresses the phosphorylation of HSF1 at Ser326 [145]. |
| Cyclosporin A (CsA) | 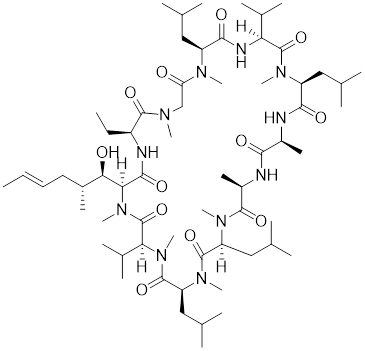 | Tolypocladiuminflatum | Enhances the phosphorylation of HSF1 at Ser303 and Ser307 [147]. |
| Dorsomorphin | 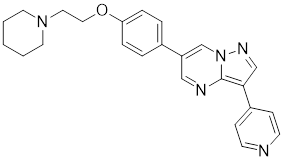 | A synthesized compound that inhibits AMPK | Reduces heat-induced Ser320 phosphorylation and nuclear translocation of HSF1 [149]. |
| EX527 | 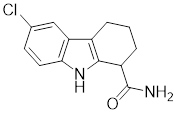 | A SIRT1 inhibitor | Promotes HSF1 deacetylation and ubiquitination [44]. |
| AGK2 | 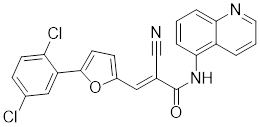 | A SIRT2 inhibitor | Promotes HSF1 deacetylation and ubiquitination [44]. |
| Targeting trimerization and nuclear translocation of HSF1 | |||
| Vitexin | 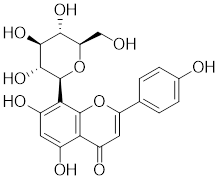 | Vitex agnus-castus | Impedes HSF1 oligomerization by interacting with its DNA-binding domain [151]. |
| Chelerythrine | 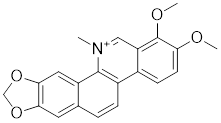 | Chelidonium maju | Suppresses the nucleus translocation of HSF1 and expression of HSP70 [153]. |
| DTHIB | 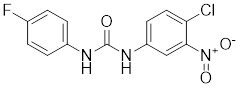 | In vitro screen targeting recombinant human HSF1 DBD | Specifically targets the DBD of HSF1 and accelerates nuclear HSF1 degradation [154]. |
| Schizandrin A (Sch A) | 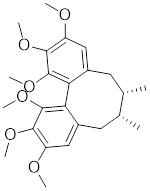 | Schisandra chinensis | Suppresses HSF1-regulated transcription and expression of HSPs [157]. |
| Quercetin | 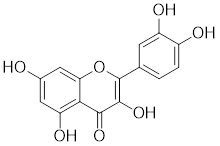 | A plant pigment | Prevents HSF1-HSE binding and inhibits heat-induced HSP70 expression [158]. |
| Fisetin | 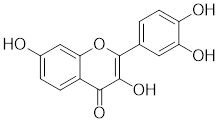 | A natural flavonoid | Blocks the binding of HSF1 to the HSP70 promoter [160]. |
| Targeting HSF1-dependent recruitment of transcription factors | |||
| IHSF115 | 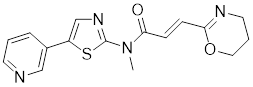 | A structure-activity analysis couple with computational screening | Interferes with ATF1-containing complex assembly, and dampens HSF1 activity [162]. |
| KRIBB11 | 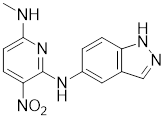 | A synthetic chemical library screening | Inhibits HSF1-dependent recruitment of p-TEFb to the HSP70 promoter [163]. |
| SNS-032 | 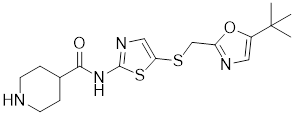 | Inhibitor of CDK9 | Inhibits HSF1 transcriptional activity and HSP70 expression [170, 171]. |
| 4,6-pyrimidine 25 | 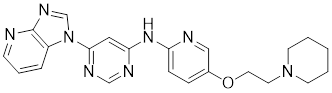 | Inhibitor of CDK9 | Inhibits HSF1 transcriptional activity and HSP70 expression [167]. |
| Cantharidin | 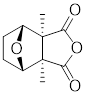 | Secreted by many species of blister beetles | Blocks HSF1 binding to HSP70 promoter and prevents p-TEFb recruitment [173]. |
| Targeting the transactivation function of HSF1 | |||
| Triptolide | 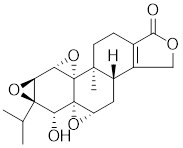 | Tripterygium wilfordii | Interferes with the activity of the C-terminal transactivation domain of HSF1 [174]. |
| HSF1 inhibitors with unclear mechanisms | |||
| Sulphoraphane | 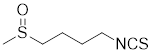 | A naturally isothiocyanate | Suppresses the expression of HSF1, HSP70 and HSP90 [175]. |
| CL-43 | 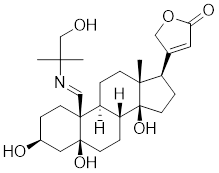 | A member of the cardenolide family | Suppresses the expression of HSP70, HSP90 and HSP40 [176]. |
| Stresgenin B | 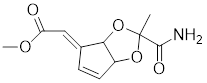 | Streptomyces sp. AS-9 | Suppresses the expression of HSPs [177]. |
| Ginsenoside Rg3 | 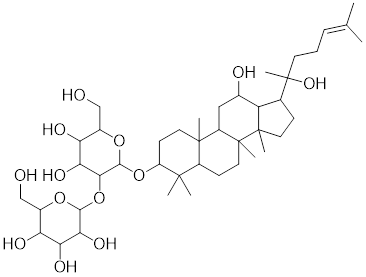 | Panax ginseng | Suppresses the expression of HSF1 and FUT4 [178]. |
| Rocaglamide A (Roc A) | 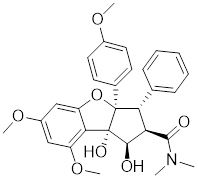 | Aglaia genus | Inhibits HSE reporter system with an IC50 of approximately 50 nM [179]. |
| Homoharringtonine | 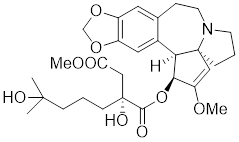 | Cephalotoxus fortunei | Directly binds to HSF1 and suppresses expression of HSF1 and HSPs [183]. |
| Rohinitib (RHT) | 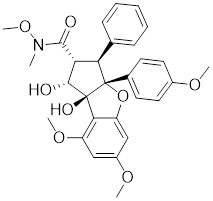 | Rocaglamide A analogs | Inhibits HSE reporter system with an IC50 of approximately 20 nM [179]. |
| KNK437 | 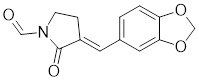 | A benzylidene lactam compound | Suppresses the expression of HSPs [184, 185]. |
| VM1 | 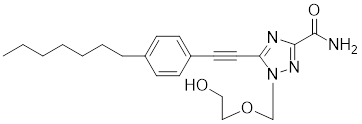 | A triazole nucleoside analogue | Suppresses the expression of HSF1 and HSPs [186]. |
| Bisamide 26 (CCT251236) | 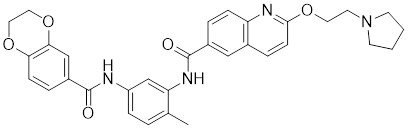 | A chemical probe | Suppresses the expression of HSP70 and HSP27 [141]. |
| NZ28 | 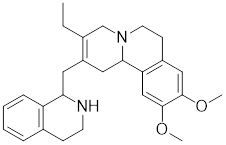 | A dehydroemetine derivative | Suppresses the expression of HSP70 and HSP27 [170]. |
| Emunin (NZ71) | 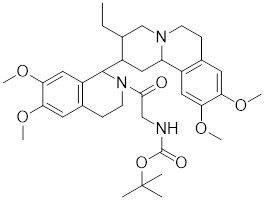 | A glycine amide conjugate of emetine | Suppresses the expression of HSP70 and HSP27 [170]. |
Additionally, through a comprehensive HSR pathway phenotypic screen of a chemical diversity library with over 100,000 compounds, PW3405, CB14649 and CB01587 are emerged as the most potent HSF1 inhibitors, effectively blocking HSF1 activity by suppressing the phosphorylation at Ser326 (Table 1) [145].
Ser303 and Ser307 are recognized as inhibitory phosphorylation sites on HSF1, and cyclosporin A (CsA) can affect these sites to alleviate HSR [33]. CsA, an immunosuppressive agent derived from the fungal species Tolypocladium inflatum, exerts its therapeutic effects by binding and inhibiting the activity of calcineurin, thereby disrupting the downstream signaling cascade critical for T cell activation. [146]. It enhances the phosphorylation of HSF1 at Ser303 and Ser307 through the ERK1/2, GSK3β and CK2 pathways in heat-treated HeLa cells. Consequently, the transcriptional activity of HSF1 is reduced and the formation of the HSF1-SSBP1 complex is disrupted, leading to downregulated expression of HSPs. This cascade of events ultimately results in a significant decrease in cancer cell survival under hyperthermia or chemotherapy (Table 1) [147]. This study establishes a conceptual basis for devising innovative therapeutic approaches for cervical cancer, proposing the integration of CsA in hyperthermic or chemotherapeutic protocols.
Ser320 is an additional target site for HSF1 inhibitors [148]. Dorsomorphin, also known as compound C, is a synthesized compound extensively used to inhibit adenosine monophosphate-activated protein kinase (AMPK) [148]. Previous studies have indicated that dorsomorphin effectively reduces heat-induced Ser320 phosphorylation and the subsequent nuclear translocation of HSF1 in cancer cells, leading to a reduction of heat-induced HSPs expression in an AMPK-independent manner [149]. Moreover, dorsomorphin sensitizes cancer cells to HSP90 inhibitor, downregulates the expression of HSP70, and induces apoptosis of cancer cells, positioning it as a potential therapeutic candidate in oncology (Table 1) [149].
The acetylation and ubiquitination of HSF1 are also strategic targets for inhibitors. For instance, EX527 and AGK2 are synthetic chemical compounds designed to inhibit the deacetylases SIRT1 and SIRT2, respectively [44]. Treatment with EX527 or AGK2 decreases the expression of HSF1 and HSP27 through HSF1 deacetylation under both non-stress and heat stress conditions [150]. Additionally, these compounds are capable of promoting the ubiquitination of HSF1, which ultimately suppresses cancer cell growth and migration (Table 1) [150]. Together, these discoveries suggest that destabilizing the HSF1 protein represents a promising therapeutic approach to inhibit HSF1 activation.
4.1.1.3 Targeting trimerization and nuclear translocation of HSF1
Vitexin, a natural flavone glucoside apigenin found in plants such as Vitex agnus-castus, Phyllostachys nigra, and pearl millet, has been observed to promote nuclear localization of HSF1 while simultaneously impeding HSF1 oligomerization through interaction with its DNA-binding domain (Table 1). Computational modeling indicates that the vitexin-HSF1 complex is stabilized by hydrophobic interactions, which initiates the autophagic cascade and, in turn, suppresses the growth of CRC cell [151]. Chelerythrine, a benzophenanthridine alkaloid derived from Chelidonium majus, serves as a selective inhibitor of protein kinase C (PKC) [152]. Chelerythrine inhibits the nuclear translocation of HSF1 and the expression of HSP70, thereby inducing apoptosis in Dalton's lymphoma cells (Table 1) [153]. These results indicate that HSF1 is a viable target for vitexin and chelerythrine in cancer therapy, offering the potential to enhance the effectiveness of chemotherapy.
Summary of HSF1 activators
| Compounds | Chemical structure | Source | Mechanism of action |
|---|---|---|---|
| Targeting HSF1-interacting proteins | |||
| HSF1A | 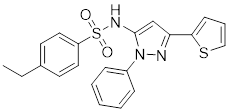 | A cell-permeable pyrazolylsulfonamide compound | Disrupts HSF1-TRiC complex through directly binding to TRiC [16, 115]. |
| Geldanamycin | 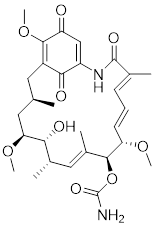 | HSP90 inhibitor | Efficiently disrupts the stable HSP90/HSF1 complex by targeting HSP90 [53, 189]. |
| Phenethyl isothiocyanate (PEITC) | 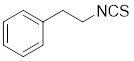 | Nasturtium officinal | Triggers the dissociation of HSF1 from HSP90 and promotes HSF1 phosphorylation at Ser326 [190]. |
| Targeting PTMs of HSF1 | |||
| SYSU-3d | 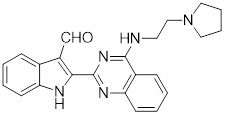 | 2-pyrimidinylindole derivative | Promotes the phosphorylation of HSF1 at Ser326 and its nuclear translocation [191]. |
| Englerin A (EA) | 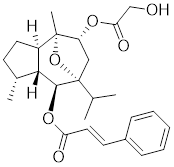 | Phyllanthus engleri | Triggers PKCθ-mediated phosphorylation of HSF1 at Ser333, causing HSF1 dissociation from HSP90 [193]. |
| Resveratrol | 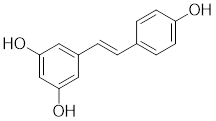 | A natural phenol or polyphenol | Promotes SIRT1-mediated deacetylation of HSF1 and upregulates the expression of HSP25 and HSP70 [195]. |
| Targeting trimerization and nuclear translocation of HSF1 | |||
| Celastrol | 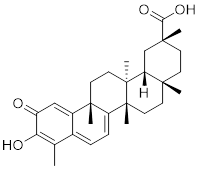 | Tripterygium wilfordiiand Tripterygium regelii | Activating HSF1-PGC1αaxis to regulate energy metabolism [196]. |
| Curcumin | 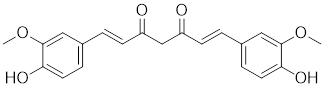 | Curcuma longa | Enhances HSF1 polymerization, nuclear translocation, and DNA binding ability [200, 201] |
| Oridonin | 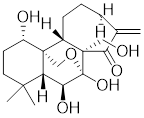 | Isodonrubescens | Directly binding to the Cys153 of HSF1 and promotes HSF1 trimerization [203]. |
| Metformin | 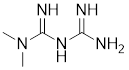 | Galega officinalis | Promotes HSF1 nuclear translocation and activates HSF1-UPRmt pathway [205]. |
| Calycosin | 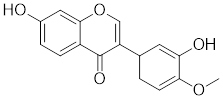 | Astragalus membranaceus Bge | Promotes HSF1 translocate to nucleus [207]. |
| Activate HSF1 at the transcriptional level | |||
| Bimoclomol | 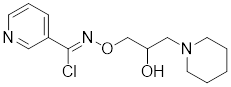 | A non-toxic hydroxylamine derivative | Enhances HSF1's binding to HSE and induces moderate HSF1 phosphorylation [208-210]. |
| FLZ | 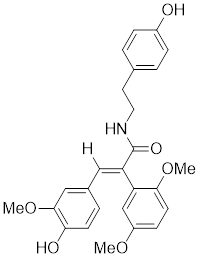 | Annona glabra | Enhances HSF1 nuclear translocation and DNA binding [211]. |
| Activate HSF1 at post-translational level | |||
| MG132 | 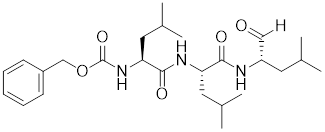 | Proteasome inhibitor | Stabilizes HSF1 by inhibiting its proteasomal degradation [212]. |
| bortezomib | 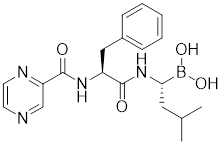 | Proteasome inhibitor | Stabilizes HSF1 by inhibiting its proteasomal degradation [212]. |
| The activators with unclear mechanisms | |||
| Tanshinone IIA (TIIA) | 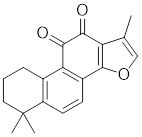 | Salvia miltiorrhiza | Increases the expression of HSF1 and the phosphorylation of HSP27 at Ser82 [213]. |
| Paeoniflorin | 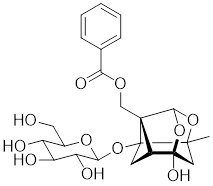 | Paeonia lactiflora | Increases the expression of HSF1 and HSP70 [214]. |
| Ginsenoside Rd | 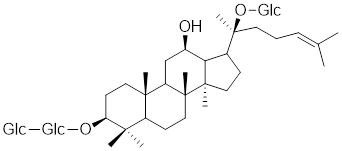 | Panax ginseng. | Increases the expression of HSF1 and HSP [215]. |
| U-133 | 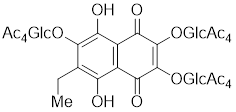 | An acetylated tris-O-glucoside echinochrome | Mildly activates HSF1 and enhances HSP70 expression [216]. |
| Taurine | 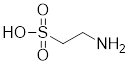 | An amino sulfonic acid in fish | Increases the myocardial expression of HSF1 and HSP70 [6]. |
| TRC051384 | 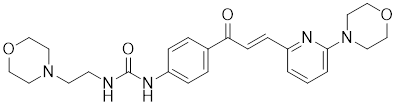 | A derivative of 2-propen-1-one | Increased the expression of HSP70 [217]. |
| Astragaloside IV | 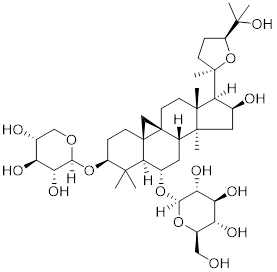 | Astragalus membranaceu Bge | Upregulates the expression of HSF1 and HSP70 [218]. |
4.1.2 Inhibiting HSF1 transcriptional activity
4.1.2.1 Targeting DNA binding ability of HSF1
The identification of Direct Targeted HSF1 Inhibitor (DTHIB) through an in vitro differential scanning fluorimetry (DSF)-based platform marks a milestone. DTHIB specifically targets the DNA binding domain of HSF1 with a high affinity, selectively accelerating the degradation of nuclear HSF1, while sparing the inactive cytosolic pool. As a result, DTHIB effectively arrested tumor progression in therapy-resistant prostate cancer animal models [154]. The selectivity of DTHIB extends to the suppression of HSF1, HSP90 and SDHC, thereby impeding the leukemia stem cell self-renewal in an AML animal model (Table 1) [155]. These findings reveal that pharmacological targeting HSF1 holds promise for a broad-spectrum anti-leukemic effect.
The RNA aptamer iaRNAHSF1, designed in 2011, binds to HSF1 with an affinity of dissociation constant (Kd) ∼ 8 nM, impeding the binding of HSF1 to HSE (Table 1). In Drosophila, iaRNAHSF1 induced a reduction in HSP83 level in a HSF1-dependent manner, leading to developmental abnormalities [156]. This exemplifies the promise of RNA aptamer technology as a cutting-edge chemical genetic approach for designing small molecules to investigate biological mechanisms.
Several natural compounds possess the capacity to regulate the transcription process of HSF1. Schizandrin A (Sch A), derived from the Chinese medicinal plant Schisandra chinensis, exhibits a moderate affinity towards HSF1 through hydrogen bonding, which effectively suppresses HSF1 transcription and the expression of HSPs, thereby inducing apoptosis in CRC cells (Table 1) [157]. Quercetin, a plant pigment with potent antioxidant properties, inhibits the heat-induced HSP70 expression by preventing HSF1 from binding to the HSE [158]. Additionally, when quercetin is combined with tumor radiofrequency ablation (RFA), a synergistic effect on cancer cell death is observed (Table 1) [159]. Fisetin, a natural flavonoid known for its antioxidant and anti-inflammatory properties, blocks the binding of HSF1 to the HSP70 promoter (Table 1) [160]. This leads to the suppression of HSP70/BAG3 complex formation and a downregulation of HSP70 expression, culminating in the induction of apoptosis in cancer cells [161].
4.1.2.2 Targeting the recruitment of transcription factors and transactivation function of HSF1
A structure-activity analysis coupled with computational screening led to the discovery of IHSF115, a novel HSF1 inhibitor that suppresses HSF1 transcriptional activity by interfering with the assembly of ATF1-containing transcription complexes (Table 1) [162]. Moreover, KRIBB11 was discovered by using a synthetic chemical library screening in 2011. Specifically, KRIBB11 inhibits HSF1-dependent recruitment of p-TEFb to the HSP70 promoter to suppress the process of transcription activation and induce apoptosis in cancer cells (Table 1) [163]. Notably, KRIBB11 treatment significantly suppresses the lymphatic metastasis in bladder cancer without apparent toxicity [164]. Both of SNS-032 and 4,6-pyrimidine series are inhibitors of CDK9 [165-167]. Because CDK9 is a component of the p-TEFb, SNS-032 and 4,6-pyrimidine series may affect the HSF1-regulated transcription [168, 169]. Consistently, recent studies have demonstrated that both compounds inhibit HSF1 transcriptional activity and effectively suppress HSP70 expression in U2OS cells [170, 171]. Additionally, 4,6-pyrimidine 25 effectively blocks the expression of HSP70, representing a promising therapeutic strategy for cancer treatment (Table 1) [167]. Cantharidin, a compound derived from traditional Chinese medicine [172], not only restrains the binding of HSF1 to the HSP70 promoter, but also hinders the recruitment of HSF1-dependent p-TEFb and blocks the p-TEFb-dependent phosphorylation of C-terminal domain (CTD) of Pol II, thereby arresting the transcriptional elongation and inducing apoptotic cell death in CRC cells (Table 1) [173].
Triptolide, derived from the traditional Chinese herb Tripterygium wilfordii, selectively interfered with the proper activity of the C-terminal transactivation domain of HSF1 without affecting its trimerization, hyperphosphorylation, or DNA-binding capacity (Table 1). Additionally, triptolide reversibly inhibits HSP70 mRNA expression and enhances stress-induced cell death in Hela cells [174].
4.1.3 HSF1 inhibitors with unclear mechanisms
To date, the intricate mechanisms underlying the functional exertion of certain inhibitors remain incompletely understood. Our current understanding is limited to the observation that these inhibitors cause a reduction in the expression levels of HSF1 and/or HSPs. While this observation provides some insights into the potential mode of action of these inhibitors, it falls short of offering a comprehensive explanation of their detailed mechanisms. Further researches are necessary to elucidate the full range of interactions and pathways through which these inhibitors exert their effects, potentially involving additional molecular targets and regulatory networks. Such insights could lead to the development of more effective and targeted therapeutic strategies.
4.1.3.1 Natural HSF1 Inhibitors
Sulphoraphane, a naturally isothiocyanate prevalent in cruciferous vegetables, suppresses the expression of HSF1 and its target genes HSP70 and HSP90, ultimately induces apoptosis in breast cancer cells (Table 1) [175]. CL-43, a member of the cardenolide family, effectively inhibits the expression of HSP70, HSP90 and HSP40 in HCT-116 cells, potentially through HSF1-mediated transcriptional mechanism. This suppression diminishes the motility and colony formation ability of HCT-116 cells, without exhibiting toxicity towards several normal cell lines (Table 1) [176]. Stresgenin B, isolated from a culture supernatant of Streptomyces spp., causes the decreased expression of HSPs, including HSP70, HSP90 and HSP110, and confers moderate cytotoxic effects on several cancer cell lines (Table 1) [177]. Ginsenoside Rg3, a steroidal saponin isolated from Panax ginseng, downregulates the expression of HSF1 and its downstream target gene fucosyltransferase IV (FUT4) (Table 1) [178]. As FUT4 is an essential enzyme in glycolipids fucosylation that promotes cancer cells growth, Rg3 treatment ultimately induces the death of gastric cancer cells [178]. Rocaglamide A (Roc A), a natural product isolated from Aglaia genus, potently inhibits HSE reporter system with an IC50 of approximately 50 nM (Table 1) [179]. Rohinitib (RHT), an analog of Roc A, exhibits greater potency than Roc A in inhibiting the HSE reporter, with an IC50 of around 20 nM [179]. RHT abolishes HSF1 binding throughout the genome, resulting in decreased mRNA levels of HSP40 and HSP70 without affecting HSF1 protein levels, ultimately leading to the suppression of tumor growth (Table 1) [179]. Given that Roc A also suppresses the activity of the translation initiation factor eIF4A [179, 180], it is suggested that rocaglates including Roc A and RHT may hinder HSF1 function by inhibiting translational flux, including protein synthesis and energy metabolism [179, 180]. Homoharringtonine, a natural plant alkaloid derived from Cephalotoxus fortune, is a protein synthesis inhibitor and has emerged as a critical therapeutic agent in hematologic malignancies [181, 182]. Homoharringtonine directly binds to HSF1, and suppresses its expression and transcription of HSF1 target genes, leading to the selective inhibition of pancreatic cancer cell viability with high HSF1 expression (Table 1) [183].
4.1.3.2 Synthetic HSF1 Inhibitors
KNK437, a benzylidene lactam compound, is recognized as a potent experimental inhibitor of HSF1. KNK437 effectively inhibits the expression of heat-induced HSPs in human CRC cells, including a range of proteins such as HSP27, HSP40, HSP70 and HSP105 (Table 1) [184, 185]. VM1, a triazole nucleoside analogue, downregulates the expression of HSF1, HSPs (HSP27, HSP70 and HSP90α) and androgen receptor (AR), offering potential benefits for prostate cancer therapy (Table 1) [186]. NZ28, a dehydroemetine derivative, was identified as a potent and non-toxic inhibitor of the HSR through high-throughput chemical screening [170]. NZ28 effectively suppresses the induction of HSPs, including HSP70 and HSP27, in various cell lines under various stress conditions (Table 1) [170]. Emunin (NZ71), a glycine amide conjugate of emetine, also displays non-toxicity while reduces the expression of HSP70 and HSP27, and significantly increases the sensitivity of myeloma and prostate carcinoma cells to inhibitors of proteasome and HSP90 (Table 1) [170].
4.2 Activators of HSF1
4.2.1 Activate HSF1 at the pre-transcriptional level
4.2.1.1 Targeting HSF1-interacting proteins
HSF1A, a potent small-molecule activator of HSF1, mitigates the suppressive effects of the HSF1-TRiC interaction though its direct binding to the TRiC/CCT complex (Table 2). This intervention enhances the expression of HSPs, thereby providing cellular defense mechanisms against protein misfolding and stress-induced apoptosis [16, 115]. Additionally, the synergistic application of HSF1A alongside 17-AAG has demonstrated the ability to augment the reconstituting activity of ex-vivo-cultured hematopoietic stem cells (HSCs), ensuring proteostasis and bolstering the regenerative capacity of HSCs resilience during periods of culture-induced stress and senescence [187]. Geldanamycin, noted for its specific affinity to HSP90 [188], effectively dismantles the stable complex of HSP90/HSF1, thereby activating HSF1 in mammalian cells (Table 2) [53, 189]. This activation is linked to neuroprotective effects, offering therapeutic promise in the treatment of HD [53, 189]. Phenethyl isothiocyanate (PEITC), abundant in watercress (Nasturtium officinale), triggers the dissociation of HSF1 from HSP90 and further induces HSF1 phosphorylation at Ser326 (Table 2) [190]. This dual action drives the nuclear accumulation of HSF1 and triggers a marked induction of HSP70, highlighting its potential as a therapeutic agent [190].
4.2.1.2 Targeting PTMs of HSF1
SYSU-3d, a 2-pyrimidinylindole derivative, facilitates the phosphorylation of HSF1 at Ser326 and its nuclear translocation in HCC cells (Table 2) [191]. Subsequently, SYSU-3d activates the HSF1/PPARγ coactivator-1α (PGC-1α) pathway, increases mitochondrial biogenesis, combats oxidative stress, and alleviates non-alcoholic steatohepatitis (NASH) [191]. Englerin A (EA), derived from the stem bark extract of Phyllanthus engleri [192], stimulates the protein kinase C-θ (PKCθ)-dependent phosphorylation of HSF1 at Ser333, leading to the dissociation of HSF1 from HSP90, and enhancing the transcriptional activity of HSF1 (Table 2). EA induces tumor cell glucose dependency and triggers glycolytic cell death under glucose deprivation conditions [193]. Resveratrol, a natural phenol synthesized by various plants in response to stress [194], promotes SIRT1-mediated deacetylation of HSF1 and upregulates the expression of HSP25 and HSP70 (Table 2) [195]. These actions protect motor neurons from mutant superoxide dismutase 1 (SOD1)-induced neurotoxicity, significantly extend the lifespan of mutant SOD1-bearing mice. These findings suggest that resveratrol may be a prospective therapeutic for amyotrophic lateral sclerosis (ALS) [195].
4.2.1.3 Targeting trimerization and nuclear translocation of HSF1
Celastrol, a chemical compound derived from the root extracts of Tripterygium wilfordii and Tripterygium regelii, regulates energy metabolism through activating HSF1-PGC1α transcriptional axis [196]. Additionally, celastrol is frequently combined with other therapeutic drugs to achieve synergistic antitumor efficacy. For instance, the combination of nanoparticle-encapsulated celastrol with DOX can enhance drug accumulation in multidrug-resistant cells. This is achieved by inducing HSF1 trimerization and nuclear translocation, which in turn promotes autophagy and apoptosis in DOX-resistant cells, pointing to a promising strategy for overcoming DOX resistance via synergistic chemotherapy [197]. Similarly, our recent study also reveals that the combination of celastrol with erastin, well-known ferroptosis inducer, synergistically triggers cell death of lung cancer cells via the ROS-mitochondiral fission-mitophagy pathway. Interestingly, the co-treatment of celastrol and erastin also significantly boosts the expression of HSPs by promoting the phosphorylation and nuclear translocation of HSF1. Knockdown of HSF1 further enhances the cytotoxic of combination of erastin and celastrol in vitro and in vivo [198]. Collectively, these observations clearly highlight the role of celastrol as a potential activator of HSF1, with significant implications for cancer therapy (Table 2).
Curcumin, a natural product extracted from the rhizome of the turmeric plant (Curcuma longa) [199], induces HSF1 trimerization [200], promotes its nuclear translocation [201], and enhances the DNA binding capacity, leading to increased HSP70 expression in human colorectal carcinoma [200] and HeLa cells (Table 2) [201] . Additionally, curcumin specifically accelerates HSP90 mRNA degradation in C. albicans [202], which may contribute to the activation of HSF1. Oridonin, a natural terpenoids derived from the traditional Chinese medicinal herb Isodon rubescens, triggers oxidative stress in cancer cells by directly binding to the C153 residue of HSF1 [203]. This interaction prevents the formation of intramolecular disulfide covalent bonds and instead promotes the formation of intermolecular disulfide covalent bond formation, facilitating HSF1 trimerization and increasing the expression of HSP70 and ubiquitin proteins (Table 2) [203]. Metformin, a biguanide that is extracted from the herb Galega officinalis, is the first-line therapy for type 2 diabetes mellitus due to its efficacy, safety profile, and cost-effectiveness [204]. Metformin promotes the nuclear translocation of HSF1 and activation of HSF1-UPRmt signaling pathway, providing a cardioprotective effect under pressure overload conditions [205]. Calycosin is an O-methylated isoflavone isolated from Astragalus membranaceus Bge [206]. A recent study indicates that HSF1 can translocate to the nucleus to enhance cell survival following calycosin treatment (Table 2) [207].
4.2.2 Activate HSF1 at the transcriptional level
Bimoclomol, a non-toxic hydroxylamine derivative with therapeutic potential in diabetes and cardiac dysfunction, exhibits a subtle yet significant binding affinity to HSF1. This interaction prolongs HSF1's residence time on the HSE and triggers a moderate phosphorylation of HSF1 in response to cellular stress, thereby activating the HSR (Table 2) [208-210]. FLZ, synthesized from the Chinese herb Annona glabra, facilitates the nuclear translocation of HSF1 and promotes its binding to the HSEs (Table 2) [211]. Importantly, FLZ treatment significantly upregulates the expression of HSP27 and HSP70, conferring a robust neuroprotective effect in experimental models of PD [211].
4.2.3 Activate HSF1 at post-translational level
Proteasome inhibitors, such as MG132 and bortezomib, are used as clinical drugs for treatment of myeloma. These drugs stabilize HSF1 by inhibiting its proteasomal degradation (Table 2) [212]. Moreover, these agents promote the differentiation of CD69+ regulatory T cells (Tregs), resulting in the efficient production of Tregs for the treatment of colitis and other autoimmune diseases characterized by Tregs deficiency [212].
4.2.4 The activators with unclear mechanisms
Tanshinone IIA (TIIA) is a compound derived from the traditional medicinal herb Salvia miltiorrhiza with prominent anticancer properties. TIIA induces the expression of HSF1 and phosphorylation of HSP27 at Ser82 in gastric cancer cells (Table 2) [213]. This triggers a cascade that includes the accumulation of ROS, the activation of UPR, and the cell death in gastric cancer [213]. Paeoniflorin, derived from herb Paeonia lactiflora, increases the expression of HSF1 and HSP70, suppresses protein aggregation in neuronal differentiated SH-SY5Y cells, and improves the neurodegenerative symptoms in animal models of PD and AD (Table 2) [214]. Total ginsenoside (TG), a mixture of the primary active ginsenosides from Panax ginseng, upregulates the expression of HSF1 and HSP-16.2 in C. elegans. One of its main constituents, ginsenoside Rd, extends the lifespan of C. elegans, indicating the involvement of TG in HSF1-mediated aging (Table 2) [215]. U-133, an acetylated tris-O-glucoside echinochrome and the main pigment in sea urchins, activates HSF1 to an extent akin to mild heat shock, promoting HSP70 expression (Table 2). This activation counters PD-like neurodegeneration [216]. Taurine, an amino sulfonic acid abundant in fish, upregulates the myocardial expression of HSF1 and HSP70, thereby exerting cardioprotective effects (Table 2) [6]. TRC051384, a compound from the substituted 2-propen-1-one class, is a potent inducer of HSP70. It significantly attenuates stroke-associated neuronal impairment and dysfunction in a rat model of transient ischemic stroke (Table 2) [217]. Astragaloside IV (AS-IV), a natural product isolated from Astragalus membranaceus Bge, elevates the expression of HSF1 at both the mRNA and protein levels (Table 2) [218]. Additionally, AS-IV increases the expression levels of HSP70, HIF-1α and VEGF, collectively contributing to an amelioration of heart failure induced by pressure overload [218].
5. Discussion
HSF1 stands as a pivotal transcriptional regulator that orchestrates the induction of HSPs, serving as a protective mechanism for proteomic quality control (PQC) and maintenance of protein homeostasis in response to stress [219, 220]. The successful isolation, cloning and identification of HSF1 have marked significant milestones, propelling research in this domain to rapidly progress over the past three decades [221]. The elaborate assembly of multi-protein complexes and HSF1-mediated transcriptional activation are cornerstones of cellular PQC mechanisms [221, 222]. Disruptions or aberrant activation of HSF1 pathways can precipitate a cascade of events that destroy the physiological and pathological balance, leading to dysfunction or abnormality in protein PQC [101, 221]. Such imbalances are implicated in the etiology of cancer [98], neurodegenerative diseases [19, 115] and a spectrum of other human diseases [5, 130, 131, 133, 134].
The discovery of HSF1 has not only deepened our understanding of the cellular stress response but has also unveiled potential therapeutic targets for various diseases. The activation of HSF1 and the subsequent upregulation of HSPs can protect cells from stress-induced damage, offering a preventive or mitigating effect against the progression of certain pathologies [19, 115]. Conversely, the dysregulation of HSF1 activity, such as sustained overactivation or insufficient activation, contributes to pathological outcomes [19, 115]. For instance, in neurodegenerative diseases, the failure to mount an adequate stress response leads to the accumulation of misfolded proteins and cellular toxicity [19, 115]. Moreover, the intricate interplay between HSF1 and other cellular pathways adds layers of complexity to its role in disease [95, 100, 223]. The modulation of HSF1 activity, therefore, needs to be finely balanced to harness its potential therapeutic benefits without exacerbating pathological conditions. Ongoing research is focused on elucidating these complex mechanisms to develop targeted therapies that can modulate HSF1 activity in a controlled manner.
In the current landscape of pharmacological research, a multitude of inhibitors and activators that target different stages of HSF1 activation have emerged, demonstrating significant therapeutic potential in experimental settings. As research progresses, the development of HSF1 modulators will likely benefit from advances in structural biology, computational modeling, and high-throughput screening technologies. These tools facilitate the design of molecules with improved binding affinity, selectivity, and pharmacokinetic properties. Despite their promise, the majority of small molecules face the challenge of lacking specificity and potency due to their inability to bind directly to HSF1. However, a select group of HSF1 inhibitors has been identified that can interact directly with HSF1, including vitexin [151], DTHIB [154], Sch A [157], IHSF115 [162], bimoclomol [208], oridonin [42] and HHT [181]. These selective inhibitors have opened new avenues in the development of HSF1 modulators, offering more precise control over HSF1 activity. Additionally, the RNA aptamer technology, exemplified by iaRNAHSF1, represents a promising chemical genetic approach to discovering novel HSF1 inhibitors. This technology harnesses the specificity of nucleic acid hybridization to modulate protein function, providing a new frontier in the search for HSF1 inhibitors. In addition to direct HSF1 modulators, many inhibitors of HSP70 and HSP90, such as Timosaponin AIII (Tim-AIII) [224], C0818 [225], and VER-155008 [226-228], have been identified. While these compounds were initially discovered for their effects on HSPs, their potential to modulate HSF1 activity warrants further exploration, as they may indirectly influence the HSR by affecting the expression or function of HSPs. The exploration of these diverse compounds necessitates a comprehensive understanding of the complex interplay between HSF1 and other cellular pathways. It is essential to elucidate how these inhibitors and activators integrate into the intricate network of cellular responses to stress and how they might be leveraged to achieve therapeutic outcomes. Moreover, the translation of these findings from the laboratory to the clinic will require rigorous preclinical and clinical evaluation to assess the safety, efficacy and optimal dosing regimens of these compounds. The goal is to develop therapies that can be tailored to individual patient needs, taking into account factors such as disease stage, genetic background and comorbidities.
In summary, the pivotal role of HSF1 in upholding cellular proteostasis and its profound implications in a diverse array of diseases highlight the imperative for ongoing research. The quest for the discovery and refinement of HSF1 modulators stands as a dynamic and promising field, with the capacity to revolutionize therapeutic approaches for diseases characterized by protein misfolding and aggregation. The advancement of HSF1-targeted therapies is expected to benefit from interdisciplinary collaboration, merging insights from molecular biology, pharmacology, and genomics. The integration of bioinformatics and systems biology will further enhance our ability to predict the effects of HSF1 modulation and to identify potential off-target effects, thereby facilitating the design of safer and more effective drugs.
Abbreviations
3'-UTR: 3'-untranslated region; ACC: Adrenocortical carcinoma; AD: Alzheimer's disease; AMPK: adenosine monophosphate-activated protein kinase; APOJ: apolipoprotein J; AS-IV: Astragaloside IV; ATF1: activating transcription factor 1; Aβ: amyloid-β; BAP1: BRCA1-associated protein-1; BDNF: brain-derived neurotrophic factor; BLCA: Bladder urothelial carcinoma; BRCA: Breast invasive carcinoma; CCT: chaperonin containing TCP-1; CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHIP: carboxyl-terminus of HSP70 interacting protein; CHOL: Cholangio carcinoma; CHOP: C/EBP homologous protein; COAD: Colon adenocarcinoma; CRC: colorectal cancer; CTD: C-terminal domain; DLBC: Lymphoid neoplasm diffuse large B-cell lymphoma; DOX: Doxorubicin; DSF: differential scanning fluorimetry; DSIF: DRB sensitivity-inducing factor; DTHIB: Direct Targeted HSF1 Inhibitor; EA: Englerin A; EGFR: epidermal growth factor receptor; EMT: epithelial-mesenchymal transition; ESCA: Esophageal carcinoma; FTO: fat mass and obesity-associated protein; FUT4: fucosyltransferase IV; FOXO3a: forkhead box protein O3a; GBM: glioblastoma; GBM: Glioblastoma multiforme; HBx: hepatitis B X protein; HBV: hepatitis B virus; HCC: hepatocellular carcinoma; HD: Huntington's disease; Hnrnpa2b1: heterogeneous nuclear ribonucleoprotein A2/B1; HNSC: Head and neck squamous cell carcinoma; HSCs: hematopoietic stem cells; HSF: human heat shock factor; HSF1: heat shock factor 1; HSE: heat shock element; HSPs: heat shock proteins; HSR: Heat shock response; IGF-IIR: insulin-like growth factor receptor II; JMJD6: Jumonji domain-containing protein 6; Kd: dissociation constant; KICH: Kidney chromophobe; KIRC: Kidney renal clear cell carcinoma; KIRP: Kidney renal papillary cell carcinoma; LAML: Acute myeloid leukemia; LGG: Brain lower grade glioma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; miRNAs: microRNAs; MESO: Mesothelioma; NEDD4: neuronally expressed downregulated protein 4; NELF: negative elongation factor; NFAT: nuclear factor of activated T cells; NRF1: nuclear respiratory factor 1; OS: overall survival; OV: Ovarian serous cystadenocarcinoma; PAAD: Pancreatic adenocarcinoma; PARP1: poly (ADP)-ribose polymerase 1; PCPG: Pheochromocytoma and paraganglioma; PD: Parkinson's disease; PDSM: phosphorylation-dependent SUMOylation motif; PEITC: Phenethyl isothiocyanate; PGC-1α: PPARγ coactivator-1α; PKC: protein kinase C; Pol II: RNA polymerase II; polyQ: polyglutamine; PQC: proteomic quality control; PRAD: Prostate adenocarcinoma; PTMs: post-translational modifications; READ: Rectum adenocarcinoma; RFA: radiofrequency ablation; RHT: Rohinitib; RIPK3: receptor-interacting protein kinase 3; Roc A: Rocaglamide A; RPA: replication protein A; SARC: Sarcoma; Sch A: Schizandrin A; SDHC: succinate dehydrogenase C; SGO2: shugoshin 2; SIRT1: sirtuin 1; SKCM: Skin cutaneous melanoma; SOD1: superoxide dismutase 1; SSBP1: single-stranded DNA binding protein 1; STAD: Stomach adenocarcinoma; TAD: transactivation domain; TG: Total ginsenoside; TGCT: Testicular germ cell tumors; THCA: Thyroid carcinoma; THYM: Thymoma; TIIA: Tanshinone IIA; Tim-AIII: Timosaponin AIII; TME: tumor microenvironment; TRiC: TCP-1 ring complex; TRIM11: tripartite motif containing 11; Tregs: regulatory T cells; UCEC: Uterine corpus endometrial carcinoma; UCS: Uterine carcinosarcoma; UPS: ubiquitin-proteasome system; UPRmt: mitochondrial unfolded protein response; UVM: Uveal melanoma; ZBP1: Z-DNA binding protein 1; ΔNp63α: p63 isoform in the p53 family.
Acknowledgements
This work was supported by the Guiding Funds of Central Government for Supporting the Development of the Local Science and Technology (236Z3003G), the Hebei Province Higher Education Scientific Research Special Task Project (JZX2024020), Key Development Fund of Hebei Normal University (L2024ZD16), and Interdisciplinary Research Fund of Hebei Normal University (L2024J04).
Author contributions
Bingwei Zhang: Writing-review & editing, Writing-original draft, Methodology, Conceptualization. Yumei Fan: Writing-review & editing, Writing-original draft. Ke Tan: Writing-review & editing, Writing-original draft, Methodology, Investigation, Conceptualization, Funding acquisition.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Molecular cell. 2010;40:253-66
2. Nakai A. Heat shock factor: Springer; 2016
3. Hu C, Yang J, Qi Z, Wu H, Wang B, Zou F. et al. Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm. 2022;3:e161
4. Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA. et al. Guidelines for the nomenclature of the human heat shock proteins. Cell stress & chaperones. 2009;14:105-11
5. Wang N, Ma H, Li J, Meng C, Zou J, Wang H. et al. HSF1 functions as a key defender against palmitic acid-induced ferroptosis in cardiomyocytes. Journal of molecular and cellular cardiology. 2021;150:65-76
6. Setyarani M, Zinellu A, Carru C, Zulli A. High dietary taurine inhibits myocardial apoptosis during an atherogenic diet: association with increased myocardial HSP70 and HSF-1 but not caspase 3. European journal of nutrition. 2014;53:929-37
7. Qin T, Chen K, Li J, Qian W, Xiao Y, Wu E. et al. Heat shock factor 1 inhibition sensitizes pancreatic cancer to gemcitabine via the suppression of cancer stem cell-like properties. Biomedicine & Pharmacotherapy. 2022;148:112713
8. Neudegger T, Verghese J, Hayer-Hartl M, Hartl FU, Bracher A. Structure of human heat-shock transcription factor 1 in complex with DNA. Nature structural & molecular biology. 2016;23:140-6
9. Anckar J, Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annual review of biochemistry. 2011;80:1089-115
10. Hentze N, Le Breton L, Wiesner J, Kempf G, Mayer MP. Molecular mechanism of thermosensory function of human heat shock transcription factor Hsf1. eLife. 2016;5:e11576
11. Rabindran SK, Haroun RI, Clos J, Wisniewski J, Wu C. Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science (New York, NY). 1993;259:230-4
12. Green M, Schuetz TJ, Sullivan EK, Kingston RE. A heat shock-responsive domain of human HSF1 that regulates transcription activation domain function. Molecular and cellular biology. 1995;15:3354-62
13. Piskacek M. Common Transactivation Motif 9aaTAD recruits multiple general co-activators TAF9, MED15, CBP and p300. Nature Precedings. 2009
14. Kmiecik SW, Mayer MP. Molecular mechanisms of heat shock factor 1 regulation. Trends in biochemical sciences. 2022;47:218-34
15. Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes & development. 1998;12:3788-96
16. Neef DW, Jaeger AM, Gomez-Pastor R, Willmund F, Frydman J, Thiele DJ. A direct regulatory interaction between chaperonin TRiC and stress-responsive transcription factor HSF1. Cell reports. 2014;9:955-66
17. Vujanac M, Fenaroli A, Zimarino V. Constitutive nuclear import and stress-regulated nucleocytoplasmic shuttling of mammalian heat-shock factor 1. Traffic (Copenhagen, Denmark). 2005;6:214-29
18. Peteranderl R, Rabenstein M, Shin YK, Liu CW, Wemmer DE, King DS. et al. Biochemical and biophysical characterization of the trimerization domain from the heat shock transcription factor. Biochemistry. 1999;38:3559-69
19. Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nature reviews Molecular cell biology. 2010;11:545-55
20. Littlefield O, Nelson HC. A new use for the 'wing' of the 'winged' helix-turn-helix motif in the HSF-DNA cocrystal. Nature structural biology. 1999;6:464-70
21. Zhang B, Fan Y, Cao P, Tan K. Multifaceted roles of HSF1 in cell death: A state-of-the-art review. Biochimica et biophysica acta Reviews on cancer. 2021;1876:188591
22. Chowdhary S, Kainth AS, Pincus D, Gross DS. Heat Shock Factor 1 Drives Intergenic Association of Its Target Gene Loci upon Heat Shock. Cell reports. 2019;26:18-28.e5
23. Chowdhary S, Kainth AS, Paracha S, Gross DS, Pincus D. Inducible transcriptional condensates drive 3D genome reorganization in the heat shock response. Molecular cell. 2022;82:4386-99.e7
24. Ray J, Munn PR, Vihervaara A, Lewis JJ, Ozer A, Danko CG. et al. Chromatin conformation remains stable upon extensive transcriptional changes driven by heat shock. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:19431-9
25. Kmiecik SW, Drzewicka K, Melchior F, Mayer MP. Heat shock transcription factor 1 is SUMOylated in the activated trimeric state. The Journal of biological chemistry. 2021;296:100324
26. Niskanen EA, Palvimo JJ. Chromatin SUMOylation in heat stress: To protect, pause and organise?: SUMO stress response on chromatin. BioEssays: news and reviews in molecular, cellular and developmental biology. 2017 39 (6)
27. Zelin E, Zhang Y, Toogun OA, Zhong S, Freeman BC. The p23 molecular chaperone and GCN5 acetylase jointly modulate protein-DNA dynamics and open chromatin status. Molecular cell. 2012;48:459-70
28. Raychaudhuri S, Loew C, Körner R, Pinkert S, Theis M, Hayer-Hartl M. et al. Interplay of acetyltransferase EP300 and the proteasome system in regulating heat shock transcription factor 1. Cell. 2014;156:975-85
29. Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic acids research. 2015;43:D512-20
30. Budzyński MA, Puustinen MC, Joutsen J, Sistonen L. Uncoupling Stress-Inducible Phosphorylation of Heat Shock Factor 1 from Its Activation. Molecular and cellular biology. 2015;35:2530-40
31. Zheng X, Krakowiak J, Patel N, Beyzavi A, Ezike J, Khalil AS. et al. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. eLife. 2016;5:e18638
32. Lu WC, Omari R, Ray H, Wang J, Williams I, Jacobs C. et al. AKT1 mediates multiple phosphorylation events that functionally promote HSF1 activation. The FEBS journal. 2022;289:3876-93
33. Jin X, Qiao A, Moskophidis D, Mivechi NF. Modulation of Heat Shock Factor 1 Activity through Silencing of Ser303/Ser307 Phosphorylation Supports a Metabolic Program Leading to Age-Related Obesity and Insulin Resistance. Molecular and cellular biology. 2018;38:e00095-18
34. Ishikawa Y, Kawabata S, Sakurai H. HSF1 transcriptional activity is modulated by IER5 and PP2A/B55. FEBS letters. 2015;589:1150-5
35. Asano Y, Kawase T, Okabe A, Tsutsumi S, Ichikawa H, Tatebe S. et al. IER5 generates a novel hypo-phosphorylated active form of HSF1 and contributes to tumorigenesis. Scientific reports. 2016;6:19174
36. Fujimoto M, Takii R, Matsumoto M, Okada M, Nakayama KI, Nakato R. et al. HSF1 phosphorylation establishes an active chromatin state via the TRRAP-TIP60 complex and promotes tumorigenesis. Nature Communications. 2022;13:4355
37. Niskanen EA, Malinen M, Sutinen P, Toropainen S, Paakinaho V, Vihervaara A. et al. Global SUMOylation on active chromatin is an acute heat stress response restricting transcription. Genome biology. 2015;16:153
38. Hietakangas V, Ahlskog JK, Jakobsson AM, Hellesuo M, Sahlberg NM, Holmberg CI. et al. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Molecular and cellular biology. 2003;23:2953-68
39. Liebelt F, Sebastian RM, Moore CL, Mulder MPC, Ovaa H, Shoulders MD. et al. SUMOylation and the HSF1-Regulated Chaperone Network Converge to Promote Proteostasis in Response to Heat Shock. Cell reports. 2019;26:236-49.e4
40. Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A. et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:45-50
41. Li X, Wang Z, Gao B, Dai K, Wu J, Shen K. et al. Unveiling the impact of SUMOylation at K298 site of heat shock factor 1 on glioblastoma malignant progression. Neoplasia (New York, NY). 2024;57:101055
42. Huang C, Hu W, Wang J, Tong L, Lu X, Wu F. et al. Methylene blue increases the amount of HSF1 through promotion of PKA-mediated increase in HSF1-p300 interaction. The international journal of biochemistry & cell biology. 2017;84:75-88
43. Yuan W, Zhang Q, Zhao Y, Xia W, Yin S, Liang X. et al. BAP1 regulates HSF1 activity and cancer immunity in pancreatic cancer. Journal of experimental & clinical cancer research: CR. 2024;43:275
44. Westerheide SD, Anckar J, Stevens SM Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science (New York, NY). 2009;323:1063-6
45. Kourtis N, Moubarak RS, Aranda-Orgilles B, Lui K, Aydin IT, Trimarchi T. et al. FBXW7 modulates cellular stress response and metastatic potential through HSF1 post-translational modification. Nature cell biology. 2015;17:322-32
46. Kim E, Wang B, Sastry N, Masliah E, Nelson PT, Cai H. et al. NEDD4-mediated HSF1 degradation underlies α-synucleinopathy. Human molecular genetics. 2016;25:211-22
47. Shi Y, Mosser DD, Morimoto RI. Molecular chaperones as HSF1-specific transcriptional repressors. Genes & development. 1998;12:654-66
48. Baler R, Zou J, Voellmy R. Evidence for a role of Hsp70 in the regulation of the heat shock response in mammalian cells. Cell stress & chaperones. 1996;1:33-9
49. Kmiecik SW, Le Breton L, Mayer MP. Feedback regulation of heat shock factor 1 (Hsf1) activity by Hsp70-mediated trimer unzipping and dissociation from DNA. The EMBO journal. 2020;39:e104096
50. Alasady MJ, Koeva M, Takagishi SR, Segal D, Amici DR, Smith RS. et al. An HSF1-JMJD6-HSP feedback circuit promotes cell adaptation to proteotoxic stress. Proceedings of the National Academy of Sciences of the United States of America. 2024;121:e2313370121
51. Alderson TR, Kim JH, Markley JL. Dynamical Structures of Hsp70 and Hsp70-Hsp40 Complexes. Structure (London, England: 1993). 2016;24:1014-30
52. Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nature reviews Molecular cell biology. 2010;11:579-92
53. Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471-80
54. Bharadwaj S, Ali A, Ovsenek N. Multiple components of the HSP90 chaperone complex function in regulation of heat shock factor 1 In vivo. Molecular and cellular biology. 1999;19:8033-41
55. Conde R, Belak ZR, Nair M, O'Carroll RF, Ovsenek N. Modulation of Hsf1 activity by novobiocin and geldanamycin. Biochemistry and cell biology. 2009;87:845-51
56. Trott A, West JD, Klaić L, Westerheide SD, Silverman RB, Morimoto RI. et al. Activation of heat shock and antioxidant responses by the natural product celastrol: transcriptional signatures of a thiol-targeted molecule. Molecular biology of the cell. 2008;19:1104-12
57. Westerheide SD, Bosman JD, Mbadugha BN, Kawahara TL, Matsumoto G, Kim S. et al. Celastrols as inducers of the heat shock response and cytoprotection. The Journal of biological chemistry. 2004;279:56053-60
58. Schulte TW, Akinaga S, Soga S, Sullivan W, Stensgard B, Toft D. et al. Antibiotic radicicol binds to the N-terminal domain of Hsp90 and shares important biologic activities with geldanamycin. Cell stress & chaperones. 1998;3:100-8
59. Lawson B, Brewer JW, Hendershot LM. Geldanamycin, an hsp90/GRP94-binding drug, induces increased transcription of endoplasmic reticulum (ER) chaperones via the ER stress pathway. Journal of cellular physiology. 1998;174:170-8
60. Kijima T, Prince TL, Tigue ML, Yim KH, Schwartz H, Beebe K. et al. HSP90 inhibitors disrupt a transient HSP90-HSF1 interaction and identify a noncanonical model of HSP90-mediated HSF1 regulation. Scientific reports. 2018;8:6976
61. Gestaut D, Limatola A, Joachimiak L, Frydman J. The ATP-powered gymnastics of TRiC/CCT: an asymmetric protein folding machine with a symmetric origin story. Current opinion in structural biology. 2019;55:50-8
62. Rossi A, Riccio A, Coccia M, Trotta E, La Frazia S, Santoro MG. The proteasome inhibitor bortezomib is a potent inducer of zinc finger AN1-type domain 2a gene expression: role of heat shock factor 1 (HSF1)-heat shock factor 2 (HSF2) heterocomplexes. The Journal of biological chemistry. 2014;289:12705-15
63. Shinkawa T, Tan K, Fujimoto M, Hayashida N, Yamamoto K, Takaki E. et al. Heat shock factor 2 is required for maintaining proteostasis against febrile-range thermal stress and polyglutamine aggregation. Molecular biology of the cell. 2011;22:3571-83
64. Alasady MJ, Mendillo ML. The heat shock factor code: Specifying a diversity of transcriptional regulatory programs broadly promoting stress resilience. Cell stress & chaperones. 2024;29:735-49
65. Sandqvist A, Björk JK, Akerfelt M, Chitikova Z, Grichine A, Vourc'h C. et al. Heterotrimerization of heat-shock factors 1 and 2 provides a transcriptional switch in response to distinct stimuli. Molecular biology of the cell. 2009;20:1340-7
66. Jaeger AM, Pemble CWt, Sistonen L, Thiele DJ. Structures of HSF2 reveal mechanisms for differential regulation of human heat-shock factors. Nature structural & molecular biology. 2016;23:147-54
67. Smith RS, Takagishi SR, Amici DR, Metz K, Gayatri S, Alasady MJ. et al. HSF2 cooperates with HSF1 to drive a transcriptional program critical for the malignant state. Science advances. 2022;8:eabj6526
68. Santopolo S, Riccio A, Rossi A, Santoro MG. The proteostasis guardian HSF1 directs the transcription of its paralog and interactor HSF2 during proteasome dysfunction. Cellular and molecular life sciences: CMLS. 2021;78:1113-29
69. Syafruddin SE, Ling S, Low TY, Mohtar MA. More Than Meets the Eye: Revisiting the Roles of Heat Shock Factor 4 in Health and Diseases. Biomolecules. 2021;11:523
70. Cui X, Xie PP, Jia PP, Lou Q, Dun G, Li S. et al. Hsf4 counteracts Hsf1 transcription activities and increases lens epithelial cell survival in vitro. Biochimica et biophysica acta. 2015;1853:746-55
71. Zhang Y, Frejtag W, Dai R, Mivechi NF. Heat shock factor-4 (HSF-4a) is a repressor of HSF-1 mediated transcription. Journal of cellular biochemistry. 2001;82:692-703
72. Fujimoto M, Izu H, Seki K, Fukuda K, Nishida T, Yamada S. et al. HSF4 is required for normal cell growth and differentiation during mouse lens development. The EMBO journal. 2004;23:4297-306
73. Fujimoto M, Oshima K, Shinkawa T, Wang BB, Inouye S, Hayashida N. et al. Analysis of HSF4 binding regions reveals its necessity for gene regulation during development and heat shock response in mouse lenses. The Journal of biological chemistry. 2008;283:29961-70
74. Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E. et al. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes & development. 2003;17:1402-14
75. Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S. et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41-51
76. Ni Z, Saunders A, Fuda NJ, Yao J, Suarez JR, Webb WW. et al. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Molecular and cellular biology. 2008;28:1161-70
77. Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Molecular cell. 2004;13:55-65
78. Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes & development. 2000;14:792-803
79. Ouararhni K, Hadj-Slimane R, Ait-Si-Ali S, Robin P, Mietton F, Harel-Bellan A. et al. The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity. Genes & development. 2006;20:3324-36
80. Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74-84
81. Petesch SJ, Lis JT. Activator-induced spread of poly(ADP-ribose) polymerase promotes nucleosome loss at Hsp70. Molecular cell. 2012;45:64-74
82. Fujimoto M, Takii R, Takaki E, Katiyar A, Nakato R, Shirahige K. et al. The HSF1-PARP13-PARP1 complex facilitates DNA repair and promotes mammary tumorigenesis. Nature Communications. 2017;8:1638
83. Tan K, Fujimoto M, Takii R, Takaki E, Hayashida N, Nakai A. Mitochondrial SSBP1 protects cells from proteotoxic stresses by potentiating stress-induced HSF1 transcriptional activity. Nature communications. 2015;6:6580
84. Zhou Z, Fan Y, Zong R, Tan K. The mitochondrial unfolded protein response: A multitasking giant in the fight against human diseases. Ageing research reviews. 2022;81:101702
85. Zhang S, Guo H, Wang H, Liu X, Wang M, Liu X. et al. A novel mitochondrial unfolded protein response-related risk signature to predict prognosis, immunotherapy and sorafenib sensitivity in hepatocellular carcinoma. Apoptosis: an international journal on programmed cell death. 2024;29:768-84
86. Wang G, Fan Y, Cao P, Tan K. Insight into the mitochondrial unfolded protein response and cancer: opportunities and challenges. Cell & bioscience. 2022;12:18
87. Zhang X, Fan Y, Tan K. A bird's eye view of mitochondrial unfolded protein response in cancer: mechanisms, progression and further applications. Cell death & disease. 2024;15:667
88. Fujimoto M, Takaki E, Takii R, Tan K, Prakasam R, Hayashida N. et al. RPA assists HSF1 access to nucleosomal DNA by recruiting histone chaperone FACT. Molecular cell. 2012;48:182-94
89. Chen L, Yang X. TRIM11 cooperates with HSF1 to suppress the anti-tumor effect of proteotoxic stress drugs. Cell cycle (Georgetown, Tex). 2019;18:60-8
90. Takii R, Fujimoto M, Matsumoto M, Srivastava P, Katiyar A, Nakayama KI. et al. The pericentromeric protein shugoshin 2 cooperates with HSF1 in heat shock response and RNA Pol II recruitment. The EMBO journal. 2019;38:e102566
91. Takii R, Fujimoto M, Tan K, Takaki E, Hayashida N, Nakato R. et al. ATF1 Modulates the Heat Shock Response by Regulating the Stress-Inducible Heat Shock Factor 1 Transcription Complex. Molecular and Cellular Biology. 2015;35:11-25
92. Carpenter RL, Gökmen-Polar Y. HSF1 as a Cancer Biomarker and Therapeutic Target. Current cancer drug targets. 2019;19:515-24
93. Alasady MJ, Mendillo ML. The Multifaceted Role of HSF1 in Tumorigenesis. Advances in experimental medicine and biology. 2020;1243:69-85
94. Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM. et al. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150:549-62
95. Scherz-Shouval R, Santagata S, Mendillo ML, Sholl LM, Ben-Aharon I, Beck AH. et al. The reprogramming of tumor stroma by HSF1 is a potent enabler of malignancy. Cell. 2014;158:564-78
96. Prince TL, Lang BJ, Guerrero-Gimenez ME, Fernandez-Muñoz JM, Ackerman A, Calderwood SK. HSF1: Primary Factor in Molecular Chaperone Expression and a Major Contributor to Cancer Morbidity. Cells. 2020;9:1046
97. Chen F, Fan Y, Cao P, Liu B, Hou J, Zhang B. et al. Pan-Cancer Analysis of the Prognostic and Immunological Role of HSF1: A Potential Target for Survival and Immunotherapy. Oxidative medicine and cellular longevity. 2021;2021:5551036
98. Dai C, Sampson SB. HSF1: guardian of proteostasis in cancer. Trends Cell Biol. 2016;26:17-28
99. Wang G, Cao P, Fan Y, Tan K. Emerging roles of HSF1 in cancer: Cellular and molecular episodes. Biochimica et biophysica acta Reviews on cancer. 2020;1874:188390
100. Dong B, Jaeger AM, Thiele DJ. Inhibiting Heat Shock Factor 1 in Cancer: A Unique Therapeutic Opportunity. Trends in pharmacological sciences. 2019;40:986-1005
101. Gomez-Pastor R, Burchfiel ET, Thiele DJ. Regulation of heat shock transcription factors and their roles in physiology and disease. Nature reviews Molecular cell biology. 2018;19:4-19
102. Björk JK, Ahonen I, Mirtti T, Erickson A, Rannikko A, Bützow A. et al. Increased HSF1 expression predicts shorter disease-specific survival of prostate cancer patients following radical prostatectomy. Oncotarget. 2018;9:31200-13
103. Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005-18
104. Solimini NL, Luo J, Elledge SJ. Non-oncogene addiction and the stress phenotype of cancer cells. Cell. 2007;130:986-8
105. Moreno R, Banerjee S, Jackson AW, Quinn J, Baillie G, Dixon JE. et al. The stress-responsive kinase DYRK2 activates heat shock factor 1 promoting resistance to proteotoxic stress. Cell death and differentiation. 2021;28:1563-78
106. Xu A, Zhang J, Zuo L, Yan H, Chen L, Zhao F. et al. FTO promotes multiple myeloma progression by posttranscriptional activation of HSF1 in an m(6)A-YTHDF2-dependent manner. Molecular therapy: the journal of the American Society of Gene Therapy. 2022;30:1104-18
107. Cheng H, Wang S, Huang A, Ma J, Gao D, Li M. et al. HSF1 is involved in immunotherapeutic response through regulating APOJ/STAT3-mediated PD-L1 expression in hepatocellular carcinoma. Cancer biology & therapy. 2023;24:1-9
108. Shaashua L, Ben-Shmuel A, Pevsner-Fischer M, Friedman G, Levi-Galibov O, Nandakumar S. et al. BRCA mutational status shapes the stromal microenvironment of pancreatic cancer linking clusterin expression in cancer associated fibroblasts with HSF1 signaling. Nature communications. 2022;13:6513
109. Wang Y, Zhu Q, Guo S, Ao J, Zhang W, Fei J. et al. HSF1 activates the FOXO3a-ΔNp63α-CDK4 axis to promote head and neck squamous cell carcinoma cell proliferation and tumour growth. FEBS letters. 2023;597:1125-37
110. Qian W, Chen K, Qin T, Xiao Y, Li J, Yue Y. et al. The EGFR-HSF1 axis accelerates the tumorigenesis of pancreatic cancer. Journal of experimental & clinical cancer research: CR. 2021;40:25
111. Liang W, Liao Y, Li Z, Wang Y, Zheng S, Xu X. et al. MicroRNA-644a promotes apoptosis of hepatocellular carcinoma cells by downregulating the expression of heat shock factor 1. Cell communication and signaling: CCS. 2018;16:30
112. Wang C, Zhang D, Wang L, Wang W. MicroRNA-455-3p inhibits osteosarcoma progression via HSF1 downregulation. Journal of orthopaedic science: official journal of the Japanese Orthopaedic Association. 2023;28:1157-64
113. Shen Q, Wang R, Liu X, Song P, Zheng M, Ren X. et al. HSF1 Stimulates Glutamine Transport by Super-Enhancer-Driven lncRNA LINC00857 in Colorectal Cancer. Cancers. 2022;14:3855
114. Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nature reviews Molecular cell biology. 2013;14:630-42
115. Neef DW, Turski ML, Thiele DJ. Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS biology. 2010;8:e1000291
116. Tambasco N, Romoli M, Calabresi P. Levodopa in Parkinson's Disease: Current Status and Future Developments. Current neuropharmacology. 2018;16:1239-52
117. Fujimoto M, Takaki E, Hayashi T, Kitaura Y, Tanaka Y, Inouye S. et al. Active HSF1 significantly suppresses polyglutamine aggregate formation in cellular and mouse models. The Journal of biological chemistry. 2005;280:34908-16
118. Kim E, Sakata K, Liao FF. Bidirectional interplay of HSF1 degradation and UPR activation promotes tau hyperphosphorylation. PLoS genetics. 2017;13:e1006849
119. Shimura H, Schlossmacher MG, Hattori N, Frosch MP, Trockenbacher A, Schneider R. et al. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson's disease. Science (New York, NY). 2001;293:263-9
120. Ho MS. Microglia in Parkinson's Disease. Advances in experimental medicine and biology. 2019;1175:335-53
121. Tiefensee Ribeiro C, Peixoto DO, Santos L, Saibro-Girardi C, Brum PO, Carazza-Kessler FG. et al. Intranasal HSP70 administration protects against dopaminergic denervation and modulates neuroinflammatory response in the 6-OHDA rat model. Brain, Behavior, & Immunity - Health. 2021;14:100253
122. Gong X, Tan Z, Xu H, Jiang X, Chen L. Paeoniflorin Attenuates Oxidative Stress and Inflammation in Parkinson's Disease by Activating the HSF1-NRF1 Axis. The American journal of Chinese medicine. 2024;52:2131-59
123. Landles C, Bates GP. Huntingtin and the molecular pathogenesis of Huntington's disease. Fourth in molecular medicine review series. EMBO reports. 2004;5:958-63
124. Hayashida N, Fujimoto M, Tan K, Prakasam R, Shinkawa T, Li L. et al. Heat shock factor 1 ameliorates proteotoxicity in cooperation with the transcription factor NFAT. The EMBO journal. 2010;29:3459-69
125. Franks H, Wang R, Li M, Wang B, Wildmann A, Ortyl T. et al. Heat shock factor HSF1 regulates BDNF gene promoters upon acute stress in the hippocampus, together with pCREB. Journal of neurochemistry. 2023;165:131-48
126. Mansky RH, Greguske EA, Yu D, Zarate N, Intihar TA, Tsai W. et al. Tumor suppressor p53 regulates heat shock factor 1 protein degradation in Huntington's disease. Cell reports. 2023;42:112198
127. Muralidar S, Ambi SV, Sekaran S, Thirumalai D, Palaniappan B. Role of tau protein in Alzheimer's disease: The prime pathological player. International Journal of Biological Macromolecules. 2020;163:1599-617
128. Tang Z, Su KH, Xu M, Dai C. HSF1 physically neutralizes amyloid oligomers to empower overgrowth and bestow neuroprotection. Science advances. 2020;6:eabc6871
129. Cheng Y, Saville L, Gollen B, Veronesi AA, Mohajerani M, Joseph JT. et al. Increased Alu RNA processing in Alzheimer brains is linked to gene expression changes. EMBO reports. 2021;22:e52255
130. Li Y, Wang D, Ping X, Zhang Y, Zhang T, Wang L. et al. Local hyperthermia therapy induces browning of white fat and treats obesity. Cell. 2022;185:949-66.e19
131. Shi X, Li T, Liu Y, Yin L, Xiao L, Fu L. et al. HSF1 Protects Sepsis-Induced Acute Lung Injury by Inhibiting NLRP3 Inflammasome Activation. Frontiers in immunology. 2022;13:781003
132. He YF, Hu XM, Khan MA, Yu BY, Sheng YC, Xiao XZ. et al. HSF1 Alleviates Brain Injury by Inhibiting NLRP3-Induced Pyroptosis in a Sepsis Model. Mediators of inflammation. 2023;2023:2252255
133. Yuan F, Cai J, Wu J, Tang Y, Zhao K, Liang F. et al. Z-DNA binding protein 1 promotes heatstroke-induced cell death. Science (New York, NY). 2022;376:609-15
134. Ling X, Lu J, Wang X, Liu L, Liu L, Wang Y. et al. Ovarian tumorB1-mediated heat shock transcription factor 1 deubiquitination is critical for glycolysis and development of endometriosis. iScience. 2022;25:105363
135. Wang Y, Zhao M, Zhao L, Geng Y, Li G, Chen L. et al. HBx-Induced HSPA8 Stimulates HBV Replication and Suppresses Ferroptosis to Support Liver Cancer Progression. Cancer research. 2023;83:1048-61
136. Velayutham M, Cardounel AJ, Liu Z, Ilangovan G. Discovering a Reliable Heat-Shock Factor-1 Inhibitor to Treat Human Cancers: Potential Opportunity for Phytochemists. Frontiers in oncology. 2018;8:97
137. Rawat PS, Jaiswal A, Khurana A, Bhatti JS, Navik U. Doxorubicin-induced cardiotoxicity: An update on the molecular mechanism and novel therapeutic strategies for effective management. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2021;139:111708
138. Kciuk M, Gielecińska A, Mujwar S, Kołat D, Kałuzińska-Kołat Ż, Celik I. et al. Doxorubicin-An Agent with Multiple Mechanisms of Anticancer Activity. Cells. 2023;12:659
139. Huang CY, Kuo WW, Lo JF, Ho TJ, Pai PY, Chiang SF. et al. Doxorubicin attenuates CHIP-guarded HSF1 nuclear translocation and protein stability to trigger IGF-IIR-dependent cardiomyocyte death. Cell death & disease. 2016;7:e2455
140. Yoon T, Kang GY, Han AR, Seo EK, Lee YS. 2,4-Bis(4-hydroxybenzyl)phenol inhibits heat shock transcription factor 1 and sensitizes lung cancer cells to conventional anticancer modalities. Journal of natural products. 2014;77:1123-9
141. Cheeseman MD, Chessum NE, Rye CS, Pasqua AE, Tucker MJ, Wilding B. et al. Discovery of a Chemical Probe Bisamide (CCT251236): An Orally Bioavailable Efficacious Pirin Ligand from a Heat Shock Transcription Factor 1 (HSF1) Phenotypic Screen. Journal of medicinal chemistry. 2017;60:180-201
142. Fok JHL, Hedayat S, Zhang L, Aronson LI, Mirabella F, Pawlyn C. et al. HSF1 Is Essential for Myeloma Cell Survival and A Promising Therapeutic Target. Clinical cancer research: an official journal of the American Association for Cancer Research. 2018;24:2395-407
143. Menezes K, Aram G, Mirabella F, Johnson DC, Sherborne AL, Houlston RS. et al. The Novel Protein HSF1 Stress Pathway Inhibitor Bisamide CCT361814 Demonstrates Pre-Clinical Anti-Tumor Activity in Myeloma. Blood. 2017;130:3072
144. Pasqua AE, Sharp SY, Chessum NEA, Hayes A, Pellegrino L, Tucker MJ. et al. HSF1 Pathway Inhibitor Clinical Candidate (CCT361814/NXP800) Developed from a Phenotypic Screen as a Potential Treatment for Refractory Ovarian Cancer and Other Malignancies. Journal of medicinal chemistry. 2023;66:5907-36
145. Zhang D, Zhang B. Selective killing of cancer cells by small molecules targeting heat shock stress response. Biochemical and biophysical research communications. 2016;478:1509-14
146. Liu J, Farmer JD Jr, Lane WS, Friedman J, Weissman I, Schreiber SL. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807-15
147. Shao J, Han B, Cao P, Zhang B, Liu M, Li D. et al. HSF1 phosphorylation by cyclosporin A confers hyperthermia sensitivity through suppression of HSP expression. Biochimica et biophysica acta Gene regulatory mechanisms. 2019;1862:846-57
148. Dasgupta B, Seibel W. Compound C/Dorsomorphin: Its Use and Misuse as an AMPK Inhibitor. Methods in molecular biology (Clifton, NJ). 2018;1732:195-202
149. Li N, Wang T, Li Z, Ye X, Deng B, Zhuo S. et al. Dorsomorphin induces cancer cell apoptosis and sensitizes cancer cells to HSP90 and proteasome inhibitors by reducing nuclear heat shock factor 1 levels. Cancer biology & medicine. 2019;16:220-33
150. Kim HW, Kim SA, Ahn SG. Sirtuin inhibitors, EX527 and AGK2, suppress cell migration by inhibiting HSF1 protein stability. Oncology reports. 2016;35:235-42
151. Bhardwaj M, Paul S, Jakhar R, Khan I, Kang JI, Kim HM. et al. Vitexin confers HSF-1 mediated autophagic cell death by activating JNK and ApoL1 in colorectal carcinoma cells. Oncotarget. 2017;8:112426-41
152. Herbert JM, Augereau JM, Gleye J, Maffrand JP. Chelerythrine is a potent and specific inhibitor of protein kinase C. Biochemical and biophysical research communications. 1990;172:993-9
153. Kumar S, Deepak P, Kumar S, Gautam PK, Acharya A. A benzophenanthridine alkaloid, chelerythrine induces apoptosis in vitro in a Dalton's lymphoma. Journal of cancer research and therapeutics. 2013;9:693-700
154. Dong B, Jaeger AM, Hughes PF, Loiselle DR, Hauck JS, Fu Y. et al. Targeting therapy-resistant prostate cancer via a direct inhibitor of the human heat shock transcription factor 1. Science translational medicine. 2020;12:eabb5647
155. Dong Q, Xiu Y, Wang Y, Hodgson C, Borcherding N, Jordan C. et al. HSF1 is a driver of leukemia stem cell self-renewal in acute myeloid leukemia. Nature communications. 2022;13:6107
156. Salamanca HH, Fuda N, Shi H, Lis JT. An RNA aptamer perturbs heat shock transcription factor activity in Drosophila melanogaster. Nucleic acids research. 2011;39:6729-40
157. Chen BC, Tu SL, Zheng BA, Dong QJ, Wan ZA, Dai QQ. Schizandrin A exhibits potent anticancer activity in colorectal cancer cells by inhibiting heat shock factor 1. Bioscience reports. 2020;40:BSR20200203
158. Wang RE, Kao JL, Hilliard CA, Pandita RK, Roti Roti JL, Hunt CR. et al. Inhibition of heat shock induction of heat shock protein 70 and enhancement of heat shock protein 27 phosphorylation by quercetin derivatives. Journal of medicinal chemistry. 2009;52:1912-21
159. Yang W, Cui M, Lee J, Gong W, Wang S, Fu J. et al. Heat shock protein inhibitor, quercetin, as a novel adjuvant agent to improve radiofrequency ablation-induced tumor destruction and its molecular mechanism. Chinese journal of cancer research = Chung-kuo yen cheng yen chiu. 2016;28:19-28
160. Rodríguez-García C, Sánchez-Quesada C, J JG. Dietary Flavonoids as Cancer Chemopreventive Agents: An Updated Review of Human Studies. Antioxidants (Basel, Switzerland). 2019;8:137
161. Kim JA, Lee S, Kim DE, Kim M, Kwon BM, Han DC. Fisetin, a dietary flavonoid, induces apoptosis of cancer cells by inhibiting HSF1 activity through blocking its binding to the hsp70 promoter. Carcinogenesis. 2015;36:696-706
162. Vilaboa N, Boré A, Martin-Saavedra F, Bayford M, Winfield N, Firth-Clark S. et al. New inhibitor targeting human transcription factor HSF1: effects on the heat shock response and tumor cell survival. Nucleic acids research. 2017;45:5797-817
163. Yoon YJ, Kim JA, Shin KD, Shin DS, Han YM, Lee YJ. et al. KRIBB11 inhibits HSP70 synthesis through inhibition of heat shock factor 1 function by impairing the recruitment of positive transcription elongation factor b to the hsp70 promoter. The Journal of biological chemistry. 2011;286:1737-47
164. Huang M, Dong W, Xie R, Wu J, Su Q, Li W. et al. HSF1 facilitates the multistep process of lymphatic metastasis in bladder cancer via a novel PRMT5-WDR5-dependent transcriptional program. Cancer communications (London, England). 2022;42:447-70
165. Conroy A, Stockett DE, Walker D, Arkin MR, Hoch U, Fox JA. et al. SNS-032 is a potent and selective CDK 2, 7 and 9 inhibitor that drives target modulation in patient samples. Cancer chemotherapy and pharmacology. 2009;64:723-32
166. Chen R, Wierda WG, Chubb S, Hawtin RE, Fox JA, Keating MJ. et al. Mechanism of action of SNS-032, a novel cyclin-dependent kinase inhibitor, in chronic lymphocytic leukemia. Blood. 2009;113:4637-45
167. Rye CS, Chessum NE, Lamont S, Pike KG, Faulder P, Demeritt J. et al. Discovery of 4,6-disubstituted pyrimidines as potent inhibitors of the heat shock factor 1 (HSF1) stress pathway and CDK9. MedChemComm. 2016;7:1580-6
168. Krystof V, Baumli S, Fürst R. Perspective of cyclin-dependent kinase 9 (CDK9) as a drug target. Current pharmaceutical design. 2012;18:2883-90
169. Canduri F, Perez PC, Caceres RA, de Azevedo WF Jr. CDK9 a potential target for drug development. Medicinal chemistry (Shariqah (United Arab Emirates)). 2008;4:210-8
170. Zaarur N, Gabai VL, Porco JA Jr, Calderwood S, Sherman MY. Targeting heat shock response to sensitize cancer cells to proteasome and Hsp90 inhibitors. Cancer research. 2006;66:1783-91
171. Acquaviva J, He S, Sang J, Smith DL, Sequeira M, Zhang C. et al. mTOR inhibition potentiates HSP90 inhibitor activity via cessation of HSP synthesis. Molecular cancer research: MCR. 2014;12:703-13
172. Naz F, Wu Y, Zhang N, Yang Z, Yu C. Anticancer Attributes of Cantharidin: Involved Molecular Mechanisms and Pathways. Molecules (Basel, Switzerland). 2020;25:3279
173. Kim JA, Kim Y, Kwon BM, Han DC. The natural compound cantharidin induces cancer cell death through inhibition of heat shock protein 70 (HSP70) and Bcl-2-associated athanogene domain 3 (BAG3) expression by blocking heat shock factor 1 (HSF1) binding to promoters. The Journal of biological chemistry. 2013;288:28713-26
174. Westerheide SD, Kawahara TL, Orton K, Morimoto RI. Triptolide, an inhibitor of the human heat shock response that enhances stress-induced cell death. The Journal of biological chemistry. 2006;281:9616-22
175. Sarkar R, Mukherjee S, Biswas J, Roy M. Sulphoraphane, a naturally occurring isothiocyanate induces apoptosis in breast cancer cells by targeting heat shock proteins. Biochemical and biophysical research communications. 2012;427:80-5
176. Nikotina AD, Koludarova L, Komarova EY, Mikhaylova ER, Aksenov ND, Suezov R. et al. Discovery and optimization of cardenolides inhibiting HSF1 activation in human colon HCT-116 cancer cells. Oncotarget. 2018;9:27268-79
177. Akagawa H, Takano Y, Ishii A, Mizuno S, Izui R, Sameshima T. et al. Stresgenin B, an inhibitor of heat-induced heat shock protein gene expression, produced by Streptomyces sp. AS-9. The Journal of antibiotics. 1999;52:960-70
178. Aziz F, Wang X, Liu J, Yan Q. Ginsenoside Rg3 induces FUT4-mediated apoptosis in H. pylori CagA-treated gastric cancer cells by regulating SP1 and HSF1 expressions. Toxicology in vitro: an international journal published in association with BIBRA. 2016;31:158-66
179. Santagata S, Mendillo ML, Tang YC, Subramanian A, Perley CC, Roche SP. et al. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science (New York, NY). 2013;341:1238303
180. Iwasaki S, Iwasaki W, Takahashi M, Sakamoto A, Watanabe C, Shichino Y. et al. The Translation Inhibitor Rocaglamide Targets a Bimolecular Cavity between eIF4A and Polypurine RNA. Molecular cell. 2019;73:738-48.e9
181. Kantarjian HM, O'Brien S, Cortes J. Homoharringtonine/omacetaxine mepesuccinate: the long and winding road to food and drug administration approval. Clinical lymphoma, myeloma & leukemia. 2013;13:530-3
182. Yakhni M, Briat A, El Guerrab A, Furtado L, Kwiatkowski F, Miot-Noirault E. et al. Homoharringtonine, an approved anti-leukemia drug, suppresses triple negative breast cancer growth through a rapid reduction of anti-apoptotic protein abundance. American journal of cancer research. 2019;9:1043-60
183. Li GH, Miao Y, Chen H, Xue MF, Wang JY, Zhang JW. et al. The small-molecule drug homoharringtonine targets HSF1 to suppress pancreatic cancer progression. American journal of cancer research. 2024;14:2072-87
184. Ohnishi K, Takahashi A, Yokota S, Ohnishi T. Effects of a heat shock protein inhibitor KNK437 on heat sensitivity and heat tolerance in human squamous cell carcinoma cell lines differing in p53 status. International journal of radiation biology. 2004;80:607-14
185. Yokota S, Kitahara M, Nagata K. Benzylidene lactam compound, KNK437, a novel inhibitor of acquisition of thermotolerance and heat shock protein induction in human colon carcinoma cells. Cancer research. 2000;60:2942-8
186. Xia Y, Wang M, Beraldi E, Cong M, Zoubeidi A, Gleave M. et al. A Novel Triazole Nucleoside Suppresses Prostate Cancer Cell Growth by Inhibiting Heat Shock Factor 1 and Androgen Receptor. Anti-cancer agents in medicinal chemistry. 2015;15:1333-40
187. Kruta M, Sunshine MJ, Chua BA, Fu Y, Chawla A, Dillingham CH. et al. Hsf1 promotes hematopoietic stem cell fitness and proteostasis in response to ex vivo culture stress and aging. Cell stem cell. 2021;28:1950-65.e6
188. Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239-50
189. Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7841-6
190. Dayalan Naidu S, Sutherland C, Zhang Y, Risco A, de la Vega L, Caunt CJ. et al. Heat Shock Factor 1 Is a Substrate for p38 Mitogen-Activated Protein Kinases. Molecular and cellular biology. 2016;36:2403-17
191. Rao Y, Li C, Hu YT, Xu YH, Song BB, Guo SY. et al. A novel HSF1 activator ameliorates non-alcoholic steatohepatitis by stimulating mitochondrial adaptive oxidation. British journal of pharmacology. 2022;179:1411-32
192. Ratnayake R, Covell D, Ransom TT, Gustafson KR, Beutler JA. Englerin A, a Selective Inhibitor of Renal Cancer Cell Growth, from Phyllanthus engleri. Organic Letters. 2009;11:57-60
193. Sourbier C, Scroggins BT, Ratnayake R, Prince TL, Lee S, Lee MJ. et al. Englerin A stimulates PKCθ to inhibit insulin signaling and to simultaneously activate HSF1: pharmacologically induced synthetic lethality. Cancer cell. 2013;23:228-37
194. Frémont L. Biological effects of resveratrol. Life sciences. 2000;66:663-73
195. Han S, Choi JR, Soon Shin K, Kang SJ. Resveratrol upregulated heat shock proteins and extended the survival of G93A-SOD1 mice. Brain research. 2012;1483:112-7
196. Ma X, Xu L, Alberobello AT, Gavrilova O, Bagattin A, Skarulis M. et al. Celastrol Protects against Obesity and Metabolic Dysfunction through Activation of a HSF1-PGC1α Transcriptional Axis. Cell metabolism. 2015;22:695-708
197. Xiao Y, Liu J, Guo M, Zhou H, Jin J, Liu J. et al. Synergistic combination chemotherapy using carrier-free celastrol and doxorubicin nanocrystals for overcoming drug resistance. Nanoscale. 2018;10:12639-49
198. Liu M, Fan Y, Li D, Han B, Meng Y, Chen F. et al. Ferroptosis inducer erastin sensitizes NSCLC cells to celastrol through activation of the ROS-mitochondrial fission-mitophagy axis. Molecular oncology. 2021;15:2084-105
199. Forouzanfar F, Barreto G, Majeed M, Sahebkar A. Modulatory effects of curcumin on heat shock proteins in cancer: A promising therapeutic approach. BioFactors (Oxford, England). 2019;45:631-40
200. Chen YC, Tsai SH, Shen SC, Lin JK, Lee WR. Alternative activation of extracellular signal-regulated protein kinases in curcumin and arsenite-induced HSP70 gene expression in human colorectal carcinoma cells. European journal of cell biology. 2001;80:213-21
201. Dunsmore KE, Chen PG, Wong HR. Curcumin, a medicinal herbal compound capable of inducing the heat shock response. Critical care medicine. 2001;29:2199-204
202. Lee YS, Chen X, Widiyanto TW, Orihara K, Shibata H, Kajiwara S. Curcumin affects function of Hsp90 and drug efflux pump of Candida albicans. Frontiers in cellular and infection microbiology. 2022;12:944611
203. Huang H, Weng H, Dong B, Zhao P, Zhou H, Qu L. Oridonin Triggers Chaperon-mediated Proteasomal Degradation of BCR-ABL in Leukemia. Scientific reports. 2017;7:41525
204. Viollet B, Guigas B, Sanz Garcia N, Leclerc J, Foretz M, Andreelli F. Cellular and molecular mechanisms of metformin: an overview. Clinical science (London, England: 1979). 2012;122:253-70
205. Xu M, Li LP, He X, Lu XZ, Bi XY, Li Q. et al. Metformin induction of heat shock factor 1 activation and the mitochondrial unfolded protein response alleviate cardiac remodeling in spontaneously hypertensive rats. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2024;38:e23654
206. Deng M, Chen H, Long J, Song J, Xie L, Li X. Calycosin: a Review of its Pharmacological Effects and Application Prospects. Expert review of anti-infective therapy. 2021;19:911-25
207. Lai PF, Mahendran R, Tsai BC, Lu CY, Kuo CH, Lin KH. et al. Calycosin Enhances Heat Shock Related-Proteins in H9c2 Cells to Modulate Survival and Apoptosis against Heat Shock. The American journal of Chinese medicine. 2024;52:1173-93
208. Hargitai J, Lewis H, Boros I, Rácz T, Fiser A, Kurucz I. et al. Bimoclomol, a heat shock protein co-inducer, acts by the prolonged activation of heat shock factor-1. Biochemical and biophysical research communications. 2003;307:689-95
209. Polakowski JS, Wegner CD, Cox BF. Bimoclomol elevates heat shock protein 70 and cytoprotects rat neonatal cardiomyocytes. European journal of pharmacology. 2002;435:73-7
210. Bíró K, Pálhalmi J, Tóth AJ, Kukorelli T, Juhász G. Bimoclomol improves early electrophysiological signs of retinopathy in diabetic rats. Neuroreport. 1998;9:2029-33
211. Kong XC, Zhang D, Qian C, Liu GT, Bao XQ. FLZ, a novel HSP27 and HSP70 inducer, protects SH-SY5Y cells from apoptosis caused by MPP(+). Brain research. 2011;1383:99-107
212. Yu L, Zhou B, Zhu Y, Li L, Zhong Y, Zhu L. et al. HSF1 promotes CD69(+) Treg differentiation to inhibit colitis progression. Theranostics. 2023;13:1892-905
213. Yin CF, Kao SC, Hsu CL, Chang YW, Cheung CHY, Huang HC. et al. Phosphoproteome Analysis Reveals Dynamic Heat Shock Protein 27 Phosphorylation in Tanshinone IIA-Induced Cell Death. Journal of proteome research. 2020;19:1620-34
214. Chang KH, Chen WL, Lee LC, Lin CH, Kung PJ, Lin TH. et al. Aqueous Extract of Paeonia lactiflora and Paeoniflorin as Aggregation Reducers Targeting Chaperones in Cell Models of Spinocerebellar Ataxia 3. Evidence-based complementary and alternative medicine: eCAM. 2013;2013:471659
215. Yu X, Li H, Lin D, Guo W, Xu Z, Wang L. et al. Ginsenoside Prolongs the Lifespan of C. elegans via Lipid Metabolism and Activating the Stress Response Signaling Pathway. International journal of molecular sciences. 2021;22:9668
216. Ekimova IV, Plaksina DV, Pastukhov YF, Lapshina KV, Lazarev VF, Mikhaylova ER. et al. New HSF1 inducer as a therapeutic agent in a rodent model of Parkinson's disease. Experimental neurology. 2018;306:199-208
217. Mohanan A, Deshpande S, Jamadarkhana PG, Kumar P, Gupta RC, Chauthaiwale V. et al. Delayed intervention in experimental stroke with TRC051384-a small molecule HSP70 inducer. Neuropharmacology. 2011;60:991-9
218. Du P, Xu L, Wang Y, Jiao T, Cheng J, Zhang C. et al. Astragaloside IV ameliorates pressure overload-induced heart failure by enhancing angiogenesis through HSF1/VEGF pathway. Heliyon. 2024;10:e37019
219. Morán Luengo T, Mayer MP, Rüdiger SGD. The Hsp70-Hsp90 Chaperone Cascade in Protein Folding. Trends in cell biology. 2019;29:164-77
220. Ma FH, Li C, Liu Y, Shi L. Mimicking Molecular Chaperones to Regulate Protein Folding. Advanced materials (Deerfield Beach, Fla). 2020;32:e1805945
221. Liu AY, Minetti CA, Remeta DP, Breslauer KJ, Chen KY. HSF1, Aging, and Neurodegeneration. In: Turksen K, editor. Cell Biology and Translational Medicine, Volume 18: Tissue Differentiation, Repair in Health and Disease. Cham: Springer Nature Switzerland. 2023;18:23-49
222. Li J, Labbadia J, Morimoto RI. Rethinking HSF1 in Stress, Development, and Organismal Health. Trends in cell biology. 2017;27:895-905
223. Kijima T, Prince T, Neckers L, Koga F, Fujii Y. Heat shock factor 1 (HSF1)-targeted anticancer therapeutics: overview of current preclinical progress. Expert opinion on therapeutic targets. 2019;23:369-77
224. Zhou C, Yu T, Zhu R, Lu J, Ouyang X, Zhang Z. et al. Timosaponin AIII promotes non-small-cell lung cancer ferroptosis through targeting and facilitating HSP90 mediated GPX4 ubiquitination and degradation. International journal of biological sciences. 2023;19:1471-89
225. Fan Y, Liu Y, Zhang L, Cai F, Zhu L, Xu J. C0818, a novel curcumin derivative, interacts with Hsp90 and inhibits Hsp90 ATPase activity. Acta pharmaceutica Sinica B. 2017;7:91-6
226. Wen W, Liu W, Shao Y, Chen L. VER-155008, a small molecule inhibitor of HSP70 with potent anti-cancer activity on lung cancer cell lines. Experimental biology and medicine (Maywood, NJ). 2014;239:638-45
227. Schlecht R, Scholz SR, Dahmen H, Wegener A, Sirrenberg C, Musil D. et al. Functional analysis of Hsp70 inhibitors. PloS one. 2013;8:e78443
228. Massey AJ, Williamson DS, Browne H, Murray JB, Dokurno P, Shaw T. et al. A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer chemotherapy and pharmacology. 2010;66:535-45
Author contact
![]() Corresponding author: Ke Tan, E-mail: tankeedu.cn.
Corresponding author: Ke Tan, E-mail: tankeedu.cn.

 Global reach, higher impact
Global reach, higher impact