Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2025; 21(8):3666-3688. doi:10.7150/ijbs.112061 This issue Cite
Review
Omics-based Approach Towards Macrophages: New Perspectives of Biology and Function in the Normal and Diseased Heart
1. Department of Cardiology, The Second Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou 310009, China
2. State Key Laboratory of Transvascular Implantation Devices, Hangzhou 310009, China
3. Heart Regeneration and Repair Key Laboratory of Zhejiang province, Hangzhou 310009, China
4. Transvascular Implantation Devices Research Institute (TIDRI), Hangzhou 310053, China
* These authors contributed equally to this work
Received 2025-2-12; Accepted 2025-5-7; Published 2025-5-27
Abstract
Macrophages play a crucial role not only in maintaining homeostasis but also in initiating inflammatory responses to various forms of stress or injury, thereby contributing to tissue damage while concurrently promoting recovery. Furthermore, the diversity of macrophage subtypes, their spatial distribution, and distinct cellular functions are closely linked to the pathogenesis and severity of cardiovascular diseases such as myocardial infarction, atherosclerosis, heart failure, and myocarditis. This association underscores the importance of investigating macrophage heterogeneity in different pathological contexts. Recent advances in multi-omics technologies—including single-cell RNA sequencing, spatial transcriptomics, and metabolomics—have elucidated the heterogeneity of macrophages, their intercellular interactions, underlying functional mechanisms, and spatial organization. In this review, we systematically summarize the diverse phenotypes and functional plasticity of macrophages in the regulation of cardiovascular diseases, with particular emphasis on the novel insights afforded by multi-omics approaches. We focus on the characteristics of macrophages in both physiological and pathological states, thereby providing reference points for clinical macrophage-targeted strategies and their translational significance.
Keywords: macrophage, cardiovascular disease, multi-omics, single-cell, spatial transcriptomics, metabolomics
Introduction
Macrophages have long been recognized for their complex interactions within the cardiovascular system, serving as the most abundant immune cells in cardiac tissue. They play pivotal roles in both maintaining homeostasis and contributing to the development of cardiovascular diseases (CVD) [1-3]. As key regulators of post-injury inflammation and the local microenvironment, their residency and polarization are closely associated with disease progression [4]. Macrophages are essential in orchestrating phagocytosis, immune surveillance, inflammation, and cardiovascular remodeling. Following injury, they are actively recruited to damaged areas, where they become the predominant immune cells, clearing tissue debris through phagocytosis and releasing substantial amounts of pro-inflammatory cytokines and proteases [5,6]. Furthermore, macrophages secrete a diverse array of mediators that promote extracellular matrix (ECM) deposition, cell proliferation, and angiogenesis, while also modulating immune responses and fibrosis via interactions with other cell types [7].
Although macrophages share common features, their functional phenotypes vary according to specific disease contexts. Distinct macrophage subpopulations are distributed across various cardiovascular compartments—including myocardial and vascular tissues—demonstrating remarkable adaptability to different microenvironments. Therefore, effective treatment of CVD requires the precise identification and selective targeting of macrophage phenotypes that mediate distinct pathological processes. Recent advancements in investigative technologies—particularly multi-omics approaches such as single-cell RNA sequencing (scRNA-seq), spatial transcriptomics (ST-seq), proteomics, and metabolomics—have revealed numerous macrophage subsets [8-12]. These methodologies provide unprecedented insights into the spatiotemporal heterogeneity and specialized functions of cardiovascular macrophages. Multi-omics analyses have further characterized the unique attributes of each macrophage subset under both physiological and pathological conditions [13,14]. By elucidating the intricate diversity among macrophages, researchers can achieve a deeper understanding of their functional roles, advancing from broad concepts of immune processes to precise characterization of individual cellular components.
This review offers, from a multi-omics perspective, a comprehensive analysis of recent advancements in cell clustering, spatial localization, and functional heterogeneity of macrophages under homeostatic and pathological conditions—including ischemic heart injury (most commonly myocardial infarction), non-ischemic cardiac insults leading to heart failure such as myocarditis, and vascular diseases including atherosclerosis and diabetic vascular complications. By illuminating the complex biological processes mediated by macrophages, this review aims to discuss emerging therapeutic targets and novel strategies for macrophage-focused interventions in cardiovascular disease.
Please note that human genes referenced in this review follow the HUGO Gene Nomenclature Committee guidelines, with gene names represented as capitalized abbreviations. Mouse gene nomenclature adheres to the Mouse Genome Informatics conventions.
Steady-State Macrophages
Macrophage Metabolism and Physiological Functions
Macrophage metabolism plays a pivotal role in determining their physiological functions, as demonstrated by extensive research in immunometabolism [15]. Under steady-state conditions, glucose, lipids, and glutamine constitute the principal metabolic substrates for macrophages. In response to diverse stimuli, macrophages exhibit metabolic flexibility, shifting substrate utilization and activating specific metabolic pathways [16]. The accumulation of metabolic end-products and intermediates regulates macrophage phenotypes, thereby facilitating tailored responses to dynamic microenvironmental signals [17]. In addition to the cell-intrinsic effects of metabolism, intercellular influences are also significant [18]. Studies have shown that macrophages modulate their microenvironment and regulate organ function through the uptake and secretion of various metabolites [19-21]. Therefore, investigating macrophage metabolic kinetics yields valuable insights into the regulation of their phenotypes and functional roles.
M1/M2 Macrophage Polarization
Over recent decades, the in vitro M1/M2 macrophage polarization model has been widely used to investigate the interplay between immune functions and metabolism [22]. Bone marrow-derived macrophages (BMDMs) are considered to be in the M0 state after treatment with colony-stimulating factors [23]. Upon stimulation of Toll-like receptors (TLRs) by agonists such as lipopolysaccharide (LPS) and/or cytokines like interferon-γ (IFN-γ), macrophages polarize into the classically activated, pro-inflammatory M1 phenotype. These M1 macrophages are characterized by high expression levels of markers such as CD80, CD86, and inducible nitric oxide synthase (iNOS) [24]. By contrast, alternatively activated, anti-inflammatory M2 macrophages differentiate in response to interleukin-4 (IL-4) or interleukin-13 (IL-13). M2 macrophages express high levels of markers such as CD163, CD206, ARG1, FIZZ1, and YM1, reflecting their reparative and immunoregulatory functions. M2 macrophages can be further subdivided into four subpopulations: M2a, M2b, M2c, and M2d [25]. Among these, the M2b subset uniquely secretes both pro-inflammatory and anti-inflammatory factors to regulate immune responses, whereas the other subsets predominately exhibit reparative phenotypes by producing anti-inflammatory and profibrotic factors [26].
The M1 and M2 phenotypes not only perform opposing immune functions but also depend on distinct metabolic pathways. Pro-inflammatory M1 macrophages exhibit enhanced glycolysis and activation of the pentose phosphate pathway, which supports biosynthetic demands and enhances antibacterial activity [27]. Conversely, M2 macrophages primarily rely on fatty acid oxidation and glutamine metabolism, along with increased mitochondrial respiration, supporting their role in inflammation resolution [28]. Upon IL-4 stimulation, M2 macrophages upregulate genes associated with fatty acid metabolism [29] and polyamine synthesis from arginine [30], further contributing to their reparative functions. Although the M1/M2 polarization model offers certain advantages, it does not sufficiently reflect the complexity and functional diversity of macrophages in vivo, due to the influence of numerous factors, especially the broad range of changes in the microenvironment. Recent advances have redefined macrophages according to their developmental origins, classifying them as either tissue-resident or monocyte-derived, each exhibiting distinct phenotypes and functions [22,31]. Differences in macrophage origin result in significant variations at the epigenetic and transcriptomic levels, which are shaped by the availability of metabolic substrates within specific organs and conditions [32]. Thus, a more nuanced characterization of macrophage metabolic profiles can advance our understanding of their diverse functional roles.
Cardiovascular macrophages, like other tissue-resident macrophages, are relatively few in number yet are critical for maintaining tissue homeostasis [33-35]. Importantly, metabolic changes during development or under pathological conditions influence macrophage phenotypes, which in turn modulate the tissue microenvironment and function. The advent of high-throughput technologies has enabled omics-based investigations into the metabolic characteristics of cardiovascular macrophages. Whereas earlier studies focused on whole-tissue metabolic activity, recent investigations have identified macrophage-specific metabolic pathways that are closely linked to systemic alterations [36-38].
Single-Cell Sequencing of Cardiac Macrophages
The development of single-cell RNA sequencing (scRNA-seq) has transformed our understanding of macrophage biology. Previously, macrophages were simplistically classified as either pro-inflammatory or anti-inflammatory agents within the immune system [2]; they are now recognized as a highly heterogeneous population exhibiting diverse phenotypes and functions [32,39,40]. scRNA-seq has become an indispensable tool for analyzing macrophage heterogeneity at single-cell resolution. This technology has surpassed the traditional binary M1/M2 classification, revealing a complex spectrum of activation states, and enabling detailed analyses of macrophage dynamics as they adapt to intricate microenvironments [41,42].
Single-cell sequencing approaches have allowed researchers to identify distinct subsets of cardiac macrophages involved in homeostasis [43,44], without solely relying on highly specific markers or lineage tracing [45,46]. Dick et al. conducted pioneering single-cell analyses of cardiac macrophages, classifying them based on CD45 and high CD64 expression into three major resident cardiac macrophage clusters [47]: (1) cells with high levels of Timd4, Lyve1, and Folr2 (collectively termed TLF+), (2) macrophages with high Ccr2 expression (Ccr2hi), and (3) those with elevated MHCII expression (MHCIIhi). These clusters exhibit functional parallels across diverse tissues. The TLF+ cluster is enriched for cellular transport and endocytosis pathways; the Ccr2hi cluster is associated with cellular activation, degranulation, and immune responses; and the MHCIIhi cluster is involved in antigen presentation and other immune processes. These findings corroborate previous work by Chakarov et al. [46], who classified tissue-resident macrophages mainly into Lyve1hi and MHCIIhi subsets. Notably, Lyve1hi macrophages share a gene expression profile with the TLF+ macrophages identified by Dick et al.
Further characterization of human cardiac tissue has refined immune cell profiles across six anatomical regions of the adult heart [48], identifying resident cardiac macrophages with distinct inflammatory and protective transcriptional signatures. These include the LYVE1+FOLR2+ cluster, analogous to the murine TLF+ macrophages, and antigen-presenting macrophages expressing HLA-related genes such as HLA-DRA, HLA-DMA, HLA-DMB, and HLA-DPA1, resembling the MHCIIhi cluster in mice. Additionally, a unique DOCK4+ macrophage cluster expressing IL4R, STAT3, and ITGAM—but lacking C1QA or FOLR2—has also been described.
Spatial Transcriptomics of Cardiac Macrophages
The application of spatial transcriptomics has addressed the lack of structural context inherent to single-cell technologies, enabling insights into tissue niches and specialized cell populations with distinct functions [49,50]. Analysis of eight anatomical regions in the heart using spatial transcriptomics identified tissue-resident macrophages (LYVE1+) not only in the sinoatrial node (SAN), but also in the atrioventricular node (AVN) [50]. Notably, the human SAN exhibits a compartmentalized structure, consisting of a central region where functionally essential P cells are intermingled with activated fibroblasts and glial cells, surrounded by a peripheral region containing immune cells—including LYVE1+ macrophages—and fibroblast populations, a configuration not observed in the AVN. In the epicardium across all four cardiac chambers, LYVE1+IGF1+ macrophages, as well as plasma B cells, are present alongside other cell types such as mesothelial cells, fibroblasts, lymphatic endothelial cells, and adipocytes. Niche analysis of immune cells in the epicardium revealed that LYVE1+IGF1+ macrophages have a pivotal role in recruiting and supporting plasma B cells, which secrete immunoglobulins and function as an immune barrier. Dysregulation of these macrophages may contribute to autoimmune mechanisms underlying heart disease [51]. Another study indicated that Lyve1loMHCIIhi macrophages predominantly localize adjacent to nerves, while Lyve1hiMHCIIlo macrophages are preferentially found near blood vessels [52].
Origin and Development of Cardiac Macrophage Subsets
The origins of steady-state macrophage subsets have long been of interest in immunological research. Early studies indicate that resident macrophages—especially the TLF+ subset and the MHCIIhi / HLA-DRhi CCR2- population—are initially derived primarily from the yolk sac, followed by the fetal liver during embryogenesis [53,54]. After birth, adult tissue macrophages begin to receive input from, or are progressively replenished by, circulating monocytes, which primarily differentiate into CCR2+ macrophages [13,55]. Transitional stages in macrophage development can be tracked using markers such as Ccr2, Csf1r, and Cx3cr1[56]. Circulating monocytes express Ccr2, a chemokine receptor important for migration, and are enriched for components of the NOD-like receptor protein 3 (NLRP3) inflammasome. Ccr2 expression reflects dynamic changes in macrophage phenotype and origin, making it a key marker for classifying cardiac macrophages. Both resident and recruited cardiac macrophages exhibit variable MHC-II/HLA-DR expression, which is crucial for antigen presentation and T-cell activation [46]. Notably, MHC-II expression in embryonically derived macrophages is gradually upregulated after birth, initially appearing in the Ccr2+ subset and subsequently in the Ccr2- subset [47,57]. As a result, most cardiac macrophages in neonatal mice are CCR2- MHC-IIlo, whereas the adult mouse heart contains three distinct resident macrophage subsets [58].
Macrophage Subsets in Various Heart Diseases
Myocardial Infarction
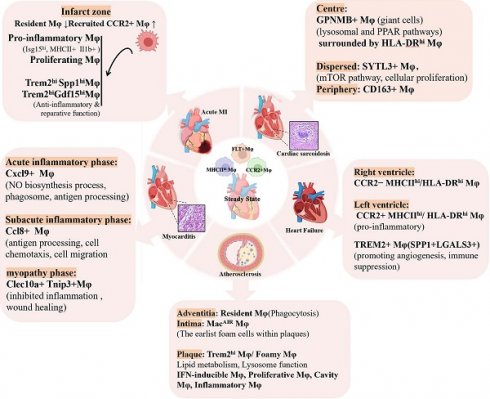
Myocardial infarction (MI) causes ischemic injury, which triggers a robust inflammatory response, the influx of immune cells, and cardiac remodeling [59]. Acute MI leads to the diversification, mobilization, and recruitment of both innate and adaptive immune cells to the infarcted region [60]. Among these, macrophages play a central role in clearing necrotic tissue, facilitating wound healing [61,62], and orchestrating remodeling processes [63]. The functions of macrophages depend on the type and proportion of subsets present at different stages—such as the acute inflammatory and chronic repair phases. Moreover, the distribution and roles of macrophage subsets differ between the infarct zone and peri-infarct region, underscoring their spatial and temporal heterogeneity during cardiac injury and repair.
Metabolic changes of macrophages in MI
In the acute phase, MI can be divided into two stages: the early inflammatory phase and the late resolution phase [64]. Initially, hypoxia in the infarcted area leads to a metabolic switch toward glycolysis and increased lactate production. This glycolytic reprogramming is associated with the predominance and expansion of CCR2+ monocyte-derived macrophage subsets. Traditionally, glycolysis provides rapid ATP production and contributes to the generation of ribose sugars and NADPH via the pentose phosphate pathway [18], which, in turn, promotes the secretion of pro-inflammatory cytokines. This has been corroborated by the upregulation of glycolytic and hypoxia-response genes in cardiac macrophages within 24 hours post-MI [65]. During ischemia, enhanced glycolysis is accompanied by suppressed oxidative phosphorylation. Concurrent inhibition of the tricarboxylic acid (TCA) cycle leads to the accumulation of succinate [66] and citrate [67]. Succinate stabilizes HIF1α, further activating glycolysis [68], and upon oxidation by succinate dehydrogenase during reperfusion, promotes IL-1β production in macrophages [69]. Citrate can be exported to the cytoplasm and converted to acetyl-CoA for histone acetylation [70], thereby regulating inflammatory gene expression [71].
Accompanying MI is widespread apoptosis of both myocardial and non-myocardial cells, phagocytosis of apoptotic cells triggers metabolic switching to oxidative phosphorylation, mediated by engulfed metabolites especially lipids [72]. Lysosomal hydrolysis of cholesterol esters within endocytic compartments leads to the formation of anti-inflammatory oxysterols [73]. Additionally, increased fatty acids promote anti-inflammatory macrophage responses by enhancing IL-10 synthesis [19] and activating PPARγ coactivator-1β (PGC-1β) [29]. These metabolic shifts underlie macrophage phenotypic plasticity, resulting in the emergence of non-steady-state subsets as MI progresses [74,75]. As inflammation subsides, the infarcted area is increasingly infiltrated by fibroblasts and extracellular matrix components. Restoration of blood flow via angiogenesis steers cardiac metabolism toward fatty acid oxidation, favoring the expansion and activity of anti-inflammatory or reparative macrophage subsets such as Trem2hi and Bhlhe41+ macrophages [76,77]. In this environment, macrophages downregulate glycolytic genes and increasingly utilize fatty acids to generate pro-resolving mediators, thereby supporting anti-inflammatory functions and tissue repair [78].
Spatiotemporal Dynamics of Cardiac Macrophages in MI
Following MI, there is a rapid influx of CCR2+ Ly6Chi monocytes and CCR2+ monocyte-derived macrophages, which displace resident macrophages (including TLF+ and MHC-IIhi subsets) from ischemic regions within 48 hours [31,33]. Between day 4 and day 28 post-MI, the absolute number of resident macrophages gradually increases, while the proportion of recruited macrophages declines. However, even at four weeks post-MI, the ratio of resident to recruited macrophages does not return to baseline. Jung et al. showed that, in MI mice, the proportion of resident macrophages sharply declines but begins to recover by day 3 for the MHC-IIhi cluster and day 5 for the TLF+ cluster, stabilizing around day 7 post-MI [79]. Spatial transcriptomics sequencing (ST-seq) revealed that, on the first day after MI, macrophages are distributed throughout the heart rather than localized to the infarct region. By day 3, macrophages begin accumulating in the infarct zone, with peak abundance during the late phase [79]. These data highlight the spatial and temporal heterogeneity and redistribution of macrophages throughout MI progression.
Functional Macrophage Subsets and Fibrotic Crosstalk
Dynamic changes in macrophage populations during the late acute phase of MI are closely linked to post-infarction repair processes [80]. Distinct macrophage clusters emerge during this period [79], including interferon-responsive (IFN) clusters and proliferating clusters. Rizzo et al. identified that the infarcted heart features two pro-inflammatory macrophage clusters—Isg15hi (also termed IFNIC) and MHCII+Il1b+—as well as a non-inflammatory Trem2hi cluster [81]. The infiltration of MHCII+Il1b+ and Isg15hi macrophages peaks between days 3 and 7 post-MI, whereas Trem2hi macrophages peak between days 3 and 5. Trem2hi macrophages are localized to the ischemic area but are absent from remote, viable myocardium, reflecting their likely role in phagocytosis.
Amrute et al. demonstrated distinct spatial relationships between macrophage subsets and fibroblast populations in infarcted human hearts [82]. CCR2+ macrophages preferentially co-localize with fibrotic FAP+/POSTN+ fibroblasts, thereby establishing an immune-fibroblast niche within the infarct core, while resident macrophages are associated with APOE+/AGT+ fibroblasts. Notably, regions enriched in FAP+/POSTN+ fibroblasts show greater infiltration of CCR2+ macrophages compared to remote areas, suggesting that local microenvironmental cues drive recruitment. Mechanistically, CCR2+ macrophages promote fibroblast activation through two principal pathways: the TGF-β/Smad3 pathway, which facilitates fibroblast migration, transdifferentiation, and extracellular matrix synthesis; and macrophage-derived IL-6, which triggers autocrine STAT3 activation in fibroblasts, further enhancing TGF-β/Smad3 signaling and promoting fibrotic remodeling. Additionally, the IL-1β/NF-κB axis drives fibroblast proliferation [83]. Targeting this axis with IL-1β-neutralizing antibodies significantly reduces the abundance of FAP+ fibroblasts and improves cardiac function in experimental models, underscoring its translational therapeutic potential [82]. Collectively, multi-omics findings elucidated the distribution, function, and intercellular communication of distinct macrophage subgroups within the infarcted, border, and remote zones of the heart.
Macrophage subsets as potential therapeutic targets for MI
Advances in single-cell sequencing have revealed distinct macrophage subsets that represent promising therapeutic targets for MI repair. In early ischemic cardiac tissue, SPP1+ macrophage clusters increase significantly. These macrophages promote inflammation and fibrosis by remodeling the extracellular matrix and activating fibroblasts through SPP1/CD44 and SPP1-αvβ3 integrin signaling pathways [82,83]. Their essential role in cardiac remodeling has been validated in zebrafish studies [86]. Within these SPP1+ macrophage clusters, CD36—a key receptor for phagocytosis—is upregulated and is crucial for binding and clearing apoptotic and necrotic neutrophils, thus playing a unique role in cardiac remodeling post-MI [87]. Trem2hi macrophages are primarily active during the later stages of MI [81]. By engaging the TREM2/SYK signaling axis, these macrophages promote tissue repair and immunomodulation [88]. They secrete anti-inflammatory cytokines such as IL-10 and TGF-β, reduce neutrophil infiltration, and maintain cardiomyocyte homeostasis by clearing dysfunctional mitochondria. Loss of the Trem2 gene exacerbates post-MI remodeling, while administration of soluble Trem2 improves cardiac recovery through enhanced anti-inflammatory activity [33]. In human myocardial infarction samples, about half of TREM2-expressing macrophages also co-express SPP1, indicating a shared phenotype involved in tissue repair and remodeling. Rizzo et al. further identified two populations among Trem2hi macrophages: the Trem2hiSpp1hi subset, representing an intermediate state between monocytes and macrophages, and the Trem2hiGdf15hi subset, corresponding to differentiated macrophages. These populations peak sequentially in the infarcted heart. Bhlhe41+ resident macrophages appear transiently in the "developing" infarct zone, which is characterized by abundant monocytes, macrophages, and myofibroblasts, while monocyte-derived Trem2hi Spp1hi macrophages predominate in the "old" infarct zone, where neutrophils, endothelial cells, and fibroblasts are enriched [77]. Bhlhe41+ resident macrophages suppress myofibroblast activation and myocardial fibrosis, thus limiting infarct expansion following MI [76]. Collectively, these findings highlight the phenotypic plasticity of macrophage subsets as spatiotemporal regulators of myocardial repair and support the development of stratified, phase-specific therapeutic strategies.
In summary, these studies highlight the dynamic shifts in resident and monocyte-derived macrophage populations across cardiac regions following MI—including the ischemic zone (comprising the infarct and peri-infarct regions) and remote non-infarcted myocardium—and their distinct contributions to tissue repair and remodeling (Figure 1). We have compiled macrophage subpopulations, their markers, and functional characteristics from MI-related studies as a quick reference for readers (Table 1).
Atherosclerosis
Atherosclerosis is a chronic, lipid-driven vascular inflammatory disease characterized by the formation of plaques in large arteries, driven primarily by the subendothelial deposition of lipoproteins in regions of disturbed blood flow [89]. Deposited lipoproteins generate pro-inflammatory derivatives, recruit leukocytes [90], and drive the polarization of resident immune cells [91]. The balance between pro-inflammatory and inflammation-resolving processes within plaques determines whether lesions remain stable or regressive, or instead progress toward instability and rupture [92]. Macrophages, as key mediators of the inflammatory response, play critical roles throughout all stages of atherosclerosis, including plaque initiation, calcification, rupture, and regression. They accumulate through both the recruitment of circulating monocytes and the proliferation of locally differentiated macrophages. During the progression of atherosclerosis, macrophages produce a broad array of pro-inflammatory and anti-inflammatory mediators, pro-thrombotic tissue factors, and proteolytic enzymes. These secreted factors modulate the growth, cellular composition, and stability of atherosclerotic plaques, thereby significantly influencing disease outcomes [93-95].
Schematic of Changes in Major Macrophage Subsets in Myocardial Infarction.
Key macrophage subsets, their markers and function in selected published studies on MI.
| Publication | Disease | Species/Model | Macrophage subsets | Markers | Function |
|---|---|---|---|---|---|
| Dick, S. A. et al.[33] (2019) | MI | Cx3cr1CreER-YFPR26Td mice / LAD artery ligations model; Patients with end-stage cardiomyopathy during the time of implantation of the left ventricular assist device | Timd4 cluster | Timd4, Lyve1, Flor2 | Tissue repair, inflammation resolution via IGF-1 secretion and efferocytosis. |
| MHC-IIhi cluster | Cd14, Cx3cr1, Adgre1 | Antigen presentation, tissue immune homeostasis. | |||
| Ccr2+ cluster | Ccr2, Fcgr1, Plac8 | Pro-inflammatory responses, contributing to ischemic damage. | |||
| Isg cluster | Irf7, Isg20, Ifit1 | Type I interferon signaling pathways, antiviral responses, inflammation regulation. | |||
| proliferating | Mki67, Top2a | Proliferating. | |||
| 7 unique post-infarct macrophage clusters | Fn1, Il1b, Mmp14, Hif1a, Sirpb1a, Cd72, Lrg1, Trem2 | Exhibiting plasticity but failing to fully adopt reparative functions. Contribute to heterogeneous pro-inflammatory and partial repair roles. | |||
| Farbehi, N. et al.[56] (2019) | MI | PdgfraGFP/+ mice / LAD artery ligations model | M1Mo | Ifitm6, Mcemp1 | Phagocytose debris, secrete IL-1β/IL-6/TNFα, amplify acute inflammation. |
| M1 Mφ | Arg1, C1qb, Ccr2, Ly6c2 | Sustain inflammation via cytokine/chemokine secretion, promote leukocyte recruitment. | |||
| M2 Mφ | Ly6c2-, Adgre1 high | Late-stage anti-inflammatory/reparative macrophages, secrete IL-10/TGF-β, resolve inflammation, promote angiogenesis, ECM remodeling and fibrosis resolution. | |||
| MAC-TR | Timd4, Lyve1 | Antigen presentation, phagocytosis, pro-regenerative roles, maintain cardiac homeostasis and fetal coronary development. | |||
| MAC-IFNIC | Ifit3, Ifit1, Ifi47 | IFN-γ/α/β-responsive subset, amplify inflammatory signaling, impair tissue repair. | |||
| MAC6 | Csf3r, Siglecf, S100a9 | Granulocyte-enriched population, role in early inflammation unclear. | |||
| Bajpai, G. et al.[31] (2019) | MI | Cx3cr1CreER-YFP: R26Td mice / IR injury model | proliferating | Ki67, Ccnb2, Aurkb | Actively proliferate in situ, contribute to macrophage pool expansion post-injury. |
| Tnip3+ cluster | Tnip3, ltgb7, Ltb4r1, ltgax | Unique cluster with dendritic cell-like features, role in immune regulation unclear. | |||
| Lyve1+ cluster | Lyve1, CD163, Tim4, Lilra5 | Regulate tissue homeostasis and repair. | |||
| Fos+ cluster | Fos, Egr1, Hspa1a | Participate in inflammatory or stress responses. | |||
| Mgst1+ cluster | Mgst1, Gpx3, Kif3a, Anpep | Anti-inflammatory, cell protection or homeostasis maintenance. | |||
| Arg1+ cluster | Arg1, Adam8, Spp1 | Anti-inflammatory/reparative phenotype, enriched in pathways for tissue repair and fibrosis. | |||
| Cxcl1+ cluster | Cxcl1, Nlrp3, Ccrl2 | Linked to adverse ventricular remodeling. | |||
| Ifit3+ cluster | Ifit1-3, Mx1, lsg15 | Associated with type I interferon signaling | |||
| Jung, S.-H. et al.[79](2019) | MI | C57BL/6 mice / LAD artery ligations | steady-state (SS) Mφ1 | Lyve1, F13a1, Cbr2, Cd163, Folr2 | Tissue-resident macrophages, maintain homeostasis and prevent fibrosis. |
| SS-Mφ2 | H2-Eb1, H2-Aa, H2-Ab1, Cd74 | Antigen-presenting resident macrophages. | |||
| MI Early-Mφ | Cd68, Fcgr1, Itgam, Ccr2 | Monocyte-derived pro-inflammatory macrophages, clear debris via phagocytosis. | |||
| Late-Mφ 1 | Apoe, Fcrls, Rgs10, Adgre1 | Transitional macrophages with mixed inflammatory/repair functions. | |||
| Late-Mφ 2 | Trem2, Gpnmb, Fabp5, Spp1 | Anti-inflammatory repair macrophages, promote tissue remodeling and angiogenesis. | |||
| transient Mφ 1 | Saa3, Fn1, Ltc4s | Phagocytic activity during mid-phase inflammation. | |||
| transient Mφ 2 | Fabp5, Spp1, Gpnmb | Early fibrotic signaling. | |||
| transient Mφ 3 | Hmox1, Prdx1, Gclm | Oxidative stress response. | |||
| IFN- Mφ | Irf7, Isg15, Ifit2 | Anti-viral response, modulate adaptive immunity. | |||
| Proliferating Mφ | Top2a, Mki67, Hist1h1b | Self-renewal via local proliferation. | |||
| Rizzo, G. et al.[81] (2022) | MI | C57BL/6 mice / LAD artery ligations; Patients with ischemic cardiomyopathy | RTM-TIMD4 cluster | Lyve1, Timd4, Folr2 | Tissue-resident macrophages, self-renewing population, cardioprotective functions. |
| RTM-MHCII cluster | MHCII, Mgl2 | Tissue-resident macrophages, partially express CCR2, homeostatic maintenance. | |||
| MHCII + Il1b + cluster | H2-Aa, Il1b, Tnip3, Tlr2, Tnfsf9, Axl | Pro-inflammatory phenotype, associated with tissue damage, NLRP3 inflammasome activity. | |||
| Isg15hi Mφ | Isg15、Irf7、Cxcl10, Il18 | Type I interferon response, pathogenic inflammation, linked to IFNγ signaling. | |||
| Trem2hi Spp1hi Mφ | Trem2, Spp1, Hmox1, Arg1 | Monocyte-to-macrophage intermediate, profibrotic, efferocytosis activity. | |||
| Trem2hi Gdf15hi Mφ | Trem2, Gdf15, Igf1, MerTk, Timp2, Apoe | Anti-inflammatory, tissue repair, cholesterol efflux regulation. | |||
| Trem2hi Prdx1hi Mφ | Trem2, Prdx1 | Iron-handling subset, antioxidant functions. | |||
| Amrute, J. M. et al.[82] (2024) | MI | Six healthy donors, four patients with acute MI, six patients with ICM and six patients with NICM | CCR2+ Mφ | CCR2, IL-1β, CD68 | Primary source of IL-1β signaling, drive FAP/POSTN fibroblast activation via IL-1β, co-localize with fibrotic niches in infarct zones. |
| Inflammatory Mφ | SPP1, MMP9, CCL2 | Mediate early inflammatory responses, promote extracellular matrix degradation, transition to profibrotic states. | |||
| CCR2- Resident Mφ | FOLR2, LYVE1, MERTK | Associated with F4 fibroblasts (APOE/AGT), maintain tissue homeostasis, less involved in fibrotic remodeling. | |||
| Kuppe, C. et al.[84] (2022) | MI | Four non-transplanted donor hearts (controls); Samples of patients with acute MI | LYVE+ PLTP+ Mφ | MAMDC2, PDE4D, SCN9A | Tissue-resident macrophages, associated with vascular homeostasis and tissue surveillance. |
| LYVE+ FLOR+ Mφ | CD14, CX3CR1, ADGRE1 | Tissue-resident macrophages, involved in immune regulation and tissue repair. | |||
| SPP1+ Mφ | ITGAX, MMP19, SPP1 | Phagocytotic activity, lipid uptake, and pro-fibrotic signaling, dominant in ischemic zones. | |||
| CCL18+ Mφ | KCNMA1, HS3ST2, NHSL1, CPM | Linked to fibrotic remodeling and extracellular matrix deposition. | |||
| Xu, Y. et al.[77] (2023) | MI | Cx3cr1CreER-YFP: R26Td mice / LAD artery ligations model | Bhlhe41+ Mφ | Bhlhe41, Fabp5, Gpnmb, Cd36, Grn | Suppress myofibroblast activation, reduce fibrosis, limit infarct expansion. |
| LYVE1+ MHCII- CCR2-Mφ | LYVE1, CCR2- | Homeostatic surveillance, decrease post-MI. | |||
| LYVE1- MHCII+ CCR2+ Mφ | MHCII, CCR2+ | Minor population at baseline, expand during inflammation. | |||
| Trem2+ Spp1+ Mφ | Trem2+, Spp1+, IL-10+ | Associated with late-stage remodeling, may promote fibrotic processes. | |||
| Cluster 1 | Tgfbi, Cd74, Ms4a4c, Ly6a | Pro-inflammatory response. | |||
| Cluster 2 | Il10, Spp1, Arg1, Cxcl3 | Anti-inflammatory/repair functions. | |||
| Mki67+ Mφ | Stmn1, Top2a, Mki67, Birc5 | Active proliferation during early inflammation. | |||
| Ninh, V. K. et al.[76] (2024) | MI | Ccr2-/-, Irf3-/- mice; Patients with acute MI | IFNIC Mφ | ISGs (Ifit1, Rsad2, Cxcl10) | Localized to border zone IFNIC colonies; exhibit blunted pro-reparative functions due to type I IFN signaling, impairing fibroblast activation and matrix deposition. |
| Resident Mφ | Timd4, Lyve1 | Maintain tissue homeostasis, phagocytose debris, and initiate early repair responses in remote zones. | |||
| Infiltrating Pro-inflammatory Mφ | Ccr2, Ly6c2, Chil3 | Secrete pro-inflammatory cytokines, amplify sterile inflammation in the infarct zone. |
Metabolic changes of macrophages in atherosclerosis
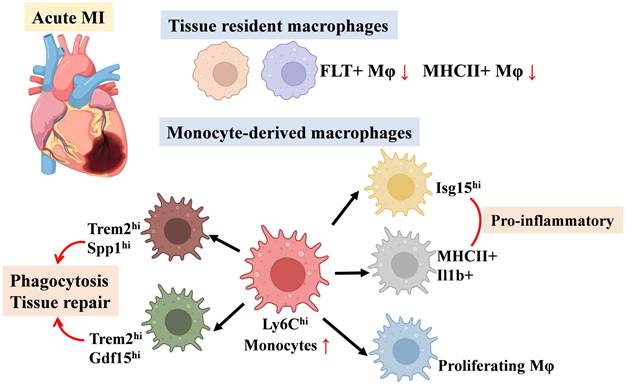
Previous studies have demonstrated that macrophages within atherosclerotic plaques undergo profound metabolic reprogramming [96]. These macrophages exhibit increased glucose uptake and enhanced glycolytic activity, likely mediated by upregulation of the HIF1α-GLUT1 axis, which promotes a pro-inflammatory phenotype [32]. Despite the abundance of fatty acids and cholesterol in plaques—which could theoretically support oxidative phosphorylation—mitochondrial dysfunction limits oxygen consumption. The accumulation of fatty acids and cholesterol instead activates TLR4 signaling, further exacerbating inflammation [97]. In contrast, during the regression phase of plaques, macrophages may shift to lipid-based metabolism under the influence of pro-resolving mediators or efferocytosis, thereby facilitating the resolution of inflammation and promoting tissue repair [98].
Resident macrophages in atherosclerosis
Resident macrophages are present in both healthy and atherosclerotic aortic adventitia. In healthy aortas, these macrophages express higher levels of Lyve1, whereas atherosclerotic aortas show increased Ccr2 expression [99]. Atherosclerotic aortas contain both true resident macrophages and newly recruited cells, which may acquire similar gene expression profiles upon infiltration. Notably, resident macrophages in atherosclerosis correspond to TLF+ macrophages. Depletion of Lyve1+ macrophages in Lyvewt/cre Csf1rflox/flox mice leads to increased arterial stiffness and collagen deposition, underscoring their essential role in maintaining vascular homeostasis [100]. Pathway analyses indicate that resident-like plaque macrophages are actively involved in receptor-mediated endocytosis [101,102].
Additionally, the aortic intima contains a distinct population of resident macrophages, termed aortic intimal resident macrophages (MacAIR) [103]. These cells originate from bone marrow progenitors that colonize the aorta at birth and maintain their population through self-renewal. Although transcriptionally similar to foam/Trem2hi macrophages, MacAIR cells display higher expression of MHCII transcripts and lower levels of foam cell-associated genes (Trem2, Spp1). MacAIR cells act as primary precursors of foam cells during early atherosclerosis, mediating initial lipid deposition via SR-A1/CD36-mediated uptake of oxidized (oxLDL) and aggregated LDL (agLDL). Their tissue-resident nature enables lipid accumulation before monocyte infiltration, and elevated baseline expression of lipid metabolism genes (e.g., CD36) primes them for rapid lipid uptake during hyperlipidemia. Brief cholesterol elevation amplifies both lipid loading and MacAIR proliferation, accelerating fatty streak formation. As atherosclerotic plaques advance, the limited proliferative capacity of MacAIR cells, combined with persistent hypercholesterolemia, overwhelms their cholesterol metabolic potential, promoting accelerated foam cell differentiation and apoptosis. Consequently, MacAIR populations peak during early lesion development but decline as monocyte-derived macrophages predominate in advanced plaques [104].
Pro-inflammatory macrophages and TREM2hi / foam macrophages in atherosclerosis
The intima of atherosclerotic aortas, compared to healthy arteries, contains distinctive macrophage populations, including pro-inflammatory and TREM2hi macrophages. Pro-inflammatory macrophages predominantly localize to the shoulder regions of plaques and are defined by high expression of pro-inflammatory chemokines, Tlr2, and Nlrp3[105]. Zernecke et al. further identified a population, termed “inflammatory-Mφ,” comprising two subsets [105]: inflammatory-Nlrp3 macrophages, which highly express Nlrp3 and Il1b, and CCR2int MHCII+ macrophages, which exhibit intermediate expression of Ccr2 and express MHCII-related transcripts such as Cd74, H2-Aa, and H2-Eb1. Inflammatory macrophages constitute the dominant macrophage population of the plaque intima, and as the principal non-foam cells exclusive to atherosclerotic aortas, act as key drivers of inflammation in advanced lesions. Kim et al. demonstrated that non-foam macrophages display even greater pro-inflammatory activity than foam macrophages, underscoring their central role in promoting plaque inflammation [101].
Recent research has redefined Trem2hi macrophages as lipid-laden foam cells, refining the traditional foam cell classification. Single-cell sequencing consistently shows that Trem2hi macrophages possess classic foam cell features [101]. These cells express high levels of Trem2, CD9, Fabp4, Apoe, and Apoc1, and are primarily localized within plaque intima and necrotic cores [99]. Zernecke et al. further identified two Trem2hi/foam macrophage subsets: the Trem2hi-Slamf9 subset, with high expression of Slamf9, Ch25h, Cd72, and related markers, and the Trem2hi-Gpnmb subset, which is marked by elevated Gpnmb, Atp6v0d2, other foam cell-associated transcripts, and Trem2-related genes such as Lpl, Lipa, Fabp5, Apoc1, and Apoe [105]. Pathway analyses reveal that Trem2hi foam macrophages are enriched for processes including organic substance metabolism, lipid metabolism, cholesterol efflux regulation, and oxidative stress pathways [101,102]. Van Kuijk et al. demonstrated that Trem2hi macrophages also highly express SPP1 and MMPs, indicating a role in promoting fibrosis, mirroring the fibrotic activity of Trem2+ hepatic macrophages [106]. These findings highlight the metabolic and functional specialization of Trem2hi macrophages within atherosclerotic plaques.
Other Macrophage Subtypes in Atherosclerosis
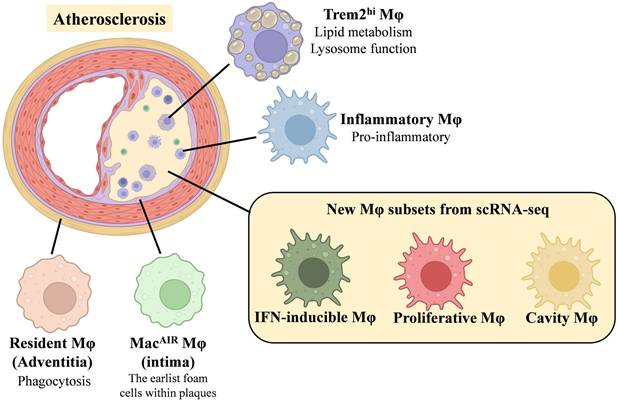
Atherosclerosis additionally involves several distinct macrophage subtypes, including proliferative macrophages, IFNICs, and intimal macrophages. Proliferative macrophages are defined by high expression of Birc5, and display elevated levels of the proliferation marker Mki67 [107]. Van Kuijk et al. [106] and Zernecke et al. [105] reported the presence of numerous IFNIC macrophages, while Cai et al. [108] identified interferon-responsive macrophages. However, it remains unclear whether interferon-activated macrophages represent a homeostatic population or arise exclusively in disease, although their association with type I interferon signaling suggests a pro-atherosclerotic role. Additionally, Zernecke et al. identified a macrophage subset resembling intimal macrophages in the aortas of atherosclerotic mice. These cells share a gene expression profile with subadventitial macrophages (CD226+CD11c+MHCII+) found in the SPM/cavity regions [109]. The origin and precise functions of these macrophages, which have not been previously identified in the aorta, remain uncertain and merit further investigation through single-cell analyses.
Human Atherosclerotic Plaque Macrophage Clusters
In analyses of human atherosclerotic plaques, Zernecke et al. identified three primary macrophage clusters: hInflammatory-Mφ, hFoamy/TREM2hi-Mφ, and hLYVE1-Mφ, along with smaller clusters such as type I interferon response macrophages (hIFNIC-Mφ) and proliferating macrophages (hProlif cluster) [105]. These findings closely parallel the macrophage subsets observed in murine atherosclerosis models. Expression of signature markers for major vascular macrophage subtypes demonstrates a conserved distribution of cell clusters across species. Winkels et al. identified two potential human macrophage subtypes [110]: CD11b+HLA-DRmed and CD11b+CD36+. Similarly, Depuydt et, al. [111] and Fernandez et, al. [112] reported the presence of both pro-inflammatory and anti-inflammatory macrophage clusters in human plaques. Of note, human foam macrophages display relatively stronger anti-inflammatory properties compared to non-foam macrophages, as they significantly suppress Il1b expression, implicating their potential role in modulating inflammatory responses within plaques.
In conclusion, the distribution of macrophage subsets involved in these atherosclerosis studies is shown in Figure 2. These macrophage subsets, their markers and functional characteristics are shown in Table 2.
Heart Failure Induced by Non-Ischemic Cardiomyopathy
Heart failure (HF) is a global health challenge, affecting over 23 million individuals and imposing a substantial burden on healthcare systems [113,114]. Cardiac overload triggers the release of pro-inflammatory cytokines, which drive monocyte infiltration into the myocardium—a critical component of the immune response and a major factor in disease progression [115,116]. In non-ischemic conditions, diverse stimuli can activate fibrotic signaling pathways in macrophages, leading to myocardial fibrosis. For instance, pressure overload (PO) induces fibrosis through mechanical stress and activation of the renin-angiotensin-aldosterone system (RAAS), whereas reactive oxygen species (ROS) play a central role in the pathogenesis of dilated cardiomyopathy (DCM). Interstitial fibrosis in non-ischemic heart injury represents a chronic and progressive process, predominantly driven by sustained, unregulated inflammation and persistent activation of profibrotic pathways. As cardiac injury advances, the heart loses its capacity to manage normal volume and/or pressure loads. The resulting microenvironment of the failing heart is characterized by extracellular matrix accumulation, impaired microcirculation, and excessive activation of immune cells, all of which further exacerbate HF progression [117].
Dynamic changes of macrophages in HF
Non-ischemic cardiomyopathy (NICM) animal models can be generated using various stressors, such as transverse aortic constriction (TAC) or angiotensin II (Ang-II) infusion. These models display characteristic pathological features, including left ventricular dysfunction, progressive interstitial fibrosis, and deteriorating cardiac function. In the Ang-II infusion-induced HF model, macrophage accumulation within the heart peaks at day 7, a pattern that mirrors the dynamic changes observed in the TAC-induced mouse HF model. After 7 days of Ang-II infusion, macrophage populations present in the steady-state heart—particularly those associated with reparative functions, such as the Timd4 and AP-1 clusters—show significant reductions in number. In contrast, pro-inflammatory macrophage subsets, including those with increased ribosome synthesis (stem cell-like) and interferon-stimulated gene (ISG)-related clusters, exhibit pronounced expansion [118,119].
Schematic of Changes in Major Macrophage Subsets in Atherosclerosis.
Key macrophage subsets, their markers and function in selected published studies on atherosclerosis.
| Publication | Disease | Species/Model | Macrophage subsets | Markers | Function |
|---|---|---|---|---|---|
| Cochain, C. et al.97 [102] (2018) | Atherosclerosis | Ldlr-/- mice / fed high-fat diet | Resident- like Mφ | Csf1r, Lyve1, Folr2, Pf4, Txnip | Exhibit anti-inflammatory/M2-like traits, may contribute to atherosclerosis via Pf4 and Txnip, potential roles in tissue homeostasis and lipid handling. |
| Inflammatory Mφ | Cxcl2, Ccl3, Ccl4, Tlr2, Nlrp3 | Proinflammatory phenotype and inflammasome activation, secrete chemokines to recruit immune cells, co-express feedback inhibitors to limit excessive inflammation. | |||
| TREM2hi Mφ | Term2, Cd9, Spp1 | Lipid metabolism/catabolism specialization, enriched in oxidative stress response pathways. | |||
| Williams, J. W. et al.[103] (2020) | Atherosclerosis | CX3CR1creER/+ Rosa26- lsl- Tomato Ldlr-/" | Adventitia Mφ | Lyve1, Mrc1 (CD206), Ccl2, Ccl6 | Maintain arterial tone via ECM regulation, exhibit interferon signatures. |
| MacAIR | Il1b, Rgs1, Cd9, Mmp12, Acp5 | First foam cells in early atherosclerosis, express IL-1β (primed inflammatory state), require CSF-1 for maintenance. | |||
| Proliferating Mφ | Mik67, Ccna2, Top2a | Contribute to local expansion but insufficient for long-term plaque maintenance. | |||
| Foamy Mφ | Trem2, SPP1, Fabp5, Gpnmb, Ctsd, Plin2 | Converge from both MacAIR and monocyte origins, metabolic/storage phenotype, associated with plaque progression. | |||
| Zernecke, A. et al.[105] (2023) | Atherosclerosis | 12 scRNA-seq datasets of atherosclerotic mouse aortas; human carotid endarterectomy specimens from two studies | TLF-Cd209hi Mφ | Lyve1, Timd4, Folr2, Cd209a/f/g | Tissue-resident macrophages, vascular homeostasis, lipid handling |
| TLF-Cd209low Mφ | Lyve1, Timd4, Folr2, Pf4, Mrc1 | Adventitial-resident macrophages, tissue repair and immune surveillance | |||
| Inflammatory Mφ | Nlrp3, Il1b, Ccl4, Ccl3 | Pro-inflammatory response, NLRP3 inflammasome activation. | |||
| CCR2intMHCII+ | Ccr2, MHC-II genes (Cd74, H2-Eb1) | Transitional state between monocytes and macrophages, antigen presentation. | |||
| Trem2hi-Slamf9 | Trem2, Slamf9, Ch25h, Tnf | Early lipid-loaded macrophages, inflammatory lipid metabolism. | |||
| Trem2hi-Gpnmb | Trem2, Gpnmb, Spp1, Apoe, Fabp5 | Foamy macrophages, lipid accumulation, cholesterol efflux, lesion progression. | |||
| MacAIR Mφ | Acp5, Gngt2, MHC-II | Intimal-resident macrophages, self-renewal, vascular barrier maintenance. | |||
| IFNIC Mφ | Isg15, Oasl2, Irf7 | Type I interferon response, antiviral defense. | |||
| hInflammator Mφ | CD74, HLA-DRB1, IL1B, CXCL8 | Pro-inflammatory cytokine secretion, plaque destabilization. | |||
| hFoamy Mφ | TREM2, SPP1, APOE, FABP5, GPNMB | Lipid metabolism, plaque core formation. | |||
| hIFNIC Mφ | ISG15、IFI6、MX1 | Type I interferon response, immune activation. | |||
| hProlif | TUBB、H2AFZ、STMN1 | Proliferating. | |||
| hLYVE1-Mφ | LYVE1, MRC1, FOLR2, SEPP1 | Tissue-resident macrophages, vascular repair, matrix remodeling. | |||
| van Kuijk, K. et al.[106] (2022) | Atherosclerosis | PHDko mic / fed high cholesterol diet; Human plaque tissue | Resident-like Mφ | Timd4, Lyve1, Flor2 | Maintain tissue homeostasis, may represent steady-state macrophages in early lesions. |
| Inflammatory Mφ | Tnip3, Nlrp3, Tnfsf9, Cxcl10 | Pro-inflammatory, associated with plaque inflammation. | |||
| Trem2hi Mφ | Term2, Mmp12, Spp1 | Foamy phenotype with enhanced fibrotic signaling, secrete Spp1 to activate fibroblasts for collagen production. | |||
| IFNIC Mφ | Isg15, Ifit3, Irf7 | Respond to interferon signaling, potential role in antiviral-like responses. | |||
| Cavity Mφ | Fn1, Gata6, Vsig4+ | Resemble peritoneal cavity macrophages, proposed role in debris clearance and lipid handling. | |||
| Bashore, A. C. et al.[107] (2024) | Atherosclerosis | Patients with carotid atherosclerosis | IL1B+ Mφ | IL1B, NLRP3, CCR2 (CD192), CD64, CD11c | Pro-inflammatory, NLRP3 inflammasome activation, key drivers of plaque inflammation, potential targets for IL-1β inhibitors. |
| C1Q+ Mφ | C1Q genes (C1QA, C1QB, C1QC), CD64, CD11c, MHCII | Complement activation, anti-inflammatory, efferocytotic capacity, enriched in STAT1/RELA-mediated immune regulation. | |||
| Apoptotic Mφ | Granzyme A, mitochondrial genes | Apoptotic cell clearance, high oxidative stress markers, phagocytic clearance functions. | |||
| Foam Mφ1 (TREM2+) | TREM2, ABCA1, LPL, CD36, APOE | Lipid metabolism specialists, cholesterol efflux, potential plaque-stabilizing functions, may represent homeostatic foam cells. | |||
| Foam Mφ2 (Inflammatory) | TREM2, APOE, C1Q genes, CCL18 | Hybrid phenotype (foamy + inflammatory), lipid processing with residual inflammation. | |||
| Proliferative Mφ | MKI67, TUBB, STMN1 | MYC-driven cell cycling, high DNA damage signaling, may contribute to macrophage persistence. | |||
| ACTA2+ Mφ | MYH11, ACTA2, MYOCD, CNN1 | Express smooth muscle cell genes, fibrotic/EMT pathway activation, possibly SMC-derived transdifferentiated cells, SMAD3/MRTF-mediated regulation. | |||
| Cai, J. et al.[108] (2020) | Atherosclerosis | allograft-induced transplant arteriosclerosis mouse | Resident- like Mφ | Mrc1, Folr2 | Anti-inflammatory, phagocytosis of apoptotic cells, tissue homeostasis. |
| Inflammatory Mφ | Eno1, Tpi1, Prdx5, Ccl2, Ccl7 | Proinflammatory response, secretion of chemokines, glycolysis and ROS production. | |||
| ISGhi Mφ | H2-Ab1, H2-Aa, Ifitm3, Ifitm6 | Strong interferon response with antigen presenting capacity. | |||
| Lgals3hi Mφ | Lgals3 | Expressed Plin2, a critical regulator in foam cell formation in atherosclerosis. | |||
| Depuydt, M. A. C. et al.[111] (2020) | Atherosclerosis | Patients with atherosclerosis | Cluster 0 (IL1B+ Mφ) | IL1B, CASP1, CASP4 | Pro-inflammatory phenotype with inflammasome activation, leukocyte transendothelial migration. |
| Cluster 1 (TNF+ Mφ) | TNF, TLR4, CCL3, CXCL1 | Pro-inflammatory macrophages driven by TNF and IFN signaling, expressed Toll-like receptors, IFNγ-driven activation via T-cell interactions. | |||
| Cluster 2 (Foam Mφ) | TREM2, ABCA1, ABCG1, MMP9 | Lipid metabolism and fibrosis promotion, anti-inflammatory via LXR/RXR pathway activation, expressed smooth muscle markers (partial), suggesting fibrotic plasticity. | |||
| Cluster 3 (Dendritic-like Mφ) | CD1C, CLEC10A, HLA-DR | Enhanced antigen presentation and IL12 production, drives T-cell activation. | |||
| Cluster 4 | CD3E, FOXP3 (misclassified) | Likely a misclustered population containing regulatory T cells. | |||
| Fernandez, D. M.[112] (2019) | Atherosclerosis | Patients with atherosclerosis | Cluster 1 | HLA-DRA, CD74 | Activated macrophages, antigen presentation and inflammatory responses. |
| Cluster 2 (Pro-inflammatory) | CYBA, LYZ, S100A9, S100A8, TIMP1 | Highly inflammatory, promotes oxidative stress and TLR signaling, reduces ECM degradation via TIMP1. | |||
| Cluster 3 | JUNB, NFKBIA, MALAT1 | Pro-inflammatory, activates NF-κB signaling, associated with cholesterol efflux (LXR/RXR pathways). | |||
| Cluster 5 (Foam cells) | APOC1, APOE, PLIN2 | Lipid-laden foam cells, cholesterol uptake, metabolism, and lipid accumulation, anti-inflammatory. |
In the TAC-induced HF mouse model, both resident macrophages (Timd4+ Ccr2-) and monocyte-derived macrophages (MoMF, Ccr2+) increase by 7 days post-TAC. By 28 days post-TAC, the numbers of most macrophage subsets return to baseline [118]. At both 7 and 28 days after TAC, the numbers of recruited M1-like CCR2+ Il1b+ macrophages are significantly elevated compared to sham controls. Resident macrophages, including CCR2- MHCIIlo M1-like and CCR2- MHCIIhi M2-like cells, also show modest increases at these time points. Martini et al. proposed that CCR2- MHCIIhi M2-like macrophages contribute to immune surveillance by promoting tissue repair and antigen presentation, whereas CCR2- MHCIIlo M1-like cells maintain homeostasis through phagocytosis of dead cardiomyocytes [118]. Conversely, the M1-like Ccr2+ Osm+ Il1b+ subset exerts pro-inflammatory effects through expression of oncostatin M (Osm), a cytokine linked to organ dysfunction [118].
Treatment with a monoclonal α-CD115 antibody, which preferentially depletes resident macrophages, was found to exacerbate fibrosis and heart failure in TAC mice [120]. These results suggest that resident macrophages are essential for attenuating cardiac deterioration and preventing fibrosis during early cardiac remodeling. In contrast, MoMFs exerted profibrotic effects in Ccr2 knockout mice. Consistent with these findings, pressure overload induced significant interactions between pro-inflammatory M1-like macrophages and activated Postn+ fibroblast subsets. CD72hi cardiac macrophages have been identified as a pro-inflammatory subset implicated in inflammation and cardiac injury following myocardial infarction [121]. This finding indicates that CD72 can serve as a marker for infiltrating monocytes/macrophages in TAC-induced heart failure. Cd72 expression, along with Ccr2, demonstrates strong similarity along the pseudo-time trajectory, and Cd72 is also highly expressed in ISG-related clusters, further linking it to pro-inflammatory Ccr2hi monocytes/macrophages. Clinically, heart failure patients with DCM exhibit increased levels of CD72hi macrophages, suggesting that the abundance of inflammatory macrophages serves as a negative predictor of cardiac recovery [121].
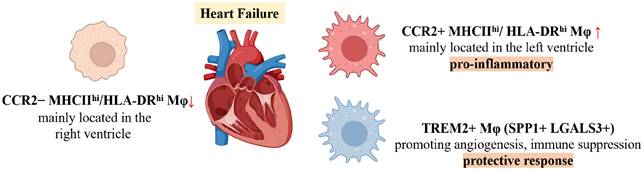
Differential Functions of Macrophage Subsets in Human Heart Failure
Using scRNA-seq, Rao et al. classified human HF macrophages into three principal subsets: CCR2-HLA-DRhi, CCR2+HLA-DRhi, and TREM2+ macrophages, which correspond to murine Ccr2-MHCIIhi, Ccr2+MHCIIhi, and Trem2+ subsets, respectively [122]. In diseased human hearts, CCR2-HLA-DRhi macrophages are more prevalent in the right ventricle (RV) of DCM patients, whereas CCR2+HLA-DRhi macrophages dominate in the left ventricle (LV), which is more severely fibrotic. The CCR2+HLA-DRhi subset displays pro-inflammatory activity, consistent with previous reports associating increased CCR2+ monocyte-derived macrophages with poor prognosis in heart failure. The specific expression of SPP1 and LGALS3 in TREM2+ macrophages suggests a potential role in promoting angiogenesis and immune suppression, contributing to protective responses under stress. Another study of human heart failure demonstrated divergent roles for monocyte-derived and resident cardiac macrophages in the disease process [123]. Monocyte-derived macrophages are enriched in hearts of patients who do not recover after left atrial volume reduction surgery (LAVD), expressing pro-inflammatory genes such as PLAUR, IL1B, TNF, and CCL4, which are associated with myocarditis and heart failure. These results emphasize that inflammatory macrophages serve as negative prognostic indicators for cardiac recovery. In contrast, CD163+ resident cardiac macrophages, which are significantly depleted during heart failure, return to normal levels in patients who achieve recovery. These macrophages exhibit transcriptional profiles closely linked to cardiac repair and remodeling, indicating their key regulatory role in heart failure recovery.
In conclusion, Figure 3 illustrates the macrophage subsets involved in heart failure, while Table 3 provides a detailed overview of these macrophage subsets, including their markers and functional characteristics.
Myocarditis
Myocarditis is characterized by dysregulated cardiac inflammation driven by dynamic macrophage heterogeneity [124]. In experimental models, cardiac macrophage expansion predominantly results from the recruitment of Ly6ChiCCR2+ monocytes, which differentiate into MHC-IIhiCCR2+ macrophages during the early stages of injury [125-127]. Using the experimental autoimmune myocarditis (EAM) model, Hua et al. [128] demonstrated that the major macrophage population present during the acute phase—distinguished by differential expression of Nos2, Arg1, and Ass1—produces nitric oxide through Ass1-mediated biosynthesis, thereby enhancing phagosomal antigen processing, IFN-γ responsiveness, and ROS metabolism. During the subacute inflammatory phase, a separate macrophage subset marked by high Ccl8 expression facilitates the processing and presentation of foreign peptide antigens via MHCII, and mediates cellular chemotaxis and monocyte migration. In the chronic myopathic phase, macrophage clusters with distinct expression of Tnf, Il-10, Vsig4, and Tnip3 contribute to the activation of mitogen-activated protein kinase (MAPK) cascades, TNF signaling, and NF-κB pathways, which collectively attenuate inflammation and promote wound healing through the synergistic effects of Il-10 and Tnip3. In giant cell myocarditis (GCM), multiple macrophage clusters have been identified [129], including monocyte-derived populations as well as mixed clusters containing both monocyte-derived and resident macrophages. Monocyte-derived macrophages express elevated levels of Prdx1 and Prdx5, genes associated with autoimmunity, while mixed clusters express members of the Ms4a gene family; other clusters express Nr4a1 and Pf4. Nr4a1 acts to inhibit macrophage polarization towards the pro-inflammatory M1 phenotype, whereas Pf4, a broad-spectrum inflammatory chemokine, functions to restrain the activation of resident macrophages.
Schematic of Changes in Major Macrophage Subsets in Heart failure.
Key macrophage subsets, their markers and function in selected published studies on HF.
| Publication | Disease | Species/Model | Macrophage subsets | Markers | Function |
|---|---|---|---|---|---|
| Martini, E. et al. [119] (2019) | HF | C57BL/6J mice / TAC model | Timd4hiMHCIIhi Mφ | Mgl2, Timd4, MHCII | Promote angiogenesis and reduce fibrosis, maintain cardiac homeostasis. |
| Timd4hi (Lyve1hi) Mφ | Timd4, Lyve1, Ccr2- | Phagocytosis of apoptotic cells, proliferate in response to stress, protect against adverse remodeling. | |||
| MHCIIhi Mφ | MHCII, Ccr2+ | Pro-inflammatory roles, increase during early stress but decline in late heart failure. | |||
| Double-Negative Mφ | Low/negative for Timd4, MHCII | Function unclear, may represent transitional or precursor states. | |||
| Revelo, X. S. et al[120] (2021) | HF | C57BL/6J mice, CCR2KO mice / TAC model | CCR2- Resident Mφ | Timd4, Lyve1, Cd163 | Promote angiogenesis, inhibit fibrosis, maintain tissue homeostasis, regulate cardiac conduction and metabolic stability. |
| CCR2+ monocyte-derived Mφ (MoMFs) | MHC-II, Ccr2, Ccl5, Cxcl10, Il1b, Spp1, Thbs1, Fn1 | Promote proinflammatory responses, drive fibrosis, recruit monocytes and T cells. | |||
| Cluster 0 (MoMFs) | Ifit1, Cxcl10 | Recruit monocytes and Th1 cells via Cxcl10. | |||
| Cluster 3 (Resident) | Myd88 pathway genes | Mediate innate immune signaling. | |||
| Cluster 4 (Resident) | Ptder4 | Regulate prostaglandin-mediated tissue repair. | |||
| Cluster 5 (MoMFs) | Spp1 | Promote fibrotic remodeling through osteopontin. | |||
| Cluster10 (MoMFs) | Ccl5, Cxcl16 | Enhance inflammation and leukocyte recruitment. | |||
| Cluster14 (MoMFs) | Thbs1, Fn1 | Drive ECM deposition and fibrosis. | |||
| Ni, S.-H. et al.[121] (2022) | HF | C57BL/6 mice; TAC and chronic Ang II infusion; patients with DCM | CD72hi Mφ | Cd72, Ccr2 | Pro-inflammatory phenotype, induce cardiomyocyte apoptosis, promote oxidative stress via ROS production, aggravate cardiac injury. |
| Rao, M. et al.[122] (2021) | HF | Patients with DCM and ICM who were undergoing heart transplantation | CCR2-HLA-DRhi C1 | LYVE1, LILRB5, MAF, SIGLEC1, SLC40A1, BLVRB, STAB1, DAB2 | Tissue-resident macrophages enriched in mild-lesion/normal hearts, negative immunomodulation, maintain tissue homeostasis. |
| CCR2-HLA-DRhi C2 | CLEC4E, CLEC7A, CLEC10A | Tissue-resident macrophages, enriched in pattern recognition receptor signaling, may mediate pathogen sensing and immune surveillance. | |||
| CCR2+HLA-DRhi C1 | C1QA, C1QB | Tissue-resident macrophages enriched in severely fibrotic regions, exhibit phagocytic activity, antigen clearance and immune regulation. | |||
| CCR2+HLA-DRhi C2 | CXCL8, NLRP3, BIRC3, EGR1 | Pro-inflammatory subset enriched in fibrotic regions, activate NLRP3 inflammasome and NF-κB signaling, recruit leukocytes and promote inflammation, interact with activated endothelial cells via CXCL8-DARC axis. | |||
| CCR2+HLA-DRhi C3 | CCR2, S100A8, S100A9 | Infiltration-derived macrophages, transition into CCR2⁺HLA-DR⁺ⁱ C2 via ATF3/KLF4 regulons, contribute to pro-inflammatory responses. | |||
| TREM2+ Mφ | TREM2, SPP1, LGALS3 | Angiogenesis and immune suppression, intermediate state with reduced inflammatory response compared to CCR2⁺HLA-DR⁺ⁱ subsets. | |||
| Amrute, J. M. et al.[123] (2023) | HF | Patients with HF who experienced either cardiac recovery or ongoing HF before and after LVAD implantation | Mφ1 | SPP1, FN1, TPRG1, PPARG | Matrix remodeling, may contribute to pathological tissue changes. |
| Mφ2 | NAMPT, NR4A1, PLAUR, FOSB | Pro-inflammatory, negative prognosis for cardiac recovery, monocyte-derived inflammatory macrophages. | |||
| Mφ3 | CXCL10, CCL4, ENOX1, GBP1, BIRC3 | Promote inflammation and respond to interferon signaling. | |||
| Mφ4 | IFI44L, MX1, EPSTI1 | Interferon-responsive state, unresolved inflammation. | |||
| Mφ5 | NAV2, SCN9, ARNF150, MAMDC2 | Resident macrophages, tissue repair and remodeling. | |||
| Proliferating Mφ | DIAPH3, ARHGAP11B, ATAD2 | Proliferating. |
The increasing use of immune checkpoint inhibitors (ICIs), including PD-1/PD-L1 and CTLA-4 inhibitors, in cancer therapy has brought greater attention to immune-related adverse events, among which myocarditis is notably severe [130]. In genetic models of ICI-induced myocarditis, a CCR2+ monocyte-derived macrophage subset characterized by Cxcl9+Cxcl10+ expression exhibits an activated phenotype and exacerbates disease progression via three principal mechanisms: (1) T-cell hyperactivation through CXCL16/CXCR6-mediated CD8+ T cell interactions and CXCL9/CXCL10-CXCR3-dependent recruitment and activation of both CD4+ and CD8+ T cells, which amplify cytotoxic attacks on myocardial cells; (2) amplification of chemokine storms via CCL2/MCP1 and CCL7/MCP3 production, recruiting peripheral immune cells and intensifying myocardial inflammation; and (3) direct myocardial injury mediated by effector T-cell activation, antibody-dependent cellular cytotoxicity (ADCC), and phagocytosis. This pathogenic cascade is orchestrated by IFN-γ-STAT1 signaling, and therapeutic inhibition of the JAK2/STAT1 axis with ruxolitinib has been shown to reduce cardiovascular mortality in patients with ICI-induced myocarditis. In comparison to Cxcl9+Cxcl10+ macrophages, Nlrp3+ macrophages are enriched for genes involved in responses to LPS, regulation of IL-1β production, and stromal cell proliferation. CD163+ resident macrophages are enriched for pathways that regulate epithelial cell proliferation and stress response.
These findings highlight the diverse roles of macrophage subpopulations in myocarditis (Table 4).
Diabetic Cardiomyopathy
Diabetic cardiomyopathy (DbCM) is defined by structural and functional abnormalities of the myocardium in diabetic patients, with metabolic dysregulation and myocardial fibrosis as hallmark features [133,134]. During early hyperglycemia, cardiac interstitial macrophage infiltration is triggered by advanced glycation end-product (AGE) accumulation, adipokine secretion, activation of the RAAS, microvascular dysfunction, and increased oxidative stress. The progression of DbCM is primarily driven by impaired insulin signaling and mitochondrial dysfunction [135], both of which disrupt oxidative phosphorylation in cardiomyocytes. Insulin resistance hampers glucose uptake via GLUT1 and GLUT4 and inhibits fatty acid oxidation by suppressing key rate-limiting enzymes [136]. The subsequent mismatch between mitochondrial dysfunction and excessive fatty acid accumulation results in elevated ROS production, which further skews macrophages toward a pro-inflammatory CCR2+ phenotype. Dectin-1, a receptor predominantly expressed by macrophage pattern-recognition receptors (PRRs), promotes this inflammatory polarization in hyperglycemic environments by activating the Syk/NF-κB pathway [137]. These pro-inflammatory macrophages secrete cytokines including TNF-α, IL-1β, and IL-6, which markedly increase the expression of resistin—an adipokine that exacerbates insulin resistance—thereby further perpetuating hyperglycemia. Moreover, resistin itself can enhance the production of inflammatory cytokines, creating a self-reinforcing vicious cycle.
Key macrophage subsets, their markers and function in selected published studies on myocarditis and CS.
| Publication | Disease | Species/Model | Macrophage subsets | Markers | Function |
|---|---|---|---|---|---|
| Hua, X. et al.[128] (2020) | Myocarditis | Balb/c mice / Autoimmune myocarditis (EAM model) | Cluster 2 | Mlr1, Cxcl9, Ly6i, Nos2, Arg1 | Oxidative phosphorylation, antigen processing/presentation, nitric oxide biosynthesis, dominant in acute inflammation. |
| Cluster 3 | Ccl8, Fbx32, Hif1a, Rxra | Antigen presentation, neutrophil degranulation, peak in subacute phase. | |||
| Cluster 7 | Ccr7, Clec10a, Tnip3 | IL-10-mediated anti-inflammatory responses, wound healing, dominant in myopathy phase. | |||
| Cluster 8 | Tnf, Il-10, Vsig4 | Immune regulation through IL-10 signaling, increased in chronic phase. | |||
| Hu, Z. et al.[129] (2023) | Myocarditis | Lewis rats / Giant cell myocarditis model | Cluster 1 | Prdx1, Prdx5, Arg1, Ass1 | High IFN-γ stimulated gene scores, enhanced phagocytosis and inflammation, autoimmunity. |
| Cluster 2 | Ms4a7, Tmem176a, Tmem176b | Expressed MS4A family genes, involved in antigen processing/presentation. | |||
| Cluster 3 | Nr4a1 | Tissue-resident macrophages, regulates immune response. | |||
| Cluster 4 | Pf4 | Pleiotropic chemokine expression, limits macrophage activation, recovery processes. | |||
| Cluster 5 | Fcnb, Plac8, Vcan | Tumor necrosis factor signaling, interferon/cytokine signaling, pro-inflammatory functions. | |||
| Pan, M. et al. [131] (2024) | Myocarditis | Ctla4+/+Pdcd1-/-, Ctla4+/-Pdcd1-/- mice | Cd163+ resident Mφ | Cd163, Lyve1, Folr2, Cbr2 | Tissue-resident macrophages, homeostatic maintenance, potential roles in stress response and tissue repair. |
| Cxcl9+Cxcl10+ Mφ | Cxcl9, Cxcl10, Gbp2b, Fcgr4, CCR2 | IFN-γ-activated inflammatory macrophages, secretion of chemokines, antigen presentation, T-cell recruitment/activation via CXCR3 signaling, antibody-dependent cytotoxicity (ADCC) potential. | |||
| Nlrp3+ Mφ | Nlrp3, Ccl4, Cd14 | Pro-inflammatory, inflammasome activation, IL-1β production, stromal interaction. | |||
| Liu, J. et al.[132] (2022) | CS | patients with cardiac sarcoidosis | GPNMB+ Mφ | GPNMB, TPRG1, SNTB1 | Lysosomal biogenesis, PPAR pathway activation, giant cell differentiation via MITF/TFEC regulation, granuloma structural organization. |
| HLA-DR+ Mφ | HLA-DR, CIITA, IL12RB1 | Antigen presentation, mTOR pathway activation, interface between giant cells and immune microenvironment. | |||
| SYTL3+ Mφ | SYTL3, FCGR3A, IL18R1 | Pro-inflammatory, mTOR-driven proliferation, intermediate differentiation state, phagosome/ECM remodeling. | |||
| Resident Mφ | CD163, F13A1, SCN9A | Tissue homeostasis, low inflammatory activity. |
Cardiac fibrosis—a defining feature of mid- to late-stage DbCM—results from macrophage-fibroblast interactions that drive interstitial and perivascular fibrosis [133,139]. In mouse models of DbCM, endothelial cell and macrophage numbers decrease while fibroblast and cardiomyocyte populations increase, reflecting aggravated fibrosis and endothelial injury [140]. In diabetic mouse hearts induced by HFD/STZ, Egfr and Pdgfra are highly expressed in cardiac fibroblasts, whereas macrophages exhibit increased Pdgfc expression [141]. These data indicate that macrophage-fibroblast crosstalk contributes to the development of myocardial fibrosis in DbCM. Notably, dynamic changes in macrophage subsets over time during DbCM progression remain understudied. Future research should employ longitudinal multi-omics and macrophage lineage tracing strategies to comprehensively characterize the evolution of macrophage phenotypes and functions, thereby providing a robust theoretical framework for developing targeted interventions in DbCM.
Cardiac Sarcoidosis
Cardiac sarcoidosis (CS) is histologically defined by granulomatous inflammation, with granulomas composed primarily of macrophages [142]. One of the main diagnostic challenges in CS is distinguishing it from other inflammatory cardiomyopathies—such as giant cell myocarditis (GCM) and lymphocytic myocarditis—due to overlapping histopathologic features. Recent spatial transcriptomic analyses have elucidated distinct cellular architectures that facilitate this differentiation [132]. Notably, GPNMB+ (glycoprotein non-metastatic melanoma protein B) multinucleated giant cells are surrounded by dense infiltrates of HLA-DRhi macrophages in CS, forming a distinctive granulomatous structure absent in other types of inflammatory heart disease. Within these granulomas, SYTL3+ macrophages are diffusely distributed, whereas CD163+ macrophages, CD1c+ dendritic cells, non-classical monocytes, and T cells localize to the periphery and external regions (Figure 4). Transcriptomic profiling reveals unique enrichment of lysosomal and PPAR signaling pathways in GPNMB+ giant cells, along with selective mTOR pathway activation—a key regulator of cellular proliferation—in HLA-DRhi and SYTL3+ macrophages. SYTL3+ macrophages may represent a transitional differentiation stage, potentially giving rise to both GPNMB+ giant cells and adjacent mature epithelioid-like histiocytes (HLA-DRhi macrophages).
Schematic of Changes in Major Macrophage Subsets in Cardiac Saroidosis.
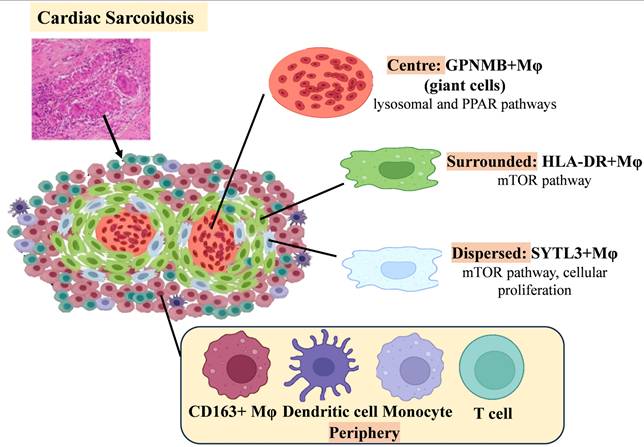
Subsequent studies have identified GPNMB as a novel marker of multinucleated giant cells in CS, with its regulation potentially mediated by the microphthalmia-associated transcription factor (MITF) family. Although GPNMB immunohistochemistry detects giant cells in both CS and GCM, the spatial organization of HLA-DRhi macrophages offers a means of diagnostic distinction: in CS, GPNMB+ giant cells are closely encircled by HLA-DRhi macrophages, whereas in GCM, these cell populations are more diffusely distributed throughout lesions. This organizational distinction provides a practical diagnostic tool. Combining GPNMB staining with HLA-DR spatial mapping thus enhances diagnostic accuracy, addressing longstanding challenges in distinguishing inflammatory cardiomyopathies. These findings underscore the value of multi-omics technologies in refining histopathological criteria.
Discussion
Decoding Macrophage Heterogeneity through scRNA-Seq
scRNA-seq has transformed our understanding of macrophage heterogeneity in cardiovascular pathology by revealing distinct functional subsets shaped by disease-specific microenvironments. Monocyte-derived CCR2+ macrophages initiate inflammatory cascades and promote adverse ventricular remodeling, whereas resident macrophage populations confer cardioprotective effects that maintain cardiac homeostasis and functional integrity [143,144]. These broad categories further differentiate into context-dependent subpopulations (Table 5): for example, TREM2hi foam cells in atherosclerotic plaques facilitate lipid uptake and enhance cholesterol efflux; post-MI TREM2hi macrophages upregulate Arg1 and IL-10 to promote reparative processes; SPP1+ macrophages secrete TGF-β and IL-10, exacerbating fibrosis; and CD72hi subsets amplify inflammatory signaling, accelerating heart failure progression. These populations are defined not only by unique surface markers, but also by specific metabolic pathways and epigenetic regulators, making them compelling therapeutic targets. Importantly, global depletion of cardiac macrophages may impair left ventricular remodeling due to the loss of resident macrophage subsets critical for myocardial homeostasis [145,146]. Therefore, selective targeting of specific macrophage subsets is essential and requires a comprehensive understanding of their functional diversity within mixed populations. In pathological settings, the inflammatory microenvironment can shift cardiac macrophages from reparative to detrimental phenotypes, meaning treatment efficacy varies depending on the timing and subsets targeted [147]. Integrating scRNA-seq with other omics technologies enables multimodal analysis of macrophage interactions with cardiomyocytes, fibroblasts, and other immune cells, helping to resolve niche-specific activation trajectories.
Major macrophage subtypes, their biomarkers, and functions in MI, Atherosclerosis and HF.
| Major Macrophage Subset | Key Markers | Functions |
|---|---|---|
| Timd4+/Lyve1+ Mφ | Timd4, Lyve1, Folr2 | In MI: Maintain cardiac homeostasis, promote repair, IGF-1 secretion, modulate inflammation |
| In atherosclerosis: Maintain vascular homeostasis, anti-inflammatory, lipid handling to reduce arterial stiffness | ||
| In HF: Phagocytosis and clearance of apoptotic cells, antifibrotic, maintain heart function | ||
| MHCIIhi Mφ | MHC-II, Cd74, H2-Ab1, H2-Eb1, Cx3cr1, Adgre1 | In MI: Antigen presentation, immune regulation, involved in repair |
| In atherosclerosis: Antigen presentation, partly pro-inflammatory | ||
| In HF: Antigen presentation, tissue repair, immune surveillance | ||
| CCR2+ Monocyte-derived Mφ | CCR2, Ly6C, FCGR1, Ccl2, Plac8, Osm | In MI: Pro-inflammatory responses, clear necrotic debris, promote fibrosis, recruit immune cells |
| In atherosclerosis: Pro-inflammatory, promote plaque progression, lipid deposition | ||
| In HF: Pro-inflammatory, drive cardiac fibrosis and dysfunction | ||
| Trem2hi Mφ | Trem2, Gpnmb, Fabp5, Apoe, Arg1, Mmp12 | In MI: Anti-inflammatory/reparative, promote remodeling, clear apoptotic and dysfunctional mitochondria |
| In atherosclerosis: Foam cells with lipid metabolism, promote cholesterol efflux and fibrosis progression | ||
| In HF: Promote angiogenesis and immune suppression, reduce inflammation | ||
| ISG+ / IFNIC Mφ | Isg15, Ifit1/2/3, Irf7, Rsad2, Cxcl10 | In MI: Regulate antiviral and inflammatory responses, partially impair repair |
| In atherosclerosis: Promote inflammation and antiviral responses in plaque | ||
| In HF: May promote persistent inflammation, negative prognosis | ||
| Proliferating Mφ | Mki67, Top2a, Ccnb2, Birc5, Tubb | In MI: Expand early inflammatory macrophage pool |
| In atherosclerosis: Support local expansion of foam cells | ||
| In HF: Expand macrophage populations early in remodeling | ||
| CD72hi Mφ | Cd72, Ccr2 | In MI: - |
| In atherosclerosis: - | ||
| In HF: Pro-inflammatory, induce cardiomyocyte apoptosis and oxidative stress, worsen cardiac injury | ||
| MacAIR Mφ | Il1b, Rgs1, Cd9, Mmp12, Acp5, MHC-II | In MI: - |
| In atherosclerosis: Foam cell precursors in early plaque, maintain vascular barrier | ||
| In HF: - |
New strategies for targeted macrophage therapy
Current therapeutic approaches targeting specific macrophage subtypes remain imprecise, typically focusing on M2-like macrophages and their associated anti-inflammatory mediators. Emerging strategies include: (1) Direct targeting of subset-specific receptors—soluble TREM2 can reprogram post-MI macrophages toward reparative phenotypes, and CCR2 antagonists (e.g., cenicriviroc) reduce inflammatory monocyte infiltration in atherosclerosis and MI; (2) Metabolic reprogramming—inhibition of glycolysis promotes oxidative metabolism and tissue repair, while PPARγ agonists enhance fatty acid oxidation and cholesterol efflux; (3) Epigenetic engineering—CRISPR-Cas9-mediated editing of genes such as TREM2 or BHLHE41 in foam cells modulates lipid metabolism and fibrotic pathways [148]; (4) Nano-targeted delivery—nanoparticle platforms selectively deliver drugs or siRNA to Lyve1+ resident macrophages or Ly6Chi monocytes, thereby preserving beneficial subsets [149]; (5) Integration of AI-driven multi-omics analyses and deep-learning-based screening facilitates the personalized design of macrophage-targeted therapies, with the potential to improve cardiovascular outcomes.
Spatial Transcriptomics Reveals Macrophage Niche in Cardiovascular Disease
Spatial transcriptomics, an emerging technique, complements scRNA-seq by retaining the spatial context of gene expression within tissues. This approach is particularly valuable in cardiovascular research, as it enables the mapping of macrophage localization in relation to other cardiac and vascular cell types, offering new insights into the tissue microenvironment that regulates macrophage function. Tissue homeostasis and cell function rely on direct interactions between neighboring cell types; spatial omics methods thus provide crucial information about how distinct macrophage subpopulations interact with their cellular milieu to coordinate tissue responses. For example, following myocardial infarction, different regions (control, peri-infarct, remote myocardium) exhibit spatially distinct enrichment of macrophage functional states, providing clarity on how cell states are modulated by their local neighborhoods. Integration of single-cell multi-omics with spatial data has uncovered new relationships between macrophage subtypes and myofibroblast differentiation during cardiac remodeling. Notably, it is important to recognize that macrophage markers identified from nuclei in human tissue differ substantially from those acquired via whole-cell sequencing. Therefore, macrophage subpopulations defined through nuclear sequencing often cannot be mapped directly onto conventional classifications, underscoring the need for more refined classification strategies in human studies.
Technical Bottlenecks in Cardiac Macrophage Metabolomics
While metabolomics has seen widespread application in macrophage research, particularly in oncology, its use in cardiovascular macrophages remains limited. Cancer studies have elucidated that tumor cells reprogram macrophage metabolism by releasing metabolites such as succinate and fatty acids into the microenvironment [150,151]. By contrast, most insights into cardiovascular macrophage metabolism are derived from transcriptomic profiling and knockout mouse models [32,152]. Since metabolic activity is regulated at multiple levels, transcriptomic data alone are insufficient to fully explain metabolic phenotypes. Although unbiased metabolomics provides critical functional insights, several technical barriers hinder its adoption. Cardiac macrophage studies face unique challenges: (1) Their low abundance (representing only ~7% of cardiac cells [153]) complicates isolation, risking tissue-level metabolome analyses that misrepresent macrophage-specific metabolic states; (2) Tissue dissociation can induce metabolic artifacts; (3) High-throughput platforms (MS/NMR) often require trade-offs between quantitative accuracy and coverage, necessitating resource-intensive targeted validation; (4) Incomplete metabolite annotation limits functional interpretation [154]. As a result, most studies rely on more readily accessible primary macrophages (e.g., BMDMs, peritoneal macrophages) rather than tissue-resident cardiac populations [34]. For instance, Tabas et al. identified apoptotic cell-derived arginine as essential for efferocytosis-induced macrophage polarization using BMDMs [155], while Thorp's group demonstrated that engulfed fatty acids drive pro-resolving functions in peritoneal macrophages through mitochondrial metabolism [19]. However, these observations may not accurately capture the metabolic dynamics of cardiac macrophages. A practical approach often involves in vitro screening of metabolic pathways, with subsequent in vivo validation. Nonetheless, such reductions risk oversimplifying the complex interplay between cardiac macrophages and their tissue niches. Future methodological advances should address the need for increased sensitivity (enabling single-cell metabolomics) and spatial resolution, while minimizing ex vivo manipulation artifacts.
The limitations of Multi - Omics
Current multi-omics approaches in cardiac macrophage research are hampered by limitations in temporal and spatial resolution, incomplete metabolome coverage, and batch effects, all of which impede mechanistic insights and translational advances. Firstly, static omics methods (like scRNA-seq) cannot adequately capture the temporal dynamics of macrophage phenotypic transitions—for example, delineating the shift from pro-inflammatory to reparative states following MI requires time-resolved analysis, which is lacking in snapshot datasets. Longitudinal multi-omics paired with lineage-tracing systems (such as Cre/Dre recombinases) offer promising solutions. Secondly, spatial transcriptomics typically aggregates multiple cells per spot, obscuring detailed macrophage-microenvironment interactions; achieving true single-cell resolution remains a priority. Furthermore, integration of distinct omics layers is complicated by platform-specific biases and batch effects, necessitating advanced computational harmonization—such as the application of multimodal variational autoencoders—to reliably elucidate cross-omic relationships [156]. Lastly, metabolomics is constrained by limited MS and NMR sensitivity, particularly for detecting atherosclerosis-relevant lipids critical to foam cell formation. Although emerging techniques like MALDI-MSI enable single-cell metabolic imaging [157], widespread adoption is limited by technical immaturity.
Future Directions
Advancements and Challenges in Single-Cell Multi-Omics
Single-cell technologies now enable multidimensional analysis of cellular processes, although integration across omics layers remains challenging. Traditional CRISPR-based screens (e.g., Perturb-seq combined with scRNA-seq) elucidate transcriptomic responses to genetic perturbations but lack spatial context. Conversely, spatial transcriptomic platforms such as multiplexed error-robust fluorescence in situ hybridization (MERFISH) provide precise tissue architecture but are generally incompatible with CRISPR perturbations [158]. Perturb-FISH bridges this gap, enabling simultaneous mapping of genetic perturbations, transcriptomes, and spatial information at single-cell resolution [159]. Validated in THP1 macrophages, this approach holds promise for dissecting macrophage heterogeneity in cardiovascular tissues. Additionally, spatial single-cell proteomics platforms (e.g., PhenoCycler-Fusion) provide visualization and quantification of protein expression at single-cell resolution, furnishing critical spatial context for understanding cellular localization and function [160]. Spatial metabolomics using imaging mass cytometry (IMC) and mass spectrometry imaging (MSI) is also increasingly employed to identify cellular biomarkers and metabolites while preserving tissue architecture. Despite these advances, single-cell metabolic imaging remains technically underdeveloped. Recent multimodal MSI workflows integrating MALDI-MSI and IMC enable single-cell analysis of metabolic heterogeneity and its relationship to specific cell populations in human tissue [161], though scalability, reproducibility, and efficiency require further validation before routine adoption. Collectively, these innovations underscore the transformative potential of single-cell multi-omics approaches, while highlighting the need for continued technical refinement to achieve seamless integration across molecular layers.
AI-Driven Multi-Omics Integration in Cardiac Macrophages
The complexity of multi-omics datasets spanning transcriptomic, epigenetic, metabolic, and spatial dimensions challenges conventional data integration approaches. Artificial intelligence (AI) and machine learning (ML) mitigate these challenges in four principal domains [162]. First, for data integration and analysis, AI/ML models—including supervised, unsupervised, and deep learning methods—facilitate harmonization across multi-omics datasets. Algorithms such as random forests, support vector machines, and transformer architectures excel at identifying cross-omic patterns and relationships often missed by standard statistical techniques. Initiatives like AtheroNET exemplify how such integration is fundamental for elucidating macrophage-driven mechanisms in cardiovascular pathology. Second, in biomarker discovery, AI/ML accelerates the identification of novel macrophage-specific markers by mining integrated multi-omics profiles. Third, in mechanistic insight, these tools help to unravel complex biological networks, clarifying macrophage-mediated pathways (e.g., inflammation, oxidative stress, mitochondrial dysfunction) underlying cardiac disease progression. Finally, in predictive modeling, ML frameworks combine multi-omics and clinical data to forecast disease progression and cardiovascular outcomes [163]. Previous studies have demonstrated the utility of integrating AI-driven models with multi-omics and clinical data to improve prediction of atherosclerotic cardiovascular disease (ASCVD) risk and inform precision diagnostic and therapeutic strategies [164].
Abbreviations
CVD: cardiovascular diseases; ECM: extracellular matrix; scRNA-seq: single-cell RNA sequencing; ST-seq: spatial transcriptomics sequencing; MI: myocardial infarction; TLRs: toll-like receptors; LPS: lipopolysaccharide; IFN-γ: interferon gamma; iNOS: inducible nitric oxide synthase; IL-4: interleukin-4; IL-13: interleukin-13; ARG1: arginase 1; YM1: chitinase-like protein 3; PGC-1β: PPARγ coactivator-1β; TCA: tricarboxylic acid cycle; HIF1α: hypoxia-inducible factor 1 alpha; TREM2: triggering receptor expressed on myeloid cells 2; Bhlhe41: basic helix-loop-helix family member e41; NLRP3: NOD-like receptor protein 3; MHCII: major histocompatibility complex class II; LYVE1: lymphatic vessel endothelial hyaluronan receptor 1; FOLR2: folate receptor 2; CCR2: C-C chemokine receptor type 2; HLA-DRA: human leukocyte antigen-DR alpha; HLA-DMA: human leukocyte antigen-DM alpha; HLA-DMB: human leukocyte antigen-DM beta; HLA-DPA1: human leukocyte antigen-DP alpha 1; IL4R: interleukin-4 receptor; STAT3: signal transducer and activator of transcription 3; ITGAM: integrin alpha M; C1QA: complement C1q A chain; SAN: sinoatrial node; AVN: atrioventricular node; IGF1: insulin-like growth factor 1; BMDMs: bone marrow-derived macrophages; TGF-β: transforming growth factor-beta; Smad3: SMAD family member 3; IL-1β: interleukin-1 beta; NF-κB: nuclear factor kappa B; SPP1: secreted phosphoprotein 1; CD44: cluster of differentiation 44; SYK: spleen tyrosine kinase; TNF: tumor necrosis factor; CCL4: C-C motif chemokine ligand 4; HLA-DR: human leukocyte antigen-DR; PD-1: programmed cell death protein 1; PD-L1: programmed death-ligand 1; CTLA-4: cytotoxic T-lymphocyte associated protein 4; CXCL9: C-X-C motif chemokine ligand 9; CXCL10: C-X-C motif chemokine ligand 10; CXCR3: C-X-C chemokine receptor type 3; CCL2: C-C motif chemokine ligand 2; MCP1: monocyte chemoattractant protein 1; CCL7: C-C motif chemokine ligand 7; MCP3: monocyte chemoattractant protein 3; ADCC: antibody-dependent cellular cytotoxicity; STAT1: signal transducer and activator of transcription 1; JAK2: janus kinase 2; GLUT1: glucose transporter 1; GLUT4: glucose transporter 4; ROS: reactive oxygen species; EGFR: epidermal growth factor receptor; PDGFRA: platelet-derived growth factor receptor alpha; PDGFC: platelet-derived growth factor C; GPNMB: glycoprotein non-metastatic melanoma protein B; MITF: microphthalmia-associated transcription factor; CRISPR: clustered regularly interspaced short palindromic repeats; Cas9: CRISPR-associated protein 9; AI: artificial intelligence, ML: machine learning; MALDI-MSI: matrix-assisted laser desorption/ionization mass spectrometry imaging; IMC: imaging mass cytometry; MSI: mass spectrometry imaging; MERFISH: multiplexed error-robust fluorescence in situ hybridization.
Acknowledgements
Figures were created under the academic license of BioRender.
Funding
This work was supported by National Natural Science Foundation of China (U22A20267 and 82030014 to J.W., 82370374 to W.Z.) and financially supported by Transvascular Implantation Devices Research Institute (TIDRI) under Grant No.KY042025003.
Author Contributions
CY and JL conceived and designed the study and wrote the manuscript. CY and YC were involved in paper collection. JW and WZ conceived the current study and reviewed the manuscript. All authors reviewed previous versions and read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Khoury MK, Yang H, Liu B. Macrophage Biology in Cardiovascular Diseases. Arterioscler Thromb Vasc Biol. 2021;41(2):e77-e81
2. Zaman R, Epelman S. Resident cardiac macrophages: Heterogeneity and function in health and disease. Immunity. 2022;55(9):1549-1563
3. Bajpai G, Lavine KJ. Isolation of Macrophage Subsets and Stromal Cells from Human and Mouse Myocardial Specimens. J Vis Exp JoVE. 2019 (154). doi: 10.3791/60015
4. Lafuse WP, Wozniak DJ, Rajaram MVS. Role of Cardiac Macrophages on Cardiac Inflammation, Fibrosis and Tissue Repair. Cells. 2020;10(1):51
5. Frodermann V, Nahrendorf M. Macrophages and Cardiovascular Health. Physiol Rev. 2018;98(4):2523-2569
6. Czubryt MP, Hale TM. Cardiac fibrosis: Pathobiology and therapeutic targets. Cell Signal. 2021;85:110066
7. Jian Y, Zhou X, Shan W. et al. Crosstalk between macrophages and cardiac cells after myocardial infarction. Cell Commun Signal CCS. 2023;21(1):109
8. Yang S, Qian L, Li Z. et al. Integrated Multi-Omics Landscape of Liver Metastases. Gastroenterology. 2023;164(3):407-423.e17
9. Zhang J, Song J, Tang S. et al. Multi-omics analysis reveals the chemoresistance mechanism of proliferating tissue-resident macrophages in PDAC via metabolic adaptation. Cell Rep. 2023;42(6):112620
10. Kloosterman DJ, Erbani J, Boon M. et al. Macrophage-mediated myelin recycling fuels brain cancer malignancy. Cell. 2024;187(19):5336-5356.e30
11. Gu Y, Zhou Y, Ju S. et al. Multi-omics profiling visualizes dynamics of cardiac development and functions. Cell Rep. 2022;41(13):111891
12. Zhang X, Xiao Y, Hu B. et al. Multi-omics analysis of human tendon adhesion reveals that ACKR1-regulated macrophage migration is involved in regeneration. Bone Res. 2024;12(1):27
13. Epelman S, Lavine KJ, Randolph GJ. Origin and Functions of Tissue Macrophages. Immunity. 2014;41(1):21-35
14. Qie J, Liu Y, Wang Y. et al. Integrated proteomic and transcriptomic landscape of macrophages in mouse tissues. Nat Commun. 2022;13(1):7389
15. Chen H-J, Sévin DC, Griffith GR. et al. Integrated metabolic-transcriptomic network identifies immunometabolic modulations in human macrophages. Cell Rep. 2024;43(9):114741
16. Baig MS, Barmpoutsi S, Bharti S. et al. Adaptor molecules mediate negative regulation of macrophage inflammatory pathways: a closer look. Front Immunol. 2024;15:1355012
17. Ma J, Wei K, Liu J. et al. Glycogen metabolism regulates macrophage-mediated acute inflammatory responses. Nat Commun. 2020;11(1):1769
18. Tabas I, Bornfeldt KE. Intracellular and Intercellular Aspects of Macrophage Immunometabolism in Atherosclerosis. Circ Res. 2020;126(9):1209-1227
19. Zhang S, Weinberg S, DeBerge M. et al. Efferocytosis Fuels Requirements of Fatty Acid Oxidation and the Electron Transport Chain to Polarize Macrophages for Tissue Repair. Cell Metab. 2019;29(2):443-456.e5
20. Yurdagul A, Subramanian M, Wang X. et al. Macrophage Metabolism of Apoptotic Cell-Derived Arginine Promotes Continual Efferocytosis and Resolution of Injury. Cell Metab. 2020;31(3):518-533.e10
21. Colegio OR, Chu N-Q, Szabo AL. et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513(7519):559-563
22. Murray PJ, Allen JE, Biswas SK. et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14-20
23. Assouvie A, Daley-Bauer LP, Rousselet G. Growing Murine Bone Marrow-Derived Macrophages. Methods Mol Biol Clifton NJ. 2018;1784:29-33
24. Orecchioni M, Ghosheh Y, Pramod AB. et al. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS-) vs. Alternatively Activated Macrophages. Front Immunol. 2019;10:1084
25. Witherel CE, Abebayehu D, Barker TH. et al. Macrophage and Fibroblast Interactions in Biomaterial-Mediated Fibrosis. Adv Healthc Mater. 2019;8(4):e1801451
26. Huang X, Li Y, Fu M. et al. Polarizing Macrophages In Vitro. Methods Mol Biol Clifton NJ. 2018;1784:119-126
27. Wang S, Liu R, Yu Q. et al. Metabolic reprogramming of macrophages during infections and cancer. Cancer Lett. 2019;452:14-22
28. Huang SC-C, Everts B, Ivanova Y. et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol. 2014;15(9):846-855
29. Vats D, Mukundan L, Odegaard JI. et al. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 2006;4(1):13-24
30. Puleston DJ, Buck MD, Klein Geltink RI. et al. Polyamines and eIF5A Hypusination Modulate Mitochondrial Respiration and Macrophage Activation. Cell Metab. 2019;30(2):352-363.e8
31. Bajpai G, Bredemeyer A, Li W. et al. Tissue Resident CCR2- and CCR2+ Cardiac Macrophages Differentially Orchestrate Monocyte Recruitment and Fate Specification Following Myocardial Injury. Circ Res. 2019;124(2):263-278
32. Wculek SK, Dunphy G, Heras-Murillo I. et al. Metabolism of tissue macrophages in homeostasis and pathology. Cell Mol Immunol. 2022;19(3):384-408
33. Dick SA, Macklin JA, Nejat S. et al. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat Immunol. 2019;20(1):29-39
34. Nicolás-Ávila JA, Lechuga-Vieco AV, Esteban-Martínez L. et al. A Network of Macrophages Supports Mitochondrial Homeostasis in the Heart. Cell. 2020;183(1):94-109.e23
35. Lim HY, Lim SY, Tan CK. et al. Hyaluronan Receptor LYVE-1-Expressing Macrophages Maintain Arterial Tone through Hyaluronan-Mediated Regulation of Smooth Muscle Cell Collagen. Immunity. 2018;49(2):326-341.e7
36. Sansbury BE, DeMartino AM, Xie Z. et al. Metabolomic analysis of pressure-overloaded and infarcted mouse hearts. Circ Heart Fail. 2014;7(4):634-642
37. Lai L, Leone TC, Keller MP. et al. Energy metabolic reprogramming in the hypertrophied and early stage failing heart: a multisystems approach. Circ Heart Fail. 2014;7(6):1022-1031
38. McGarrah RW, Crown SB, Zhang G-F. et al. Cardiovascular Metabolomics. Circ Res. 2018;122(9):1238-1258
39. Zaman R, Hamidzada H, Epelman S. Exploring cardiac macrophage heterogeneity in the healthy and diseased myocardium. Curr Opin Immunol. 2021;68:54-63
40. Mosser DM, Hamidzadeh K, Goncalves R. Macrophages and the maintenance of homeostasis. Cell Mol Immunol. 2021;18(3):579-587
41. Li S, Yu J, Huber A. et al. Metabolism drives macrophage heterogeneity in the tumor microenvironment. Cell Rep. 2022;39(1):110609
42. Park MD, Silvin A, Ginhoux F. et al. Macrophages in health and disease. Cell. 2022;185(23):4259-4279
43. Li L, Ma Q, Wang M. et al. Single-cell transcriptome sequencing of macrophages in common cardiovascular diseases. J Leukoc Biol. 2023;113(2):139-148
44. Lother A, Kohl P. The heterocellular heart: identities, interactions, and implications for cardiology. Basic Res Cardiol. 2023;118(1):30
45. Yona S, Kim K-W, Wolf Y. et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38(1):79-91
46. Chakarov S, Lim HY, Tan L. et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science. 2019;363(6432):eaau0964
47. Dick SA, Wong A, Hamidzada H. et al. Three tissue resident macrophage subsets coexist across organs with conserved origins and life cycles. Sci Immunol. 2022;7(67):eabf7777
48. Litviňuková M, Talavera-López C, Maatz H. et al. Cells of the adult human heart. Nature. 2020;588(7838):466-472
49. Asp M, Giacomello S, Larsson L. et al. A Spatiotemporal Organ-Wide Gene Expression and Cell Atlas of the Developing Human Heart. Cell. 2019;179(7):1647-1660.e19
50. Kanemaru K, Cranley J, Muraro D. et al. Spatially resolved multiomics of human cardiac niches. Nature. 2023;619(7971):801-810
51. Murphy SP, Kakkar R, McCarthy CP. et al. Inflammation in Heart Failure: JACC State-of-the-Art Review. J Am Coll Cardiol. 2020;75(11):1324-1340
52. Wei Q, Deng Y, Yang Q. et al. The markers to delineate different phenotypes of macrophages related to metabolic disorders. Front Immunol. 2023: 14: 1084636.
53. Leid J, Carrelha J, Boukarabila H. et al. Primitive Embryonic Macrophages are Required for Coronary Development and Maturation. Circ Res. 2016;118(10):1498-1511
54. Schulz C, Gomez Perdiguero E, Chorro L. et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336(6077):86-90
55. Epelman S, Lavine KJ, Beaudin AE. et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 2014;40(1):91-104
56. Farbehi N, Patrick R, Dorison A. et al. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. eLife. 2019;8:e43882
57. Bajpai G, Schneider C, Wong N. et al. The human heart contains distinct macrophage subsets with divergent origins and functions. Nat Med. 2018;24(8):1234-1245
58. Lavine KJ, Epelman S, Uchida K. et al. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc Natl Acad Sci U S A. 2014;111(45):16029-16034
59. Anderson JL, Morrow DA. Acute Myocardial Infarction. N Engl J Med. 2017;376(21):2053-2064
60. X W, Mr R, M K-K. et al. Angiogenesis after acute myocardial infarction. Cardiovasc Res. 2021;117(5):1257-1273
61. Jin K, Gao S, Yang P. et al. Single-Cell RNA Sequencing Reveals the Temporal Diversity and Dynamics of Cardiac Immunity after Myocardial Infarction. Small Methods. 2022;6(3):e2100752
62. W W, L A-H, V T. et al. Deciphering the Dynamic Transcriptional and Post-transcriptional Networks of Macrophages in the Healthy Heart and after Myocardial Injury. Cell Rep. 2018;23(2):622-636
63. Yap J, Irei J, Lozano-Gerona J. et al. Macrophages in cardiac remodelling after myocardial infarction. Nat Rev Cardiol. 2023;20(6):373-385
64. Peet C, Ivetic A, Bromage DI. et al. Cardiac monocytes and macrophages after myocardial infarction. Cardiovasc Res. 2020;116(6):1101-1112
65. Heidt T, Courties G, Dutta P. et al. Differential contribution of monocytes to heart macrophages in steady-state and after myocardial infarction. Circ Res. 2014;115(2):284-295
66. Chouchani ET, Pell VR, Gaude E. et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515(7527):431-435
67. Hassel B, Ilebekk A, Tønnessen T. Cardiac accumulation of citrate during brief myocardial ischaemia and reperfusion in the pig in vivo. Acta Physiol Scand. 1998;164(1):53-59
68. MacKenzie ED, Selak MA, Tennant DA. et al. Cell-permeating alpha-ketoglutarate derivatives alleviate pseudohypoxia in succinate dehydrogenase-deficient cells. Mol Cell Biol. 2007;27(9):3282-3289
69. Mills EL, Kelly B, Logan A. et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell. 2016;167(2):457-470.e13
70. Wellen KE, Hatzivassiliou G, Sachdeva UM. et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324(5930):1076-1080
71. Lauterbach MA, Hanke JE, Serefidou M. et al. Toll-like Receptor Signaling Rewires Macrophage Metabolism and Promotes Histone Acetylation via ATP-Citrate Lyase. Immunity. 2019;51(6):997-1011.e7
72. Thorp EB. Cardiac macrophages and emerging roles for their metabolism after myocardial infarction. J Clin Invest. 2023;133(18):e171953
73. Viaud M, Ivanov S, Vujic N. et al. Lysosomal Cholesterol Hydrolysis Couples Efferocytosis to Anti-Inflammatory Oxysterol Production. Circ Res. 2018;122(10):1369-1384
74. Langston PK, Shibata M, Horng T. Metabolism Supports Macrophage Activation. Front Immunol. 2017;8:61
75. O'Neill LAJ, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213(1):15-23
76. Ninh VK, Calcagno DM, Yu JD. et al. Spatially clustered type I interferon responses at injury borderzones. Nature. 2024;633(8028):174-181
77. Xu Y, Jiang K, Su F. et al. A transient wave of Bhlhe41+ resident macrophages enables remodeling of the developing infarcted myocardium. Cell Rep. 2023;42(10):113174
78. Mouton AJ, DeLeon-Pennell KY, Rivera Gonzalez OJ. et al. Mapping macrophage polarization over the myocardial infarction time continuum. Basic Res Cardiol. 2018;113(4):26
79. Jung S-H, Hwang B-H, Shin S. et al. Spatiotemporal dynamics of macrophage heterogeneity and a potential function of Trem2hi macrophages in infarcted hearts. Nat Commun. 2022;13(1):4580
80. Frangogiannis NG. Cardiac fibrosis: Cell biological mechanisms, molecular pathways and therapeutic opportunities. Mol Aspects Med. 2019;65:70-99
81. Rizzo G, Gropper J, Piollet M. et al. Dynamics of monocyte-derived macrophage diversity in experimental myocardial infarction. Cardiovasc Res. 2022;119(3):772-785
82. Amrute JM, Luo X, Penna V. et al. Targeting immune-fibroblast cell communication in heart failure. Nature. 2024;635(8038):423-433
83. Chen R, Zhang H, Tang B. et al. Macrophages in cardiovascular diseases: molecular mechanisms and therapeutic targets. Signal Transduct Target Ther. 2024;9(1):130
84. Kuppe C, Ramirez Flores RO, Li Z. et al. Spatial multi-omic map of human myocardial infarction. Nature. 2022;608(7924):766-777
85. Hoeft K, Schaefer GJL, Kim H. et al. Platelet-instructed SPP1+ macrophages drive myofibroblast activation in fibrosis in a CXCL4-dependent manner. Cell Rep. 2023;42(2):112131
86. Bevan L, Lim ZW, Venkatesh B. et al. Specific macrophage populations promote both cardiac scar deposition and subsequent resolution in adult zebrafish. Cardiovasc Res. 2020;116(7):1357-1371
87. Lindsey ML, Jung M, Yabluchanskiy A. et al. Exogenous CXCL4 infusion inhibits macrophage phagocytosis by limiting CD36 signalling to enhance post-myocardial infarction cardiac dilation and mortality. Cardiovasc Res. 2019;115(2):395-408
88. Gong S, Zhai M, Shi J. et al. TREM2 macrophage promotes cardiac repair in myocardial infarction by reprogramming metabolism via SLC25A53. Cell Death Differ. 2024;31(2):239-253
89. Cole JE, Park I, Ahern DJ. et al. Immune cell census in murine atherosclerosis: cytometry by time of flight illuminates vascular myeloid cell diversity. Cardiovasc Res. 2018;114(10):1360-1371
90. Libby P. Inflammation during the life cycle of the atherosclerotic plaque. Cardiovasc Res. 2021;117(13):2525-2536
91. Koelwyn GJ, Corr EM, Erbay E. et al. Regulation of macrophage immunometabolism in atherosclerosis. Nat Immunol. 2018;19(6):526-537
92. Tabas I, Bornfeldt KE. Macrophage Phenotype and Function in Different Stages of Atherosclerosis. Circ Res. 2016;118(4):653-667
93. Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. 2013;13(10):709-721
94. Blagov AV, Markin AM, Bogatyreva AI. et al. The Role of Macrophages in the Pathogenesis of Atherosclerosis. Cells. 2023;12(4):522
95. Moore KJ, Tabas I. Macrophages in the pathogenesis of atherosclerosis. Cell. 2011;145(3):341-55
96. Xiao Q, Hou R, Xie L. et al. Macrophage metabolic reprogramming and atherosclerotic plaque microenvironment: Fostering each other? Clin Transl Med. 2023;13(5):e1257
97. Carroll RG, Zasłona Z, Galván-Peña S. et al. An unexpected link between fatty acid synthase and cholesterol synthesis in proinflammatory macrophage activation. J Biol Chem. 2018;293(15):5509-5521
98. Barrett TJ. Macrophages in Atherosclerosis Regression. Arterioscler Thromb Vasc Biol. 2020;40(1):20-33
99. Willemsen L, de Winther MP. Macrophage subsets in atherosclerosis as defined by single-cell technologies. J Pathol. 2020;250(5):705-714
100. Takaoka M, Zhao X, Lim HY. et al. Early intermittent hyperlipidaemia alters tissue macrophages to fuel atherosclerosis. Nature. 2024;634(8033):457-465
101. Kim K, Shim D, Lee JS. et al. Transcriptome Analysis Reveals Nonfoamy Rather Than Foamy Plaque Macrophages Are Proinflammatory in Atherosclerotic Murine Models. Circ Res. 2018;123(10):1127-1142
102. Cochain C, Vafadarnejad E, Arampatzi P. et al. Single-Cell RNA-Seq Reveals the Transcriptional Landscape and Heterogeneity of Aortic Macrophages in Murine Atherosclerosis. Circ Res. 2018;122(12):1661-1674
103. Williams JW, Zaitsev K, Kim K-W. et al. Limited proliferation capacity of aortic intima resident macrophages requires monocyte recruitment for atherosclerotic plaque progression. Nat Immunol. 2020;21(10):1194-1204
104. Paulson KE, Zhu S-N, Chen M. et al. Resident intimal dendritic cells accumulate lipid and contribute to the initiation of atherosclerosis. Circ Res. 2010;106(2):383-390
105. Zernecke A, Erhard F, Weinberger T. et al. Integrated single-cell analysis-based classification of vascular mononuclear phagocytes in mouse and human atherosclerosis. Cardiovasc Res. 2023;119(8):1676-1689
106. van Kuijk K, Demandt JAF, Perales-Patón J. et al. Deficiency of myeloid PHD proteins aggravates atherogenesis via macrophage apoptosis and paracrine fibrotic signalling. Cardiovasc Res. 2022;118(5):1232-1246
107. Bashore AC, Yan H, Xue C. et al. High-Dimensional Single-Cell Multimodal Landscape of Human Carotid Atherosclerosis. Arterioscler Thromb Vasc Biol. 2024;44(4):930-945
108. Cai J, Deng J, Gu W. et al. Impact of Local Alloimmunity and Recipient Cells in Transplant Arteriosclerosis. Circ Res. 2020;127(8):974-993
109. Zernecke A, Winkels H, Cochain C. et al. Meta-Analysis of Leukocyte Diversity in Atherosclerotic Mouse Aortas. Circ Res. 2020;127(3):402-426
110. Winkels H, Ehinger E, Vassallo M. et al. Atlas of the Immune Cell Repertoire in Mouse Atherosclerosis Defined by Single-Cell RNA-Sequencing and Mass Cytometry. Circ Res. 2018;122(12):1675-1688
111. Depuydt MAC, Prange KHM, Slenders L. et al. Microanatomy of the Human Atherosclerotic Plaque by Single-Cell Transcriptomics. Circ Res. 2020;127(11):1437-1455
112. Fernandez DM. Single-cell immune landscape of human atherosclerotic plaques. Nat Med. 2019;25:30
113. Bozkurt B. et al. HF STATS 2024: Heart Failure Epidemiology and Outcomes Statistics. J Card Fail. 2025;31(1):66-116
114. Feng J, Zhang Y, Zhang J. Epidemiology and Burden of Heart Failure in Asia. JACC Asia. 2024;4(4):249-264
115. Bacmeister L, Schwarzl M, Warnke S. et al. Inflammation and fibrosis in murine models of heart failure. Basic Res Cardiol. 2019;114(3):19
116. Duncan SE, Gao S, Sarhene M. et al. Macrophage Activities in Myocardial Infarction and Heart Failure. Cardiol Res Pract. 2020;2020:4375127
117. Tanai E, Frantz S. Pathophysiology of Heart Failure. Compr Physiol. 2015;6(1):187-214
118. Martini E, Kunderfranco P, Peano C. et al. Single-Cell Sequencing of Mouse Heart Immune Infiltrate in Pressure Overload-Driven Heart Failure Reveals Extent of Immune Activation. Circulation. 2019;140(25):2089-2107
119. Zhang D, Wang X, Zhu L. et al. TIMD4hiMHCⅡhi Macrophages Preserve Heart Function Through Retnla. JACC Basic Transl Sci. 2024;10(1):65-84
120. Revelo XS, Parthiban P, Chen C. et al. Cardiac Resident Macrophages Prevent Fibrosis and Stimulate Angiogenesis. Circ Res. 2021;129(12):1086-1101
121. Ni S-H, Xu J-D, Sun S-N. et al. Single-cell transcriptomic analyses of cardiac immune cells reveal that Rel-driven CD72-positive macrophages induce cardiomyocyte injury. Cardiovasc Res. 2022;118(5):1303-1320
122. Rao M, Wang X, Guo G. et al. Resolving the intertwining of inflammation and fibrosis in human heart failure at single-cell level. Basic Res Cardiol. 2021;116(1):55
123. Amrute JM, Lai L, Ma P. et al. Defining cardiac functional recovery in end-stage heart failure at single-cell resolution. Nat Cardiovasc Res. 2023;2(4):399-416
124. Zhu H, Galdos FX, Lee D. et al. Identification of Pathogenic Immune Cell Subsets Associated With Checkpoint Inhibitor-Induced Myocarditis. Circulation. 2022;146(4):316-335
125. Clemente-Casares X, Hosseinzadeh S, Barbu I. et al. A CD103+ Conventional Dendritic Cell Surveillance System Prevents Development of Overt Heart Failure during Subclinical Viral Myocarditis. Immunity. 2017;47(5):974-989.e8
126. Leuschner F, Courties G, Dutta P. et al. Silencing of CCR2 in myocarditis. Eur Heart J. 2015;36(23):1478-1488
127. Mizushina Y, Shirasuna K, Usui F. et al. NLRP3 protein deficiency exacerbates hyperoxia-induced lethality through Stat3 protein signaling independent of interleukin-1β. J Biol Chem. 2015;290(8):5065-5077
128. Hua X, Hu G, Hu Q. et al. Single-Cell RNA Sequencing to Dissect the Immunological Network of Autoimmune Myocarditis. Circulation. 2020;142(4):384-400
129. Hu Z, Hua X, Mo X. et al. Inhibition of NETosis via PAD4 alleviated inflammation in giant cell myocarditis. iScience. 2023;26(7):107162
130. Axelrod ML, Meijers WC, Screever EM. et al. T cells specific for α-myosin drive immunotherapy-related myocarditis. Nature. 2022;611(7937):818-826
131. Ma P, Liu J, Qin J. et al. Expansion of Pathogenic Cardiac Macrophages in Immune Checkpoint Inhibitor Myocarditis. Circulation. 2024;149(1):48-66
132. Liu J, Ma P, Lai L. et al. Transcriptional and Immune Landscape of Cardiac Sarcoidosis. Circ Res. 2022;131(8):654-669
133. Jia G, Hill MA, Sowers JR. Diabetic Cardiomyopathy: An Update of Mechanisms Contributing to This Clinical Entity. Circ Res. 2018;122(4):624-638
134. Chiang C-E, Ueng K-C, Chao T-H. et al. 2021 Consensus Pathway of the Taiwan Society of Cardiology on Novel Therapy for Type 2 Diabetes. JACC Asia. 2021;1(2):129-146
135. Jia G, DeMarco VG, Sowers JR. Insulin resistance and hyperinsulinaemia in diabetic cardiomyopathy. Nat Rev Endocrinol. 2016;12(3):144-153
136. Xie S, Xu S-C, Deng W. et al. Metabolic landscape in cardiac aging: insights into molecular biology and therapeutic implications. Signal Transduct Target Ther. 2023;8(1):114
137. Yang N, Wang M, Lin K. et al. Dectin-1 deficiency alleviates diabetic cardiomyopathy by attenuating macrophage-mediated inflammatory response. Biochim Biophys Acta Mol Basis Dis. 2023;1869(6):166710
138. Suresh Babu S, Thandavarayan RA, Joladarashi D. et al. MicroRNA-126 overexpression rescues diabetes-induced impairment in efferocytosis of apoptotic cardiomyocytes. Sci Rep. 2016;6:36207
139. Tan Y, Zhang Z, Zheng C. et al. Mechanisms of diabetic cardiomyopathy and potential therapeutic strategies: preclinical and clinical evidence. Nat Rev Cardiol. 2020;17(9):585-607
140. Su Q, Huang W, Huang Y. et al. Single-cell insights: pioneering an integrated atlas of chromatin accessibility and transcriptomic landscapes in diabetic cardiomyopathy. Cardiovasc Diabetol. 2024;23(1):139
141. Li W, Lou X, Zha Y. et al. Single-cell RNA-seq of heart reveals intercellular communication drivers of myocardial fibrosis in diabetic cardiomyopathy. eLife. 2023;12:e80479
142. Sakthivel P, Bruder D. Mechanism of granuloma formation in sarcoidosis. Curr Opin Hematol. 2017;24(1):59-65
143. Sansonetti M, Waleczek FJG, Jung M. et al. Resident cardiac macrophages: crucial modulators of cardiac (patho)physiology. Basic Res Cardiol. 2020;115(6):77
144. Hulsmans M, Clauss S, Xiao L. et al. Macrophages Facilitate Electrical Conduction in the Heart. Cell. 2017;169(3):510-522.e20
145. van Amerongen MJ, Harmsen MC, van Rooijen N. et al. Macrophage depletion impairs wound healing and increases left ventricular remodeling after myocardial injury in mice. Am J Pathol. 2007;170(3):818-829
146. Frantz S, Hofmann U, Fraccarollo D. et al. Monocytes/macrophages prevent healing defects and left ventricular thrombus formation after myocardial infarction. FASEB J Off Publ Fed Am Soc Exp Biol. 2013;27(3):871-881
147. Gombozhapova A, Rogovskaya Y, Shurupov V. et al. Macrophage activation and polarization in post-infarction cardiac remodeling. J Biomed Sci. 2017;24(1):13
148. Lee Y-W, Mout R, Luther DC. et al. In Vivo Editing of Macrophages through Systemic Delivery of CRISPR-Cas9-Ribonucleoprotein-Nanoparticle Nanoassemblies. Adv Ther. 2019;2(10):1900041
149. Chen W, Schilperoort M, Cao Y. et al. Macrophage-targeted nanomedicine for the diagnosis and treatment of atherosclerosis. Nat Rev Cardiol. 2022;19(4):228-249
150. Wu J-Y, Huang T-W, Hsieh Y-T. et al. Cancer-Derived Succinate Promotes Macrophage Polarization and Cancer Metastasis via Succinate Receptor. Mol Cell. 2020;77(2):213-227.e5
151. Yang P, Qin H, Li Y. et al. CD36-mediated metabolic crosstalk between tumor cells and macrophages affects liver metastasis. Nat Commun. 2022;13(1):5782
152. Kopecky BJ, Lavine KJ. Cardiac macrophage metabolism in health and disease. Trends Endocrinol Metab TEM. 2024;35(3):249-262
153. Pinto AR, Ilinykh A, Ivey MJ. et al. Revisiting Cardiac Cellular Composition. Circ Res. 2016;118(3):400-409
154. Cui L, Lu H, Lee YH. Challenges and emergent solutions for LC-MS/MS based untargeted metabolomics in diseases. Mass Spectrom Rev. 2018;37(6):772-792
155. Schilperoort M. et al. The role of efferocytosis-fueled macrophage metabolism in the resolution of inflammation. Immunol Rev. 2023;319(1):65-80
156. Reel PS, Reel S, Pearson E. et al. Using machine learning approaches for multi-omics data analysis: A review. Biotechnol Adv. 2021;49:107739
157. Angel PM, Baldwin HS, Gottlieb Sen D. et al. Advances in MALDI imaging mass spectrometry of proteins in cardiac tissue, including the heart valve. Biochim Biophys Acta Proteins Proteomics. 2017;1865(7):927-935
158. Binan L, Danquah S, Valakh V. et al. Simultaneous CRISPR screening and spatial transcriptomics reveals intracellular, intercellular, and functional transcriptional circuits. BioRxiv Prepr Serv Biol. 2023. 2023 11.30.569494
159. Wang T, Chen X, Han Y. et al. scProAtlas: an atlas of multiplexed single-cell spatial proteomics imaging in human tissues. Nucleic Acids Res. 2025;53(D1):D582-D594
160. Rosenberger FA, Thielert M, Strauss MT. et al. Spatial single-cell mass spectrometry defines zonation of the hepatocyte proteome. Nat Methods. 2023;20(10):1530-1536
161. Nunes JB, Ijsselsteijn ME, Abdelaal T. et al. Integration of mass cytometry and mass spectrometry imaging for spatially resolved single-cell metabolic profiling. Nat Methods. 2024;21(10):1796-1800
162. Picard M, Scott-Boyer M-P, Bodein A. et al. Integration strategies of multi-omics data for machine learning analysis. Comput Struct Biotechnol J. 2021;19:3735-3746
163. Friedrich S, Groß S, König IR. et al. Applications of artificial intelligence/machine learning approaches in cardiovascular medicine: a systematic review with recommendations. Eur Heart J Digit Health. 2021;2(3):424-436
164. Sopic M, Vilne B, Gerdts E. et al. Multiomics tools for improved atherosclerotic cardiovascular disease management. Trends Mol Med. 2023;29(12):983-995
Author contact
![]() Corresponding authors: Jian'an Wang, M.D. Department of Cardiology, Second Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou 310009, China. E-mail: wangjianan111edu.cn; Wei Zhu Ph.D. Department of Cardiology, Second Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou 310009, China. E-mail: weizhu65edu.cn
Corresponding authors: Jian'an Wang, M.D. Department of Cardiology, Second Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou 310009, China. E-mail: wangjianan111edu.cn; Wei Zhu Ph.D. Department of Cardiology, Second Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou 310009, China. E-mail: weizhu65edu.cn

 Global reach, higher impact
Global reach, higher impact