10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2025; 21(8):3705-3725. doi:10.7150/ijbs.105550 This issue Cite
Research Paper
ITLN1 exacerbates Crohn's colitis by driving ZBP1-dependent PANoptosis in intestinal epithelial cells through antagonizing TRIM8-mediated CAPN2 ubiquitination
1. Department of Gastrointestinal Surgery, The Affiliated Changzhou No. 2 People's Hospital of Nanjing Medical University, The Third Affiliated Hospital of Nanjing Medical University, Changzhou Medical Center, Nanjing Medical University, China.
2. Department of Gastroenterology, The Affiliated Taizhou People's Hospital of Nanjing Medical University, Taizhou School of Clinical Medicine, Nanjing Medical University, China.
3. Department of Orthopedics, Changshu Hospital Affiliated to Nanjing University of Chinese Medicine, China.
4. Department of General Surgery, The First Affiliated Hospital of Nanjing Medical University, China.
5. Department of General Surgery, Affiliated Taizhou People's Hospital of Nanjing Medical University, Taizhou School of Clinical Medicine, Nanjing Medical University, China.
# These authors contributed equally to this article.
Received 2024-10-21; Accepted 2025-4-12; Published 2025-5-31
Abstract
Background: This study aimed to investigate the mechanisms by which PANoptosis of intestinal epithelial cells (IECs) promotes Crohn's disease (CD) progression.
Methods: Single-cell RNA sequencing (scRNA-seq) was performed on inflamed and uninflamed colon tissues from patients with CD. The biological functions of intelectin-1 (ITLN1) in inflammation and PANoptosis were verified through in vitro experiments. The molecular mechanisms underlying its biological functions were examined using co-immunoprecipitation (Co-IP) combined with mass spectrometry (MS) and RNA-seq and further validated with rescue experiments. Additionally, the in vivo function of ITLN1 regulation on inflammation, PANoptosis, and the intestinal mucosal barrier was explored in interleukin-10 knockout (IL-10 KO) colitis model mice.
Results: ITLN1 was significantly overexpressed in IECs from inflamed colon tissues and specifically associated with CD-related inflammatory markers. RNA-seq and in vitro experiments indicated that ITLN1 promotes inflammation, PANoptosis, and impaired tight junctions. Co-IP and MS analyses revealed that ITLN1 can bind to the PANoptosis-promoting protein calpain-2 (CAPN2) and enhance its stability. The E3 ubiquitin ligase, a tripartite motif containing 8 (TRIM8), directly interacts with CAPN2 and mediates its ubiquitination degradation. ITLN1 can bind to TRIM8, and its impact on inflammation and Z-DNA binding protein 1 (ZBP1)-induced PANoptosis can be antagonized by CAPN2. These in vivo studies indicated that short hairpin-ITLN1 improves colonic inflammation and intestinal barrier function in IL-10 KO mice.
Conclusion: We identified the ITLN1-TRIM8-CAPN2 axis that drives IEC PANoptosis in CD progression. Pharmacological inhibition of ITLN1 significantly mitigated epithelial damage and colitis both in vivo and in vitro, establishing ITLN1-targeted therapies and PANoptosis modulation as viable clinical strategies for CD treatment.
Keywords: Crohn's disease, ITLN1/TRIM8/CAPN2 axis, PANoptosis, ubiquitination
Introduction
Crohn's disease (CD) is a persistent inflammatory condition characterized by autoimmune dysregulation. As one of the principal forms of inflammatory bowel disease (IBD), this disorder is characterized by pan-enteric tropism, potentially involving any segment from the oral cavity to the anal canal [1]. CD poses a significant global public health challenge, with a notable incidence worldwide [1]. Incidence rates are exceptionally high in Western countries compared with Asian regions [2]. However, recent data indicate an increasing trend in the incidence of CD in China, especially in major cities [3]. The complex pathogenesis of CD involves genetics, immunity, and environmental factors and remains unclear [4]. The CD is currently clinically incurable and prone to recurrence, primarily relying on nutritional support, medication, and surgical interventions to alleviate symptoms and control inflammation [5,6]. This underscores the imperative for mechanistic investigations to uncover novel molecular targets for improved clinical interventions.
Emerging evidence implicates intestinal epithelial cells (IECs) dyshomeostasis as a pivotal contributor to CD pathophysiology [7]. Pathogenic cascades involve multidimensional disruptions: (1) malabsorption of essential ions and metabolites; (2) compromised mucosal integrity through junctional complex degradation; (3) ecological shifts in commensal microbiota; (4) systemic immune network destabilization [8,9]. A reduction in the number of IECs and the destruction of tight junctions are frequently observed in CD, resulting in impaired intestinal epithelium mechanical barrier function [10]. This impairment permits many bacteria and other pathogenic factors to penetrate the epithelial layer and directly interact with the intestinal immune system, leading to a persistently and excessively activated immune response and promoting CD progression [11].
PANoptosis, a novel inflammatory cell death paradigm regulated by the PANoptosome multiprotein complex, synthesizes molecular hallmarks of pyroptosis, apoptosis, and necroptosis, transcending singular classification within conventional death modalities [12,13]. This coordinated demise mechanism functions as a molecular amplifier of inflammation, orchestrating the secretion of cytokine storm initiators that propagate pathological immune activation—a critical driver of autoimmune pathogenesis [14]. Current investigations have delineated its mechanistic involvement in immune dysregulation disorders, with robust evidence in rheumatoid arthritis (RA) [15] and systemic lupus erythematosus [16]. However, to date, no reports have directly linked PANoptosis with CD.
Single-cell sequencing demonstrated that ITLN1 overexpression in colonic epithelium is associated with inflammatory severity. Our functional studies demonstrated that ITLN1 facilitates PANoptosis by stabilizing CAPN2 through competition with TRIM8, thereby establishing a mechanistic connection between PANoptosis and CD progression. This axis represents a new therapeutic target, as evidenced by ITLN1 inhibition, which mitigates epithelial damage in both in vivo and in vitro models. The ITLN1-CAPN2 interaction interface offers a framework for creating biologics to inhibit pathogenic PANoptosis, addressing a critical requirement in CD precision therapeutics.
Materials and methods
Patient samples
Colonic tissue samples (both inflamed and non-inflamed regions) were obtained from 14 CD patients undergoing colectomy or ileocolectomy. All procedures strictly complied with the ethical guidelines outlined in the Helsinki Declaration. Immediately after resection, lamina propria-containing tissue fragments (approximately soybean-sized) were flash-frozen in liquid nitrogen and stored in an ultra-low temperature freezer at -80°C for subsequent RNA extraction. This prospective, single-center observational study was conducted at the First Affiliated Hospital of Nanjing Medical University (2023-2024) with approval from the Institutional Ethics Committee (Approval No.: 2023-SR-115). The study participants' inclusion and exclusion criteria are outlined in Table 1. All participants provided informed consent. Table 2 presents the demographic data of the enrolled patients, including age, gender, disease, and location. Before surgery, all enrolled patients with CD were assessed for inflammatory markers (for instance, C-reactive protein) based on previously reported methods [17,18].
Single-cell RNA (scRNA) sequencing
ScRNA-seq and bioinformatic analysis were conducted by Genechem Co., Ltd. Sequencing libraries were generated using the BD Rhapsody WTA Pipeline (version 1.8). Raw FASTQ files were aligned to reference genomes (human GRCh38/mouse mm10) with transcriptome annotations (GENCODE version 32/Ensembl 98 for human; GENCODE version M23/Ensembl 98 for mouse). A UMI count matrix was processed through the Scanpy Python package (version 1.8). Quality control steps included the following: (1) Filtering cells with low UMI/gene counts (below set thresholds); (2) removing cells exhibiting > 10% mitochondrial gene expression. Post-QC, data underwent library size normalization (normalize_total function) and log transformation. Highly variable genes were identified using the methods described by Macosko et al. [19]. Dimensionality reduction via PCA was followed by graph-based clustering and 2D/3D Uniform Manifold Approximation and Projection (UMAP) visualization. Cluster-specific marker genes were determined using Seurat's Wilcoxon test (adjusted P < 0.05, |log2FC| > 2) [20]. Functional enrichment analysis (GO/KEGG pathways) was performed with g:Profiler2 [21] using hypergeometric testing.
Inclusion and exclusion criteria
| Inclusion criteria |
|---|
| aged 18-65 years |
| diagnosed with CD based on pathological evidence |
| underwent elective surgical resection for colonic (or ileocolonic) CD |
| with signed informed consent |
| Exclusion criteria |
| with diffuse small bowel lesions |
| combined with malignancies |
| combined with other autoimmune or autoinflammatory diseases |
| declined to participate |
CD, Crohn's disease.
Patient demographic data
| Single-cell sequencing (Discovery set) | Validation set | |
|---|---|---|
| Number | 4 | 10 |
| Age (year) | 42.0±2.2 | 42.3±5.3 |
| Gender, n (%) | - | - |
| Male | 3 (75.0) | 7 (70.0) |
| Female | 1 (25.0) | 3 (30.0) |
| Body mass index (kg/m2) | 20.9±0.9 | 21.2±1.2 |
| Disease location, n (%) | - | - |
| Colonic | 2 (50.0) | 3 (30.0) |
| Ileocolic | 2 (50.0) | 7 (70.0) |
| Duration of disease (month) | 21.3±6.5 | 21.8±6.3 |
Data are expressed as means ± S.E.M or number with percentage.
Co-immunoprecipitation (Co-IP) and protein mass spectrometry (MS)
Cells were lysed with IP lysis buffer, and total proteins were isolated via centrifugation. After verifying protein quality using Western blotting, Co-IP experiments proceeded with SureBeads magnetic beads. Resuspended beads (20 μL) were washed with phosphate buffer saline (PBS) with Tween 20, incubated with 5 μL IP antibody on a rotary shaker (1 h, RT), and subsequently mixed with lysate-containing target proteins (2 h, RT). Post-washing, protein complexes were denatured in a loading buffer (40 μL, 95 °C, and 10 min) and collected for Western blotting or MS.
Silver staining was performed using the Fast Silver Stain Kit (Beyotime). Samples were analyzed via an UltiMate 3000 RSLCnano system coupled to a Q Exactive HF MS (Thermo). Raw MS data were processed with MaxQuant (version 1.6.6.0) [22], aligned to the UniProt human database (2023-06-19 release, 20,423 entries) [23]. Significant hits required a fold change > 2 and ≥ 2 unique peptides.
Animal experiments
C57BL/6 IL-10 knockout (KO) mice (GemPharmatech Co. Ltd.) were housed under specific pathogen-free conditions and used as colitis models. Twelve-week-old male wild-type (WT) and IL-10 knockout (KO) mice were divided into four groups (n = 5 per group): (i) WT group; (ii) IL-10 KO mice treated with normal saline; (iii) IL-10 KO mice treated with a short hairpin (sh)-ITLN1 (administered 200 μL of 1e11 viral genomes adeno-associated virus (AAV) vector carrying sh-ITLN1 via enema once); (iv) IL-10 KO mice were treated with sh-ITLN1 combined with oe-CAPN2 (AAV carried sh-ITLN1 and oe-CAPN2 via enema once). Mice were euthanized via cervical dislocation one-week post-intervention, and proximal colon tissues were collected for analysis. All procedures were approved by the Ethics Committee of Nantong University (No. S20200323-289).
Disease activity index (DAI)
Based on standardized criteria, a score of 1 was allocated for each observed clinical manifestation: unkempt fur, fecal occult blood, minor rectal prolapse (< 1 mm), or loose stool consistency [24]. Severe diarrhea or rectal prolapse exceeding 1 mm warranted an additional point. The final DAI was calculated as the cumulative sum of these scores.
Histological analysis
Following euthanasia, colon length was recorded, and proximal colon segments were collected. Tissues were washed with PBS, preserved in 10% formalin, and paraffin-embedded. Sections (6 μm) were prepared for hematoxylin-eosin (HE) or Alcian Blue-Periodic Acid-Schiff (AB-PAS) staining. Two blinded pathologists independently assessed colitis severity and inflammation using established grading guidelines [25].
Determination of levels of pro-inflammatory cytokines
IL-17, tumor necrosis factor-alpha (TNF-α), and interferon-gamma (IFN-γ) levels in proximal colon homogenates were analyzed via ELISA (R&D Systems). Briefly, tissues were rinsed, homogenized in PBS, and centrifuged (5,000 ×g, 5 min). Supernatants (50 μL/well) were loaded onto 96-well plates with Assay Diluent RDI-63. After incubation and washes, absorbance at 450 nm was quantified using a BioTek EL800 reader.
Epithelial apoptosis analysis
Epithelial apoptosis was evaluated via TUNEL assay (In Situ Cell Death Detection Kit, Roche) following standard protocols [26]. Colon sections underwent permeabilization with 1% Triton X-100 and 0.1% sodium citrate, followed by incubation with TUNEL reagent in darkness. Nuclei were counterstained with DAPI (Servicebio), and slides were mounted in 50% glycerol. TUNEL-positive cells were quantified per field using confocal microscopy (Olympus).
Epithelial cells isolation
Colonic epithelial cells from human and murine tissues were isolated via chelation-based methods [27]. Murine colon segments (0.5 cm) or surgically resected human CD specimens (inflamed/uninflamed) were rinsed in cold PBS to clear debris. Tissues were treated with dithiothreitol (2 mmol/L) and EDTA (1 mmol/L) in PBS under gentle agitation (37 °C, 20 min, two cycles). Cell suspensions were purified via percoll-RPMI density gradient centrifugation (200 × g, 5 min), and pelleted epithelial cells were retained for downstream applications.
Immunofluorescence staining of colon tissues and cells
Immunofluorescence staining of colon tissues or cells was conducted to investigate protein localization and expression levels, as described by a previous study [28]. Cell coverslips or frozen sections were rinsed, followed by antigen retrieval using EDTA buffer in a microwave. After washing, the sections were dried, circled with a hydrophobic barrier pen, and soaked in 3% hydrogen peroxide to remove endogenous peroxidase. After washing again in PBS, BSA was added to the block for 30 min. The primary antibody was applied and incubated overnight at 4 °C. The sections were subsequently treated with an HRP-conjugated secondary antibody at room temperature for 50 min, washed, and CY3-TSA was added and incubated in the dark for 10 min. This process was repeated for subsequent primary and secondary antibodies with FITC-TSA, CY5-TSA, and 594-TSA. Nuclei were counterstained with DAPI, and the slides were mounted and photographed. The primary antibodies used included anti-ZO-1 (Abcam, #ab221547), anti-occludin (Abcam, #ab216327), anti-ZBP1 (Thermo Fisher, #PA5-20455), ITLN1 (Proteintech, #11770-1-AP), anti-caspase1 (Servicebio, #GB11383), anti-RIPK3 (Servicebio, #GB115458), anti-ASC (Servicebio, #GB113966, GB115270), and anti-TRIM8 (Proteintech, #27463-1-AP).
Western blotting
Protein lysates from tissues or cells were prepared using RIPA buffer (Beyotime) supplemented with phosphatase/protease inhibitors. Protein concentration was quantified via BCA assay (Pierce Biotechnology). As described by a previous study [29], equal amounts (15 µg) were resolved on 10% SDS-PAGE gels and transferred to PVDF membranes (Millipore). After blocking with 5% non-fat milk, membranes were probed with primary antibodies at 4 °C overnight, followed by secondary antibody incubation (2 h). Signals were developed using chemiluminescent reagents (Cell Signaling Technology) and analyzed with ImageJ. Loading controls included GAPDH, vinculin, or β-actin. Primary antibodies were as follows: anti-ZO-1 (Abcam, #ab221547), anti-occludin (Abcam, #ab216327), anti-CAPN2 (Abcam, #ab236650), anti-caspase1 (AdipoGen, #AG-20B-0042), anti-caspase3 (CST, #9662), anti-caspase-7 (CST, #9492), anti-pMLKL (CST, #37333), anti-MLKL (Servicebio, #GB115699), anti-RIPK3 (Servicebio, #GB115458), anti-pRIPK3 (CST, #91702), anti-GSDMD-N (Abcam, #ab215203), anti-ASC (Servicebio, #GB113966, GB115270), anti-TRIM8 (Proteintech, #27463-1-AP), anti-Ub-WT (Servicebio, #GB115700), anti-Ub-K48 (Abcam, #ab140601), and anti-Ub-K63 (Abcam, #ab179434).
RNA extraction and qRT-PCR
Total RNAs were extracted from colon tissues/cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. The extracted RNA was reverse transcribed into cDNA using the First-Strand cDNA Synthesis Kit (Invitrogen). Furthermore, qPCR was performed using SYBR Green (Applied Biosystems) on an ABI 7500 system. Relative expression levels were normalized via the 2-△△Ct method, with triplicate technical replicates. Primer sequences are detailed in Table 3.
Primers sequences for qRT-PCR
| Name | Sequence (5′-3′) |
|---|---|
| Human IGHG3 | |
| Forward | CTCTACTCCCTCAGCAGCGTG |
| Reverse | CTGGGCTTGTGATTCACGTT |
| Human ITLN1 | |
| Forward | AGTGTTGGACTGACAACGGC |
| Reverse | TACATCCGGTGACCCTCATTC |
| Human HMGCS2 | |
| Forward | GCCTCTCAGGACATGTTCGAC |
| Reverse | AGCCATAAGAGAAGGCACCA |
| Human CAPN2 | |
| Forward | CTCCACCAAGTCATCGTTGCT |
| Reverse | TTCCAGTATTCTCGGGATCCAG |
| Human TRIM8 | |
| Forward | CGGAGAATTGGAAGAACTGCT |
| Reverse | TGTAGGCCTGGTTGCACTCTG |
| Human GAPDH | |
| Forward | TCCTGGGCTACACTGAGCAC |
| Reverse | CTGTTGCTGTAGCCAAATTCGTTG |
| Mouse ITLN1 | |
| Forward | ACTCACAATGGGTACAGCAGTAG |
| Reverse | CATGCCTTGGAGCCCACAATG |
| Mouse TRIM8 | |
| Forward | TGCGGAAGATGCTAGAAGGTC |
| Reverse | GTCTCCAGGAAGCTAGCCTCA |
| Mouse CAPN2 | |
| Forward | GACATGCACACCATTGGCTT |
| Reverse | CGCGGAGGTTAATGAAGGTAT |
| Mouse GAPDH | |
| Forward | GTCAAGGCCGAGAATGGGAA |
| Reverse | CTCGTGGTTCACACCCATCA |
qRT-PCR, quantitative real time-polymerase chain reaction; ITLN1, intelectin-1; CAPN2, calpain-2; TRIM8, tripartite motif containing 8; IGHG3, immunoglobulin heavy constant gamma 3; HMGCS2, 3-hydroxy-3-methylglutaryl-CoA synthase 2.
Transmission electron microscopy (TEM) of tight junctions (TJs)
TJs were analyzed via TEM, as described previously [30]. Tissue samples were fixed in 4% glutaraldehyde, post-fixed in 1% osmium tetroxide, dehydrated, and embedded in Epon 812 resin. Ultrathin sections were stained with uranyl acetate and lead citrate and imaged using a Hitachi H-600 TEM.
Ussing chamber studies
Proximal colon segments were mounted in Lucite chambers with Ringer's buffer (37 °C) to evaluate permeability [31]. Basal mannitol flux was measured by adding 1 mM mannitol to the mucosal compartment. Transepithelial electrical resistance (TEER) and short-circuit current (Isc) were recorded via an automated voltage clamp, with TEER calculated using Ohm's law [32].
Intestinal permeability assay
In vivo intestinal permeability was assessed using FITC-dextran (60 mg/100 g body weight; Sigma-Aldrich) administered via oral gavage [33]. The serum was collected 4 h post-administration and was analyzed for FITC-dextran levels via fluorescence quantification (BMG Labtech).
Flow cytometry (FCM) of apoptosis
Epithelial cell apoptosis was assessed using propidium iodide (PI) and Annexin V-FITC staining, following a previously described method [34]. Cells were stained with 10 µL PI and 5 µL Annexin V-FITC and analyzed with an EpicsAltra flow cytometer (Beckman Coulter). Early apoptotic cells (Annexin V+/PI-) and late apoptotic cells (Annexin V+/PI+) were quantified with triplicate technical replicates.
Cell transfection
NCM460 cells were transfected with TRIM8 siRNA (Lipofectamine™2000, Thermo Fisher), ITLN1 shRNA (pLKO.1 vector, Sangon Biotech), or TRIM8/CAPN2 overexpression plasmids (pcDNA3.1), following standard protocols. siRNA/shRNA sequences are listed in Table 4.
Detection of ubiquitination levels
To detect the ubiquitination levels of CAPN2, HEK 293 cells were transfected with CAPN2 to promote its overexpression. After 24 h, CAPN2 protein was purified and separated using SDS-PAGE. An anti-ubiquitin antibody was subsequently used to identify which lysine residues in CAPN2 were more prone to ubiquitination. In an in vitro reaction, the target protein CAPN2 was mixed with HA-ubiquitin and TRIM8 (a protein hypothesized to induce CAPN2 ubiquitination) in a reaction buffer and incubated. The resulting mixture was analyzed using immunoprecipitation and Western blotting techniques to further study CAPN2 ubiquitination.
Cycloheximide (CHX) chase assay
HEK293 cells transfected with MG132, si-TRIM8, oe-TRIM8, or controls were treated with CHX (100 μg/mL) 24 h post-transfection [35]. CAPN2 protein degradation was monitored via Western blotting at designated timepoints.
Cell live/dead (Calcein/PI) staining
Live/dead (Calcein/PI) staining was used to assess the proportion of dead cells among IECs, following the manufacturer's instructions (Proteintech, #PF00007). A mixture of 30 µL of 1.5 mM PI and 5 µL of 4 mM Calcein AM was combined with 10 mL of PBS. Cells (5 × 105 /mL) were incubated with a staining solution (vortexed in PBS) for 15 min in the dark and visualized under a fluorescence microscope.
Primers for sh- and si-RNA sequences
| Name | Sequence (5′-3′) |
|---|---|
| Human sh-ITLN1-1 | Target: GCTAATACTTACTTCAAGGAA |
| Forward | CCGGGCTAATACTTACTTCAAGGAACTCGAGTTCCTTGAAGTAAGTATTAGCTTTTTG |
| Reverse | AATTCAAAAAGCTAATACTTACTTCAAGGAACTCGAGTTCCTTGAAGTAAGTATTAGC |
| Human sh-ITLN1-2 | Target: GATATGGAACTCATGTTGGTT |
| Forward | CCGGGATATGGAACTCATGTTGGTTCTCGAGAACCAACATGAGTTCCATATCTTTTTG |
| Reverse | AATTCAAAAAGATATGGAACTCATGTTGGTTCTCGAGAACCAACATGAGTTCCATATC |
| Human sh-ITLN1-3 | Target:CCAGTGAAATATGGAGAAGGA |
| Forward | CCGGCCAGTGAAATATGGAGAAGGACTCGAGTCCTTCTCCATATTTCACTGGTTTTTG |
| Reverse | AATTCAAAAACCAGTGAAATATGGAGAAGGACTCGAGTCCTTCTCCATATTTCACTGG |
| Human si-TRIM8-1 | Target: GCCAGTACTGCTGCTACTACAGC |
| Forward | UGUAGUAGCAGCAGUACUGGC |
| Reverse | CAGUACUGCUGCUACUACAGC |
| Human si-TRIM8-2 | Target: AGGATGTCAGCTTCATGAAGAAC |
| Forward | UCUUCAUGAAGCUGACAUCCU |
| Reverse | GAUGUCAGCUUCAUGAAGAAC |
| Human si-TRIM8-3 | Target: CACCAAGTCTGTGAAAATCCTGA |
| Forward | AGGAUUUUCACAGACUUGGUG |
| Reverse | CCAAGUCUGUGAAAAUCCUGA |
| Mouse sh-ITLN1-1 | Target: CTCGGAAGACAGCCTCTTATT |
| Forward | CCGG CTCGGAAGACAGCCTCTTATT CTCGAG AATAAGAGGCTGTCTTCCGAG TTTTTG |
| Reverse | AATTCAAAAA CTCGGAAGACAGCCTCTTATT CTCGAG AATAAGAGGCTGTCTTCCGAG |
| Mouse sh-ITLN1-2 | Target: CTCGGAAGACAGCCTCTTATT |
| Forward | AATTCAAAAA CTCGGAAGACAGCCTCTTATT CTCGAG AATAAGAGGCTGTCTTCCGAG |
| Reverse | AATTCAAAAA CCAGCATTACCTGTAGTCTAT CTCGAG ATAGACTACAGGTAATGCTGG |
| Mouse sh-ITLN1-3 | Target:CTCGGAAGACAGCCTCTTATT |
| Forward | CCGG CACGAAGAATGGTGTCATCTA CTCGAG TAGATGACACCATTCTTCGTG TTTTTG |
| Reverse | AATTCAAAAA CACGAAGAATGGTGTCATCTA CTCGAG TAGATGACACCATTCTTCGTG |
qRT-PCR, quantitative real time-polymerase chain reaction; ITLN1, intelectin-1; CAPN2, calpain-2.
Statistical analysis
Data were analyzed using SPSS 19.0 or GraphPad Prism 8.0. Group comparisons were conducted using Student's t-test or analysis of variance (ANOVA), while Pearson's correlation was used to assesse ITLN1 expression and inflammatory markers in Crohn's disease. Significance was set at P < 0.05.
Results
IECs were significantly reduced in CD inflammatory tissues
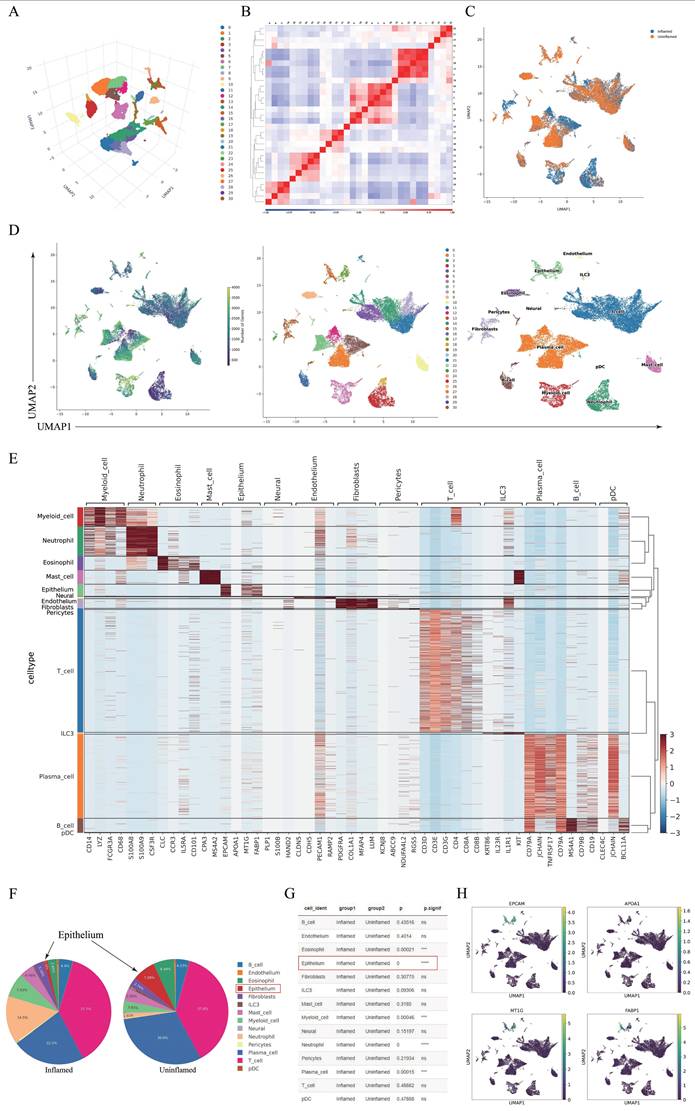
Single-cell RNA sequencing (scRNA-seq) was conducted on colon tissues from patients with CD, including four inflamed and three non-inflamed samples (one excluded due to quality control). Using the UMAP method, we clustered the colon cell populations. The clustering and 3D projection results are illustrated in Figure 1A. Pearson's correlation heatmap (Figure 1B) and cluster distribution plots (Figure 1C) highlighted intergroup heterogeneity. Cell subtypes were annotated using marker gene expression (Figures 1D-E). The comparison of cell subgroup numbers between groups indicated that the reduction in the number of IECs was most significant in the inflamed regions (P = 0.000, Figures 1F-G). Given the pivotal role of IECs in CD pathogenesis and their marked depletion in inflamed tissues, subsequent analyses focused on this population. Marker genes (EPCAM, APOA1, MT1G, and FABP1) exhibited cluster-specific expression patterns (Figure 1H).
The presence of PANoptosis in IECs under inflammatory conditions
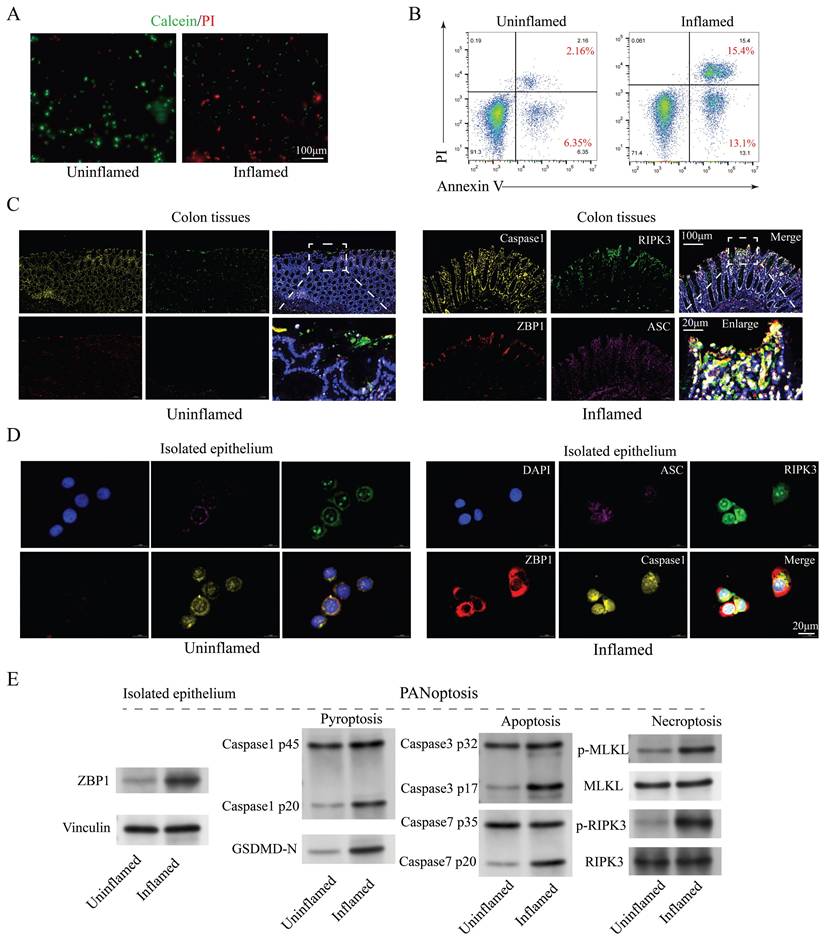
The number of IECs is influenced by various factors, including genetics, lifestyle, inflammation, environment, and medication [36]. Considering that this study compares the differences in the number of IECs between inflamed and non-inflamed regions within the same patient with CD, we can exclude these external factors. Inflammation is probably the primary cause of the significant reduction in IEC numbers. The live/dead cell staining (Figure 2A) and apoptosis flow cytometry (Figure 2B) analysis of the primary isolated IECs from colon tissues revealed markedly elevated cell death in inflamed regions, suggesting apoptosis as a key driver of IEC depletion. To delineate the modes of IEC death, markers of apoptosis (caspase3 and caspase7), pyroptosis (ASC, caspase1, and GSDMD-N), and necroptosis (pMLKL and pRIPK3) were analyzed. Western blotting demonstrated significant upregulation of these markers in inflamed colon tissues (Figure 2C) and isolated epithelial cells (Figures 2D-E), implicating multiple forms of cell death, known as PANoptosis, were present in CD inflammation. Additionally, the subcellular localization of these cell death marker proteins in IEC exhibited significant overlap (Figure 2D), indicating the possible formation of protein complexes termed PANoptosomes. These findings suggest that PANoptosis is present in IECs under CD inflammation and may play a role in CD progression.
ITLN1 in IECs is closely linked to CD inflammation
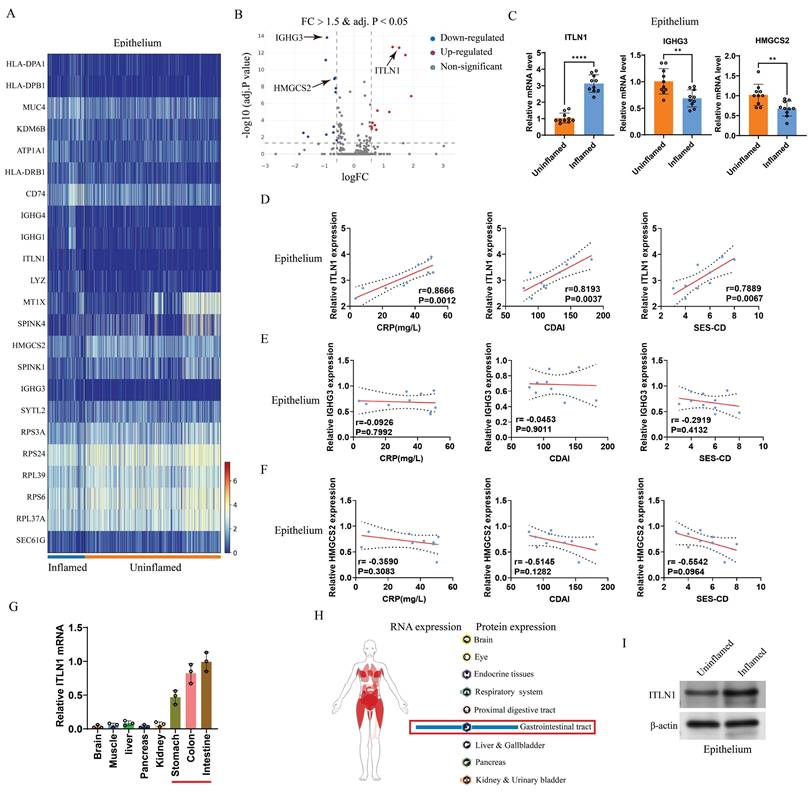
scRNA-seq analysis identified differentially expressed genes (DEGs) between inflamed and non-inflamed IECs (Figures 3A-B). The top five upregulated and downregulated genes were selected for validation with an expanded sample size (10 inflamed versus 10 uninflamed). Three differentially expressed genes (ITLN1, IGHG3, and HMGCS2) were identified (Figure 3C). The correlation between these three genes and CD-specific inflammatory markers identified ITLN1 as a core molecule (Figures 3D-F). ITLN1 is a protein that contains a carbohydrate-binding domain and can bind to specific carbohydrate structures, and it can be secreted by IECs [37]. We examined ITLN1 expression in various tissues (brain, liver, kidney, and gastrointestinal tract). We found that ITLN1 is specifically expressed in the gastrointestinal system, with very low expression in other systems (Figure 3G), consistent with the Atlas database (Figure 3H). Protein expression analysis further confirmed pronounced ITLN1 upregulation in inflamed IECs (Figure 3I). These findings suggest that ITLN1 is probably an important regulatory molecule in the progression of CD inflammation.
ITLN1 influences PANoptosis in IECs
In the lipopolysaccharides (LPS) and adenosine triphosphate (ATP) co-stimulated normal colon mucosa 460 (NCM460) in vitro inflammation model, ITLN1 expression was significantly upregulated (Figure 4A), suggesting a close relationship between ITLN1 and inflammation. Three distinct shRNA sequences targeting ITLN1 were designed to investigate ITLN1's functional role. The qRT-PCR and Western blotting (Supplemental Figures 1A-B) identified sh-ITLN1-1 as the most effective construct, selected for downstream analyses. The heatmap and volcano plot of differentially expressed genes from the RNA-seq analysis of sh-ITLN1 and sh-control are depicted in Figures 4B-C, respectively. KEGG pathway enrichment highlighted “cell growth and death” as a top-ranked pathway (Figure 4D), aligning with ITLN1's potential regulatory role in inflammatory cell death. In the LPS/ATP-induced in vitro inflammation model, no single death inhibitor could completely reverse the inflammation-induced reduction in cell proliferation activity (Figure 4E), further supporting the existence of PANoptosis. Results of apoptosis FCM (Figure 4F) live/dead (Calcein/PI) staining (Figure 4G) confirmed that sh-ITLN1 inhibited cell death. Furthermore, the proinflammatory cytokines IL-1β and IL-18, which were elevated by LPS + ATP induction, were significantly reduced under the sh-ITLN1 intervention (Figure 4H). Moreover, sh-ITLN1 significantly suppressed the expression of PANoptosis-related marker proteins (Figure 4I). These results further support that ZBP1-induced PANoptosis is probably involved in the progression of CD inflammation, and sh-ITLN1 can inhibit inflammation and ZBP1-induced PANoptosis.
ITLN1 influences tight junctions in IECs
ITLN1, a galactofuranose-binding secretory lectin, primarily localizes to gastrointestinal goblet cells and omentum [38]. To investigate the biological functions and molecular mechanism of ITLN1, we performed IP and MS (Figures 5A-C). Volcano plot analysis highlighted differentially precipitated proteins (Figure 5D), and KEGG pathway enrichment ranked "tight junction" as a top pathway under "cellular processes" (Figure 5E). Previous studies have revealed that ITLN1 can recognize and clear pathogens by binding to specific carbohydrate structures [39] and contribute to the protection of the colonic mucosa [40]. Accordingly, we further investigated the impacts of ITLN1 on tight junctions in IECs. Under LPS + ATP-induced inflammation, Western blotting (Figure 5F) and immunofluorescence (Figure 5G) revealed significant downregulation of occludin and ZO-1 in NCM460 cells, which was reversed by sh-ITLN1 knockdown.
ITLN1 regulates calpain-2 (CAPN2) ubiquitination through competitive binding with tripartite motif containing 8 (TRIM8)
Immunoprecipitation (IP) and mass spectrometry (MS) analyses identified CAPN2 as a primary interactor of ITLN1 (Figure 6A). CAPN2, a calcium-dependent protease, directly participates in apoptosis and necroptosis signaling pathways, making it directly relevant to PANoptosis [41,42].
Single-cell RNA sequencing analysis of inflamed and uninflamed colon tissues from patients with CD. (A) Three-dimensional projection of cell clustering in the colon was performed using the UMAP method. (B) Heatmap illustrating the correlation between different cell populations, calculated by Pearson's correlation coefficients. (C) Clustering analysis of cells derived from inflamed and uninflamed colon tissues, highlighting distinct cell groupings. (D) UMAP-based cell type annotation, illustrating various cell types in the colon tissue samples. (E) Expression levels of marker genes across each cell cluster. (F-G) Comparative analysis of cell subgroup distributions between inflamed and uninflamed tissues, indicating statistical significance. (H) Expression profiles of specific IEC marker genes across various cell clusters. IECs, intestinal epithelial cells; CD, Crohn's disease; UMAP, Uniform Manifold Approximation and Projection.
Evidence of PANoptosis in CD under inflammatory conditions. (A) Live/dead cell staining (Calcein/PI) of primary isolated IECs from inflamed and uninflamed colon tissues, illustrating viable (green) and dead (red) cells. Scale bar: 100 μm. (B) FCM analysis of apoptosis with Annexin V/PI staining in primary isolated IECs from inflamed and uninflamed colon tissues. (C) Immunofluorescence analysis of PANoptosis-related marker genes (caspase1, RIPK3, ZBP1, and ASC) in inflamed versus uninflamed regions of colon tissues. Scale bars: 100 and 20 μm. (D) Immunofluorescence detection of PANoptosis-related marker genes (caspase1, RIPK3, ZBP1, and ASC) in isolated IECs from inflamed and uninflamed colon tissues. Scale bar: 20 μm. (E) Western blotting analysis for PANoptosis-related marker genes (caspase1, caspase3, caspase7, GSDMD-N, MLKL, and RIPK3) in isolated IECs from inflamed and uninflamed colon tissues. IECs, intestinal epithelial cells; CD, Crohn's disease; FCM, flow cytometry.
ITLN1 in IECs closely correlates with CD inflammation. (A) Heatmap and (B) volcano plot displaying differentially expressed genes in IECs from inflamed and uninflamed colon tissues of patients with CD (threshold: FC > 1.5 and adjusted P value < 0.05). (C) qRT-PCR validation of differentially expressed genes in the isolated epithelium (10 inflamed versus 10 uninflamed), confirming the upregulation of ITLN1, IGHG3, and HMGCS2 in the inflamed epithelium. (D-F) Correlation analysis between ITLN1, IGHG3, and HMGCS2 expression in inflamed epithelium with CD-associated inflammatory markers (CRP, CDAI, and SES-CD), demonstrating significant associations. (G) Relative mRNA expression of ITLN1 in various WT mice tissues, exhibiting its highest expression in the gastrointestinal system. (H) RNA and protein expression of ITLN1 in human tissues, as provided by the Atlas online database. (I) Western blotting analysis of ITLN1 protein expression in IECs from inflamed and uninflamed colon tissues of patients with CD, highlighting the elevated expression in inflamed IECs. CD, Crohn's disease; IECs, intestinal epithelial cells; ITLN1, intelectin-1; FC, fold change; qRT-PCR, quantitative real-time-polymerase chain reaction; CRP, C-reactive protein; CDAI, Crohn's disease activity index; SES-CD, simple endoscopic score for Crohn's disease. Data are presented as mean ± standard deviation (SD). ** P < 0.01, *** P < 0.001, **** P < 0.0001 by paired t test.
While CAPN2 mRNA levels remained unchanged between inflamed and non-inflamed intestinal epithelial cells (IECs), its protein expression was markedly upregulated in inflamed regions (Figure 6B). Furthermore, the sh-ITLN1 intervention did not significantly affect CAPN2 mRNA levels but diminished its protein levels (Figure 6C). This suggests that increased protein stability is the main reason for CAPN2 upregulation under inflammatory conditions, with ITLN1 positively regulating its expression by affecting protein stability. The CHX protein stability assay results indicated that MG132 significantly increased CAPN2 protein stability (Figure 6D), suggesting that CAPN2 is mainly degraded via the ubiquitin-proteasome pathway. We re-examined the ITLN1 protein spectrum and found that TRIM8—an E3 ubiquitin ligase—was prominently listed among the interacting proteins (Figure 6A). Computational molecular docking results suggest potential binding sites between TRIM8 and CAPN2 (Figure 6E). Bidirectional Co-IP experiments confirmed that CAPN2 and TRIM8 can bind to each other (Figure 6F). Endogenous Co-IP (Figure 6G) experiments demonstrated that si-TRIM8 (Supplemental Figure 1C) could inhibit the ubiquitination level of the CAPN2 protein, while oe-TRIM8 could upregulate its ubiquitination level. Exogenous Co-IP experiments also confirmed that TRIM8 can induce dose-dependent ubiquitination modification of the CAPN2 protein (Figure 6H). Immunofluorescence results revealed that TRIM8 and ITLN1 were highly co-localized in the NCM460 cell line (Figure 6I). Besides, computational molecular docking suggested potential binding sites between the two proteins (Figure 6J). Additionally, CHX experiments demonstrated that TRIM8 can promote the degradation of the CAPN2 protein (Figure 6K), while si-TRIM8 can inhibit its degradation (Figure 6L). All these results strongly suggest that ITLN1 regulates CAPN2 ubiquitination by competitively binding with TRIM8.
ITLN1 influences PANoptosis in IECs. (A) Western blotting and (B) q-PCR analysis revealing ITLN1 expressions in NCM460 cells with or without LPS/ATP stimulation. (C-D) The heatmap and volcano plots illustrating differentially expressed genes from RNA-seq analysis comparing sh-ITLN1 versus sh-control. (E) KEGG pathway enrichment analysis of differentially expressed genes. (F) Effect of various cell death inhibitors on cell viability in LPS/ATP-induced NCM460 cells. (G) Annexin V/PI FCM analysis of apoptosis in LPS/ATP-induced NCM460 cells treated with sh-ITLN1 or control. (H) Live/dead cell staining (Calcein/PI) of LPS/ATP-induced NCM460 cells, visualizing viable (green) and dead (red) cells. Scale bar: 100 μm. (I) Influence of sh-ITLN1 or control on the proinflammatory cytokine levels in LPS/ATP-induced NCM460 cells. (J) Western blotting analysis for PANoptosis-related marker genes (caspase-1, caspase-3, GSDMD-N, MLKL, RIPK3) in LPS/ATP-induced NCM460 cells. ITLN1, Intelectin-1; IECs, intestinal epithelial cells; LPS, lipopolysaccharide; ATP, adenosine triphosphate; FCM, flow cytometry. ** P < 0.01, *** P < 0.001, **** P < 0.0001 by paired t test.
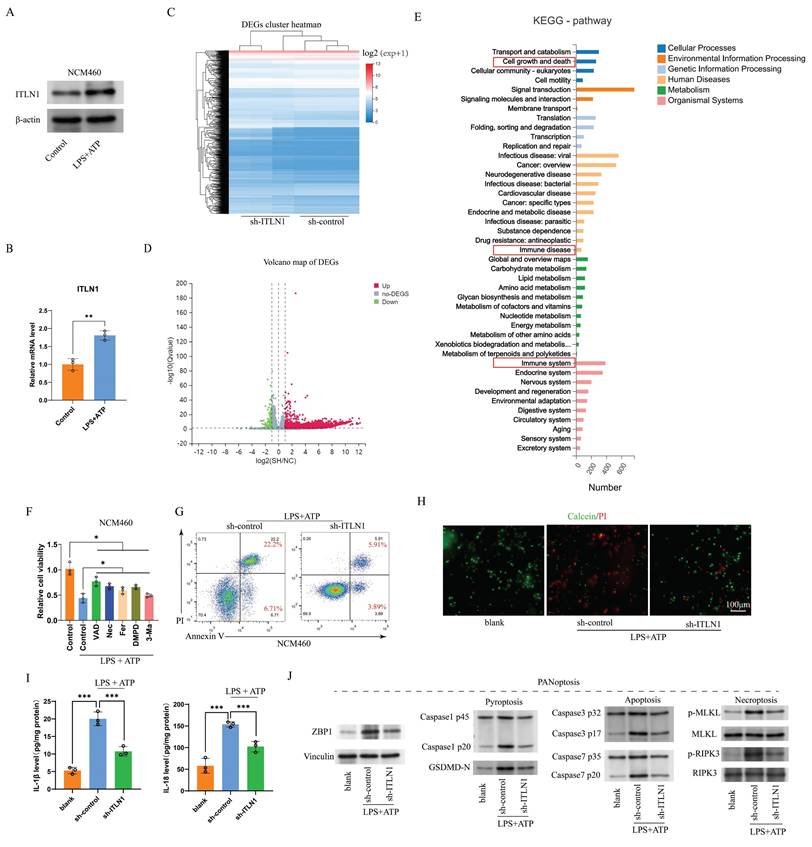
ITLN1 influences tight junctions in IECs. (A) Western blotting analysis following ITLN1 immunoprecipitation, confirming the presence of ITLN1 in the immunoprecipitated complex. (B) Distribution of peptide lengths obtained from mass spectrometry after immunoprecipitation, illustrating the size distribution of peptides associated with ITLN1. (C) Venn diagram displaying the overlap of proteins between ITLN1 and IgG immunoprecipitations. (D) Volcano plot of differentially expressed proteins (threshold: FC > 2, P < 0.05). (E) KEGG pathway analysis of differentially expressed proteins. (F) Western blotting analysis illustrating the effect of sh-ITLN1 on the expression of tight junction proteins (occludin and ZO-1) in NCM460 cells with or without LPS/ATP. (G) Immunofluorescence images illustrating the localization and expression of tight junction proteins (occludin and ZO-1) in IECs, with the impact of sh-ITLN1 treatment, indicating disruption of tight junction integrity. Scale bar: 20 μm. CD, Crohn's disease; IECs, intestinal epithelial cells; ITLN1, intelectin-1; DEGs, differentially expressed genes; LPS, lipopolysaccharide; ATP, adenosine triphosphate; FC, fold change; FCM, flow cytometry; KEGG, Kyoto encyclopedia of genes and genomes. Data are presented as mean ± SD. ** P < 0.01, *** P < 0.001, **** P < 0.0001 by one-way ANOVA test.
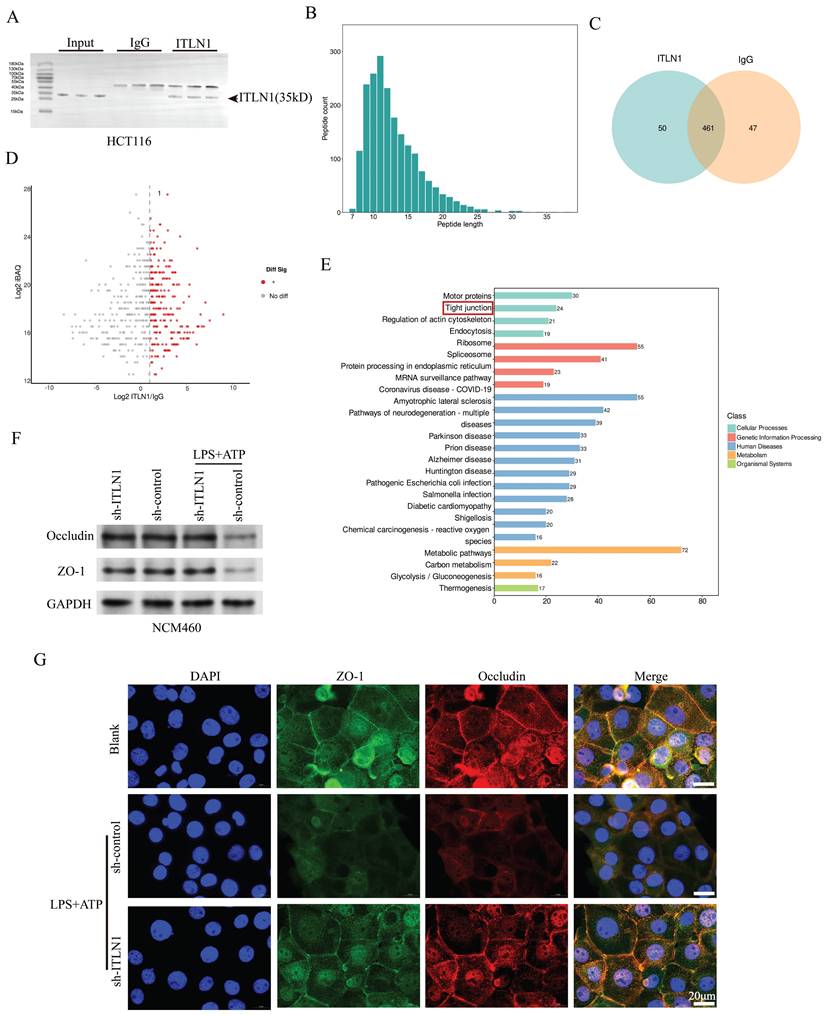
ITLN1 regulates CAPN2 ubiquitination through competitive binding with TRIM8. (A) Silver staining analysis of ITLN1 IP, identifying interacting proteins. (B) mRNA and protein expression levels of CAPN2 in isolated IECs from inflamed and uninflamed colon tissues illustrating no significant change. (C) Impact of sh-ITLN1 on CAPN2 mRNA and protein expression levels in NCM460 cell lines. (D) CHX protein stability assay evaluating the impact of ITLN1 on the stability of CAPN2 protein over time in NCM460 cells. (E) CHX protein stability assay evaluating the stability of CAPN2 protein over time in HEK293 cells. (F) Computational molecular docking analysis identifying potential binding sites between TRIM8 and CAPN2. (G) Bidirectional Co-IP between CAPN2 and TRIM8, confirming their direct interaction. (H) Endogenous and (I) exogenous assessments of TRIM8's impact on the ubiquitination level of CAPN2 protein in HEK293 cells using ubiquitin immunoblotting. (J) Immunofluorescence co-localization of ITLN1 and TRIM8 in NCM460 cells, confirming their spatial proximity. (K) Computational molecular docking analysis revealing potential binding sites between TRIM8 and ITLN1. (L-M) CHX protein stability assay evaluating the impact of TRIM8 and si-TRIM8 on the degradation of the CAPN2 protein in HEK293 cells. IECs, intestinal epithelial cells; IP, immunoprecipitation; ITLN1, intelectin-1; CAPN2, calpain-2; TRIM8, tripartite motif containing 8; CHX, cycloheximide. Data are presented as mean ± SD. ** P < 0.01, *** P < 0.001, **** P < 0.0001 by unpaired t test.
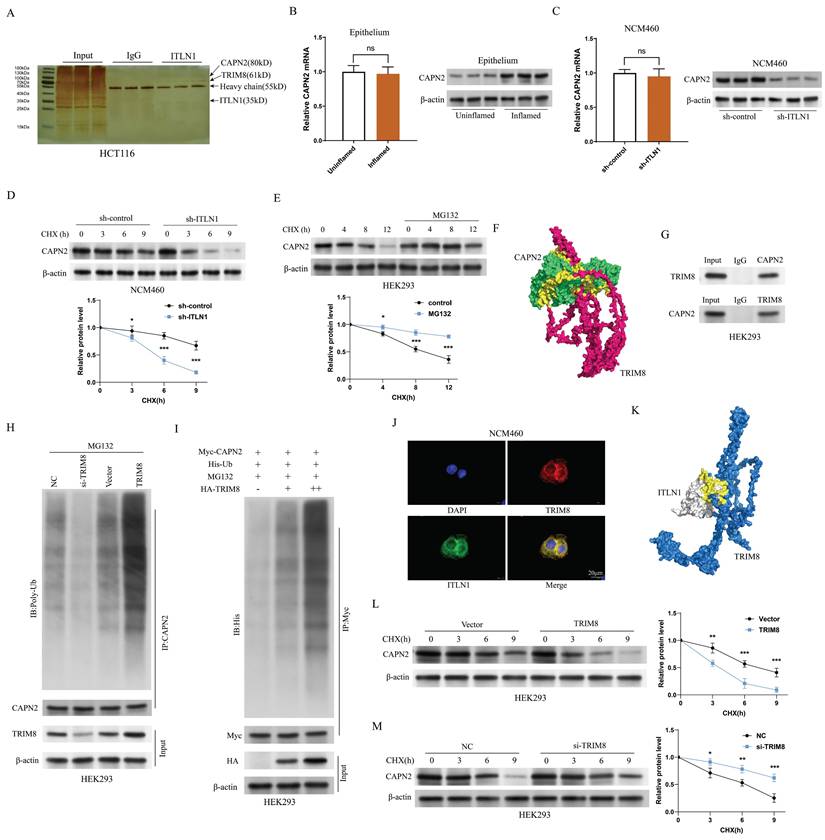
CAPN2 interacts with TRIM8. (A) The predicted ubiquitination sites of CAPN2 determined by Prop1.0, illustrating potential propeptide cleavage regions. (B) Immunoblot analysis of lysates from HEK293 cells transfected with His-tagged K48-Ub or K63-Ub, indicating the ubiquitination pattern of Myc-tagged CAPN2 in the presence or absence of HA-tagged TRIM8. (C) Schematic diagram of TRIM8 and its truncation mutants. (D) Immunoprecipitation of HA-tagged TRIM8 or its mutants and Myc-tagged CAPN2 in HEK293 cells, followed by immunoblotting with the indicated antibodies to analyze their interaction. (E) Schematic diagram of CAPN2 and its truncation mutants. (F) Immunoprecipitation of Myc-tagged CAPN2 or its mutants and HA-tagged TRIM8 in HEK293 cells, followed by immunoblotting to assess the interaction between the proteins.
TRIM8 interacts with CAPN2
Figure 7A illustrates the predicted ubiquitination sites of CAPN2 through the database Prop 1.0 [43]. To characterize TRIM8-mediated polyubiquitination, ubiquitin mutants (K48 and K63) were co-expressed in transfection assays. TRIM8-driven CAPN2 polyubiquitination was observed exclusively with K48 mutants, not K63 (Figure 7B). Structural analysis revealed TRIM8's functional domains: an N-terminal RING-finger domain (essential for E3 ligase activity), BBOX1/2 motifs, a coiled-coil domain, and an RFP-like domain (Figure 7C). Truncated TRIM8 and CAPN2 mutants were constructed to search for the domains responsible for the interaction between CAPN2 and TRIM8. The complementary binding studies using truncated TRIM8 constructs demonstrated that CAPN2 mainly interacted with RING and BBOX domains of TRIM8 (Figure 7D). Moreover, TRIM8 mainly interacted with the calpain catalytic domain of CAPN2 (Figures 7E-F).
CAPN2 antagonizes the impact of ITLN1 on inflammation and ZBP1-induced PANoptosis
We further investigated whether the effect of ITLN1 on inflammation and PANoptosis depends on CAPN2. Intervention with sh-ITLN1 (Supplemental Figure 1D) and ALLN (a CAPN2 inhibitor) significantly suppressed CAPN2 protein levels, while oe-CAPN2 rescued CAPN2 expression (Figure 8A). The inhibitory effect of sh-ITLN1 on LPS + ATP-induced inflammatory cytokines was also rescued by oe-CAPN2 (Figure 8B). Live/dead cell staining (Figure 8C) and apoptosis FCM (Figure 8D) similarly revealed that CAPN2 could counteract the inhibitory effect of sh-ITLN1 on cell death. Moreover, the inhibition of ZBP1-induced PANoptosis-related marker proteins by sh-ITLN1 could be reversed by CAPN2 (Figures 8E-F).
Sh-ITLN1 improves colonic inflammation in IL-10 KO mice
IL-10 KO mice were used as a CD colitis model and administered AAV-carried sh-ITLN1. We found that sh-ITLN1 reduced ITLN1 expression (Figure 9A) and decreased CAPN2 protein levels in colon tissue (Figure 9B). The sh-ITLN1 intervention significantly improved the DAI (Figure 9C), pathological inflammation score (disrupted intestinal integrity, reduced inflammatory cell infiltrates in the lamina propria and reduced goblet cells; Figure 9D), and inflammatory cytokine expression (Figure 9E) in IL-10 KO mice, while CAPN2 counteracted the anti-inflammatory effects of sh-ITLN1. Additionally, sh-ITLN1 significantly inhibited the expressions of ZBP1-induced PANoptosis-related marker proteins of colon tissues, which can be reversed by CAPN2 (Figures 9F-G).
Sh-ITLN1 improves intestinal barrier function
Sh-ITLN1 treatment significantly improved intestinal permeability in IL-10 KO mice, as evidenced by reduced mannitol flux (Figure 10A), lower FITC-dextran levels (Figure 10C), and increased electrical resistance (Figure 10B). The expression and distribution of tight junction (TJ) proteins, occludin, and ZO-1 were significantly improved with sh-ITLN1 treatment (Figures 10D-E). Similarly, MUC2 immunohistochemistry (Figure 10F) and AB-PAS staining (Figure 10G) revealed that sh-ITLN1 enhanced the mucus barrier in mice. Furthermore, sh-ITLN1 significantly reduced epithelial cell apoptosis in colon tissue (Figure 10H) and improved TJ morphology under TEM (Figure 10I). However, the beneficial effects of sh-ITLN1 on intestinal barrier function in IL-10 KO mice were reversed by CAPN2 (Figures 10A-I).
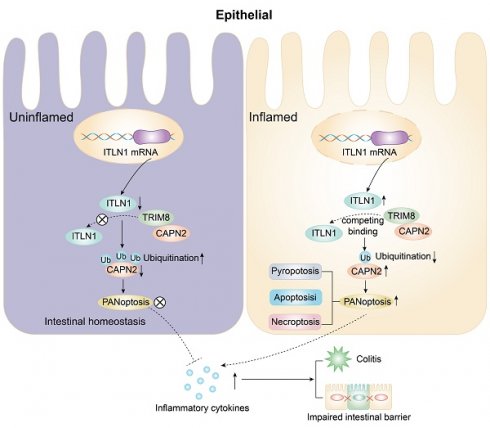
Key findings
The abnormally high expression of ITLN1 competitively binds to TRIM8, inhibiting the ubiquitination of CAPN2 and leading to its increased expression. This induces ZBP1-mediated PANoptosis in IECs, resulting in impaired intestinal mucosal barrier function and the release of a substantial number of inflammatory cytokines, promoting the progression of Crohn's colitis (Figure 11).
Discussion
Based on the single-cell sequencing, in vivo, and in vitro studies, this study demonstrated that the ITLN1/TRIM8/CAPN2 axis is involved in the colonic inflammation progression of CD via mediating PANoptosis. This novel insight adds a new dimension to understanding CD pathogenesis and identifies novel clinical therapeutic targets for CD.
Emerging evidence highlights ITLN1's involvement in diverse physiological and pathological processes [44], including inflammation, immune modulation, and metabolic regulation, with a significant association with type 2 diabetes [45]. There is some controversy over whether ITLN1 promotes or inhibits inflammation. However, ITLN1's interaction with adiponectin receptor 1 inhibits the NF-κB pathway in macrophages, suppressing LPS-induced proinflammatory cytokine production and demonstrating anti-inflammatory properties [46]. Conversely, ITLN1 has been reported to enhance allergen-induced IL-25 and IL-33 secretion in airway epithelial cells, exacerbating allergic airway inflammation [47]. Furthermore, ITLN1 has been identified as a potential genetic risk factor for IBD, with significantly elevated expression in ulcerative colitis (UC), possibly participating in UC pathogenesis by disrupting the intestinal mucus barrier and microbiota homeostasis [48]. However, the correlation between ITLN1 and CD remains unknown. The mRNA and protein levels of ITLN1 were significantly elevated in IECs in inflamed regions, and its expression was positively correlated with CD-specific inflammatory markers such as CRP, CDAI, and SES-CD. This suggests that there may be a specific correlation between ITLN1 and CD.
CAPN2 is a calcium-dependent cysteine protease involved in various cellular processes. Extensive research has demonstrated a strong connection between CAPN2 and inflammation. A previous study revealed that CAPN1 or CAPN2 depletion provided protective effects against lung inflammation caused by ventilators [49].
CAPN2 antagonizes the effects of ITLN1 on inflammation and ZBP1-induced PANoptosis. (A) Western blotting analysis illustrating the effect of sh-ITLN1 and CAPN2 rescue on CAPN2 protein expression in NCM460 cells, confirming the modulation of CAPN2 expression upon sh-ITLN1 and CAPN2 rescue treatment. (B) ELISA quantification of proinflammatory cytokines (IL-6 and IL-8) in NCM460 cells, demonstrating the influence of sh-ITLN1 and CAPN2 rescue on cytokine levels following LPS/ATP co-stimulation. (C) Live/dead (Calcein/PI) staining of NCM460 cells indicating the effect of sh-ITLN1 and CAPN2 rescue on cell viability. Scale bar: 100 μm. (D) Flow cytometry analysis of apoptosis in LPS/ATP co-stimulated NCM460 cells treated with sh-ITLN1, sh-ITLN1 + CAPN2 rescue, or ALLN. (E) Immunofluorescence analysis of PANoptosis-related marker genes (caspase-1, RIPK3, ZBP1, and ASC) in NCM460 cells, illustrating the effect of sh-ITLN1 and CAPN2 rescue on PANoptosis expression. Scale bar: 20 μm. (F) Western blotting analysis of PANoptosis-related proteins (caspase-1, caspase-3, caspase-7, GSDMD-N, p-MLKL, p-RIPK3) in LPS/ATP co-stimulated NCM460 cells, indicating the modulation of PANoptosis pathways by sh-ITLN1 and CAPN2 rescue. IECs, intestinal epithelial cells; ITLN1, intelectin-1; CAPN2, calpain-2; FCM, flow cytometry; LPS, lipopolysaccharide; ATP, adenosine triphosphate. Data are presented as mean ± SD. *** P < 0.001 by one-way ANOVA test.
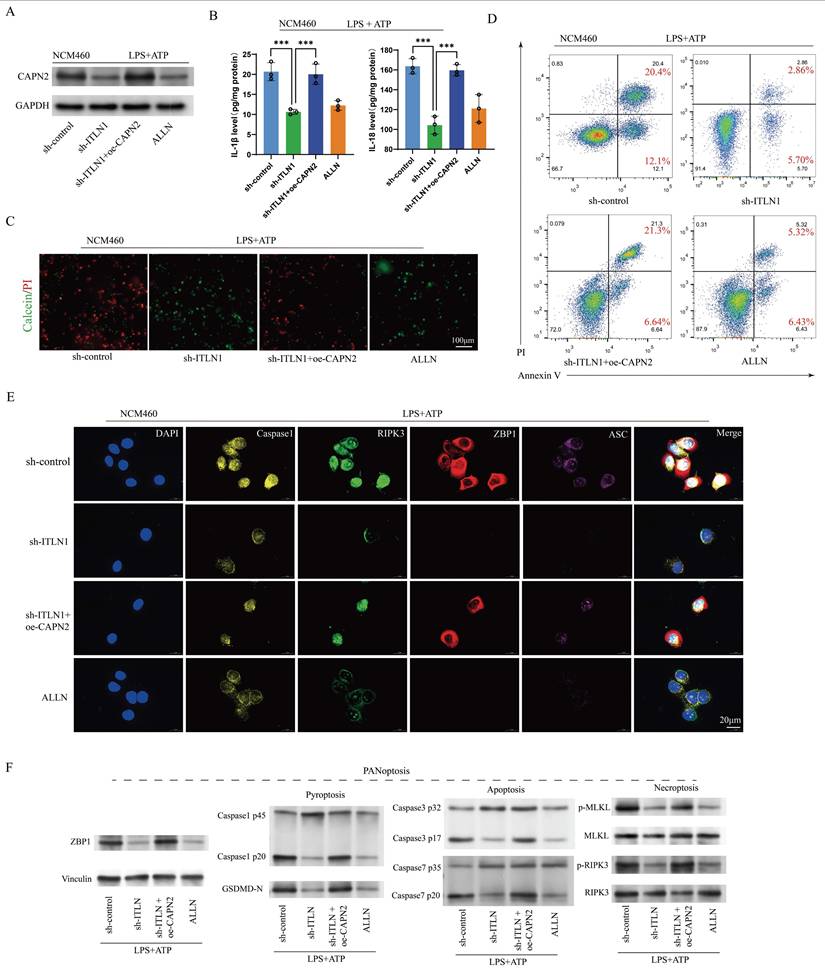
Sh-ITLN1 alleviates colonic inflammation in IL-10 KO mice. (A) Effect of sh-ITLN1 and CAPN2 rescue on ITLN1 mRNA levels in colon tissues of mice, illustrating a significant reduction in ITLN1 expression upon sh-ITLN1 treatment. (B) Western blotting analysis revealing the impact of sh-ITLN1 on ITLN1 and CAPN2 protein expression in colon tissues from IL-10 KO mice. (C) Effect of sh-ITLN1 on the disease activity index, indicating a significant reduction in disease severity in the IL-10 KO model upon ITLN1 knockdown. (D) Histological analysis of colon tissues using HE staining, revealing the influence of sh-ITLN1 on the pathological inflammation score. Scale bar: 20 μm. (E) Quantification of inflammatory cytokine levels (TNF-α, IFN-γ, and IL-17) in colon tissues, demonstrating a significant decrease following sh-ITLN1 treatment. (F) Immunofluorescence and (G) Western blotting analysis of PANoptosis-associated marker genes in colon tissues, demonstrating the effect of sh-ITLN1 and CAPN2 rescue on PANoptosis level. Scale bar: 20 μm. ITLN1, intelectin-1; CAPN2, calpain-2; IL-10 KO, IL-10 knock-out. Data are presented as mean ± SD. ** P < 0.01, *** P < 0.001 by one-way ANOVA test.
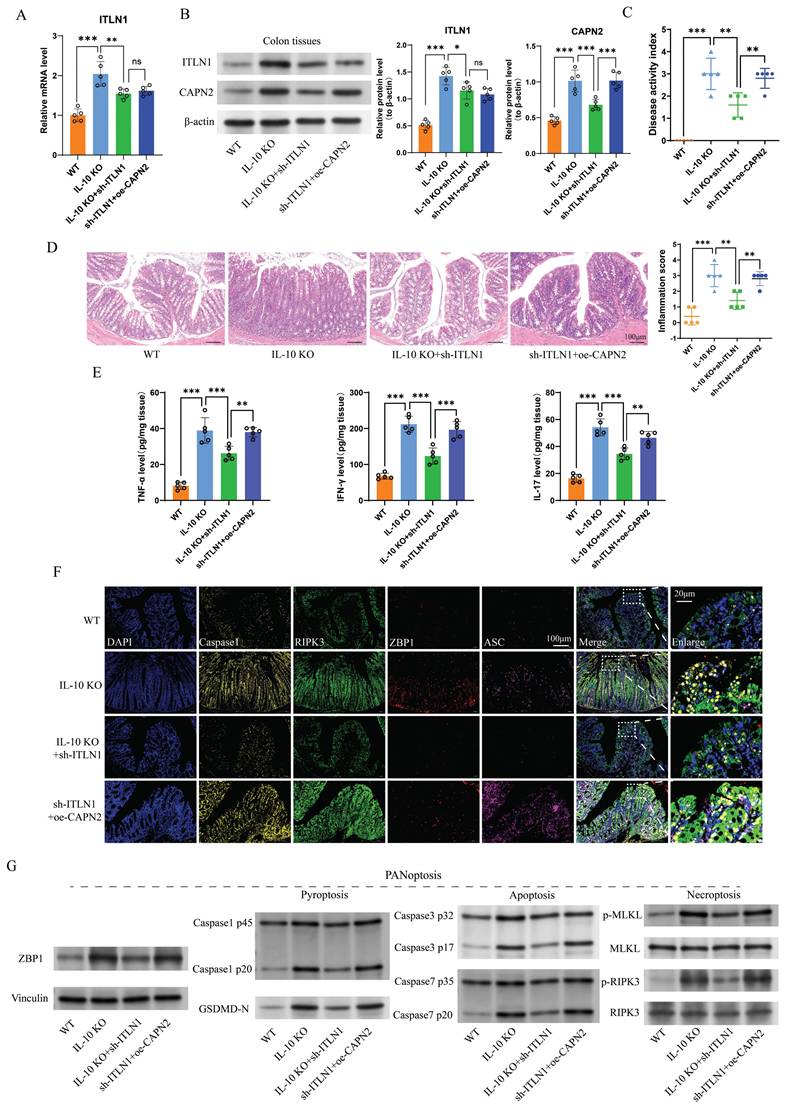
Sh-ITLN1 enhances intestinal barrier function in IL-10 KO mice. (A) Mannitol fluxes, (B) electrical resistance, and (C) FITC-dextran analyses evaluating the effect of sh-ITLN1 and CAPN2 restoration on intestinal permeability. (D) Western blotting and (E) immunofluorescence analysis (scale bar: 100 and 20 μm) illustrating the effect of sh-ITLN1 and CAPN2 rescue on tight junction proteins (occludin and ZO-1) in colon tissues. (F) MUC2 immunohistochemistry in colon tissues. (scale bar: 50 and 20 μm). (G) AB-PAS staining in colon tissues (scale bar: 100 and 20 μm). (H) TUNEL staining in colon tissues (scale bar: 100 μm). (I) Representative TEM images of tight junctions in each group (scale bar: 1 μm). ITLN1, intelectin-1; CAPN2, calpain-2; IL-10 KO, IL-10 knockout; FITC, fluorescein isothiocyanate; TJ, tight junction; AB-PAS, Alcian Blue-Periodic Acid-Schiff; TEM, transmission electron microscope; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling. Data are presented as mean ± SD. * P < 0.05, ** P < 0.01, *** P < 0.001 by one-way ANOVA test.
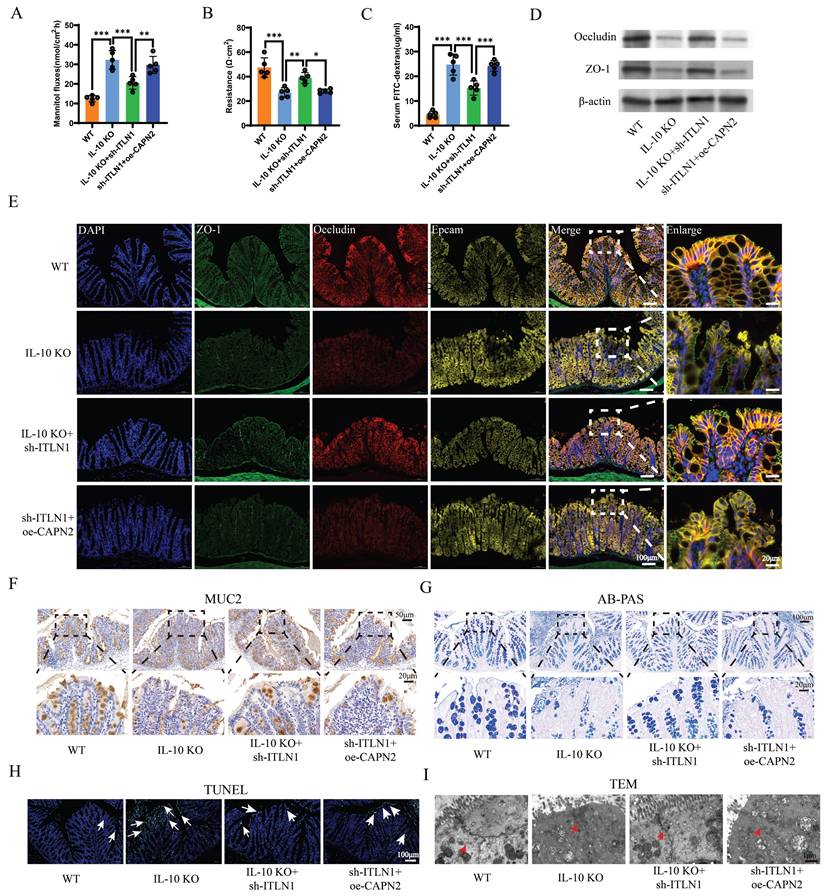
A schematic diagram illustrating how ITLN1 promotes Crohn's colitis by inducing PANoptosis. IECs, intestinal epithelial cells; ITLN1, intelectin-1; CAPN2, calpain-2; TRIM8, tripartite motif containing 8.
Additionally, calpain activation has been linked to neuronal apoptosis following spinal cord injuries and in neurodegenerative disorders [50], highlighting its dual role in inflammatory and degenerative pathologies. Another study has indicated that CAPN1/CAPN2 can facilitate cisplatin-induced pyroptosis in esophageal cancer [51]. Similarly, Bozym et al. [52] reported that CAPN2 mediates coxsackievirus B-induced cellular necrosis and is implicated in the actin cytoskeleton rearrangement and disruption of the junctional complex. These findings cumulatively suggest a close relationship between CAPN2 and cell death. Our study has found a positive correlation between CAPN2 and PANoptosis. A previous study has demonstrated the therapeutic potential of CAPN2 inhibitors in murine colitis and colitis-associated cancer, utilizing the azoxymethane/dextran sulfate sodium model to suppress macrophage activation and restrict tumor progression [53]. Our findings are consistent with and further support these observations.
Recent research has increasingly highlighted the strong association between PANoptosis and IBD. The excessive IEC death induced by PANoptosis compromises the intestinal barrier, facilitates bacterial translocation, and triggers secondary inflammation, further aggravating mucosal epithelial damage in ulcerative colitis (UC) patients [54,55]. Currently, researchers have studied PANoptosis-related genes in CD through a combination of bioinformatics, machine learning, and related experiments. They discovered that PANoptosis plays a nonnegligible role in CD via interacting with CD-associated genes and regulating the immune system [56]. Besides, numerous studies have confirmed the direct positive correlation between PANoptosis and inflammation [57]. This study demonstrated that the expressions of PANoptosis-related marker proteins were significantly elevated in patients with CD, IL-10 KO colitis mice, and LPS + ATP-induced in vitro inflammation models, which strongly suggested the presence of PANoptosis under inflammatory conditions. Furthermore, it revealed that PANoptosis is probably involved in CD pathogenesis, and targeting PANoptosis could become a novel therapeutic modality for CD.
Herein, CAPN2 is identified as one of the key mechanisms by which ITLN1 regulates PANoptosis. ITLN1 does not directly affect CAPN2 mRNA levels; instead, it regulates the protein stability of CAPN2. Protein translational modifications (PTMs) are currently among the most extensively studied mechanisms that regulate protein stability [58]. PTMs enhance the functional diversity of the proteome by adding functional groups, regulating subunit proteolysis, or degrading proteins. These modifications include phosphorylation, ubiquitination, methylation, acetylation, and glycosylation [58]. Our findings suggest that CAPN2 is primarily degraded via the ubiquitin-proteasome pathway. TRIM8, as an E3 ubiquitin ligase, transfers ubiquitin molecules to target proteins, thereby mediating ubiquitination. This is a crucial mechanism for regulating protein expression levels within cells, impacting key biological processes (autophagy, apoptosis, innate immunity, signal transduction, and others) [59,60]. We speculated that ITLN1 may competitively bind to the E3 ligase TRIM8, thereby reducing the ubiquitination level of the CAPN2 protein and promoting its increased expression.
This study elucidates a pathogenic pathway in which ITLN1 competitively binds TRIM8 to inhibit CAPN2 ubiquitination and degradation, stabilizing CAPN2 to promote ZBP1-mediated PANoptosis in IECs and exacerbate Crohn's colitis. This mechanism offers viable therapeutic options: targeted inhibition of ITLN1 (via shRNA or monoclonal antibodies) or pharmacological inhibition of CAPN2 (using calpain inhibitors such as MDL28170) significantly reduced intestinal inflammation and PANoptosis in vitro and in vivo. These effects were accompanied by the restoration of mucosal barrier integrity in colitis models. This establishes a robust basis for subsequent clinical translation. Targeting the ITLN1/CAPN2 axis reduced inflammation and PANoptosis significantly and restored mucosal barrier integrity in colitis models, thereby establishing a basis for subsequent clinical application. This corresponds with other therapeutic strategies in IBD, including polyphenols, which regulate inflammation and gut microbiota, enhancing barrier function [61]. We intend to validate ITLN1-neutralizing biologics in patient-derived organoids and evaluate ITLN1 as a biomarker for anti-PANoptosis therapies, bridging mechanistic insights to precision treatments in CD.
Supplementary Material
Supplementary figure S1.
Acknowledgements
Authors thank Home for Researchers editorial team (www.home-for-researchers.com) for improving the English language in this manuscript.
Funding
This work was supported by National Natural Science Foundation of China (82300609), National Natural Science Foundation of Jiangsu Province (BK20241775), Changzhou Sci&Tech Program (2022CZBJ066, 2022CZLJ017, CJ20245024, QY202306), Changzhou Medical Center of Nanjing Medical University Program (CMC2024PY17, CMC2024PY01), Taizhou School of Clinical Medicine, Nanjing Medical University (TZKY20230308, TZKY20230102), Social Development Plan of Taizhou (TS202213).
Author contributions
J Z, YJ L, and P Y: Study design and data analysis.
J Z, ZW X, DM W and HG W: Patient recruitment, data collection and writing up of the first draft of the paper.
J Z, and LM T: scientific advice, supervision and drafting of the manuscript.
Ethical approval and consent to participate
Consent to participate is not applicable in this study. The clinical study was approved by the ethics committee of The First Affiliated Hospital of Nanjing Medical University (No. 2023-SR-115). All the experimental protocols were approved by the Ethics Committee of the Laboratory Animal Center of Nantong University (No. S20200323-289) according to the guidelines by the Chinese Council on Animal Care.
Availability of data and material
Please contact the corresponding author upon reasonable request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Roda G, Chien Ng S, Kotze PG. et al. Crohn's disease. Nat Rev Dis Primers. 2020;6:22
2. Ng SC, Shi HY, Hamidi N. et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390:2769-2778
3. Cui G, Liu H, Xu G. et al. Exploring Links Between Industrialization, Urbanization, and Chinese Inflammatory Bowel Disease. Front Med (Lausanne). 2021;8:757025
4. Petagna L, Antonelli A, Ganini C. et al. Pathophysiology of Crohn's disease inflammation and recurrence. Biol Direct. 2020;15:23
5. Caio G, Lungaro L, Caputo F. et al. Nutritional Treatment in Crohn's Disease. Nutrients. 2021;13:1628
6. Torres J, Mehandru S, Colombel JF. et al. Crohn's disease. Lancet. 2017;389:1741-1755
7. Foerster EG, Mukherjee T, Cabral-Fernandes L. et al. How autophagy controls the intestinal epithelial barrier. Autophagy. 2022;18:86-103
8. Larabi A, Barnich N, Nguyen H. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy. 2020;16:38-51
9. Alula KM, Dowdell AS, LeBere B. et al. Interplay of gut microbiota and host epithelial mitochondrial dysfunction is necessary for the development of spontaneous intestinal inflammation in mice. Microbiome. 2023;11:256
10. Kanke M, Kennedy Ng MM, Connelly S. et al. Single-Cell Analysis Reveals Unexpected Cellular Changes and Transposon Expression Signatures in the Colonic Epithelium of Treatment-Naïve Adult Crohn's Disease Patients. Cell Mol Gastroenterol Hepatol. 2022;13:1717-1740
11. Garcia-Carbonell R, Yao SJ, Das S. et al. Dysregulation of Intestinal Epithelial Cell RIPK Pathways Promotes Chronic Inflammation in the IBD Gut. Front Immunol. 2019;10:1094
12. Sun X, Yang Y, Meng X. et al. PANoptosis: Mechanisms, biology, and role in disease. Immunol Rev. 2023
13. Zhu P, Ke ZR, Chen JX. et al. Advances in mechanism and regulation of PANoptosis: Prospects in disease treatment. Front Immunol. 2023;14:1120034
14. Wang L, Zhu Y, Zhang L. et al. Mechanisms of PANoptosis and relevant small-molecule compounds for fighting diseases. Cell Death Dis. 2023;14:851
15. Li J, Cui J, Wu L. et al. Machine learning and molecular subtype analyses provide insights into PANoptosis-associated genes in rheumatoid arthritis. Arthritis Res Ther. 2023;25:233
16. Sun W, Li P, Wang M. et al. Molecular characterization of PANoptosis-related genes with features of immune dysregulation in systemic lupus erythematosus. Clin Immunol. 2023;253:109660
17. Best WR, Becktel JM, Singleton JW. et al. Development of a Crohn's disease activity index. National Cooperative Crohn's Disease Study. Gastroenterology. 1976;70:439-444
18. Daperno M, D'Haens G, Van Assche G. et al. Development and validation of a new, simplified endoscopic activity score for Crohn's disease: the SES-CD. Gastrointest Endosc. 2004;60:505-512
19. Macosko EZ, Basu A, Satija R. et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202-1214
20. Butler A, Hoffman P, Smibert P. et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411-420
21. Raudvere U, Kolberg L, Kuzmin I. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019;47:W191-W198
22. Prianichnikov N, Koch H, Koch S. et al. MaxQuant Software for Ion Mobility Enhanced Shotgun Proteomics. Mol Cell Proteomics. 2020;19:1058-1069
23. UniProt. the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480-D489
24. Wirtz S, Neufert C, Weigmann B. et al. Chemically induced mouse models of intestinal inflammation. Nat Protoc. 2007;2:541-546
25. Singh UP, Singh S, Taub DD. et al. Inhibition of IFN-gamma-inducible protein-10 abrogates colitis in IL-10-/- mice. J Immunol. 2003;171:1401-1406
26. Zhao J, Lin Z, Ying P. et al. circSMAD4 Promotes Experimental Colitis and Impairs Intestinal Barrier Functions by Targeting Janus Kinase 2 Through Sponging miR-135a-5p. J Crohns Colitis. 2023;17:593-613
27. Yu M, Luo Y, Cong Z. et al. MicroRNA-590-5p Inhibits Intestinal Inflammation by Targeting YAP. J Crohns Colitis. 2018;12:993-1004
28. Zhao J, Zhao Z, Ying P. et al. METTL3-mediated m(6) A modification of circPRKAR1B promotes Crohn's colitis by inducing pyroptosis via autophagy inhibition. Clin Transl Med. 2023;13:e1405
29. Ji ML, Jiang H, Zhang XJ. et al. Preclinical development of a microRNA-based therapy for intervertebral disc degeneration. Nat Commun. 2018;9:5051
30. Wang H, Dong J, Shi P. et al. Anti-mouse CD52 monoclonal antibody ameliorates intestinal epithelial barrier function in interleukin-10 knockout mice with spontaneous chronic colitis. Immunology. 2015;144:254-262
31. Arrieta MC, Madsen K, Doyle J. et al. Reducing small intestinal permeability attenuates colitis in the IL10 gene-deficient mouse. Gut. 2009;58:41-48
32. Looijer-van Langen M, Hotte N, Dieleman LA. et al. Estrogen receptor-β signaling modulates epithelial barrier function. Am J Physiol Gastrointest Liver Physiol. 2011;300:G621-626
33. Ahmad R, Rah B, Bastola D. et al. Obesity-induces Organ and Tissue Specific Tight Junction Restructuring and Barrier Deregulation by Claudin Switching. Sci Rep. 2017;7:5125
34. Zhao J, Wang H, Zhou J. et al. miR-130a-3p, a Preclinical Therapeutic Target for Crohn's Disease. J Crohns Colitis. 2021;15:647-664
35. Wang L, Li H, Huang A. et al. Mutual regulation between TRIM21 and TRIM8 via K48-linked ubiquitination. Oncogene. 2023;42:3708-3718
36. Parikh K, Antanaviciute A, Fawkner-Corbett D. et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 2019;567:49-55
37. Chen L, Li J, Yang G. A comparative review of intelectins. Scand J Immunol. 2020;92:e12882
38. Washimi K, Yokose T, Yamashita M. et al. Specific expression of human intelectin-1 in malignant pleural mesothelioma and gastrointestinal goblet cells. PLoS One. 2012;7:e39889
39. Li D, Zhao X, Xiao Y. et al. Intelectin 1 suppresses tumor progression and is associated with improved survival in gastric cancer. Oncotarget. 2015;6:16168-16182
40. Cao W, Li RW, Chin Y. et al. Transcriptome analysis reveals the protective role of fructo-oligosaccharide in colonic mucosal barriers in exercise-induced stressed mice. Food Funct. 2021;12:4484-4495
41. Keller N, Ozmadenci D, Ichim G. et al. Caspase-8 function, and phosphorylation, in cell migration. Semin Cell Dev Biol. 2018;82:105-117
42. Shapovalov I, Harper D, Greer PA. Calpain as a therapeutic target in cancer. Expert Opin Ther Targets. 2022;26:217-231
43. Duckert P, Brunak S, Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel. 2004;17:107-112
44. Zhao A, Xiao H, Zhu Y. et al. Omentin-1: a newly discovered warrior against metabolic related diseases. Expert Opin Ther Targets. 2022;26:275-289
45. Nonnecke EB, Castillo PA, Akahoshi DT. et al. Characterization of an intelectin-1 (Itln1) knockout mouse model. Front Immunol. 2022;13:894649
46. Kobayashi H, Uchimura K, Ishii T. et al. Intelectin1 ameliorates macrophage activation via inhibiting the nuclear factor kappa B pathway. Endocr J. 2022;69:539-546
47. Yi L, Cheng D, Zhang K. et al. Intelectin contributes to allergen-induced IL-25, IL-33, and TSLP expression and type 2 response in asthma and atopic dermatitis. Mucosal Immunol. 2017;10:1491-1503
48. Matute JD, Duan J, Flak MB. et al. Intelectin-1 binds and alters the localization of the mucus barrier-modifying bacterium Akkermansia muciniphila. J Exp Med. 2023;220:e20211938
49. Liu D, Yan Z, Minshall RD. et al. Activation of calpains mediates early lung neutrophilic inflammation in ventilator-induced lung injury. Am J Physiol Lung Cell Mol Physiol. 2012;302:L370-379
50. Momeni HR. Role of calpain in apoptosis. Cell J. 2011;13:65-72
51. Li RY, Zheng ZY, Li ZM. et al. Cisplatin-induced pyroptosis is mediated via the CAPN1/CAPN2-BAK/BAX-caspase-9-caspase-3-GSDME axis in esophageal cancer. Chem Biol Interact. 2022;361:109967
52. Bozym RA, Patel K, White C. et al. Calcium signals and calpain-dependent necrosis are essential for release of coxsackievirus B from polarized intestinal epithelial cells. Mol Biol Cell. 2011;22:3010-3021
53. Rose AH, Huang Z, Mafnas C. et al. Calpain-2 Inhibitor Therapy Reduces Murine Colitis and Colitis-associated Cancer. Inflamm Bowel Dis. 2015;21:2005-2015
54. Ungaro R, Mehandru S, Allen PB. et al. Ulcerative colitis. Lancet. 2017;389:1756-1770
55. Wang JM, Yang J, Xia WY. et al. Comprehensive Analysis of PANoptosis-Related Gene Signature of Ulcerative Colitis. Int J Mol Sci. 2023;25:348
56. Lu J, Li F, Ye M. PANoptosis and Autophagy-Related Molecular Signature and Immune Landscape in Ulcerative Colitis: Integrated Analysis and Experimental Validation. J Inflamm Res. 2024;17:3225-3245
57. Lee E, Song CH, Bae SJ. et al. Regulated cell death pathways and their roles in homeostasis, infection, inflammation, and tumorigenesis. Exp Mol Med. 2023;55:1632-1643
58. Lee JM, Hammarén HM, Savitski MM. et al. Control of protein stability by post-translational modifications. Nat Commun. 2023;14:201
59. Hatakeyama S. TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends Biochem Sci. 2017;42:297-311
60. Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399-434
61. Boaru DL, Fraile-Martinez O, De Leon-Oliva D. et al. Harnessing the Anti-Inflammatory Properties of Polyphenols in the Treatment of Inflammatory Bowel Disease. Int J Biol Sci. 2024;20:5608-5672
Author contact
![]() Corresponding authors: Liming Tang (tanglimingedu.cn, Department of Gastrointestinal Surgery, The Affiliated Changzhou No.2 People's Hospital of Nanjing Medical University, The Third Affiliated Hospital of Nanjing Medical University, Changzhou Medical Center, Nanjing Medical University, 68 Gehu Road, Changzhou, 213000, China) or Honggang Wang (honggangtzcom, Department of General Surgery, Affiliated Taizhou People's Hospital of Nanjing Medical University, Taizhou School of Clinical Medicine, Nanjing Medical University, Taizhou, 225300, China) or Dongmei Wang (dmwangedu.cn, Department of Gastrointestinal Surgery, The Affiliated Changzhou No.2 People's Hospital of Nanjing Medical University, The Third Affiliated Hospital of Nanjing Medical University, Changzhou Medical Center, Nanjing Medical University, 68 Gehu Road, Changzhou, 213000, China).
Corresponding authors: Liming Tang (tanglimingedu.cn, Department of Gastrointestinal Surgery, The Affiliated Changzhou No.2 People's Hospital of Nanjing Medical University, The Third Affiliated Hospital of Nanjing Medical University, Changzhou Medical Center, Nanjing Medical University, 68 Gehu Road, Changzhou, 213000, China) or Honggang Wang (honggangtzcom, Department of General Surgery, Affiliated Taizhou People's Hospital of Nanjing Medical University, Taizhou School of Clinical Medicine, Nanjing Medical University, Taizhou, 225300, China) or Dongmei Wang (dmwangedu.cn, Department of Gastrointestinal Surgery, The Affiliated Changzhou No.2 People's Hospital of Nanjing Medical University, The Third Affiliated Hospital of Nanjing Medical University, Changzhou Medical Center, Nanjing Medical University, 68 Gehu Road, Changzhou, 213000, China).

 Global reach, higher impact
Global reach, higher impact