10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2025; 21(9):4270-4292. doi:10.7150/ijbs.113193 This issue Cite
Review
Deubiquitylating Enzymes in Hepatocellular Carcinoma
1. Department of Liver Surgery, Xiangya Hospital, Central South University, 110 Xiangya Road, Changsha, Hunan 410078, China.
2. Department of General Surgery, The Second Hospital of Liling City, Baitutang Town, Liling City, Zhuzhou City, Hunan 412207, China.
* Xingyu Mi and Qingfeng Li contributed equally to this work.
Received 2025-3-4; Accepted 2025-6-10; Published 2025-6-20
Abstract
Ubiquitination is a reversible and dynamic process, precisely regulated by ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), ubiquitin ligases (E3), and deubiquitinating enzymes (DUBs). Dysregulation of DUBs disrupts the dynamic balance of ubiquitination, contributing to the development of various diseases, particularly cancer. An increasing number of studies have identified dysregulation of DUBs in various tumor types and have explored their regulatory mechanisms in these contexts. Hepatocellular carcinoma (HCC), the most common form of liver cancer, is highly malignant and has limited treatment options, necessitating the exploration of additional therapeutic strategies. Current research has identified dysregulation of DUBs in HCC, but their mechanisms of action remain poorly understood. Given the large number of DUB family members, there is significant potential for further investigation. This review summarizes the DUBs associated with HCC, including their structures, functions, and the mechanisms through which they regulate HCC development. Furthermore, we provide a brief overview of the discussed DUBs, aiming to offer new perspectives for future HCC research.
Keywords: hepatocellular carcinoma, deubiquitylating enzymes
Introduction
Ubiquitination is an important post-translational modification process in which ubiquitin molecules, consisting of 76 amino acids, are covalently conjugated to target proteins[1]. The main steps of ubiquitination are mediated by three types of enzymes: ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3)[2, 3]. And deubiquitinating enzymes (DUBs) can remove ubiquitin from substrate proteins. It is noteworthy that ubiquitination is a reversible and dynamic process, precisely and orderly maintained by E1, E2, E3 and DUBs. Ubiquitin proteins are first activated by E1, a process that requires ATP. Subsequently, Ubiquitin proteins bind to E2 and then to substrate proteins through E3 to form a covalent bond[4]. While DUBs can clean ubiquitin proteins from substrate proteins to negatively regulate this process[5]. There are some current researches indicating that imbalance between ubiquitination and deubiquitination is associated with the occurrence and development of human cancer[6-10]. Some targeted drugs for the ubiquitin system have been developed and have shown progress[11]. However, considering the number and diversity of E2s, E3s, and DUBs, there is still potential for the development of more drugs. In particular, DUBs are inherently attractive as potential drug targets[11, 12].
Liver cancer is one of the most common malignant tumors of the digestive system. According to the global cancer statistics of 2022 released by the International Agency for Research on Cancer, the mortality rate of liver cancer ranks 3rd[13]. The most common subtype of liver cancer is hepatocellular carcinoma (HCC), accounting for approximately 90% of cases[14]. Curative surgery is the only treatment that has the opportunity to cure early-stage HCC, but the majority of patients are diagnosed at an advanced stage and lose the opportunity for surgery[15]. Although targeted drugs for the treatment of advanced HCC, such as sorafenib and lenvatinib, which are tyrosine kinase inhibitors, have been developed and utilized in clinical practice, there are currently no drugs targeting DUBs used in the treatment of advanced HCC.
This article will review the research progress of deubiquitinating enzymes in HCC, explore their potential roles in the occurrence, development, metastasis, and treatment of HCC, and provide new ideas and directions for the precision treatment of HCC.
The DUB Family and Structure
It has been hypothesized that approximately 100 DUBs are encoded by the human genome. According to sequence and catalytic domain, DUBs are classified into 6 families: ubiquitin-specific proteases (USPs), ovarian tumor proteases (OTUs), ubiquitin C-terminal hydrolases (UCHs), Machado-Joseph domain-containing proteases (MJDs), the motif interacting with ubiquitin (MIU)-containing novel DUB family (MINDYs), and JAMM/MPN domain-associated Zn-depend metalloproteases (JAMMs)[16].
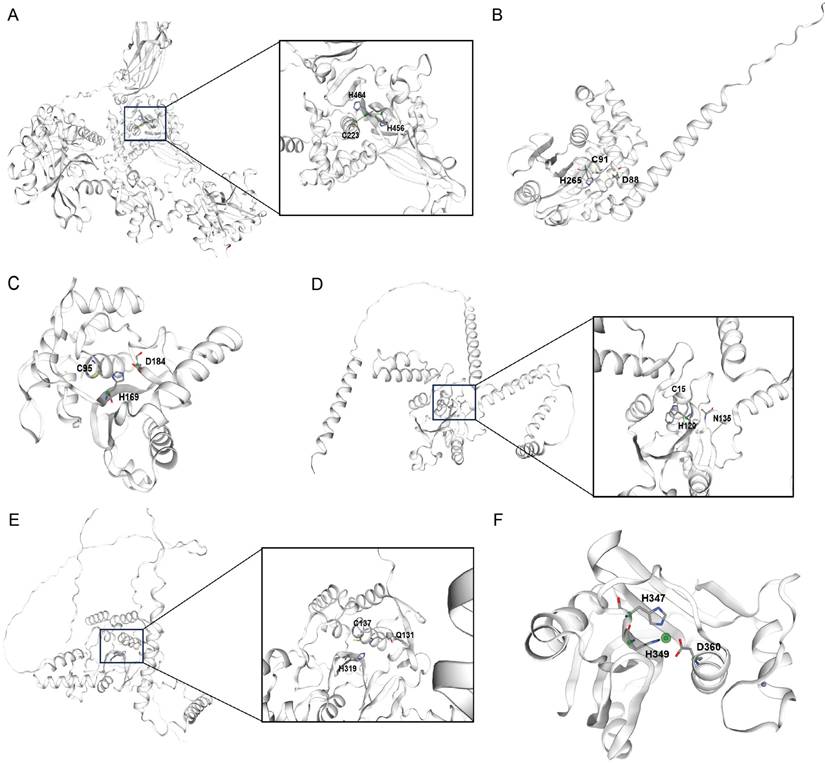
The USPs range from 50 to 300kDa, and their enzymatic functions are carried out by the thiol group of the central cysteine[17]. The catalytic region of a USP features a structure akin to that of papain, and this conserved domain allows the majority of USPs to cleave isopeptide bonds through a comparable process (Fig. 1A)[18]. USPs possess specialized cysteine protease domains that operate through distinct catalytic mechanisms; these regions are composed of three conserved subdomains, which are analogous to the anatomical components of the palm, thumb, and fingers of a human right hand, respectively[18]. The majority of USP family members consist of a central catalytic domain, along with additional N- and C-terminal extensions. These extensions can vary in composition, incorporating distinct domains or sequences that confer specialized functions[19].
The OTU domain was initially discovered through bioinformatics analysis in a gene from ovarian tumors in Drosophila melanogaster[20]. Further studies revealed that certain members of the OTU family function as deubiquitinating enzymes, featuring potential catalytic cysteine and histidine residues[21, 22]. Almost all OTUs possess an OTU catalytic domain along with an ubiquitin interaction domain. The OTU core domain's structure consists of β-strands that are sandwiched between α-helical regions, and the α-helical regions are formed by an anterior α-helical region, a central β-sandwich region, and a posterior α-helical region[21, 23]. The OTUs' active site is established at the midpoint of the OTU's surface, precisely where the helical domain intersects with the β-strands domain (Fig. 1B)[24]. Although the overall structure of OTUs varies, the arrangement of catalytic residues is similar and conserved[25].
The UCH subgroup comprises four members, which are UCH-L1, UCH-L3, UCH37, and BRCA1-associated protein-1. Each UCH featuring an N-terminal C12 peptidase domain that is shaped by a knotted peptide backbone, a C-terminal extension, and an unstructured loop that modulates substrate access to the catalytic site. The active site of a UCH typically consists of two parts: one is the central β-sheet structure, and the other is the α-helical structure surrounding the β-sheet. Furthermore, UCHs also contain an unstructured loop structure that limits the binding of substrates to the active site (Fig. 1C)[26]. In the absence of substrate, the active sites of UCHs are distorted and inactive. However, when bound to ubiquitin, the structures of UCHs active sites undergo changes, acquiring enzymatic activity, allowing the catalytic triad—composed of aspartic acid, histidine, and cysteine—to bind with the substrate[26].
The MJD family encompasses several distinct deubiquitinating enzymes, including Ataxin-3, Ataxin-3L, JOSD1, and JOSD2, all of which contain a Josephin catalytic domain composed of approximately 180 amino acids[27]. The Josephin domain of MJD family proteins is responsible for their catalytic activity, while additional structural domains are involved in regulating their substrate specificity and subcellular localization (Fig. 1D)[27].
MINDYs represent a relatively newly discovered family of deubiquitinating enzymes that contain an uncharacterized catalytic domain. Their crystal structure reveals a unique protein folding that shows no homology with any known deubiquitinating enzymes, but MINDYs still belong to the superfamily of cysteine proteases[28]. The catalytic core domain of MINDY-1 resembles a light bulb consisting of two subdomains, a central ''bulb'' subdomain that sits on a ''stalk'' subdomain, which resembles the base of the bulb (Fig. 1E)[28].
Unlike other families of deubiquitinating enzymes, the JAMM family of deubiquitinating enzymes is a class of metalloproteases with a metal catalytic center, which typically contains a zinc ion that plays a key role in the catalytic reaction. JAMMs are typically composed of several distinct structural domains, including an MPN domain that is involved in metal ion binding and the catalytic reaction, as well as a JAMM domain that participates in substrate recognition and binding. JAMMs utilize a zinc ion in the metal catalytic center to polarize water molecules, which then attack the carbonyl carbon of the ubiquitin molecule, leading to the cleavage of the isopeptide bond between ubiquitin and the target protein (Fig. 1F).
Structural representations of DUB catalytic domains across six families. Active site cysteine (for OTUs, USPs, UCHs, MJDs, MINDYs) or zinc (for JAMMs) is highlighted. Representative structures are shown for:(A) USP family (USP7, PDB: 1NB8) (B) OTU family (OTUB1, PDB: 2ZFY) (C) UCH family (UCH-L3, PDB: 1UCH) (D) MJD family (Ataxin-3, PDB: 1YZB) (E) MINDY family (MINDY1, PDB: 5JKN) (F) JAMM family (STAMBPL1, PDB: 2ZNR).
Function of the USP Family and Research Advances in Hepatocellular Carcinoma
The USP family is currently the most extensively studied family. This article will proceed to introduce the research progress of USPs in HCC.
USP1
The best-characterized function of USP1 is as a regulator of several important steps in the DNA damage response[29]. In addition, recent evidence suggests that USP1 may also contribute to regulate differentiation in specific cellular contexts[30]. Current studies have found that USP1 is overexpressed in liver cancer tissue and associated with poor survival in HCC patients[31, 32]. And it has been proven that ML-323 (a USP1 inhibitor) inhibits the growth of HCC cells and induces G1 phase cell cycle arrest by regulating the expression of cell cycle proteins. Additionally, treatment of HCC cells with ML-323 results in the accumulation of ubiquitinated proteins, induces endoplasmic reticulum stress, and triggers Noxa-dependent apoptosis regulated by Activating Transcription Factor 4[31]. The Hippo signaling pathway has been proven to be a significant suppressive pathway in the development of HCC[33]. A study's DUB siRNA screening showed that USP1 is a key regulator of Hippo signaling activity. The mechanistic research of the article proves that USP1 interacts with the WW domain of TAZ, thereby enhancing the stability of TAZ through the inhibition of K11-linked polyubiquitination[32]. In addition, USP1 induces mitochondrial fission by enhancing phosphorylation of Drp1 at Ser616 via deubiquitination and stabilization of CDK5, thereby enhancing the metabolic reprogramming of HCC and affecting the occurrence and development of HCC[34].
USP2
The dysregulation of USP2 expression is closely related to the progression of HCC. Studies have found that compared with adjacent non-tumor tissue or normal liver tissue, the protein and mRNA levels of USP2 in HCC tissue are significantly altered[35]. Among the different isoforms of USP2, USP2b is dominant in normal liver tissue but is markedly down-regulated in HCC[36]. In contrast, USP2a is up-regulated in HCC, and its increased expression is positively correlated with poor patient prognosis[35, 37]. The data of Chongqing Medical University indicate that USP2a can promote HCC progression via deubiquitination and stabilization of RAB1A[37]. Non-alcoholic steatohepatitis (NASH) is also one of the causes of HCC. USP2 is also believed to be involved in lipid metabolism, further regulating the progression of HCC[38].
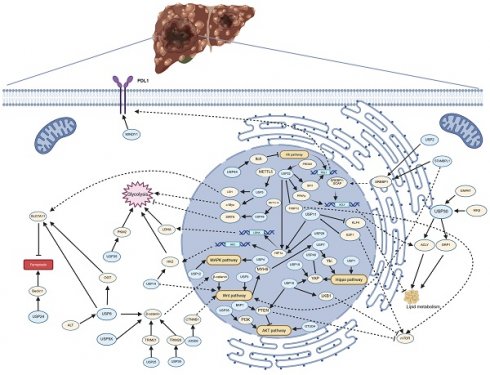
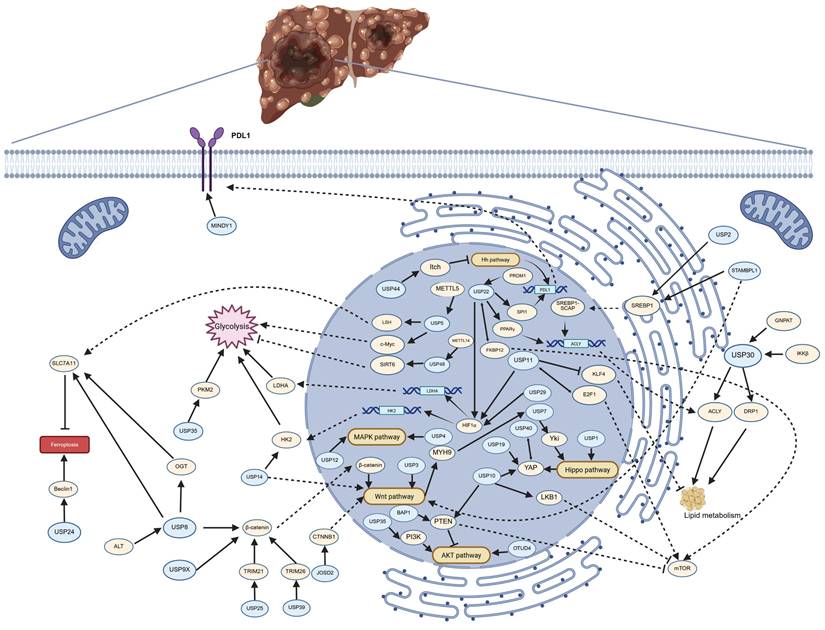
The pathways regulated by deubiquitinating enzymes (DUBs) in the progression of hepatocellular carcinoma (HCC). The figure illustrates the pathways by which DUBs regulate glycolysis, lipid metabolism, ferroptosis, PDL1 synthesis, and multiple signaling pathways in HCC. Arrows (→) indicate promotion or activation, while blocked lines (┫) denote inhibition or suppression. Created in https://BioRender.com.
USP3
USP3 protein is upregulated in HCC tissues, but its upregulation level is not related to clinical pathological staging. Knocking down USP3 inhibits the growth of HCC cells and induces apoptosis in HCC cells. It also inhibits the growth of drug-resistant HCC cells. After USP3 knockdown, the levels and activity of β-catenin in HCC cells decrease and can be reversed by Wnt activators (lithium)[39].
USP4
A study indicated that Cyclophilin A (CypA) is an important molecule in USP4-mediated oncogenic activity in HCC. USP4 interacts with CypA and inhibits CypA degradation through deubiquitination in HCC cells, thereby enhancing the stability of CypA. Subsequently, the USP4/CypA complex activates the MAPK signaling pathway and prevents the phosphorylation of CrkII, thereby promoting the progression of HCC[40]. USP4 also interacts with TGFR-1 and deubiquitinates it, thereby activating the TGF-β signaling pathway and subsequently inducing epithelial-mesenchymal transition (EMT) in HCC cells[41]. Another study has elaborated on a different pathway affecting the EMT in HCC. LOXL3 interacts with the snail family transcriptional repressor 1 (Snail1), and Snail1 binds to the promoter of USP4, while USP4 deubiquitinates and stabilizes the expression of Snail1, thus forming a regulatory loop[42].
USP5
USP5 is overexpressed in highly malignant HCC and is positively correlated with the expression of SLUG. Downregulation of USP5 inhibits the deubiquitination of SLUG, thereby suppressing the proliferation, metastasis, and invasion of HCC cells, while the overexpression of USP5 promotes the stability of SLUG and EMT both in vitro and in vivo[43]. USP5-mediated deubiquitination of LSH suppresses ferroptosis in liver cancer cells by upregulating SLC7A11, thereby promoting the occurrence of HCC. Moreover, the USP5 inhibitor can inhibit the DUB activity of USP5 towards LSH, thereby suppressing the progression of HCC[44]. The P14(ARF) -P53 signaling was activated by USP5 knockdown in HCC cells. And Hpn induces apoptosis by inhibiting USP5 expression and activating the p14(ARF)-P53 signaling pathway[45]. CREB1/P300 serves as a key transcriptional regulator of METTL5, which controls the translation process of USP5, thereby regulating the ubiquitination of c-Myc and subsequently modulating the glycolytic metabolism in HCC[46].
USP7
USP7 expression in HCC tissues is significantly higher than in the surrounding non-tumor tissues, and it is associated with shorter overall survival and a higher cumulative recurrence rate of HCC[47-49]. USP7 modulates the Hippo pathway by regulating the ubiquitination status of Yes-associated protein (YAP). The transcriptional coactivator Yorkie, a key effector of the Hippo pathway, has its binding with USP7 diminished upon pathway activation, yet this activation promotes the phosphorylation of Yorkie[50]. Transmembrane protein 43 (TMEM43) plays a significant role in cancer, with a study revealing its high expression in HCC. Knockdown of TMEM43 has been shown to inhibit tumor progression. The research further discovered that USP7 regulates the ubiquitination levels of TMEM43 through deubiquitination, thereby affecting the progression of HCC[51]. Yin-yang-1 (YY1) is a DNA-binding protein with a zinc finger structure and is an important transcription factor that regulates the proliferation, migration, and EMT of tumor cells. USP7 can prevent the ubiquitin-dependent degradation of YY1, stabilizing the expression of YY1, thereby promoting the proliferation, migration, and EMT of HCC cells[52]. Mass spectrometry analysis revealed BTF3 as a potential substrate of USP7. Consequently, Hu and colleagues conducted a series of studies confirming the interaction between USP7 and BTF3, and that USP7 downregulates the ubiquitination levels of BTF3, thereby stabilizing BTF3 protein. Overexpression of BTF3 partially restored the inhibitory effects of USP7 depletion on the malignant phenotype and stemness characteristics of HCC cells[53]. USP7 can also form complexes with certain target proteins, thereby regulating downstream pathways. USP7 forms a complex with thyroid hormone receptor-interacting protein 12 (TRIP12), thereby stabilizing TRIP12. TRIP12, an E3 ubiquitin ligase, further induces the ubiquitination of constitutive p14(ARF) and promotes the progression of HCC[48]. TRIM27 is also an E3 ubiquitin ligase that is stabilized through binding to USP7. The USP7-TRIM27 complex regulates tumor progression in HCC by activating STAT3[54]. USP7 also has upstream regulatory factors. MiR-205 may negatively regulate the protein levels of USP7 in HCC cells by targeting the 3'-untranslated region (3'-UTR) of USP7, thereby modulating the p53 signaling pathway and cellular proliferation levels[47]. ENKUR inhibits the nuclear translocation of MYH9 by binding to β-catenin, reducing the expression of MYH9. The low expression of MYH9 suppresses the recruitment of the deubiquitinating enzyme USP7, promotes the degradation of c-Myc, and inhibits the proliferation, metastasis, and sorafenib resistance of HCC[55]. The results of qRT-PCR showed that METTL3 and USP7 are both highly expressed in HCC tissues and cell lines, and they are positively correlated. Further experiments indicated that METTL3 may regulate the expression of USP7 through m6A methylation, promoting the invasion, migration, and proliferation of HCC cells[56]. Another study demonstrated that flap endonuclease 1 (FEN1) can prevent the ubiquitination and degradation of mouse double minute 2 (MDM2) by recruiting USP7, thereby inactivating the p53 signaling pathway[57]. In addition to the specific USP7 inhibitor P22077, arsenic can also inhibit the expression of USP7, playing a role in inhibiting tumor development[58, 59]. However, arsenic is cytotoxic, and targeted delivery of arsenic drugs to liver tumors may be one of the research directions for the future.
USP8
USP8 was initially discovered in breast cancer and is associated with the regulation of the epidermal growth factor receptor (EGFR)[60]. Subsequent studies have found that the upregulation or mutation of USP8 stabilizes numerous oncogenes or proto-oncogenes, thereby activating multiple signaling pathways, leading to cancer progression and survival[61]. Compared with normal liver tissue, USP8 is significantly upregulated in liver cancer tissue[62-64]. A study found that inhibiting USP8 can lead to the suppression of sensitive and resistant to doxorubicin HCC cell growth, as well as induce apoptosis. Meanwhile, the inhibition of USP8 resulted in a 90% reduction in the levels of various receptor tyrosine kinases (RTKs), including EGFR and c-Met[62]. Our research group conducted some studies and found that USP8 regulates ferroptosis in HCC. One of our studies indicated that USP8 stabilizes β-catenin through deubiquitination, thereby promoting the growth, invasion, tumor stem-like characteristics, and ferroptosis resistance of HCC[63]. Our other study found that targeting USP8 can inhibit the stability of O-GlcNAc transferase (OGT), subsequently suppressing the O-GlcNAcylation of SLC7A11, ultimately promoting ferroptosis in HCC[65]. Our research has also found that USP8 stabilizes OGT through deubiquitination, thereby promoting the progression of intrahepatic cholangiocarcinoma (ICC). At the same time, the absence of USP8 enhances the response of ICC to pemigatinib[64].
However, another study has presented an opposing conclusion, suggesting that USP8 inhibits the progression of liver cancer by regulating TRAF6-mediated signaling to activate NF-κB and induce autophagy[66]. The researchers incorporated lipopolysaccharide (LPS), a key component of Gram-negative bacteria and potent TLR4 activator known to modulate the tumor immune microenvironment, in their investigation of USP8-mediated hepatocellular carcinoma regulation. This experimental design focusing on immune microenvironment mechanisms may account for the divergent conclusions observed in this study.
In tumor tissues of other organ systems (including lung, gastric, and pancreatic cancers), USP8 functions as a tumor suppressor. Targeted modulation of USP8 not only inhibits tumor progression but also enhances sensitivity to anti-tumor agents.
USP9X
USP9X is highly expressed in liver cancer tissues and is an independent risk factor for poor prognosis. Knockdown of USP9X significantly inhibits the proliferation of HCC cells. Mechanistically, USP9X regulates the expression of β-catenin to promote HCC cell proliferation[67]. In HCC cell lines, miR-26b targets the 3'UTR of USP9X, thereby affecting EMT through the Smad4 and TGF-β signaling pathways[68]. In HCC, the expression level of miR-26b is reduced and negatively correlated with HCC grading[69]. Another study has shown that in HCC, miR-26b inhibits USP9X-mediated p53 deubiquitination, enhancing the sensitivity of HCC cells to doxorubicin[70]. Long non-coding LNC473 can recruit the deubiquitinating enzyme USP9X to inhibit the ubiquitination level of survivin, thereby increasing the expression of survivin, which in turn promotes the progression of HCC[71].
USP10
Current research has demonstrated that USP10 regulates the development of HCC, but the mechanisms are complex. Regarding its role as an oncogene or a tumor suppressor, some studies have reached contradictory conclusions. An article published by Dalian Medical University in 2018 stated that USP10 inhibits the progression of HCC by suppressing the activation of mTOR[72]. The team continued their research and found that in HCC, USP10 removes the ubiquitination of LKB1, thereby inhibiting the activation of mTOR[73]. The continuous activation of Smad4 and TGF-β is closely related to the metastasis of advanced HCC, while USP10 can maintain the protein levels of Smad4 and activate TGF-β signaling, thereby promoting the metastasis of HCC[74]. The small molecule inhibitor of USP10 can suppress the development of HCC by inhibiting the deubiquitination of YAP, thereby promoting the degradation of YAP and downregulating P53[75]. Growing evidence suggests that chronic stress, such as depression, is associated with poor prognosis in cancer patients, and the upregulation of PLAGL2 is related to chronic stress[76-78]. USP10 forms a signaling loop with PLAGL2, and stress-induced secretion of adrenaline activates this loop, thereby promoting the progression of HCC[79].
USP11
A study evaluated the expression of USP11 in 71 clinical samples of HCC by using immunohistochemical and analyzed the relationship between USP11 expression and clinical pathological characteristics and overall survival time. The study has found that the expression level of USP11 in tumor tissue is higher than that in non-tumor tissue, and is associated with vascular invasion, differentiation, number of tumors, recurrence rate, and shorter overall survival time[80]. The research team further discovered that USP11 interacts with nuclear factor 90 (NF90), promoting its deubiquitination, thereby stabilizing its function in HCC cells[81]. A study from Jinan University has reported that USP11 interacts with E2F1, maintaining the stability of E2F1 protein by deubiquitination, while E2F1 regulates the expression of USP11 at the transcriptional level, thus forming a positive feedback loop between E2F1/USP11 that promotes the proliferation and migration of HCC cells[82]. Another study demonstrated that USP11 promotes EMT and metastasis in HCC via eEF1A1/SP1/HGF dependent-EMT[83]. USP11 has been reported to regulate the metabolism of liver cancer cells. Elevated levels of USP11 have been detected in both in vitro models of hepatic steatosis and in public clinical data from patients with non-alcoholic fatty liver disease and HCC[84]. Under in vitro steatotic conditions, the absence of USP11 results in reduced lipid content and downregulation of genes involved in fatty acid metabolism[84]. Furthermore, USP11 modulates glucose metabolism in HCC through the HIF1α/LDHA axis, thereby affecting proliferation and metastasis[85].
USP12
Compared with adjacent normal tissues, USP12 is specifically upregulated in liver cancer tissues. After knocking down USP12, the cell cycle protein-dependent kinase 1/cyclinB1 axis was activated, inducing G2/M arrest, and triggering apoptosis through the p38/ MAPK pathway[86]. Another study found that USP12 can remove the ubiquitination of c-Myc, thereby regulating the progression of HCC[87].
USP13
Nanjing medical university investigated the function of USP13 in HCC, they found that USP13 was significantly upregulated in both of primary HCC tumor tissues and cell lines. And mechanism studies indicated that knocking down USP13 can significantly downregulate the expression levels of c-Myc and overexpression of c-Myc could significantly attenuate the effects of shUSP13 on HCC cell growth inhibition[88]. However, the study did not clarify whether USP13 regulates c-myc through the deubiquitination pathway or other pathways. The activation of the Toll-like receptor 4/Myeloid differentiation primary response gene 88/Nuclear factor-kappa B (TLR4/MyD88/NF-κB) pathway is involved in the occurrence and development of HCC. Moreover, studies have shown that TLR4 in melanoma is regulated by the ubiquitin-proteasome system. Gao et al. found that USP13 inhibits ubiquitin-mediated TLR4 degradation, thereby enhancing the proliferation, EMT, migration, and invasion of HCC[89].
USP14
USP14 exhibits significantly higher expression in tumor tissues of HCC patients compared to para-cancerous and normal liver tissues. Moreover, patients with higher USP14 expression have a markedly poorer prognosis after surgery than those with lower levels of USP14 expression[90]. Researchers have begun to explore the mechanisms by which USP14 influences the occurrence and development of HCC. A study from Wuhan University has found that the selective USP14 inhibitor, b-AP15, induces cytotoxicity in HCC cells by increasing endoplasmic reticulum (ER) stress and inhibiting the Wnt/Notch1 signaling pathway[91]. This suggests that USP14 may affect the tumor activity of HCC by activating the Wnt/Notch1 signaling pathway. China Medical University demonstrated that USP14 is involved in the maintenance of HIF1α stability to activate HIF1α-induced transactivation via its deubiquitinase activity. And USP14 depletion or its specific inhibitor treatment decreased cell proliferation, invasion, migration, and Vascular Mimicry formation even under hypoxia conditions in HCC cell lines[92]. Another study of Affiliated Hospital of Nantong University indicated that knocking down USP14 significantly inhibits the proliferation, invasion, and metastasis of liver cancer cells, promotes apoptosis, and simultaneously suppresses the expression of hexokinase 2 (HK2). Further experimental research has found that USP14 affects EMT through the HK2/AKT/P62 axis. At the same time, since HK2 is a downstream target of USP14, USP14 can mediate glucose metabolism in HCC cells[93]. USP14 also plays a significant role in the resistance mechanism of HCC to lenvatinib; silencing USP14 can markedly reduce lenvatinib resistance both in vitro and in vivo. The specific mechanism involves USP14 removing the ubiquitination of Calcium and Integrin-Binding Protein 1 (CIB1), thereby promoting the P21-activated kinase 1 (PAK1)-ERK1/2 signaling axis[94].
USP15
Current literature indicates that USP15 promotes the progression of HCC and is associated with the prognosis of HCC patients. A system biology analysis suggests that USP15 may suppress tumorigenesis in HCC by regulating gene expression, cell cycle, and signal transduction pathways involved in DNA repair[95]. After silencing the expression of USP15 in HCC cells, cell proliferation is inhibited and apoptosis is induced[96]. However, there is no further mechanistic research.
USP16
Hepatitis B virus X protein (HBx) has been shown to accelerate HCC progression by promoting tumor growth and metastasis[97-99]. Carboxyl-terminal truncated HBx (Ct-HBx) proteins are frequently present in HCC tumour tissues, but not in non-tumorous tissues[100, 101]. The expression of USP16 is significantly suppressed by Ct-HBX, thereby inhibiting tumor proliferation. Conversely, ectopic expression of USP16 suppresses the tumor-promoting activity of Ct-HBx[102].
USP18
Currently, only one article has elaborated on the role of USP18 in HCC. The article points out that the expression of USP18 in HCC tissue is higher than that in adjacent non-cancerous tissue, and it is even more highly expressed in HCC associated with hepatitis B virus infection. Moreover, it may promote the growth of HCC cells by stabilizing the BCL2L2 protein[103].
USP19
USP19 is upregulated in HCC tissues and is associated with poor prognosis. In vivo and in vitro experiments have also demonstrated that USP19 promotes the proliferation and migration of HCC. USP19 is a specific deubiquitinating enzyme for YAP, and knockdown of USP19 can reduce the expression of YAP protein and its target genes[104].
USP20
Our research group has identified that USP20 plays a role in inhibiting ferroptosis in HCC cells and enhancing their resistance to oxaliplatin. This effect is mediated by USP20's ability to remove K48-linked polyubiquitination at lysine residues K30 and K37 of the SLC7A11 protein, thereby stabilizing SLC7A11. The stabilization of SLC7A11 subsequently suppresses ferroptosis, which contributes to the development of oxaliplatin resistance. And the phosphorylation of USP20 at Ser132 and Ser368 requires the activation of ATR induced by DNA damage[105].
USP21
Li's team conducted an analysis of multiple HCC datasets and discovered that the USP21 gene, which encodes a member of the ubiquitin-specific protease family, is highly amplified and overexpressed in HCC. This upregulation is significantly associated with poor clinical prognosis. Inhibition of USP21 in HCC cell lines was found to reduce tumorigenic properties, while ectopic expression enhanced oncogenicity. Further mechanistic studies revealed that USP21 stabilizes MEK2 by removing its ubiquitination, thereby activating the ERK signaling pathway[106]. Circular RNAs (circRNAs) play a pivotal role in the oncogenesis of HCC, and the expression of USP21 is regulated by hsa_circ_0039053. Studies have shown that hsa_circ_0039053 can interact with miR-637 to promote the expression of USP21, thereby facilitating the progression of HCC[107].
USP22
A study of Ling found that USP22 promotes hypoxia-induced stemness and glycolysis in HCC cells by deubiquitination and stabilization of HIF1α. Moreover, USP22 and HIF1α formed a positive feedback loop and facilitating the development of HCC[108]. Researchers at the First People's Hospital of Hangzhou have identified another positive feedback loop involving USP22. In this loop, USP22 promotes the activation of mammalian target of rapamycin complex 1 (mTORC1) by deubiquitinating FK506-binding protein 12 (FKBP12), and the activated mTORC1 further stabilizes USP22 by inhibiting autophagic degradation. This discovery highlights the complex regulatory network of USP22 in the context of HCC and suggests that targeting this feedback loop could be a potential therapeutic strategy for HCC[109]. Elevated de novo lipogenesis is considered a crucial factor in the development of HCC[38], and USP22 has been shown to promote the de novo synthesis of fatty acids and HCC tumorigenesis. The specific mechanism involves USP22 stabilizing peroxisome proliferator-activated receptor gamma (PPARγ) through deubiquitination, which in turn increases the expression of acetyl-CoA carboxylase (ACC) and ATP citrate lyase (ACLY)[110]. USP22 has been implicated in the regulation of the immunological microenvironment in HCC. The specific mechanism involves PRDM1 enhancing the transcription of USP22, which then reduces the degradation of SPI1 protein through deubiquitination, thereby enhancing the transcription of Programmed Cell Death Ligand 1 (PDL1). This ultimately leads to the exhaustion of infiltrating CD8+ T cells[111].
USP24
The function of USP24 in tumors is intricate, with various studies presenting contrasting views on whether USP24 serves as a promoter or suppressor of cancer. In the context of neuroblastoma, a deficiency in USP24 is noted, and the overexpression of USP24 has been demonstrated to suppress the proliferation of neuroblastoma cells[112]. Conversely, in lung and gastric cancers, USP24 behaves as an oncogenic factor[113, 114]. Similar to these findings, research on HCC has also produced inconsistent conclusions. Zhou's research discovered that USP24 binds to Tumor Necrosis Factor Receptor-Associated Factor 2 (TRAF2), inhibiting its degradation and thereby promoting the accumulation of TRAF2. The upregulation of TRAF2 activates the AKT/NF-κB signaling pathway, which enhances the survival of HCC cells. Additionally, USP24 expression is positively correlated with the expression of PDL1 in HCC[115]. Hu et al. discovered that in sorafenib-resistant HCC cells, miR-21-5p is upregulated, and USP24 is a downstream target of miR-21-5p. Further mechanistic exploration revealed that the upregulation of miR-21-5p increases USP24, which in turn removes the ubiquitination level of SIRT7 through USP24, thereby inhibiting cellular autophagy[116]. Researchers at Zhengzhou University believe that USP24 is a tumor suppressor. They have discovered that USP24 can stabilize Beclin1, one of the key regulators of autophagy, thereby promoting tumor cell autophagy and ferroptosis, and increasing the susceptibility of HCC to sorafenib[117].
USP25
The USP25 protein plays a crucial role in the development of various types of cancer. For instance, in pancreatic cancer, USP25 stabilizes HIF1α through deubiquitination, thereby promoting glycolysis in cancer cells and consequently driving tumor progression[118]. In tissues and cell lines of HCC, USP25 is also highly expressed. Research has confirmed that in HCC, USP25 interacts with TRIM21 to regulate EMT and the Wnt/β-catenin signaling pathway. These findings reveal the significant role of USP25 in the development of liver cancer and may provide new targets for future therapeutic strategies[119].
USP26
USP26 plays a regulatory role in the development and progression of colorectal, cervical, breast, and liver cancers. In HBV-positive HCC, USP26 is strongly induced, and its levels are associated with poor prognosis. Mechanistically, the HBV-encoded protein HBx binds to the promoter and induces the production of USP26, which in turn stabilizes SIRT1 through deubiquitination, thereby promoting cell proliferation, inhibiting apoptosis, and accelerating the occurrence of HCC[120].
USP27
Cyclin E is frequently dysregulated in tumor cells and this may contribute to the development of various types of cancers[121-124], and USP27 promoted Cyclin E stability by negatively regulating its ubiquitination. In addition, suppression of USP27 expression results in the inhibition of the growth, migration, and invasion of hepatocellular carcinoma, and the absence of USP27 makes Hep3B cells sensitive to 5-FU-induced apoptosis, indicating that USP27 plays a role in the drug resistance of HCC[125]. USP27 can also remove the ubiquitination of the histone methyltransferase SETD3, thereby enhancing its stability and promoting the growth of tumor cells[126].
USP28
USP28 is overexpressed in the majority of tumors and holds prognostic value across various cancer types. Moreover, there is a significant correlation between USP28 and immune modulatory factors, clinical staging, checkpoint inhibitor responses, MSI (microsatellite instability), TMB (tumor mutational burden), CNV (copy number variation), MMR (mismatch repair) defects, and DNA methylation[127]. Among these, patients with liver cancer and high expression of USP28 generally have a poorer prognosis[128]. In HCC, it has been reported in literature that USP28 regulates tumor progression by removing the ubiquitination of c-Myc[128, 129]. USP28 is regulated by microRNAs and circular RNAs; miR-363-3p and miR-216b can inhibit the expression of USP28, while circMED27 upregulates USP28[128-130]. However, no further targets of USP28 have been identified in HCC.
USP29
Aberrant expression of HIF1α and increased aerobic glycolysis metabolism are drivers of resistance to Sorafenib[131, 132]. USP29 is a key deubiquitinating enzyme of HIF1α, which directly deubiquitinates and stabilizes HIF1α, thereby promoting its transcriptional activity. HK2 is one of the transcriptional targets of HIF1α and is also a key enzyme in aerobic glycolysis. Knocking down USP29 reduces the level of aerobic glycolysis in sorafenib-resistant HCC cells, restoring their sensitivity to sorafenib[133].
USP30
The GNPAT, which encodes the enzyme glyceronephosphate O-acyltransferase, exhibits amplification, upregulation, and a strong correlation with poor clinical outcomes in patients with HCC. USP30 is recruited by GNPAT, which encodes the enzyme glyceronephosphate O-acyltransferase. Subsequently, USP30 deubiquitylates and stabilizes dynamin-related protein 1 (DRP1), thereby playing a crucial role in the regulation of mitochondrial morphology, lipid metabolism, and the initiation of HCC[134]. USP30 is prominently expressed in HCC that develops in mice subjected to a high-fat diet[135]. This indicates that USP30 is also involved in the regulation of lipid metabolism in HCC cells. IKKβ phosphorylates and stabilizes USP30, which in turn promotes the deubiquitination of ACLY and fatty acid synthase (FASN) by USP30. IKKβ also directly phosphorylates ACLY, facilitating the interaction between USP30 and ACLY, as well as the deubiquitination of the latter[135].
USP32
Several studies have found that USP32 can regulate the growth and development of different tumors through the catalytic activity of deubiquitinating enzymes. Analysis of the TCGA database has yielded the expression patterns of USP32 in various malignant tumors. Among them, six types of cancer, including cholangiocarcinoma, esophageal cancer, head and neck squamous cell carcinoma, hepatocellular carcinoma, and gastric adenocarcinoma, show elevated USP32 mRNA expression levels[136]. Xiu et al. conducted CCK8, migration assays, and clonogenic assays on HCC cell lines and established a nude mouse model of HCC to verify the carcinogenic role of USP32. The experiments demonstrated that the knockdown of USP32 can inhibit the proliferation, colony formation, and migration of HCC cells, as well as suppress tumor growth in vivo[137]. In gastric cancer, USP32 regulates the expression of SMAD2 in a ubiquitin proteasome-dependent manner[138]. Additionally, USP32 promotes the progression of non-small cell lung cancer by deubiquitinating BAG3 and activating the RAF-MEK-ERK signaling pathway[139]. However, the specific mechanisms by which USP32 regulates the development of HCC have not been further explored.
USP33
Gan et al. first discovered the abnormal increase in USP33 expression in HCC tissues, and USP33 may serve as a prognostic biomarker for HCC patients. Furthermore, as a deubiquitinating enzyme for SP1, USP33 participates in the invasion and metastasis of HCC by activating the SP1/c-Met axis[140].
USP35
Immunohistochemical analysis comparing the expression of USP35 in HCC tumors and peritumoral tissues revealed a significant upregulation of USP35 in tumor tissues. Prognostic statistical analysis indicated that HCC patients with higher USP35 expression had significantly shorter overall survival times[141]. Mechanistically, USP35 promotes the development of HCC by deubiquitinating and stabilizing PKM2 and ABHD17C proteins, while also activating the PI3K/AKT signaling pathway[141, 142].
USP39
USP39 plays a regulatory role in the proliferation of HCC cells. Upon USP39 knockdown, cell proliferation is significantly suppressed, and apoptosis is induced[143, 144]. A study has observed that USP39 stabilizes the protein SP1 by promoting its deubiquitination, and USP39 facilitates the proliferation of HCC cells in an SP1-dependent manner[145]. Zinc-finger E-box-binding homeobox 1 (ZEB1) and β-catenin are associated with the proliferation and metastasis of HCC, and their protein levels are regulated by the ubiquitination system. Specifically, the E3 ubiquitin ligase TRIM26 suppresses HCC by ubiquitinating ZEB1 and β-catenin, while USP39 removes the ubiquitination. Additionally, USP39 inhibits the maturation and splicing of TRIM26 pre-mRNA, thereby further increasing ZEB1 and β-catenin levels[146, 147]. Knockdown of USP39 leads to the downregulation of the transcription factor Forkhead box M1 (FoxM1). The expression of FoxM1 target genes, including Polo-like kinase 1, Cyclin B1, and Centromere protein A, also decreases following USP39 knockout. These results suggest that USP39 may regulate the progression of HCC by modulating the pre-mRNA splicing of FoxM1[148]. A study has demonstrated that USP39 can be acetylated by the histone acetyltransferase MYST1, which leads to its degradation by the proteasome. Conversely, the lysine deacetylase sirtuin 7 (SIRT7) removes the acetylation of USP39, promoting its stability and consequently accelerating the proliferation and tumorigenesis of HCC cells both in vitro and in vivo[149]. USP39 is recruited by dynein axonemal assembly factor 5 (DNAAF5), and stabilize phosphofructokinase L (PFKL) through the deubiquitination pathway, thereby promoting HCC proliferation and resistance to sorafenib[150].
USP40
USP40 is upregulated in HCC tissues and predicts poor prognosis for HCC patients. Knockdown of USP40 inhibits the proliferation, migration, and stemness of HCC cells. Mechanistically, USP40 interacts with Claudin1 and suppresses its polyubiquitination, thereby stabilizing Claudin1 protein[151]. YAP is a crucial driver of HCC progression, and the ubiquitin-proteasome system governs its levels. USP40 interacts with YAP to remove the lysine 48 (K48)-linked polyubiquitination at K252 and K315 sites, thereby maintaining YAP stability. In turn, YAP transcriptionally activates the expression of USP40 in HCC cells. This reciprocal regulation forms a positive feedback loop between USP40 and YAP, which contributes to the progression of HCC[152].
USP44
In various types of cancer, the role of USP44 in tumorigenesis and development varies. In breast and prostate cancer, USP44 acts as an oncogenic factor. In contrast, in pancreatic cancer, thyroid cancer, colorectal cancer, and liver cancer, USP44 inhibits tumor progression. The expression of USP44 is diminished in HCC tissues, and clinical pathological analysis further indicates that low levels of USP44 expression are associated with poorer prognosis and advanced tumor stages in HCC patients[153]. A study investigating the mechanism by which USP44 suppresses tumors in HCC found that USP44 inhibits PDL1 expression by stabilizing Itch, which downregulates the Hedgehog (Hh) signaling pathway. Additionally, the Gli1 inhibitor GANT61 was found to synergize with anti-PDL1 therapy, suggesting a potential combined therapeutic approach for HCC treatment[154].
USP46
USP46 is found to be downregulated in HCC tissues, and low levels of USP46 are correlated with a poor prognosis for HCC patients. USP46 stabilizes MST1 protein by directly binding to it and reducing its ubiquitination, and MST1 antagonizes the expression of YAP, thereby inhibiting the development of HCC[155]. A series of experiments from Nanchang University have demonstrated that USP46 is a target gene of miR-27a-3p. MiR-27a-3p promotes the proliferation of cancer cells and accelerates the occurrence and development of HCC by inhibiting the expression of USP46[156].
USP48
Pyroptosis is a form of programmed cell death characterized by the activation of inflammatory caspases and the cleavage of gasdermin proteins. It can suppress tumor development and induce antitumor immunity, making the activation of pyroptosis a potential therapeutic strategy for cancer. USP48, by stabilizing Gasdermin E (GSDME), promotes pyroptosis and enhances the therapeutic efficacy of programmed cell death protein 1 inhibitors in mouse tumor models[157]. Additionally, in mouse liver tumor induced by diethylnitrosamine and human HCC, the expression of USP48 is downregulated. In HCC, USP48 stabilizes SIRT6 by deubiquitination, which hinders metabolic reprogramming and thus inhibits HCC tumorigenesis. Furthermore, the m6A modification induced by methyltransferase-like 14 (Mettl14) is involved in the regulation of USP48 in HCC by maintaining the stability of USP48 mRNA[158].
USP49
USP49 is a downstream molecule of HLNC1, which is a long non-coding RNA. HLNC1 interacts with USP49 and significantly destabilizes it, thereby promoting the viability and migration of HCC cells[159]. USP49 is also a tumor suppressor in pancreatic cancer[160]. However, in gastric cancer, USP49 stabilizes YAP1 through deubiquitination, and YAP1 in turn promotes the transcription of USP49, forming a positive feedback loop that drives the malignant progression of gastric cancer[161].
USP53
The antitumor effect of USP53 has been confirmed in lung and breast cancer, and USP53 expression is downregulated in HCC tissues as well as HCC cell lines. Furthermore, the ectopic expression of USP53 inhibits the proliferation, migration, and invasion of HCC cells and induces apoptosis in HCC cells. Immunoprecipitation (CO-IP) assays and mass spectrometry analysis identified cytochrome c (CYCS) as a target protein of USP53, and USP53 stabilizes CYCS to induce apoptosis in HCC cells[162].
CYLD
CYLD (Cylindromatosis) contains a USP catalytic domain and belongs to the USP family, capable of cleaving K63 and M1 linked polyubiquitin chains. Initially associated with the rare disease familial cylindromatosis, subsequent research has found that CYLD is involved in the regulation of the occurrence and development of various tumors. Compared to the surrounding non-malignant tissues, the protein and mRNA levels of CYLD in HCC are both lower, and it is believed that downregulating CYLD in HCC cell lines increases the resistance of HCC cells to treatment with doxorubicin, 5-fluorouracil, and cisplatin. Further findings show that the downregulation of CYLD reduces the apoptosis induced by tumor necrosis factor-α, while also leading to the degradation of the NF-κB inhibitor IκB-α, thereby enhancing NF-κB activity in HCC cells[163]. And Pannem's research found that CYLD negatively controls the expression of c-MYC in HCC through the JNK-dependent signaling pathway, thereby regulating the proliferation of HCC cells[164]. Existing research suggests that multiple microRNAs regulate the expression of CYLD, such as miR-362[165], miR-501[166, 167], and miR-922[168]. Lu's research team found in the liver cancer cell line HepG2 that SOX2 increases the expression of EGFR, and EGFR upregulates miR-222-5p, leading to the downregulation of CYLD[169].
Function of the OTU Deubiquitinase Family and Research Advances in Hepatocellular Carcinoma
Current research indicates that OTU deubiquitinating family members, including OTUB1, OTUD3, OTUD4, OTUD5, and TRABID, are associated with HCC. This article will further elaborate on the relationship between these OTUs and the occurrence and progression of HCC.
OTUB1
Chen's research team found that the expression level of OTUB1 is significantly correlated with clinical pathological parameters such as TNM staging, histological staging, metastasis/recurrence by analyzing 115 clinical HCC tissue samples. Survival analysis showed that patients with high expression of OTUB1 had a lower overall survival time compared to those with low expression of OTUB1. In vitro experiments revealed that knocking down OTUB1 inhibited the proliferation, migration, and invasion capabilities of HCC cells[170]. Overexpression of OTUB1 in HCC could be a novel, effective, and supplementary biomarker for HCC. However, the specific mechanisms by which OTUB1 regulates the progression of HCC still require further investigation.
OTUD3
OTUD3 is also significantly overexpressed in HCC. Furthermore, mass spectrometry analysis revealed that α-actinin 4 (ACTN4) is a downstream target of OTUD3, and the protein level of ACTN4 is significantly correlated with the expression of OTUD3. Further studies found that OTUD3 directly binds to ACTN4, leading to its deubiquitination and stabilization. Both in vitro and in vivo experiments confirmed that ACTN4 is crucial for OTUD3-mediated HCC proliferation and metastasis[171].
Another article, however, revealed that OTUD3 acts as a tumor suppressor in hepatocellular carcinoma while simultaneously enhancing the sensitivity of liver cancer cells to sorafenib treatment. The study found that OTUD3 is a deubiquitinating enzyme for the eukaryotic initiation factor 2α (eIF2α), which can antagonize the integrated stress response (ISR) and inhibit liver cancer[172]. ISR is a response activated in response to intrinsic and extrinsic stimuli and plays a role in tumor progression and drug resistance[173-175]. Moreover, the activation of ISR due to reduced OTUD3 expression confers resistance of liver cancer cells to sorafenib, and the combined use of the ISR inhibitor ISRIB significantly enhances the sensitivity of liver cancer cells to sorafenib[172].
OTUD3, a deubiquitinating enzyme, demonstrates functional pleiotropy in tumorigenesis through its regulation of multiple substrate targets. While its roles in HCC have been partially characterized, accumulating evidence reveals that OTUD3 can function as either a tumor suppressor or oncoprotein in a context-dependent manner across various malignancies, including lung, breast, and colorectal cancers. These seemingly contradictory findings underscore the complexity of OTUD3's mechanistic involvement in HCC pathogenesis and highlight the need for further investigation to fully elucidate its molecular functions.
OTUD4
OTUD4 mRNA expression is significantly downregulated in HCC tissues and is associated with poor prognosis. Through gene set enrichment analysis (GSEA), OTUD4 is related to apoptosis signaling pathways and the AKT signaling pathway. Therefore, the overexpression of OTUD4 may inhibit the proliferation, migration, and invasion of HCC cells by suppressing the AKT signaling pathway[176]. The specific mechanisms by which OTUD4 regulates the development of HCC cells remain unclear and warrant further investigation.
OTUD5
The research team from Xi'an Jiaotong University found that OTUD5 is significantly upregulated in HCC tissues, and high levels of OTUD5 are also detected in most HCC cell lines. Analysis of TCGA data showed that high expression of OTUD5 indicates a poorer overall survival in HCC patients. Mass spectrometry analysis revealed that solute carrier family 38 member 1 (SLC38A1) is a candidate downstream target protein of OTUD5. Further research results indicate that OTUD5 stabilizes SLC38A1 by preventing its ubiquitin-mediated proteasomal degradation, thereby promoting the proliferation of HCC cells[177].
OTUD7B
Recent studies have revealed that the deubiquitinase OTUD7B, which is significantly downregulated in HCC, directly binds to and removes lysine-linked polyubiquitin chains from both wild-type and mutant p53, thereby inhibiting its proteasomal degradation and enhancing p53 protein stability. Functional experiments demonstrated that OTUD7B suppresses HCC cell and xenograft growth by activating the p53-dependent mitochondrial apoptosis pathway (inducing PUMA/BAX expression), an effect that is abolished upon p53 depletion. Notably, OTUD7B and p53 form a bidirectional regulatory loop—OTUD7B expression is itself transcriptionally repressed by p53. Clinical data analysis showed a positive correlation between OTUD7B and p53 protein levels in HCC tissues, suggesting that the OTUD7B-p53 axis may represent a potential therapeutic target for HCC[178].
TRABID
TRAF-binding domain-containing protein (TRABID) also is a member of the OTU deubiquitinase family. Proteomic analysis based on mass spectrometry has found that TRABID associates with damaged DNAAF5 (DDB2) in HepG2 cells. TRABID causes deubiquitination of DDB2, and the proliferation of HCC cells mediated by TRABID is attenuated by DDB2 knockdown[179].
Function of the UCH Family and Research Advances in Hepatocellular Carcinoma
Despite growing evidence suggesting that UCH enzymes are closely related to human malignant tumors, there have been few studies on the role of UCH enzymes in HCC. Moreover, there are currently no studies that demonstrate the impact of UCH-L1 and UCH-L3 on the development of HCC.
UCH37
Zhongshan Hospital of Fudan University performed a functional proteomic analysis to screen UCH37-interacting proteins in hepatocellular carcinoma, and glucose-regulated protein 78 (GRP78) was identified as one interacting with UCH37[180]. GRP78 may be a target protein of UCH37, but there is a lack of research on its specific mechanisms. Zhongshan Hospital has conducted further research on UCH37 and found that UCH37 is overexpressed in HCC, making it an important indicator for predicting time to recurrence. Concurrently, in vitro experiments have confirmed that UCH37 can promote cell migration and invasion in HCC cell lines by interacting with and deubiquitinating the RNA splicing factor PRP19[181].
BRCA1-associated protein-1 (BAP1)
Compared with the adjacent non-tumor tissues, the levels of BAP1 mRNA and protein in HCC are significantly downregulated. Low expression of BAP1 is positively correlated with aggressive tumor phenotypes, and is independently associated with poorer recurrence-free survival and overall survival following curative hepatectomy. Wild-type BAP1, but not mutant BAP1, significantly inhibits the proliferation, invasion, EMT of HCC cells in vitro, and tumor progression and metastasis in vivo. Mechanistically, BAP1 interacts with PTEN and stabilizes PTEN through deubiquitination, and further negatively regulates HCC cell EMT by inactivating the AKT/GSK-3β/Snail pathway. However, these tumor-suppressive effects of BAP1 are abolished by inactivating mutations[182].
Function of the MJD Family and Research Advances in Hepatocellular Carcinoma
Machado-Joseph disease, also referred to as spinocerebellar ataxia type 3, is a neurological disorder that is widely recognized as the most common form of spinocerebellar ataxia. MJDs are closely associated with such neurological conditions[183].
As a member of MJDs and a therapeutic target in neurodegenerative disease, Ataxin-3 has been well characterized in the progression of MachadoJoseph disease[184]. As for other members of MJD such as Ataxin-3L, JOSD1, and JOSD2, there is no evidence to suggest their role in MJD. However, a growing body of evidence indicates that Ataxin-3, Ataxin-3L, JOSD1, and JOSD2 are also associated with cancer progression. Recent studies have indicated that non-small cell lung cancer is associated with Ataxin-3, testicular cancer and human colorectal cancer are also related to Ataxin-3[185-187]. Buss R et al. found that Ataxin-3L promotes the migration of NSCLC cells, and high levels of are associated with lower overall survival rates in breast cancer patients[188, 189]. But the mechanism is not yet clear. JOSD1 is frequently overexpressed in lung adenocarcinoma, head and neck squamous cell carcinoma, colon cancer, uterine cancer, melanoma, ovarian cancer, and bladder cancer[189-193]. Amplification of JOSD2 is more common in tumors such as uterine cancer, cholangiocarcinoma, lung cancer, adrenocortical carcinoma, diffuse large B-cell lymphoma of the lymphoid type, bladder cancer, breast cancer, gastric cancer, and pancreatic cancer[189, 194, 195]. Zhou et al. conducted a study and found that the expression levels of Ataxin-3, JOSD1, and JOSD2 were significantly elevated in HCC tissues and cell lines. These elevated expressions correlated with histological grading, specimen types, TP53 mutations, lymph node metastasis, gender, and age of HCC patients[196]. A study conducted by Fujian Medical University has delved into the mechanism by which JOSD2 regulates HCC. The findings indicate that JOSD2 interacts with and reduces the ubiquitination levels of catenin β 1 (CTNNB1), thereby enhancing the transmission of the Wnt pathway[197].
Function of the MINDYs and Research Advances in Hepatocellular Carcinoma
As a newly identified family of deubiquitinating enzymes, the MIDNYs have not been extensively studied and applied in cancer research. Current research indicates that MINDY1 is associated with breast cancer, bladder cancer, and HCC, and promotes their progression[198-201].
In human breast cancer tissues, there is a positive correlation between estrogen receptor α (ERα) and MINDY1 protein levels, and high expression of MINDY1 is associated with poor prognosis. Further mechanistic research has found that MINDY1 mediates the deubiquitination of ERα and increases its stability in a deubiquitination activity-dependent manner[198]. In bladder cancer, MINDY1 has been shown to interact with YAP in a deubiquitination activity-dependent manner, deubiquitinating and stabilizing YAP[199]. Xia et al. have discovered that the expression level of MINDY1 in HCC tissues is higher than that in adjacent tumor tissues and is closely related to the progression of the tumor. High expression of MINDY1 is an independent risk factor for poor prognosis in HCC patients[201]. Song and colleagues have found that MINDY1 inhibits the malignant progression of HCC by suppressing the ubiquitination of PDL1 and mediating immune escape[200].
Function of the JAMM Family and Research Advances in Hepatocellular Carcinoma
Members of the JAMM deubiquitinating enzyme family have been implicated in various cancers. Specifically, PSMD14 has been associated with glioblastoma, where it promotes tumorigenesis by deubiquitinating and stabilizing β-catenin, a key component of the Wnt signaling pathway[202]. And PSMD14 also is a novel prognostic marker and therapeutic target in osteosarcoma[203]. STAMBP has been linked to breast cancer and melanoma, potentially contributing to the progression of these cancers through its deubiquitinating activities[204-206]. Mutations in BRCC3 have been correlated with myeloid neoplasms, suggesting a role in the development of blood-related cancers[207]. Lastly, STAMBPL1 has been identified as being associated with HCC. The clinical results of the study show that STAMBPL1 is significantly increased in tumor tissues of HCC patients, and its expression is closely related to tumor size and TNM staging. Cells with overexpression of STAMBPL1 exhibit a marked enhancement in proliferation both in vitro and in vivo, while STAMBPL1 knockdown demonstrates the completely opposite effect. Mechanistically, STAMBPL1 activates the Wnt/β-catenin pathway, increasing the expression of downstream oncogenes. In addition to this, STAMBPL1 is transcriptionally regulated by Sterol Regulatory Element-Binding Protein 1 (SREBP1), and overexpression of STAMBPL1 increases lipid accumulation in HCC cells and xenografted tumors. These findings suggest that STAMBPL1 plays a role in the malignant behavior of HCC cells by modulating the Wnt/β-catenin pathway and lipid metabolism[208].
Conclusions and Future Perspectives
This article introduces and analyzes some DUBs related to HCC, indicating that DUBs play a unique regulatory role in the occurrence and development of HCC. The development of targeted drugs has become an important treatment modality for patients with high expression of deubiquitinating enzymes in HCC, and some molecular targeted drugs and small molecule inhibitors related to ubiquitination and deubiquitination enzymes have also shown therapeutic effects in clinical treatment[209-211].
Due to the involvement of multiple pathways and targets, the regulatory mechanisms of deubiquitinating enzymes in HCC are complex. Most DUBs associated with HCC tend to promote tumor activity, However, OTUD4 and BAP1 have been shown to exert inhibitory effects on tumorigenesis. The roles of USP8 and OTUD3 in HCC are less clear, with some studies yielding contradictory results regarding their oncogenic or tumor-suppressive functions. This suggests that the mechanisms by which USP8 and OTUD3 regulate HCC are more intricate and require further elucidation.
The regulatory mechanisms involving DUBs in HCC are extremely complex, with DUBs possessing multiple downstream targets. To provide a more intuitive understanding of the process by which DUBs regulate HCC, this article offers a table and a schematic diagram to illustrate the complex processes involved in the occurrence and development of HCC regulated by DUBs.
USP2, USP11, USP22, USP30 and STAMBPL1 play a role in tumor activity by regulating lipid metabolism of HCC cells. These enzymes are crucial in the development of HCC associated with NASH, as they are involved in the regulation of disease progression and, due to their role in tumorigenesis, serve as potential diagnostic biomarkers and therapeutic targets.
Tumor cell glycolytic reprogramming is an important field in cancer research, a process often associated with the Warburg effect, whereby tumor cells tend to produce energy through glycolysis rather than oxidative phosphorylation, even in the presence of ample oxygen[212]. In the tumor microenvironment, glycolytic reprogramming is related to the proliferation and invasiveness of tumor cells. USP5, USP11, USP14, USP22, USP29 and USP48 promote tumor development by regulating glycolysis in HCC, and targeting these three deubiquitinating enzymes to inhibit glycolysis in HCC presents a new approach to suppress tumor growth.
PDL1 plays a crucial role in the tumor microenvironment, where tumor cells express PDL1 to inhibit the activity of T cells, thereby achieving immune evasion[213, 214]. USP22 and MINDY1 maintain the stability of PDL1 through the process of deubiquitination. The expression levels of USP22 and MINDY1 may indicate the sensitivity to PDL1 inhibitors, and inhibitors of USP22 or MINDY1 may enhance the efficacy of PD-1/PDL1 inhibitors and reduce drug resistance.
The MAPK, Hh, Wnt, Hippo and mTOR signaling pathways play crucial roles in the occurrence, development, invasion, and metastasis of HCC. The abnormal activation of these pathways not only promotes the proliferation and survival of tumor cells but also affects tumor invasion and metastasis by regulating the tumor microenvironment. Some DUBs are involved in the regulation of the activation of these signaling pathways. USP4 and USP12 promote the activation of the MAPK signaling pathway. USP44 inhibits the progression of hepatocellular carcinoma by suppressing the Hh signaling pathway. β-catenin, as a core regulatory factor of the canonical Wnt signaling pathway, has its protein levels regulated by USP8, USP9X, USP25, and USP39. In contrast, USP3, USP14, and JOSD2 do not regulate the Wnt signaling pathway by modulating β-catenin. USP1 and USP7 are involved in the regulation of the Hippo pathway, while USP10, USP19, and USP40 regulate the protein levels of YAP, a downstream effector of the Hippo pathway. The protein stability of mTOR is regulated by USP10, USP11, and USP22. Therefore, targeting these deubiquitinating enzymes represents an important therapeutic strategy with significant clinical implications.
EMT is a dynamic process wherein epithelial cells lose their polarity and cell-cell adhesion while gaining migratory and invasive properties characteristic of mesenchymal cells. This transformation plays a pivotal role in tumor invasion and metastasis[215]. EMT regulation involves a complex interplay of multiple signaling pathways, which are increasingly recognized as therapeutic targets. In this review, we summarize the mechanistic roles of USP4, USP5, USP9X, USP11, USP14, USP39, and BAP1 in modulating EMT and discuss their potential as novel targets for suppressing HCC metastasis.
From the existing research results, it can be seen that the abnormal expression of DUBs is closely related to drug resistance. Sorafenib and lenvatinib are currently first-line treatment drugs for HCC, but their therapeutic effects are limited and prone to drug resistance. In HCC cells and tumors resistant to lenvatinib, USP14 is significantly upregulated, and mutations in OTUD3 are associated with sorafenib resistance. The abnormal expression of USP8, USP29, and USP39 is also associated with sorafenib resistance. Additionally, the abnormal expression of USP8 and USP9 may be one of the reasons for HCC cells' resistance to doxorubicin. Furthermore, knocking down USP27 makes HCC cells more sensitive to 5-FU. The development of DUB inhibitors and their combination with current antitumor drugs is one of the promising directions for future research.
Ferroptosis is a form of programmed cell death that is dependent on iron and reactive oxygen species (ROS), characterized by the accumulation of lipid peroxides within cells, which ultimately leads to the disruption of cell membrane integrity and cell death[216]. Among DUBs, USP5 and USP8 inhibit ferroptosis in HCC, thereby promoting the progression and drug resistance of HCC. Developing specific inhibitors of USP5 and USP8 represents a potential anti-tumor strategy, especially for HCC that is resistant to conventional treatments.
This review summarizes 12 DUBs associated with drug resistance in HCC. Among these, USP3, USP8, USP9X, USP20, and CYLD promote HCC resistance to conventional chemotherapeutic agents (e.g., doxorubicin, platinum-based drugs). Additionally, USP7, USP8, USP24, USP29, and USP39 confer resistance to sorafenib, while USP14 and USP28 contribute to lenvatinib resistance. To better illustrate the roles of these DUBs in HCC drug resistance mechanisms, we have provided a schematic summary in Table 2.
DUBs play a crucial role in the pathogenesis of tumors. The development of targeted drugs and small-molecule inhibitors is advancing rapidly, representing a key focus of current and future research. Although most inhibitors remain in preclinical research, some drugs have already entered clinical trials. Significant progress has been made in the development of USP1 inhibitors, with two compounds advancing to Phase I clinical studies: KSQ-4279 (NCT05240898) and ISM3091 (NCT05932862), both targeting advanced solid tumors. VLX1570, a small-molecule inhibitor targeting USP14 and UCHL5, was the only DUB inhibitor to reach Phase II clinical trials (NCT02372240) for the treatment of multiple myeloma. However, the trial was terminated due to drug toxicity issues, highlighting the challenges in developing deubiquitinase inhibitors and underscoring the need for further research. In Table 1, we systematically summarize the current progress in the development of small-molecule inhibitors directed against DUBs.
Current research suggests that OTUB1, ATXN3, and JOSD1 promote the development of HCC, but the regulatory mechanisms of these three in HCC have not yet been elucidated in the literature. The research on the role of DUBs in the pathogenesis and treatment of HCC still has significant room for development. The development of targeted drugs and the clinical application of small molecule inhibitors will also become hot topics in future research. These can provide new ideas and research directions for the treatment of HCC in the future.
Roles of deubiquitinating enzymes (DUBs) in hepatocellular carcinoma (HCC) and their inhibitors.
| DUBs family | DUBs | Mechanism pathway | Action | Article | Inhibitors |
|---|---|---|---|---|---|
| USP | USP1 | USP1/Hippo pathway | Progression | [32] | KSQ-4279 |
| USP1/CDK5 | Progression | [34] | ISM3091 FT-3171 SP-002 | ||
| USP2 | USP2a/RAB1A | Progression | [37] | Q15 | |
| USP2/SREPB1/2 | Lipid metabolism | [38] | COH29 LCAHA 6TG | ||
| USP3 | USP3/β-catenin | Proliferation/Apoptosis | [39] | NC114 U4-I05 | |
| USP4 | USP4/CypA/MAPK pathway | Growth/Migration/Invasion | [40] | Vialinin A | |
| USP4/TGFR-1/TGF-β pathway | EMT/Metastasis | [41] | U4-I05 | ||
| USP4/Snail1 | EMT | [42] | |||
| USP5 | USP5/SLUG | Proliferation/Metastasis/Invasion/EMT | [43] | WP1130 | |
| Hpn/USP5/p14(ARF)-p53 pathway | Apoptosis | [45] | PYR-41 Formononetin | ||
| USP5/LSH/SLC7A11 | Ferroptosis | [44] | Mebendazole | ||
| CREB1/P300/METTL5/USP5/c-Myc | Glycolysis | [46] | USP5-IN-1 | ||
| USP7 | USP7/Yki/Hippo pathway | Progression | [50] | Compound 19 | |
| USP7/TMEM43/VDAC1 | Progression | [51] | HBX28528 | ||
| USP7/YY1 | Proliferation/Metastasis/EMT | [52] | HBX41108 | ||
| USP7/BTF3 | Proliferation/Metastasis/Invasion | [53] | GNE-6676 | ||
| USP7/TRIP12/p14 | Progression | [48] | I-56/57 | ||
| USP7/TRIM27/STAT3 | Progression | [54] | Compound-23/25/28 | ||
| miR‑205/USP7 | Proliferation | [47] | |||
| ENKUR/β-catenin/c-Jun/MYH9/USP7 | Proliferation/EMT | [55] | FT827/671 | ||
| METTL3/USP7 | Proliferation/Metastasis/Invasion | [56] | |||
| FEN1/USP7/MDM2/p53 | Metastasis/Invasion/EMT | [57] | |||
| USP8 | USP8/RTKs | Apoptosis | [62] | ML364 Compound 22c DC-U43-10 DC-U4106 | |
| USP8/β-catenin | Tumorigenesiss | [63] | |||
| USP8/OGT/SLC7A11 | Ferroptosis | [65] | |||
| USP8/TRAF6/ NF-ΚB | Autophagy | [66] | |||
| USP9X | USP9X/β-catenin | Proliferation | [67] | WP1130 | |
| MiR26b/USP9X/Smad4/TGF-β | EMT | [68] | EOAI3402143 | ||
| MiR-26b/USP9X/p53 | Autophagy | [70] | FT709 | ||
| LNC473/USP9X/survivin | Proliferation/Invasion | [71] | |||
| USP10 | USP10/PTEN/mTORC1 | Progression | [72] | Spautin-1 | |
| USP10/LKB1/mTOR | Progression | [73] | Quercetin | ||
| USP10/Smad4 | Metastasis | [74] | DZNep | ||
| USP10/YAP/p53 | Proliferation | [75] | HBX19818 | ||
| USP10/PLAGL2 | Progression | [79] | Wu-5 | ||
| USP11 | USP11/NF90 | Proliferation/Metastasis | [81] | Mitoxantrone | |
| USP11/E2F1/ERK/mTOR | Metastasis/Autophagy | [82] | |||
| USP11/Eef1A1/SP1/HGF | EMT/Metastasis | [83] | |||
| USP11/KLF4 | Lipid metabolism | [84] | |||
| USP11/HIF-1α/LDHA | Glycolysis/Proliferation/Metastasis | [85] | |||
| USP12 | USP12/The cyclin dependent kinase 1/cyclinB1 | Proliferation | [86] | — | |
| USP12/p38/MAPK pathway | Autophagy | [86] | |||
| USP12/c-Myc | Progression | [87] | |||
| USP13 | USP13/c-Myc | Progression | [88] | Spadin-1 | |
| USP13/TLR4/MyD88/NF-κB | Proliferation/EMT/Migration/Invasion | [89] | BK50118C | ||
| USP14 | USP14/Wnt/Notch1 signaling pathway | Endoplasmic reticulum/ Progression | [91] | VLX1570 IU1 | |
| USP14/ HIF1α | Proliferation/Invasion/Migration/Vascular Mimicry formation | [92] | b-AP15 PtPT | ||
| USP14/HK2/AKT/P62 | Proliferation/Invasion/Autophagy/Glycolysis | [93] | |||
| USP15 | - | Gene expression/Cell cycle/DNA repair | [95] | USP15-IN-1 | |
| Proliferation/Autophagy | [96] | ||||
| USP16 | Ct-HBX/USP16 | Progression | [102] | — | |
| USP18 | USP18/BCL2L1 | Proliferation | [103] | — | |
| USP19 | USP19/YAP | Proliferation/Migration | [104] | Example 142 | |
| USP20 | ATR/USP20/SLC7A11 | Ferroptosis | [105] | GSK2643943A | |
| USP21 | USP21/MEK2 | Progression | [106] | BAY-805 ZINC02422616 | |
| hsa_circ_0039053/USP21 | Progression | [107] | |||
| USP22 | USP22/HIF1α | Stemness/Glycolysis | [108] | Baohuoside I | |
| USP22/ FKBP12/ mTORC1 | Tumorigenesis/Progression/Autophagy | [109] | Morusin | ||
| USP22/PPARγ/ACLY and ACC | Tumorigenesis/Lipid metabolism | [110] | Chrysin | ||
| PRDM1 /USP22/SPI1/PDL1 | Tumor immunity | [111] | USP22i-S02 Pirarubicin | ||
| USP24 | USP24/TRAF2 | Progression | [115] | WP1130 EOAI3402143 NCI677397 | |
| MiR-21-5p/USP24/SIRT7 | Progression | [116] | |||
| USP24/Beclin1 | Proliferation/Migration/Ferroptosis/ Autophagy | [117] | |||
| USP25 | USP25/TRIM21/Wnt/β-catenin signaling pathway | Proliferation/Migration/Invasion/EMT | [119] | PR619 AZ1 Vismodegib Compound 9p FT206 | |
| USP26 | HBx/USP26/SIRT1 | Proliferation/Apoptosis | [120] | — | |
| USP27 | USP27/Cyclin E | Growth/Migration/Invasion | [125] | — | |
| USP27/SETD3 | Proliferation/Progression | [126] | |||
| USP28 | miR-363-3p/USP28/c-Myc | Progression | [129] | CT113 AZ1 | |
| miR-216b/USP28 | Progression | [128] | |||
| USP29 | USP29/HIF1α/HK2 | Glycolysis | [133] | — | |
| USP30 | GNPAT/USP30/DRP1 | Lipid metabolism/Tumorigenesis | [134] | MTX652 | |
| IKKβ/USP30/ACLY | Lipid metabolism/Tumorigenesis | [135] | MF094 ST-539 USP30inh | ||
| USP33 | USP33/SP1/c-Met | Invasion/Metastasis | [140] | — | |
| USP35 | USP35/PKM2 | Proliferation/Migration/Invasion | [141] | — | |
| USP35/ABHD17C | Proliferation/Apoptosis/Migration/Invasion | [142] | |||
| USP35/PI3K/AKT signaling pathway | Proliferation/Apoptosis/Migration/Invasion | [142] | |||
| USP39 | USP39/SP1 | Proliferation | [145] | — | |
| USP39/TRIM26/ZEB1 | EMT/ Proliferation/Metastasis | [146] | |||
| USP39/TRIM26/β-catenin | Proliferation/Metastasis | [147] | |||
| USP39/FoxM1/ PLK1 and cyclin B1 | Proliferation | [143] | |||
| SIRT7/USP39 | Proliferation/Tumorigenesis | [149] | |||
| DNAAF5/USP39/PFKL | Proliferation | [150] | |||
| USP40 | USP40/Claudin1 | Proliferation/Migration/Stemness | [151] | — | |
| USP40/YAP | Progression | [152] | |||
| USP44 | USP44/Itch/Gli1/PDL1 | Progression | [154] | — | |
| USP46 | USP46/MST1 | Proliferation/Metastasis | [155] | — | |
| MiR-27a-3p/USP46 | Proliferation | [156] | |||
| USP48 | USP48/Gasdermin E | Pyroptosis | [157] | — | |
| Mettl14/USP48/SIRT6 | Glycolysis/Malignancy | [158] | |||
| USP49 | HSF1/HLNC1/USP49 | Progression | [159] | — | |
| USP53 | USP53/CYCS | Apoptosis | [162] | — | |
| CYLD | CYLD/IκB-α | Apoptosis | [163] | Dihydromyricetin | |
| CYLD/JNK/c-MYC | Proliferation | [164] | |||
| miR-362/CYLD | Growth/Metastasis | [165] | |||
| miR-501/CYLD | Proliferation | [167] | |||
| miR-922/CYLD | Proliferation | [168] | |||
| SOX2/EGFR/miR-222-5p/CYLD | Proliferation/Migration/Invasion | [169] | |||
| OTU | OTUB1 | - | Progression | [170] | Compound 61 |
| OTUD3 | OTUD3/ACTN4 | Proliferation/Metastasis | [171] | — | |
| OTUD3/ eIF2α/ISR | Progression | [172] | |||
| OTUD4 | OTUD4/AKT signaling pathway | Proliferation/Metastasis/Invasion/Autophagy | [176] | — | |
| OTUD5 | OTUD5/ SLC38A1 | Proliferation | [177] | — | |
| OTUD7B | OTUD7B/p53/PUMA/BAX | Progression/Autophagy | [178] | — | |
| TRABID | TRABID/DDB2 | Proliferation | [179] | — | |
| UCH | UCH37 | UCH37/GRP78 | Progression | [180] | b-AP15 |
| UCH37/PRP19 | Metastasis/Invasion | [181] | |||
| BAP1 | BAP1/PTEN/AKT/GSK-3β/Snail pathway | Proliferation/Invasion/EMT | [182] | iBap | |
| JOSD2 | JOSD2 | JOSD2/CTNNB1/Wnt pathway | Progression | [197] | HY041004 |
| MINDY | MINDY1 | MINDY1/PDL1 | Tumor immunity | [200] | — |
| JAMM | STAMBPL1 | STAMBPL1/Wnt/β-catenin | Proliferation | [208] | CSN5i-3 |
| STAMBPL1/SREBP1 | Lipid metabolism | [208] |
DUBs Involved in HCC Drug Resistance.
| DUBs | Resistant Drug(s) | Experimental Validation | Article |
|---|---|---|---|
| USP3 | Conventional chemotherapeutic drugs | In vitro | [39] |
| USP7 | Sorafenib | In vitro/ Patient-derived xenograft | [217] |
| USP8 | Doxorubicin/ Sorafenib | In vitro/ Cell Line-Derived Xenograft | [62] |
| USP9X | Doxorubicin | In vitro/ Cell Line-Derived Xenograft | [70, 218] |
| USP14 | Lenvatinib | In vitro/ Cell Line-Derived Xenograft | [94] |
| USP20 | Oxaliplatin | In vitro/ Cell Line-Derived Xenograft | [105] |
| USP24 | Sorafenib | In vitro/ Cell Line-Derived Xenograft | [117] |
| USP28 | Lenvatinib | Cell Line-Derived Xenograft | [130] |
| USP29 | Sorafenib | In vitro/ Patient-derived xenograft | [133] |
| USP39 | Sorafenib | In vitro | [150] |
| CYLD | Doxorubicin/ 5-Fluorouracil/ Cisplatin | In vitro | [163] |
| OTUD3 | Sorafenib | In vitro/ Cell Line-Derived Xenograft | [172] |
Abbreviations
FEN1: Flap endonuclease 1
FoxM1: Forkhead box M1
HBx: Hepatitis B virus X protein
HCC: Hepatocellular carcinoma
HK2: Hexokinase 2
ICC: Intrahepatic cholangiocarcinoma
ISR: Integrated stress response
JAMM: JAMM/MPN domain-associated Zn-depend metalloproteases
MINDY: The motif interacting with ubiquitin (MIU)-containing novel DUB family
MJD: Machado-Joseph domain-containing protease
mTORC1: Mammalian target of rapamycin complex 1
NASH: Non-alcoholic steatohepatitis
OGT: O-GlcNAc transferase
OUT: Ovarian tumor protease
PAK1: P21-activated kinase 1
PDL1: Programmed Cell Death Ligand 1
PFKL: Phosphofructokinase L
RTK: Receptor tyrosine kinase
SIRT7: Sirtuin 7
TMEM43: Transmembrane protein 43
TRABID: TRAF-binding domain-containing protein
TRAF2: Tumor Necrosis Factor Receptor-Associated Factor 2
TRIP12: Thyroid hormone receptor-interacting protein 12
UCH: Ubiquitin C-terminal hydrolase
USP: Ubiquitin-specific protease
YAP: Yes-associated protein
YY1: Yin-yang-1
ZEB1: Zinc-finger E-box-binding homeobox 1
Acknowledgements
Funding
This work was supported by the National Natural Science Foundation of China [82203160, Jianing Tang], China Postdoctoral Science Foundation [2022M713531, 2024T171061, Jianing Tang], the Natural Science Foundation of Hunan Province [2024JJ4088, Jianing Tang].
Author contributions
All authors made substantial, direct and intellectual contribution to the review. Under the guidance of the three corresponding authors, Xingyu Mi and Qingfeng Li wrote this review, with Xingyu Mi preparing the tables and figures in the manuscript. Guo Long, Yilin Pan, Yulin Xie, and Shiqi Lu collected the data. All authors reviewed and approved the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Cockram PE, Kist M, Prakash S, Chen SH, Wertz IE, Vucic D. Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ. 2021;28:591-605
2. Sharma B, Bhatt TK. Genome-wide identification and expression analysis of E2 ubiquitin-conjugating enzymes in tomato. Sci Rep. 2017;7:8613
3. Buetow L, Huang DT. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol. 2016;17:626-42
4. Zheng N, Shabek N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu Rev Biochem. 2017;86:129-57
5. Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550-63
6. Wang Z, Shen N, Wang Z, Yu L, Yang S, Wang Y. et al. TRIM3 facilitates ferroptosis in non-small cell lung cancer through promoting SLC7A11/xCT K11-linked ubiquitination and degradation. Cell Death Differ. 2024;31:53-64
7. Zhang S, Jia X, Dai H, Zhu X, Song W, Bian S. et al. SERPINE2 promotes liver cancer metastasis by inhibiting c-Cbl-mediated EGFR ubiquitination and degradation. Cancer Commun (Lond). 2024;44:384-407
8. He Y, Shi Q, Ling Y, Guo H, Fei Y, Wu R. et al. ABLIM1, a novel ubiquitin E3 ligase, promotes growth and metastasis of colorectal cancer through targeting IĸBα ubiquitination and activating NF-ĸB signaling. Cell Death Differ. 2024;31:203-16
9. Gong DA, Zhou P, Chang WY, Yang JY, Zhang YL, Huang AL. et al. SPOP promotes CREB5 ubiquitination to inhibit MET signaling in liver cancer. Biochim Biophys Acta Mol Cell Res. 2024;1871:119642
10. Zhang Y, Huang Z, Li K, Xie G, Feng Y, Wang Z. et al. TrkA promotes MDM2-mediated AGPS ubiquitination and degradation to trigger prostate cancer progression. J Exp Clin Cancer Res. 2024;43:16
11. Cohen P, Tcherpakov M. Will the ubiquitin system furnish as many drug targets as protein kinases? Cell. 2010;143:686-93
12. Huang X, Dixit VM. Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res. 2016;26:484-98
13. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-63
14. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301-14
15. Feng R, Su Q, Huang X, Basnet T, Xu X, Ye W. Cancer situation in China: what does the China cancer map indicate from the first national death survey to the latest cancer registration? Cancer Commun (Lond). 2023;43:75-86
16. Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018;17:57-78
17. Yuan T, Yan F, Ying M, Cao J, He Q, Zhu H. et al. Inhibition of Ubiquitin-Specific Proteases as a Novel Anticancer Therapeutic Strategy. Front Pharmacol. 2018;9:1080
18. Shen J, Lin X, Dai F, Chen G, Lin H, Fang B. et al. Ubiquitin-specific peptidases: Players in bone metabolism. Cell Prolif. 2023;56:e13444
19. Chen R, Pang X, Li L, Zeng Z, Chen M, Zhang S. Ubiquitin-specific proteases in inflammatory bowel disease-related signalling pathway regulation. Cell Death Dis. 2022;13:139
20. Makarova KS, Aravind L, Koonin EV. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem Sci. 2000;25:50-2
21. Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363-97
22. Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 2003;4:517-22
23. Nanao MH, Tcherniuk SO, Chroboczek J, Dideberg O, Dessen A, Balakirev MY. Crystal structure of human otubain 2. EMBO Rep. 2004;5:783-8
24. Lin SC, Chung JY, Lamothe B, Rajashankar K, Lu M, Lo YC. et al. Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20. J Mol Biol. 2008;376:526-40
25. Du J, Fu L, Sui Y, Zhang L. The function and regulation of OTU deubiquitinases. Front Med. 2020;14:542-63
26. Bishop P, Rocca D, Henley JM. Ubiquitin C-terminal hydrolase L1 (UCH-L1): structure, distribution and roles in brain function and dysfunction. Biochem J. 2016;473:2453-62
27. Weeks SD, Grasty KC, Hernandez-Cuebas L, Loll PJ. Crystal structure of a Josephin-ubiquitin complex: evolutionary restraints on ataxin-3 deubiquitinating activity. J Biol Chem. 2011;286:4555-65
28. Abdul Rehman SA, Kristariyanto YA, Choi SY, Nkosi PJ, Weidlich S, Labib K. et al. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol Cell. 2016;63:146-55
29. Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD. et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331-9
30. Williams SA, Maecker HL, French DM, Liu J, Gregg A, Silverstein LB. et al. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell. 2011;146:918-30
31. Wang L, Hu T, Shen Z, Zheng Y, Geng Q, Li L. et al. Inhibition of USP1 activates ER stress through Ubi-protein aggregation to induce autophagy and apoptosis in HCC. Cell Death Dis. 2022;13:951
32. Liu D, Li Q, Zang Y, Li X, Li Z, Zhang P. et al. USP1 modulates hepatocellular carcinoma progression via the Hippo/TAZ axis. Cell Death Dis. 2023;14:264
33. Misra JR, Irvine KD. The Hippo Signaling Network and Its Biological Functions. Annu Rev Genet. 2018;52:65-87
34. Bian S, Ni W, Zhou L, Tong Y, Dai C, Zhao X. et al. Ubiquitin-specific protease 1 facilitates hepatocellular carcinoma progression by modulating mitochondrial fission and metabolic reprogramming via cyclin-dependent kinase 5 stabilization. Cell Death Differ. 2024;31:1202-18
35. Zhang X, Nadolny C, Chen Q, Ali W, Hashmi SF, Deng R. Dysregulation and oncogenic activities of ubiquitin specific peptidase 2a in the pathogenesis of hepatocellular carcinoma. Am J Cancer Res. 2023;13:2392-409
36. Nadolny C, Zhang X, Chen Q, Hashmi SF, Ali W, Hemme C. et al. Dysregulation and activities of ubiquitin specific peptidase 2b in the pathogenesis of hepatocellular carcinoma. Am J Cancer Res. 2021;11:4746-67
37. Xiong B, Huang J, Liu Y, Zou M, Zhao Z, Gong J. et al. Ubiquitin-specific protease 2a promotes hepatocellular carcinoma progression via deubiquitination and stabilization of RAB1A. Cell Oncol (Dordr). 2021;44:329-43
38. Calvisi DF, Wang C, Ho C, Ladu S, Lee SA, Mattu S. et al. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology. 2011;140:1071-83
39. Xu J, Gui GSW, Yang C, Zhu S, Chen Z, Ma SLB. et al. USP3 inhibition is Active Against Chemo-resistant Hepatocellular Carcinoma Anchorage-independent Growth via Suppressing Wnt/β-catenin. Curr Mol Med. 2024;24:667-75
40. Li T, Yan B, Ma Y, Weng J, Yang S, Zhao N. et al. Ubiquitin-specific protease 4 promotes hepatocellular carcinoma progression via cyclophilin A stabilization and deubiquitination. Cell Death Dis. 2018;9:148
41. Qiu C, Liu Y, Mei Y, Zou M, Zhao Z, Ye M. et al. Ubiquitin-specific protease 4 promotes metastasis of hepatocellular carcinoma by increasing TGF-β signaling-induced epithelial-mesenchymal transition. Aging (Albany NY). 2018;10:2783-99
42. Li R, Shang R, Li S, Ren Y, Shen L, Yang L. et al. LOXL3-promoted hepatocellular carcinoma progression via promotion of Snail1/USP4-mediated epithelial-mesenchymal transition. Environ Toxicol. 2022;37:2540-51
43. Meng J, Ai X, Lei Y, Zhong W, Qian B, Qiao K. et al. USP5 promotes epithelial-mesenchymal transition by stabilizing SLUG in hepatocellular carcinoma. Theranostics. 2019;9:573-87
44. Yan B, Guo J, Wang Z, Ning J, Wang H, Shu L. et al. The ubiquitin-specific protease 5 mediated deubiquitination of LSH links metabolic regulation of ferroptosis to hepatocellular carcinoma progression. MedComm (2020). 2023;4:e337
45. Liu Y, Wang WM, Zou LY, Li L, Feng L, Pan MZ. et al. Ubiquitin specific peptidase 5 mediates Histidine-rich protein Hpn induced cell apoptosis in hepatocellular carcinoma through P14-P53 signaling. Proteomics. 2017 17
46. Xia P, Zhang H, Lu H, Xu K, Jiang X, Jiang Y. et al. METTL5 stabilizes c-Myc by facilitating USP5 translation to reprogram glucose metabolism and promote hepatocellular carcinoma progression. Cancer Commun (Lond). 2023;43:338-64
47. Zhu L, Liu R, Zhang W, Qian S, Wang JH. MicroRNA-205 regulates ubiquitin specific peptidase 7 protein expression in hepatocellular carcinoma cells. Mol Med Rep. 2015;12:4652-6
48. Cai JB, Shi GM, Dong ZR, Ke AW, Ma HH, Gao Q. et al. Ubiquitin-specific protease 7 accelerates p14(ARF) degradation by deubiquitinating thyroid hormone receptor-interacting protein 12 and promotes hepatocellular carcinoma progression. Hepatology. 2015;61:1603-14
49. Wang X, Zhang Q, Wang Y, Zhuang H, Chen B. Clinical Significance of Ubiquitin Specific Protease 7 (USP7) in Predicting Prognosis of Hepatocellular Carcinoma and its Functional Mechanisms. Med Sci Monit. 2018;24:1742-50
50. Sun X, Ding Y, Zhan M, Li Y, Gao D, Wang G. et al. Usp7 regulates Hippo pathway through deubiquitinating the transcriptional coactivator Yorkie. Nat Commun. 2019;10:411
51. Zhang N, Wang F, Yang X, Wang Q, Chang R, Zhu L. et al. TMEM43 promotes the development of hepatocellular carcinoma by activating VDAC1 through USP7 deubiquitination. Transl Gastroenterol Hepatol. 2024;9:9
52. Liu H, Han J, Lv Y, Zhao Z, Zheng S, Sun Y. et al. Isorhamnetin and anti-PD-L1 antibody dual-functional mesoporous silica nanoparticles improve tumor immune microenvironment and inhibit YY1-mediated tumor progression. J Nanobiotechnology. 2023;21:208
53. Hu M, Dai C, Sun X, Chen Y, Xu N, Lin Z. et al. Ubiquitination-specific protease 7 enhances stemness of hepatocellular carcinoma by stabilizing basic transcription factor 3. Funct Integr Genomics. 2024;24:28
54. Sakamoto T, Kuboki S, Furukawa K, Takayashiki T, Takano S, Yoshizumi A. et al. TRIM27-USP7 complex promotes tumour progression via STAT3 activation in human hepatocellular carcinoma. Liver Int. 2023;43:194-207
55. Hou R, Li Y, Luo X, Zhang W, Yang H, Zhang Y. et al. ENKUR expression induced by chemically synthesized cinobufotalin suppresses malignant activities of hepatocellular carcinoma by modulating β-catenin/c-Jun/MYH9/USP7/c-Myc axis. Int J Biol Sci. 2022;18:2553-67
56. Li Y, Cheng X, Chen Y, Zhou T, Li D, Zheng WV. METTL3 facilitates the progression of hepatocellular carcinoma by modulating the m6A level of USP7. Am J Transl Res. 2021;13:13423-37
57. Bian S, Ni W, Zhu M, Zhang X, Qiang Y, Zhang J. et al. Flap endonuclease 1 Facilitated Hepatocellular Carcinoma Progression by Enhancing USP7/MDM2-mediated P53 Inactivation. Int J Biol Sci. 2022;18:1022-38
58. Yi J, Gong X, Yin XY, Wang L, Hou JX, Chen J. et al. Parthenolide and arsenic trioxide co-trigger autophagy-accompanied apoptosis in hepatocellular carcinoma cells. Front Oncol. 2022;12:988528
59. Gao M, Qi Z, Deng M, Huang H, Xu Z, Guo G. et al. The deubiquitinase USP7 regulates oxidative stress through stabilization of HO-1. Oncogene. 2022;41:4018-27
60. Cao Z, Wu X, Yen L, Sweeney C, Carraway KL 3rd. Neuregulin-induced ErbB3 downregulation is mediated by a protein stability cascade involving the E3 ubiquitin ligase Nrdp1. Mol Cell Biol. 2007;27:2180-8
61. Islam MT, Chen F, Chen H. The oncogenic role of ubiquitin specific peptidase (USP8) and its signaling pathways targeting for cancer therapeutics. Arch Biochem Biophys. 2021;701:108811
62. Zhu Y, Xu J, Hu W, Wang F, Zhou Y, Gong W. et al. Inhibiting USP8 overcomes hepatocellular carcinoma resistance via suppressing receptor tyrosine kinases. Aging (Albany NY). 2021;13:14999-5012
63. Tang J, Long G, Xiao L, Zhou L. USP8 positively regulates hepatocellular carcinoma tumorigenesis and confers ferroptosis resistance through β-catenin stabilization. Cell Death Dis. 2023;14:360
64. Long G, Wang D, Tang J, Hu K, Zhou L. USP8 promotes the tumorigenesis of intrahepatic cholangiocarcinoma via stabilizing OGT. Cancer Cell Int. 2024;24:238
65. Tang J, Long G, Hu K, Xiao D, Liu S, Xiao L. et al. Targeting USP8 Inhibits O-GlcNAcylation of SLC7A11 to Promote Ferroptosis of Hepatocellular Carcinoma via Stabilization of OGT. Adv Sci (Weinh). 2023;10:e2302953
66. Kim MJ, Choi B, Kim JY, Min Y, Kwon DH, Son J. et al. USP8 regulates liver cancer progression via the inhibition of TRAF6-mediated signal for NF-κB activation and autophagy induction by TLR4. Transl Oncol. 2022;15:101250
67. Chen MY, Li ZP, Sun ZN, Ma M. USP9X promotes the progression of hepatocellular carcinoma by regulating beta-catenin. Ir J Med Sci. 2020;189:865-71
68. Shen G, Lin Y, Yang X, Zhang J, Xu Z, Jia H. MicroRNA-26b inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting USP9X. BMC Cancer. 2014;14:393
69. Trompeter HI, Dreesen J, Hermann E, Iwaniuk KM, Hafner M, Renwick N. et al. MicroRNAs miR-26a, miR-26b, and miR-29b accelerate osteogenic differentiation of unrestricted somatic stem cells from human cord blood. BMC Genomics. 2013;14:111
70. Chen E, Li E, Liu H, Zhou Y, Wen L, Wang J. et al. miR-26b enhances the sensitivity of hepatocellular carcinoma to Doxorubicin via USP9X-dependent degradation of p53 and regulation of autophagy. Int J Biol Sci. 2021;17:781-95
71. Chen H, Yang F, Li X, Gong ZJ, Wang LW. Long noncoding RNA LNC473 inhibits the ubiquitination of survivin via association with USP9X and enhances cell proliferation and invasion in hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2018;499:702-10
72. Lu C, Ning Z, Wang A, Chen D, Liu X, Xia T. et al. USP10 suppresses tumor progression by inhibiting mTOR activation in hepatocellular carcinoma. Cancer Lett. 2018;436:139-48
73. Ma C, Lin Z, Yao J, Qin W, Wang X, Li Q. et al. Loss of USP10 promotes hepatocellular carcinoma proliferation by regulating the serine synthesis pathway through inhibition of LKB1 activity. Cancer Sci. 2024;115(12):3902-3914
74. Yuan T, Chen Z, Yan F, Qian M, Luo H, Ye S. et al. Deubiquitinating enzyme USP10 promotes hepatocellular carcinoma metastasis through deubiquitinating and stabilizing Smad4 protein. Mol Oncol. 2020;14:197-210
75. Lu Y, Gao J, Wang P, Chen H, He X, Luo M. et al. Discovery of potent small molecule ubiquitin-specific protease 10 inhibitors with anti-hepatocellular carcinoma activity through regulating YAP expression. Eur J Med Chem. 2024;272:116468
76. Pinquart M, Duberstein PR. Depression and cancer mortality: a meta-analysis. Psychol Med. 2010;40:1797-810
77. Polityńska B, Pokorska O, Wojtukiewicz AM, Sawicka M, Myśliwiec M, Honn KV. et al. Is depression the missing link between inflammatory mediators and cancer? Pharmacol Ther. 2022;240:108293
78. Sekiya R, Maeda M, Yuan H, Asano E, Hyodo T, Hasegawa H. et al. PLAGL2 regulates actin cytoskeletal architecture and cell migration. Carcinogenesis. 2014;35:1993-2001
79. Wang C, Ni J, Zhai D, Xu Y, Wu Z, Chen Y. et al. Stress-induced epinephrine promotes hepatocellular carcinoma progression via the USP10-PLAGL2 signaling loop. Exp Mol Med. 2024;56:1150-63
80. Zhang S, Xie C, Li H, Zhang K, Li J, Wang X. et al. Ubiquitin-specific protease 11 serves as a marker of poor prognosis and promotes metastasis in hepatocellular carcinoma. Lab Invest. 2018;98:883-94
81. Zhang C, Xie C, Wang X, Huang Y, Gao S, Lu J. et al. Aberrant USP11 expression regulates NF90 to promote proliferation and metastasis in hepatocellular carcinoma. Am J Cancer Res. 2020;10:1416-28
82. Qiao L, Zhang Q, Sun Z, Liu Q, Wu Z, Hu W. et al. The E2F1/USP11 positive feedback loop promotes hepatocellular carcinoma metastasis and inhibits autophagy by activating ERK/mTOR pathway. Cancer Lett. 2021;514:63-78
83. Chen J, Ning D, Du P, Liu Q, Mo J, Liang H. et al. USP11 potentiates HGF/AKT signaling and drives metastasis in hepatocellular carcinoma. Oncogene. 2024;43:123-35
84. Yang H, Park D, Ryu J, Park T. USP11 degrades KLF4 via its deubiquitinase activity in liver diseases. J Cell Mol Med. 2021;25:6976-87
85. Qiao L, Hu W, Li L, Chen X, Liu L, Wang J. USP11 promotes glycolysis by regulating HIF-1α stability in hepatocellular carcinoma. J Cell Mol Med. 2024;28:e18017
86. Liu C, Li X, Feng G, Cao M, Liu F, Zhang G. et al. Downregulation of USP12 inhibits tumor growth via the p38/MAPK pathway in hepatocellular carcinoma. Mol Med Rep. 2020;22:4899-908
87. Jiang L, Qi X, Lai M, Zhou J, Yuan M, You J. et al. WDR20 prevents hepatocellular carcinoma senescence by orchestrating the simultaneous USP12/46-mediated deubiquitination of c-Myc. Proc Natl Acad Sci U S A. 2024;121:e2407904121
88. Huang J, Gu ZL, Chen W, Xu YY, Chen M. Knockdown of ubiquitin-specific peptidase 13 inhibits cell growth of hepatocellular carcinoma by reducing c-Myc expression. Kaohsiung J Med Sci. 2020;36:615-21
89. Gao S, Chen T, Li L, Liu X, Liu Y, Zhao J. et al. Hypoxia-Inducible Ubiquitin Specific Peptidase 13 Contributes to Tumor Growth and Metastasis via Enhancing the Toll-Like Receptor 4/Myeloid Differentiation Primary Response Gene 88/Nuclear Factor-κB Pathway in Hepatocellular Carcinoma. Front Cell Dev Biol. 2020;8:587389
90. Huang G, Li L, Zhou W. USP14 activation promotes tumor progression in hepatocellular carcinoma. Oncol Rep. 2015;34:2917-24
91. Ding Y, Chen X, Wang B, Yu B, Ge J. Deubiquitinase inhibitor b-AP15 activates endoplasmic reticulum (ER) stress and inhibits Wnt/Notch1 signaling pathway leading to the reduction of cell survival in hepatocellular carcinoma cells. Eur J Pharmacol. 2018;825:10-8
92. Lv C, Wang S, Lin L, Wang C, Zeng K, Meng Y. et al. USP14 maintains HIF1-α stabilization via its deubiquitination activity in hepatocellular carcinoma. Cell Death Dis. 2021;12:803
93. Zhang N, Zhang H, Yang X, Xue Q, Wang Q, Chang R. et al. USP14 exhibits high expression levels in hepatocellular carcinoma and plays a crucial role in promoting the growth of liver cancer cells through the HK2/AKT/P62 axis. BMC Cancer. 2024;24:237
94. Xu MH, Zheng YM, Liang BG, Xu WX, Cao J, Wang P. et al. Deubiquitination of CIB1 by USP14 promotes lenvatinib resistance via the PAK1-ERK1/2 axis in hepatocellular carcinoma. Int J Biol Sci. 2024;20:3269-84
95. Ren Y, Song Z, Rieser J, Ackermann J, Koch I, Lv X. et al. USP15 Represses Hepatocellular Carcinoma Progression by Regulation of Pathways of Cell Proliferation and Cell Migration: A System Biology Analysis. Cancers (Basel). 2023;15:1371
96. Yao XQ, Li L, Piao LZ, Zhang GJ, Huang XZ, Wang Y. et al. Overexpression of Ubiquitin-Specific Protease15 (USP15) Promotes Tumor Growth and Inhibits Apoptosis and Correlated With Poor Disease-Free Survival in Hepatocellular Carcinoma. Technol Cancer Res Treat. 2020;19:1533033820967455
97. Qiu X, Dong S, Qiao F, Lu S, Song Y, Lao Y. et al. HBx-mediated miR-21 upregulation represses tumor-suppressor function of PDCD4 in hepatocellular carcinoma. Oncogene. 2013;32:3296-305
98. Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang Y, Tang GN. et al. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology. 2013;57:1882-92
99. You X, Liu F, Zhang T, Li Y, Ye L, Zhang X. Hepatitis B virus X protein upregulates oncogene Rab18 to result in the dysregulation of lipogenesis and proliferation of hepatoma cells. Carcinogenesis. 2013;34:1644-52
100. Ma NF, Lau SH, Hu L, Xie D, Wu J, Yang J. et al. COOH-terminal truncated HBV X protein plays key role in hepatocarcinogenesis. Clin Cancer Res. 2008;14:5061-8
101. Tu H, Bonura C, Giannini C, Mouly H, Soussan P, Kew M. et al. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res. 2001;61:7803-10
102. Qian Y, Wang B, Ma A, Zhang L, Xu G, Ding Q. et al. USP16 Downregulation by Carboxyl-terminal Truncated HBx Promotes the Growth of Hepatocellular Carcinoma Cells. Sci Rep. 2016;6:33039
103. Cai J, Liu T, Jiang X, Guo C, Liu A, Xiao X. Downregulation of USP18 inhibits growth and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma cells by suppressing BCL2L1. Exp Cell Res. 2017;358:315-22
104. Tian Z, Xu C, He W, Lin Z, Zhang W, Tao K. et al. The deubiquitinating enzyme USP19 facilitates hepatocellular carcinoma progression through stabilizing YAP. Cancer Lett. 2023;577:216439
105. Tang J, Long G, Xiao D, Liu S, Xiao L, Zhou L. et al. ATR-dependent ubiquitin-specific protease 20 phosphorylation confers oxaliplatin and ferroptosis resistance. MedComm (2020). 2023;4:e463
106. Li W, Cui K, Prochownik EV, Li Y. The deubiquitinase USP21 stabilizes MEK2 to promote tumor growth. Cell Death Dis. 2018;9:482
107. Yang TB, Yi F, Liu WF, Yang YH, Yang C, Sun J. Identification of hsa_circ_0039053 as an up-regulated and oncogenic circRNA in hepatocellular carcinoma via the miR-637-mediated USP21 activation. J Cancer. 2020;11:6950-9
108. Ling S, Shan Q, Zhan Q, Ye Q, Liu P, Xu S. et al. USP22 promotes hypoxia-induced hepatocellular carcinoma stemness by a HIF1α/USP22 positive feedback loop upon TP53 inactivation. Gut. 2020;69:1322-34
109. Ye Q, Zhou W, Xu S, Que Q, Zhan Q, Zhang L. et al. Ubiquitin-specific protease 22 promotes tumorigenesis and progression by an FKBP12/mTORC1/autophagy positive feedback loop in hepatocellular carcinoma. MedComm (2020). 2023;4:e439
110. Ning Z, Guo X, Liu X, Lu C, Wang A, Wang X. et al. USP22 regulates lipidome accumulation by stabilizing PPARγ in hepatocellular carcinoma. Nat Commun. 2022;13:2187
111. Li Q, Zhang L, You W, Xu J, Dai J, Hua D. et al. PRDM1/BLIMP1 induces cancer immune evasion by modulating the USP22-SPI1-PD-L1 axis in hepatocellular carcinoma cells. Nat Commun. 2022;13:7677
112. Bedekovics T, Hussain S, Zhang Y, Ali A, Jeon YJ, Galardy PJ. USP24 Is a Cancer-Associated Ubiquitin Hydrolase, Novel Tumor Suppressor, and Chromosome Instability Gene Deleted in Neuroblastoma. Cancer Res. 2021;81:1321-31
113. Wang YC, Wang SA, Chen PH, Hsu TI, Yang WB, Chuang YP. et al. Variants of ubiquitin-specific peptidase 24 play a crucial role in lung cancer malignancy. Oncogene. 2016;35:3669-80
114. Zhi X, Jiang S, Zhang J, Qin J. Ubiquitin-specific peptidase 24 accelerates aerobic glycolysis and tumor progression in gastric carcinoma through stabilizing PLK1 to activate NOTCH1. Cancer Sci. 2023;114:3087-100
115. Zhou N, Guo C, Li X, Tu L, Du J, Qian Q. et al. USP24 promotes hepatocellular carcinoma tumorigenesis through deubiquitinating and stabilizing TRAF2. Biochem Pharmacol. 2024;229:116473
116. Hu Z, Zhao Y, Mang Y, Zhu J, Yu L, Li L. et al. MiR-21-5p promotes sorafenib resistance and hepatocellular carcinoma progression by regulating SIRT7 ubiquitination through USP24. Life Sci. 2023;325:121773
117. Cao J, Wu S, Zhao S, Wang L, Wu Y, Song L. et al. USP24 promotes autophagy-dependent ferroptosis in hepatocellular carcinoma by reducing the K48-linked ubiquitination of Beclin1. Commun Biol. 2024;7:1279
118. Nelson JK, Thin MZ, Evan T, Howell S, Wu M, Almeida B. et al. USP25 promotes pathological HIF-1-driven metabolic reprogramming and is a potential therapeutic target in pancreatic cancer. Nat Commun. 2022;13:2070
119. Liu Y, Ma J, Lu S, He P, Dong W. USP25 promotes hepatocellular carcinoma progression by interacting with TRIM21 via the Wnt/β-catenin signaling pathway. Chin Med J (Engl). 2023;136:2229-42
120. Ma M, Yi L, Pei Y, Zhang Q, Tong C, Zhao M. et al. USP26 as a hepatitis B virus-induced deubiquitinase primes hepatocellular carcinogenesis by epigenetic remodeling. Nat Commun. 2024;15:7856
121. Wołowiec D, Mekki Y, Ffrench P, Manel AM, Bertrand Y, Rimokh R. et al. Differential expression of cell proliferation regulatory proteins in B- and T-lineage acute lymphoblastic leukaemias. Br J Haematol. 1996;95:518-23
122. Dong Y, Sui L, Tai Y, Sugimoto K, Hirao T, Tokuda M. Prognostic significance of cyclin E overexpression in laryngeal squamous cell carcinomas. Clin Cancer Res. 2000;6:4253-8
123. Fukuse T, Hirata T, Naiki H, Hitomi S, Wada H. Prognostic significance of cyclin E overexpression in resected non-small cell lung cancer. Cancer Res. 2000;60:242-4
124. Erlanson M, Landberg G. Prognostic implications of p27 and cyclin E protein contents in malignant lymphomas. Leuk Lymphoma. 2001;40:461-70
125. Dong L, Yu L, Bai C, Liu L, Long H, Shi L. et al. USP27-mediated Cyclin E stabilization drives cell cycle progression and hepatocellular tumorigenesis. Oncogene. 2018;37:2702-13
126. Zou T, Wang Y, Dong L, Che T, Zhao H, Yan X. et al. Stabilization of SETD3 by deubiquitinase USP27 enhances cell proliferation and hepatocellular carcinoma progression. Cell Mol Life Sci. 2022;79:70
127. Zhou W, Chen J, Wang J. Comprehensive prognostic and immunological analysis of Ubiquitin Specific Peptidase 28 in pan-cancers and identification of its role in hepatocellular carcinoma cell lines. Aging (Albany NY). 2023;15:6545-76
128. Zhang JF. MicroRNA-216b suppresses the cell growth of hepatocellular carcinoma by inhibiting Ubiquitin-specific peptidase 28 expression. Kaohsiung J Med Sci. 2020;36:423-8
129. Han H, Sun D, Li W, Shen H, Zhu Y, Li C. et al. A c-Myc-MicroRNA functional feedback loop affects hepatocarcinogenesis. Hepatology. 2013;57:2378-89
130. Zhang P, Sun H, Wen P, Wang Y, Cui Y, Wu J. circRNA circMED27 acts as a prognostic factor and mediator to promote lenvatinib resistance of hepatocellular carcinoma. Mol Ther Nucleic Acids. 2022;27:293-303
131. Feng J, Dai W, Mao Y, Wu L, Li J, Chen K. et al. Simvastatin re-sensitizes hepatocellular carcinoma cells to sorafenib by inhibiting HIF-1α/PPAR-γ/PKM2-mediated glycolysis. J Exp Clin Cancer Res. 2020;39:24
132. Bao MH, Wong CC. Hypoxia, Metabolic Reprogramming, and Drug Resistance in Liver Cancer. Cells. 2021;10:1715
133. Gao R, Buechel D, Kalathur RKR, Morini MF, Coto-Llerena M, Ercan C. et al. USP29-mediated HIF1α stabilization is associated with Sorafenib resistance of hepatocellular carcinoma cells by upregulating glycolysis. Oncogenesis. 2021;10:52
134. Gu L, Zhu Y, Lin X, Li Y, Cui K, Prochownik EV. et al. Amplification of Glyceronephosphate O-Acyltransferase and Recruitment of USP30 Stabilize DRP1 to Promote Hepatocarcinogenesis. Cancer Res. 2018;78:5808-19
135. Gu L, Zhu Y, Lin X, Lu B, Zhou X, Zhou F. et al. The IKKβ-USP30-ACLY Axis Controls Lipogenesis and Tumorigenesis. Hepatology. 2021;73:160-74
136. Li S, Song Y, Wang K, Liu G, Dong X, Yang F. et al. USP32 deubiquitinase: cellular functions, regulatory mechanisms, and potential as a cancer therapy target. Cell Death Discov. 2023;9:338
137. Xiu M, Bao W, Wang J, Chen J, Li Y, Hai Y. High USP32 expression contributes to cancer progression and is correlated with immune infiltrates in hepatocellular carcinoma. BMC Cancer. 2023;23:1105
138. Dou N, Hu Q, Li L, Wu Q, Li Y, Gao Y. USP32 promotes tumorigenesis and chemoresistance in gastric carcinoma via upregulation of SMAD2. Int J Biol Sci. 2020;16:1648-57
139. Li S, Yang L, Ding X, Sun H, Dong X, Yang F. et al. USP32 facilitates non-small cell lung cancer progression via deubiquitinating BAG3 and activating RAF-MEK-ERK signaling pathway. Oncogenesis. 2024;13:27
140. Gan Q, Shao J, Cao Y, Lei J, Xie P, Ge J. et al. USP33 regulates c-Met expression by deubiquitinating SP1 to facilitate metastasis in hepatocellular carcinoma. Life Sci. 2020;261:118316
141. Lv T, Zhang B, Jiang C, Zeng Q, Yang J, Zhou Y. USP35 promotes hepatocellular carcinoma progression by protecting PKM2 from ubiquitination-mediated degradation. Int J Oncol. 2023;63:113
142. Wang L, Wang J, Ma X, Ju G, Shi C, Wang W. et al. USP35 promotes HCC development by stabilizing ABHD17C and activating the PI3K/AKT signaling pathway. Cell Death Discov. 2023;9:421
143. Yuan X, Sun X, Shi X, Jiang C, Yu D, Zhang W. et al. USP39 promotes the growth of human hepatocellular carcinoma in vitro and in vivo. Oncol Rep. 2015;34:823-32
144. Pan Z, Pan H, Zhang J, Yang Y, Liu H, Yang Y. et al. Lentivirus mediated silencing of ubiquitin specific peptidase 39 inhibits cell proliferation of human hepatocellular carcinoma cells in vitro. Biol Res. 2015;48:18
145. Dong X, Liu Z, Zhang E, Zhang P, Wang Y, Hang J. et al. USP39 promotes tumorigenesis by stabilizing and deubiquitinating SP1 protein in hepatocellular carcinoma. Cell Signal. 2021;85:110068
146. Li X, Yuan J, Song C, Lei Y, Xu J, Zhang G. et al. Deubiquitinase USP39 and E3 ligase TRIM26 balance the level of ZEB1 ubiquitination and thereby determine the progression of hepatocellular carcinoma. Cell Death Differ. 2021;28:2315-32
147. Wang W, Lei Y, Zhang G, Li X, Yuan J, Li T. et al. USP39 stabilizes β-catenin by deubiquitination and suppressing E3 ligase TRIM26 pre-mRNA maturation to promote HCC progression. Cell Death Dis. 2023;14:63
148. Yuan X, Sun X, Shi X, Jiang C, Yu D, Zhang W. et al. USP39 regulates the growth of SMMC-7721 cells via FoxM1. Exp Ther Med. 2017;13:1506-13
149. Dong L, Yu L, Li H, Shi L, Luo Z, Zhao H. et al. An NAD(+)-Dependent Deacetylase SIRT7 Promotes HCC Development Through Deacetylation of USP39. iScience. 2020;23:101351
150. Liu Y, Wu Q, Sun T, Huang J, Han G, Han H. DNAAF5 promotes hepatocellular carcinoma malignant progression by recruiting USP39 to improve PFKL protein stability. Front Oncol. 2022;12:1032579
151. Wu Q, Qiu Y, Guo J, Yuan Z, Yang Y, Zhu Q. et al. USP40 promotes hepatocellular carcinoma cell proliferation, migration and stemness by deubiquitinating and stabilizing Claudin1. Biol Direct. 2024;19:13
152. Mo H, Li R, Yang N, Han J, Xiao X, Zhang Y. et al. USP40 promotes hepatocellular carcinoma progression through a YAP/USP40 positive feedback loop. Cancer Lett. 2024;589:216832
153. Zhou H, Yang L, Lin X, Chan TF, Lee NP, Tse WKF. et al. Integrated network findings reveal ubiquitin-specific protease 44 overexpression suppresses tumorigenicity of liver cancer. Aging (Albany NY). 2023;15:4304-18
154. Chen S, Zhou B, Huang W, Li Q, Yu Y, Kuang X. et al. The deubiquitinating enzyme USP44 suppresses hepatocellular carcinoma progression by inhibiting Hedgehog signaling and PDL1 expression. Cell Death Dis. 2023;14:830
155. Qiu Y, Huang D, Sheng Y, Huang J, Li N, Zhang S. et al. Deubiquitinating enzyme USP46 suppresses the progression of hepatocellular carcinoma by stabilizing MST1. Exp Cell Res. 2021;405:112646
156. Wen M, Xu H, Peng H, Sheng Y, Yang W, Yan J. MiR-27a-3p targets USP46 to inhibit the cell proliferation of hepatocellular carcinoma. Chem Biol Drug Des. 2022;100:280-9
157. Ren Y, Feng M, Hao X, Liu X, Li J, Li P. et al. USP48 Stabilizes Gasdermin E to Promote Pyroptosis in Cancer. Cancer Res. 2023;83:1074-93
158. Du L, Li Y, Kang M, Feng M, Ren Y, Dai H. et al. USP48 Is Upregulated by Mettl14 to Attenuate Hepatocellular Carcinoma via Regulating SIRT6 Stabilization. Cancer Res. 2021;81:3822-34
159. Qian X, Li S, Yang Z, Zhang J. The long non-coding RNA HLNC1 potentiates hepatocellular carcinoma progression via interaction with USP49. J Clin Lab Anal. 2020;34:e23462
160. Wu L, Yu K, Chen K, Zhu X, Yang Z, Wang Q. et al. Fbxo45 facilitates pancreatic carcinoma progression by targeting USP49 for ubiquitination and degradation. Cell Death Dis. 2022;13:231
161. Liu Z, Li J, Ding Y, Ma M, Chen J, Lei W. et al. USP49 mediates tumor progression and poor prognosis through a YAP1-dependent feedback loop in gastric cancer. Oncogene. 2022;41:2555-70
162. Yao Y, Ma W, Guo Y, Liu Y, Xia P, Wu X. et al. USP53 plays an antitumor role in hepatocellular carcinoma through deubiquitination of cytochrome c. Oncogenesis. 2022;11:31
163. Urbanik T, Köhler BC, Boger RJ, Wörns MA, Heeger S, Otto G. et al. Down-regulation of CYLD as a trigger for NF-κB activation and a mechanism of apoptotic resistance in hepatocellular carcinoma cells. Int J Oncol. 2011;38:121-31
164. Pannem RR, Dorn C, Ahlqvist K, Bosserhoff AK, Hellerbrand C, Massoumi R. CYLD controls c-MYC expression through the JNK-dependent signaling pathway in hepatocellular carcinoma. Carcinogenesis. 2014;35:461-8
165. Ni F, Zhao H, Cui H, Wu Z, Chen L, Hu Z. et al. MicroRNA-362-5p promotes tumor growth and metastasis by targeting CYLD in hepatocellular carcinoma. Cancer Lett. 2015;356:809-18
166. Liu Y, Chai Y, Zhang J, Tang J. A Function Variant at miR-501 Alters Susceptibility to Hepatocellular Carcinoma in a Chinese Han Population. Cell Physiol Biochem. 2016;38:2500-8
167. Huang DH, Wang GY, Zhang JW, Li Y, Zeng XC, Jiang N. MiR-501-5p regulates CYLD expression and promotes cell proliferation in human hepatocellular carcinoma. Jpn J Clin Oncol. 2015;45:738-44
168. Liu J, Su Z, Zeng Y, Zhang H, Yang S, Liu G. miR-922 regulates CYLD expression and promotes the cell proliferation of human hepatocellular carcinoma. Oncol Rep. 2017;37:1445-50
169. Pu J, Wu X, Wu Y, Shao Z, Luo C, Tang Q. et al. Anti-oncogenic effects of SOX2 silencing on hepatocellular carcinoma achieved by upregulating miR-222-5p-dependent CYLD via the long noncoding RNA CCAT1. Aging (Albany NY). 2021;13:12207-23
170. Ni Q, Chen J, Li X, Xu X, Zhang N, Zhou A. et al. Expression of OTUB1 in hepatocellular carcinoma and its effects on HCC cell migration and invasion. Acta Biochim Biophys Sin (Shanghai). 2017;49:680-8
171. Xie P, Chen Y, Zhang H, Zhou G, Chao Q, Wang J. et al. The deubiquitinase OTUD3 stabilizes ACTN4 to drive growth and metastasis of hepatocellular carcinoma. Aging (Albany NY). 2021;13:19317-38
172. Dai H, Wu B, Ge Y, Hao Y, Zhou L, Hong R. et al. Deubiquitylase OTUD3 regulates integrated stress response to suppress progression and sorafenib resistance of liver cancer. Cell Rep. 2024;43:114487
173. Tian X, Zhang S, Zhou L, Seyhan AA, Hernandez Borrero L, Zhang Y. et al. Targeting the Integrated Stress Response in Cancer Therapy. Front Pharmacol. 2021;12:747837
174. Kalkavan H, Chen MJ, Crawford JC, Quarato G, Fitzgerald P, Tait SWG. et al. Sublethal cytochrome c release generates drug-tolerant persister cells. Cell. 2022;185:3356-74.e22
175. Calvo V, Zheng W, Adam-Artigues A, Staschke KA, Huang X, Cheung JF. et al. A PERK-Specific Inhibitor Blocks Metastatic Progression by Limiting Integrated Stress Response-Dependent Survival of Quiescent Cancer Cells. Clin Cancer Res. 2023;29:5155-72
176. Zhao X, Su X, Cao L, Xie T, Chen Q, Li J. et al. OTUD4: A Potential Prognosis Biomarker for Multiple Human Cancers. Cancer Manag Res. 2020;12:1503-12
177. Yang YN, Jia SY, Zhu N, Xiao XL, Ma Y, Tu KS. et al. OTUD5 promotes the growth of hepatocellular carcinoma by deubiquitinating and stabilizing SLC38A1. Biol Direct. 2024;19:12
178. Ding C, Cao L, Wang R, Wu Q, Li M, Zhang J. et al. OTUD7B is a new deubiquitinase targeting p53. Theranostics. 2025;15:2121-38
179. Chen YP, Zhang XF. TRABID targets DDB2 for deubiquitination to promote proliferation of hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2022;625:23-30
180. Fang Y, Mu J, Ma Y, Ma D, Fu D, Shen X. The interaction between ubiquitin C-terminal hydrolase 37 and glucose-regulated protein 78 in hepatocellular carcinoma. Mol Cell Biochem. 2012;359:59-66
181. Fang Y, Fu D, Tang W, Cai Y, Ma D, Wang H. et al. Ubiquitin C-terminal Hydrolase 37, a novel predictor for hepatocellular carcinoma recurrence, promotes cell migration and invasion via interacting and deubiquitinating PRP19. Biochim Biophys Acta. 2013;1833:559-72
182. Chen X, Huang A, Wang Y, Chen F, Hu B, Zhang X. et al. BRCA1-associated protein 1 serves as a tumor suppressor in hepatocellular carcinoma by deubiquitinating and stabilizing PTEN. Am J Cancer Res. 2021;11:2044-61
183. Haberhausen G, Damian MS, Leweke F, Müller U. Spinocerebellar ataxia, type 3 (SCA3) is genetically identical to Machado-Joseph disease (MJD). J Neurol Sci. 1995;132:71-5
184. Paulson HL, Das SS, Crino PB, Perez MK, Patel SC, Gotsdiner D. et al. Machado-Joseph disease gene product is a cytoplasmic protein widely expressed in brain. Ann Neurol. 1997;41:453-62
185. Sacco JJ, Yau TY, Darling S, Patel V, Liu H, Urbé S. et al. The deubiquitylase Ataxin-3 restricts PTEN transcription in lung cancer cells. Oncogene. 2014;33:4265-72
186. Shi Z, Chen J, Zhang X, Chu J, Han Z, Xu D. et al. Ataxin-3 promotes testicular cancer cell proliferation by inhibiting anti-oncogene PTEN. Biochem Biophys Res Commun. 2018;503:391-6
187. Li D, Zhang T, Lai J, Zhang J, Wang T, Ling Y. et al. MicroRNA-25/ATXN3 interaction regulates human colon cancer cell growth and migration. Mol Med Rep. 2019;19:4213-21
188. Buus R, Faronato M, Hammond DE, Urbé S, Clague MJ. Deubiquitinase activities required for hepatocyte growth factor-induced scattering of epithelial cells. Curr Biol. 2009;19:1463-6
189. Zeng C, Zhao C, Ge F, Li Y, Cao J, Ying M. et al. Machado-Joseph Deubiquitinases: From Cellular Functions to Potential Therapy Targets. Front Pharmacol. 2020;11:1311
190. Ma X, Qi W, Yang F, Pan H. Deubiquitinase JOSD1 promotes tumor progression via stabilizing Snail in lung adenocarcinoma. Am J Cancer Res. 2022;12:2323-36
191. Jing C, Liu DD, Lai QC, Li LQ, Zhou MQ, Ye BB. et al. JOSD1 promotes proliferation and chemoresistance of head and neck squamous cell carcinoma under the epigenetic regulation of BRD4. Cancer Cell Int. 2021;21:12
192. Sun Y, Liu D, Zhang X, Su P, Li X, Li Z. et al. Regulation of Hippo/YAP axis in colon cancer progression by the deubiquitinase JOSD1. Cell Death Discov. 2024;10:365
193. Wu X, Luo Q, Zhao P, Chang W, Wang Y, Shu T. et al. JOSD1 inhibits mitochondrial apoptotic signalling to drive acquired chemoresistance in gynaecological cancer by stabilizing MCL1. Cell Death Differ. 2020;27:55-70
194. Qian M, Yan F, Wang W, Du J, Yuan T, Wu R. et al. Deubiquitinase JOSD2 stabilizes YAP/TAZ to promote cholangiocarcinoma progression. Acta Pharm Sin B. 2021;11:4008-19
195. Yuan T, Zeng C, Liu J, Zhao C, Ge F, Li Y. et al. Josephin domain containing 2 (JOSD2) promotes lung cancer by inhibiting LKB1 (Liver kinase B1) activity. Signal Transduct Target Ther. 2024;9:11
196. Zhou L, Chen G, Liu T, Liu X, Yang C, Jiang J. MJDs family members: Potential prognostic targets and immune-associated biomarkers in hepatocellular carcinoma. Front Genet. 2022;13:965805
197. Huang Y, Zeng J, Liu T, Xu Q, Song X, Zeng J. Deubiquitinating enzyme JOSD2 promotes hepatocellular carcinoma progression through interacting with and inhibiting CTNNB1 degradation. Cell Biol Int. 2022;46:1089-97
198. Tang J, Luo Y, Long G, Zhou L. MINDY1 promotes breast cancer cell proliferation by stabilizing estrogen receptor α. Cell Death Dis. 2021;12:937
199. Luo Y, Zhou J, Tang J, Zhou F, He Z, Liu T. et al. MINDY1 promotes bladder cancer progression by stabilizing YAP. Cancer Cell Int. 2021;21:395
200. Song XC, Li WJ, Tian CY, Ma X, Yang WB, Zhou JH. Study on the mechanism of liver cancer immune escape mediated by MINDY1 through regulation of PD-L1 ubiquitination level. Biomol Biomed. 2024;25:144-154
201. Xia BL, Liu KW, Huang HX, Shen MM, Wang B, Gao J. Deubiquitinating enzyme MINDY1 is an independent risk factor for the maintenance of stemness and poor prognosis in liver cancer cells. Zhonghua gan zang bing za zhi = Zhonghua ganzangbing zazhi = Chinese journal of hepatology. 2023;31:518-23
202. Wang Y, Liu Y, Ma C, Liu C, Tang Q, Wang Z. et al. Deubiquitinase PSMD14 promotes tumorigenicity of glioblastoma by deubiquitinating and stabilizing β-catenin. Biofactors. 2024;50:1134-1147
203. Lai JB, Kong WK, Fu QC, Jiang ZC, Sun BH, Ye X. et al. PSMD14 is a novel prognostic marker and therapeutic target in osteosarcoma. Diagn Pathol. 2024;19:8
204. Li L, Yang XM, He MF, Xu XC, Xuan XF, Zhang JR. et al. The expression and clinical significance of STAMBP in breast cancer. Mol Biol Rep. 2023;50:899-906
205. Yang Q, Yan D, Zou C, Xue Q, Lin S, Huang Q. et al. The deubiquitinating enzyme STAMBP is a newly discovered driver of triple-negative breast cancer progression that maintains RAI14 protein stability. Exp Mol Med. 2022;54:2047-59
206. Iwakami Y, Yokoyama S, Watanabe K, Hayakawa Y. STAM-binding protein regulates melanoma metastasis through SLUG stabilization. Biochem Biophys Res Commun. 2018;507:484-8
207. Huang D, Nagata Y, Grossmann V, Radivoyevitch T, Okuno Y, Nagae G. et al. BRCC3 mutations in myeloid neoplasms. Haematologica. 2015;100:1051-7
208. Jin J, Wang Y, Hu Y. STAMBPL1, transcriptionally regulated by SREBP1, promotes malignant behaviors of hepatocellular carcinoma cells via Wnt/β-catenin signaling pathway. Mol Carcinog. 2024;63:2158-73
209. Wang D, Ma L, Wang B, Liu J, Wei W. E3 ubiquitin ligases in cancer and implications for therapies. Cancer Metastasis Rev. 2017;36:683-702
210. Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov. 2011;10:29-46
211. Deng L, Meng T, Chen L, Wei W, Wang P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther. 2020;5:11
212. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029-33
213. Cha JH, Chan LC, Li CW, Hsu JL, Hung MC. Mechanisms Controlling PD-L1 Expression in Cancer. Mol Cell. 2019;76:359-70
214. Gou Q, Dong C, Xu H, Khan B, Jin J, Liu Q. et al. PD-L1 degradation pathway and immunotherapy for cancer. Cell Death Dis. 2020;11:955
215. Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M. et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2020;21:341-52
216. Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE. et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060-72
217. Sun L, He M, Li F, Wu D, Zheng P, Zhang C. et al. Oxyberberine sensitizes liver cancer cells to sorafenib via inhibiting NOTCH1-USP7-c-Myc pathway. Hepatol Commun. 2024;8:e0405
218. Liu H, Chen W, Liang C, Chen BW, Zhi X, Zhang S. et al. WP1130 increases doxorubicin sensitivity in hepatocellular carcinoma cells through usp9x-dependent p53 degradation. Cancer Lett. 2015;361:218-25
Author contact
![]() Corresponding authors: Liang Xiao, Xingyu Mi, Jianing Tang and Ledu Zhou; Department of Liver Surgery, Xiangya Hospital, Central South University, 110 Xiangya Road, Changsha, Hunan 410078, China; xiaoliangrickedu.cn; mixingyu1998com; tangjnedu.cn; zhouldedu.cn.
Corresponding authors: Liang Xiao, Xingyu Mi, Jianing Tang and Ledu Zhou; Department of Liver Surgery, Xiangya Hospital, Central South University, 110 Xiangya Road, Changsha, Hunan 410078, China; xiaoliangrickedu.cn; mixingyu1998com; tangjnedu.cn; zhouldedu.cn.

 Global reach, higher impact
Global reach, higher impact