10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2025; 21(11):4701-4718. doi:10.7150/ijbs.115097 This issue Cite
Research Paper
TSG6 promotes epithelial-mesenchymal transition and tumor-associated macrophage polarization through Smad2/3 and MAPK signaling by facilitating TSG6-CD44-TGFβR1 or EGFR complex formation
Department of Pharmacy, College of Pharmacy, Institute of Pharmaceutical Science and Technology, Hanyang University, Ansan, Gyeonggi-do 15588, Republic of Korea.
*The authors equally contributed.
Received 2025-4-4; Accepted 2025-7-2; Published 2025-7-24
Abstract
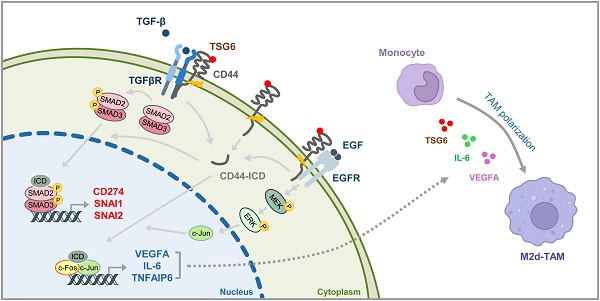
TSG6 is highly expressed during PLK1-induced epithelial-mesenchymal transition (EMT). However, the role of TSG6 in the tumor microenvironment (TME) remains poorly understood. We investigate the function and regulatory mechanisms of TSG6 in immune plasticity within the TME of lung adenocarcinoma (LUAD). The simultaneous high expression of TSG6 and PLK1 in LUAD patients was associated with lower survival rates. TSG6 and CD44 were markedly upregulated during EMT driven by TGF-b or active PLK1 in A549 and HCC827 cells. TSG6 treatment enhanced EMT by increasing N-cadherin and phosphorylated Smad2 levels. TSG6 depletion blocked the effects, which was restored upon TSG6 retreatment. Additionally, TSG6 treatment induced polarization of THP-1 monocytes into M2d tumor-associated macrophages (TAMs). In cocultures of THP-1 monocytes with A549 cells expressing TSG6, M2d-inducing factors in A549 cells and M2d markers in THP-1 cells were upregulated. Immunoprecipitation showed that TSG6 binds CD44, enhancing CD44's interaction with TGFbR or EGFR. In TSG6-treated LUAD cells, both total CD44 and its cleaved intracellular domain increased by activating TGFβR1-Smad2/3 and MAPK-ERK1/2-AP-1 pathways. Thus, TSG6 promotes EMT and M2d-TAMs polarization by activating TGFβR1/Smad and MAPK/ERK pathway through direct interaction between CD44 and TGFβR1 or EGFR.
Keywords: TSG6, invasiveness, tumor-associated macrophages, CD44
Introduction
The tumor microenvironment (TME) is a complex environment consisting of cancer cells, blood vessels, extracellular matrix, and immune cells including T cells, B cells, monocytes and dendritic cells [1-3]. The interactions between cancer cells and immune cells or blood vessel cells have been studied to understand the cell plasticity in TME [1-3]. The composition of TME depends on the tumor type but typically includes immune cells, blood vessels, and extracellular matrix (ECM) [1]. During the early stages of tumor growth, mutual relationships between cancer cells and TME components support the survival of cancer cells and development of malignancy [1, 4]. Especially, in tumor malignancy, monocytes are closely involved by differentiating into tumor-associated macrophages (TAMs) [5]. In the TME, M2-like TAMs reside and functions as anti-inflammatory and tumor-supporting macrophages [5]. M2 macrophages are classified into M2a, M2b, M2c, and M2d macrophages, which are stimulated by IL-4/IL-13, lipopolysaccharide/IL-1 receptor, IL-10, and VEGF, respectively [2]. Polarized M2 phenotype plays a crucial role in tumor proliferation, metastasis, and immune evasion by expressing anti-inflammatory factors such as IL-10 and transforming growth factor (TGF)-β [2]. Mononuclear cells are attracted by cytokines including CCL-2, CSF1, and VEGF and they are typically polarized into TAMs. The polarized M2-like macrophage markers and cytokines such as mannose receptors (CD206), scavenger receptors (CD163), VEGF, and IL-10, exhibit tumor-supporting effects [2].
Tumor necrosis factor (TNF)-stimulated gene-6 (TSG6) is the hyaluronan (HA)-binding protein through its LINK domains [6, 7]. HA synthesized by HAS releases into the ECM, which undergoes macromolecular reconstruction through its interacts with HA-binding proteins, including CD44 [8]. The interaction between HA and CD44 is an instrumental event in regulating inflammation and cancer [9]. TSG6 also forms an oligomer with HA to regulate the cross-linking of the HA and modify the ECM [10]. Moreover, both full-length TSG6 and its LINK domain enhance the binding of HA to CD44 on the cell surface, which results in leukocyte rolling and their recruitment to sites of inflammation [11]. The precise role of TSG6 in TME has remained vague, because it is upregulated in the pro-inflammatory environment [6]. Interestingly, it is also upregulated in several cancer patients including lung cancer [12], colorectal cancer (CRC) [13], head and neck cancer [14], and urothelial carcinomas [15] with poor prognosis. Notably, its overexpression significantly increased in metastatic CRC patients, activating JAK2-STAT3 signals for metastasis [13]. Furthermore, we recently found that TNFAIP6 (encoding TSG6) is the most highly expressed gene in active PLK1-driven EMT of non-small cell lung cancer (NSCLC) cells [12].
PLK1, a mitotic serine/threonine kinase, serves as a master regulator of mitosis, controlling processes from centrosome maturation at mitotic entry to mitotic exit through phosphorylation activity [16]. Because of its pivotal involvement in cell division, PLK1 contributes to cell proliferation, and its overexpression is positively associated with various cancers and with advanced stages of several malignancies [16-19] including those of the breast [17], prostate [18], and stomach [19]. In the context of lung adenocarcinoma, PLK1 promotes the EMT by phosphorylating key factors such as β-catenin [20], vimentin [21], and FoxM1 [4]. Notably, our previous work demonstrated that the active form of PLK1, phosphorylated at Thr210, drives EMT in NSCLC [12]. During this process, NSCLC cells exhibited elevated expression of TNFAIP6. Depleting TNFAIP6 showed its pivotal role in PLK1-mediated EMT, especially in cell migration and invasion [12]. Then, we observed that its expression was transcriptionally regulated by phosphorylated β-catenin by PLK1 in NSCLC cells [20]. However, the functions of TSG6 in the TME are not clearly understood. Here, we have found that TSG6 facilitates M2d-like macrophage polarization and EMT in the TME of LUAD by activating TGFβR1/Smad and MAPK/ERK signaling through its direct interaction with its receptor CD44, which promotes the complex formation with TGFβR1 or EGFR.
Materials and methods
Bioinformatics analysis
The patient data were extracted from an online database (www.kmplot.com) following previous reports [4, 21, 22]. In the database, all cancer patients were identified from the Cancer Genome Atlas (http://cancergenome.nih.gov) or the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). To establish the clinical relevance of PLK1 and TNFAIP6 expression to the survival of patients, the database was established using these genes expression data and survival information from 1,152 patients after excluding biased arrays. The samples were split into low and high groups using the median expression of each factor. Hazard ratios (HRs) with 95% confidence intervals. Log-rank P were calculated according to the online formulas provided by each database. A log-rank P value of < 0.05 was considered statistically significant. In survival analysis, HR represents the ratio of the hazard rates between two levels of an explanatory variable as shown in Table S1.
Cell culture and treatment
Human monocyte THP-1, lung fibroblast MRC5, lung adenocarcinoma (LUAD) A549, and HCC827 cells were purchased from the KCLB (Seoul, Korea). HEK293T cells were purchased from ATCC (Manassas, VA, USA). A549, HCC827, and THP-1 cells were cultured in RPMI-1640 (Corning Cellgro; Manassas, VA, USA), MRC5 cells were cultured in MEM (Corning Cellgro), and HEK293T cells were cultured in DMEM (Corning Cellgro), supplemented with 10% FBS in the presence of penicillin and streptomycin (Corning Cellgro) in a humidified 5% CO2 incubator at 37°C. For TGF-β treatment, cells were treated with 2.5 ng/ml TGF-β for 48 hours based on a previous study [12]. Cells were seeded at 2.5×104 cells/ml for 16 hours. For recombinant human TSG6 (rhTSG6) treatment, THP-1 cells were seeded 1.5×105 cells/ml and treated with 200 ng/ml with rhTSG6 for two hours or the indicated duration after 24 hours based on previous studies [23, 24]. 10 μM trametinib was applied for 48 hours based on the GI50 value in A549 cells [25]. All other chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA). For the coculture system, human monocytes THP-1 cells and A549 cells expressing TSG6 (TSG6-WT) were cultured in RPMI-1640 (Corning Cellgro) supplemented with 10% FBS in the presence of penicillin and streptomycin (Corning Cellgro) in a humidified 5% CO2 incubator at 37 °C.
Immunoblotting
For cell lysis, lysis buffer (1 mM dithiothreitol, 1 mM EGTA, 50 mM β-glycerophosphate, 0.5% Triton X-100, 20 mM Tris, pH 7.5, 2 mM MgCl2, 25 mM NaF, 1 mM Na3VO4, 100 mg/ml PMSF, and a protease inhibitor cocktail (Roche; Indianapolis, IN, USA)) was used. Proteins were resolved by SDS-PAGE after adjustment of the protein concentration, and subjected to immunoblot analysis with appropriate antibodies as follows: TSG6 (R&D systems, MAB2104; Minneapolis, USA); CD44 (Santa Cruz Biotechnology, sc-9960); PLK (Santa Cruz Biotechnology, sc-17783); phospho-PLK1T210 (Cell Signaling, 5472; Danvers, USA); N-cadherin (Sigma, C3865); E-cadherin (Cell Signaling, 4065); Smad2/3 (Cell Signaling, 8685); p-Smad2S465/S467 (Cell Signaling, 18338); β-actin (Santa Cruz Biotechnology, sc-47772); GAPDH (Santa Cruz Biotechnology, sc-47724); and IgG (Santa Cruz Biotechnology, sc-2027). Immune complexes were detected using an Odyssey infrared imaging system (LI-COR Biosciences; Lincoln, NE, USA). The values of intensity were determined using Photoshop software.
Quantitative real time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells at 48 h after exposure to TGF-β and quantified by Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA). Next, cDNA was generated by using a First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). The synthesized cDNA was mixed with SYBR Green Master Mix (Bio-Rad Laboratories, Hercules, CA, USA) and gene-specific primers, and then qRT-PCR was conducted using a CFX96 Real-Time PCR system (Bio-Rad Laboratories). The levels of mRNA were normalized using GAPDH as an internal control and all normalized values within the dataset were calculated relative to untreated control cells with the 2-ΔΔCT method [26]. The primer sequences used are shown in Table S2.
Lentivirus-based plasmid expression
For the lentiviral expression of TNFAIP6 (gene ID no. 7130) or PLK1 (gene ID no. 18817), we used pLVX-TRE3G-eRFP and pLVX-Tet3G vectors (Clontech #631351; CA, USA). TNFAIP6 and various versions of PLK1, including wild-type, T210D, and W414F/V415A mutants, were as described previously [12]. The various versions of PLK1 were subcloned into pLVX-TRE3G-eRFP and a lentivirus expressing PLK1 was generated according to the manufacturer's guide and previous reports [20]. For viral infection, cells were treated with viral particles from pLVX-Tet3G and pLVX-TRE3G-eRFP-PLK1 or pLVX-TRE3G-eRFP-TNFAIP6 with 10 µg/ml polybrene and 10 mM HEPES. G418 and puromycin were treated to select the infected cells at a concentration of 500 µg/ml or and 2 µg/ml for 5 days or 2 days, respectively. The expression of PLK1 or TSG6 was induced with 2 µg/ml of doxycycline.
Lentivirus-based shRNA preparation
To deplete TNFAIP6, we designed lentivirus-based shRNA transfer plasmids to target human TNFAIP6 (gene access no. NM_007115) at positions 530-550 (CAAATGAGTACGAAGATAA, shTNFAIP6#1) or positions 693-713 (GGGAAGATACTGTGGAGATGA, shTNFAIP6#2), and then the lentivirus was generated. The infected cells were selected using 2 µg/ml puromycin for 2 days.
Immunohistochemistry analyses from animal studies
Four-week-old BALB/c nude mice (male, Orient Bio Inc., Seoul, Korea) were injected with 1 × 106 A549 cells (in 100 μl PBS) expressing pLVX-TRE3G-eRFP-Tet3G-Mock, wild-type PLK1 (PLK1-WT), T210D mutant PLK1 (PLK1-TD), and W414F/V415A mutant PLK1 (PLK1-FA) through the tail vein as described previously [12]. To induce PLK1, doxycycline was administrated in their drinking water at a concentration of 1 mg/ml. All animal experiments were approved and managed by the guidelines of the Institutional Animal Care and Use Committee, Hanyang University (HY-IACUC-2017-0115A), as described previously [12]. Twelve weeks after the injection, mice were sacrificed. The lungs were separated and fixed in 4% paraformaldehyde for the staining of TSG6, CD44, CD68, and CD163. Briefly, tumor tissue sections were deparaffinized and rehydrated with xylene and graded alcohol, respectively. The sections were permeabilized with PBS containing 0.1% Triton X-100 (PBST) at room temperature (RT) and treated with peroxide blocking solution at RT. After washing two times with PBS, the sections were incubated with anti-TSG6 (R&D systems, MAB2104, 1:200), anti-CD44 (Santa Cruz Biotechnology, sc-9960, 1:200), anti-CD68 (Santa Cruz Biotechnology Inc. sc-20060, 1:200), and anti-CD163 (ABclonal Inc., Gyeonggi, Korea, A8383, 1:200) antibodies overnight at 4℃. The next day, the slides were washed three times with Tris-buffered saline and incubated with secondary antibody-conjugated peroxidase. After further washing, immunoperoxidase staining was developed using a DAB chromogen kit (K5007, Dako, Glostrup, Denmark) and counterstained with Meyer's hematoxylin (SH-002, KP&T, Osong, Korea). Images of cells were obtained and evaluated with a confocal microscope FW3000 (Olympus; Tokyo, Japan).
Immunoprecipitation assay
Cell lysates were incubated with anti-CD44 (Santa Cruz Biotechnology, sc-9960) and normal IgG (Santa Cruz Biotechnology) antibodies for 16 h at 4 °C with end-over-end mixing, followed by incubation with protein G agarose (Santa Cruz Biotechnology) for 4 h at 4°C. Immunoprecipitants were divided from the supernatants by centrifugation and washed four times with lysis buffer. Proteins were resolved by SDS-PAGE and then analyzed by immunoblotting.
Chromatin immunoprecipitation assays
We examined the interaction between c-Jun (AP-1) and the promoters of IL6, TNFAIP6, and VEGFA in A549 cells treated with rhTSG6. Cross-linking reaction was performed with 1.4% formaldehyde. Cells were lysed with IP buffer. Chromatin was sheared by sonication and incubated with antibody to c-Jun (Santa Cruz Biotechnology, sc-74543) or normal IgG for 16 hours. Sheared chromatin was incubated with a specific protein A/G bead for 4 hours and then washed five times with IP buffer. To the washed beads, Chelex 100 slurry (Bio-Rad; Hercules, CA, USA) was added and then the samples were boiled and incubated with Proteinase K (Invitrogen; Carlsbad, CA, USA) at 55°C for 30 min. The samples cleared by centrifugation, and the supernatants were taken for real-time PCR. The bound chromatin fractions were amplified with promoter-specific primers of human IL6, TNFAIP6, and VEGFA (Table S3) for 40 cycles. For the interaction between Smad2/3 and the promoters of CD274 (encoding PD-L1), SNAI1, and SNAI2 in rhTSG6-treated A549 cells, anti-Smad2/3 antibody (#8685, Cell signaling) and promoter-specific primers of human CD274, SNAI1, and SNAI2 (Table S3) were used. Real-time PCR was performed using SYBR Green Master Mix (Bio-Rad, #1708880) with a CFX96 Real-Time PCR system (Bio-Rad). The data were analyzed using the comparative CT (ΔΔCT) method.
Statistical analysis
All data are expressed as the means ± SDs of at least three independent experiments, each conducted in triplicate. Statistical analysis was performed using the Student's t-test and statistical significance was set at p < 0.05.
Results
Concurrent expression of TNFAIP6 and anti-inflammatory cytokines correlate with the low survival rate of lung adenocarcinoma patients
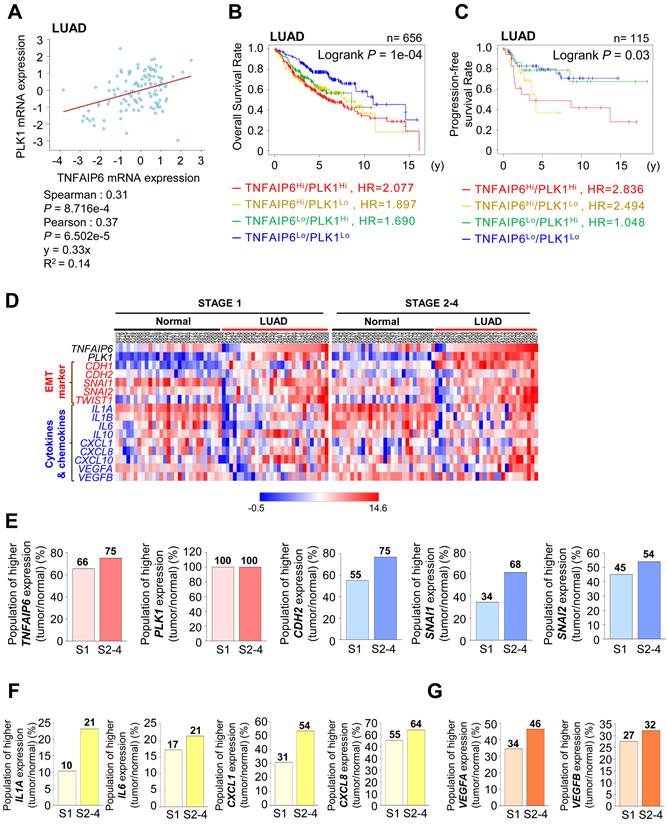
TNFAIP6 was identified as the most highly expressed gene during active PLK1-driven EMT of NSCLC [12] (Fig. S1A). Because of this, we investigated the correlation between TNFAIP6 and PLK1 expression in LUAD and squamous cell carcinoma (LUSQ) (Fig.1A, Fig. S1B). In LUAD, a positive correlation was observed between TNFAIP6 and PLK1 mRNA expression (Spearman: 0.31, p=8.716e-4; Pearson: 0.37, p=6.502e-5) (Fig. 1A). In contrast, a negative correlation was identified in LUSQ (Spearman: -0.43, p=1.539e-4; Pearson: -0.37, p=1.484e-3) (Fig. S1B). A clinical relevance between the expression of TNFAIP6/PLK1 and overall survival rate (OS) was observed in patients with lung cancer (Fig. S1C), LUAD (Fig. 1B), and LUSQ (Fig. S1D) using a Kaplan-Meier (KM) plot database [22]. The OS of LUAD patients (n = 656) with high TNFAIP6Hi/PLK1Hi expression was significantly shorter than those with low TNFAIP6Lo/PLK1Lo expression (red line, HR=2.077, Log-rank p =0.0001) (Fig. 1B, Table S1). Additionally, the progression-free survival rate (PFS) of LUAD patients (n = 115) with TNFAIP6Hi/PLK1Hi expression was much shorter than those with low TNFAIP6Lo/PLK1Lo expression (red line, HR=2.836, Log-rank p =0.03) (Fig. 1C). However, in LUSQ patients (n = 300), no significant correlation was observed between TNFAIP6 and PLK1 expression and OS (Fig. S1D, Table S1). Therefore, the high expression of TNFAIP6 and PLK1 is associated with poor prognosis in LUAD patients, but not in LUSQ.
We then determined the correlation of expression among TNFAIP6, PLK1, and mesenchymal markers depending on stages. A heatmap analysis was conducted using a TCGA dataset of LUAD patients for TNFAIP6, PLK1, and mesenchymal markers including CDH2, SNAI1, and SNAI2 in paired normal tissue (Fig. 1D-E, left, normal) and adjacent tumor tissues (Fig. 1D-E, right, LUAD) depending on stages. The expression levels and frequencies of TNFAIP6, PLK1, CDH2, SNAI1, and SNAI2 were higher in stages 2-4 than in those of stage 1 (Fig. 1D-E). In the genomic analysis of LUAD, the levels of TNFAIP6 were higher in patients with advanced stages 2-4 (75%) than in those with tumor stage 1 (66%) (Fig. 1D-E), indicating that TNFAIP6 and PLK1 were markedly expressed in advanced LUAD. Previously we found that the immune system process and inflammatory response were ranked within the top 5 pathways in KEGG pathway analysis using invasive cells expressing active PLK1 [12]. Given that TNFAIP6 is related to inflammation and cancer [9], the expression of cytokines and chemokines was observed in both normal tissues and adjacent tumor tissues of LUAD patients. The levels of IL1A and IL6 were higher in patients with advanced stages than in those with tumor stage 1 (Fig. 1D-F). In addition, the levels of CXCL1, CXCL8, VEGFA, and VEGFB related to monocyte recruitment and angiogenesis were higher in patients with advanced stages than in those with tumor stage 1 (Fig. 1D-G). The expression of these cytokines correlated with the expression of TNFAIP6 in advanced LUAD. Therefore, advanced LUAD showed a higher expression of TNFAIP6, PLK1, and cytokines related to monocyte recruitment, macrophage polarization, and angiogenesis.
Clinical correlation between the survival rate of patients and the expression of TSG6 and PLK1 in lung adenocarcinoma (LUAD). (A) Analysis of Spearman's and Pearson's correlation coefficients between the expression of TNFAIP6 and PLK1 in LUAD patients using cBioportal. (B-C) The overall survival (OS) of LUAD patients (n = 656, Log-rank P = 1e-04) (B) and the progression-free survival (PFS) of LUAD patients (n = 115, Log-rank P = 0.03) (C) were analyzed according to the expression of TNFAIP6 and PLK1 using KM PLOTTER. High (Hi) and low (Lo) were generated by dividing patients according to their expression at the median cut-off. (D) A heatmap analysis was performed using a dataset of LUAD patients from TCGA for the expression of TNFAIP6, PLK1, epithelial-mesenchymal transition markers (EMT), and cytokines and chemokines markers in paired normal (left) and tumor tissues (right) depending on stages. (E) The ratio of increased expression of TNFAIP6, PLK1, and EMT markers (CDH2, SNAI1, and SNAI2) and (F-G) cytokine and chemokine markers (IL1A, IL6, CXCL1, CXCL8, VEGFA, and VEGFB) in tumor tissue compared to adjacent normal tissue were plotted.
TSG6 and its receptor CD44 are highly expressed in TGF-β-induced EMT of LUAD cells
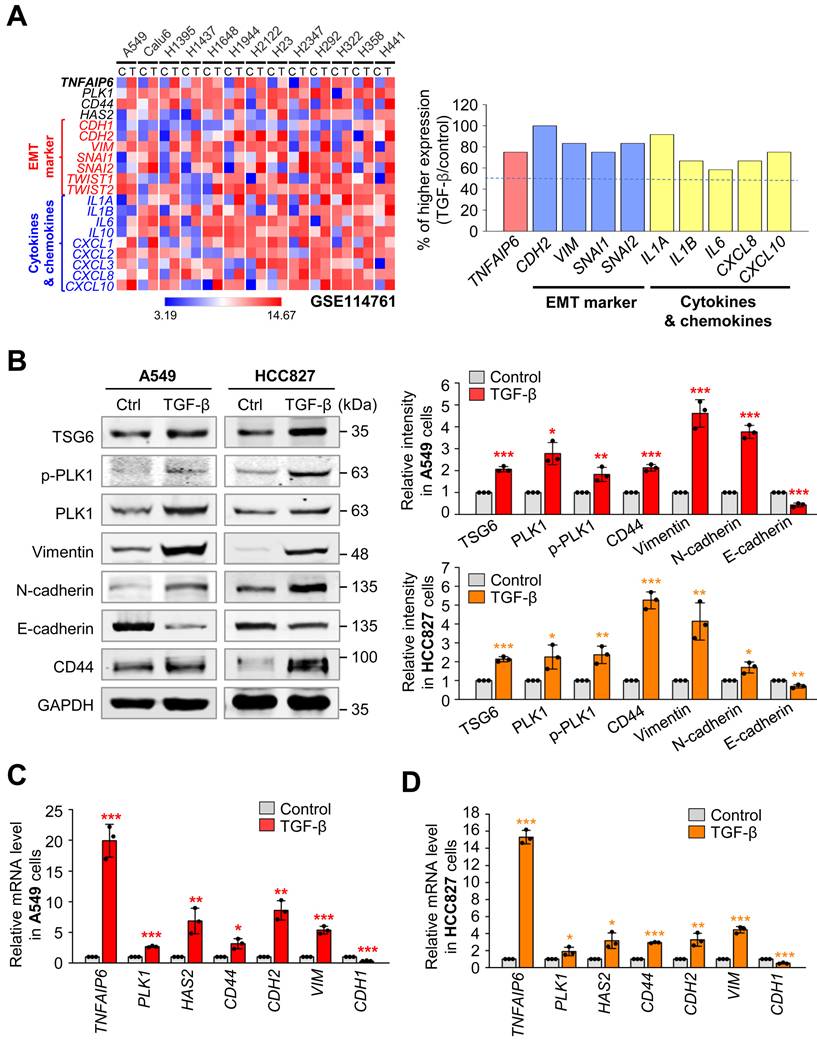
To investigate the expression of TNFAIP6 (which encodes TSG6) during LUAD metastasis, we analyzed data from the published transcriptome (GSE114761) of TGF-β-treated LUAD cells including A549, Calu6, NCI-H1395, NCI-H1437, NCI-H1648, NCI-H1944, NCI-2122, NCI-H23, NCI-H2347, NCI-H292, NCI-H322, NCI-H358, and NCI-H441 (Fig. 2A). Heatmap analysis revealed that TNFAIP6 expression after treatment with TGF-β was significantly higher in approximately 80% of TGF-β-treated LUAD cells compared to the control, when the mesenchymal markers CDH2 and VIM were high, indicating that TNFAIP6 was upregulated in TGF-β-induced EMT cells. In addition, the levels of its receptor CD44 [27] were upregulated in TGF-β-treated LUAD cells.
To confirm these observations, primary LUAD A549 and HCC827 cells were treated with TGF-β to induce the EMT and analyze the levels of TSG6 and CD44 (Fig. 2B-D). As expected, TGF-β treatment reduced the protein and mRNA levels of E-cadherin (encoded by CDH1), whereas it increased the levels of mesenchymal markers N-cadherin (encoded by CDH2) and vimentin (encoded by VIM). Under these conditions, immunoblotting revealed that the levels of TSG6 and CD44 were upregulated in TGF-β-treated A549 and HCC827 cells (Fig. 2B). Furthermore, qRT-PCR analysis showed that TNFAIP6 expression increased by approximately 15~20-fold in TGF-β-treated A549 and HCC827 cells (Fig. 2C-D). The changes in CD44, CDH2, VIM, and PLK1 in TGF-β-treated NSCLC cells, were consistent with the data from the transcriptome (GSE114761) of the TGF-β-treated cells. Thus, TSG6 and CD44 were upregulated during TGF-β-induced EMT in LUAD cells.
Inhibition of PLK1 activity reduces the expression of TSG6 and CD44 in the tail-vein injection mouse model
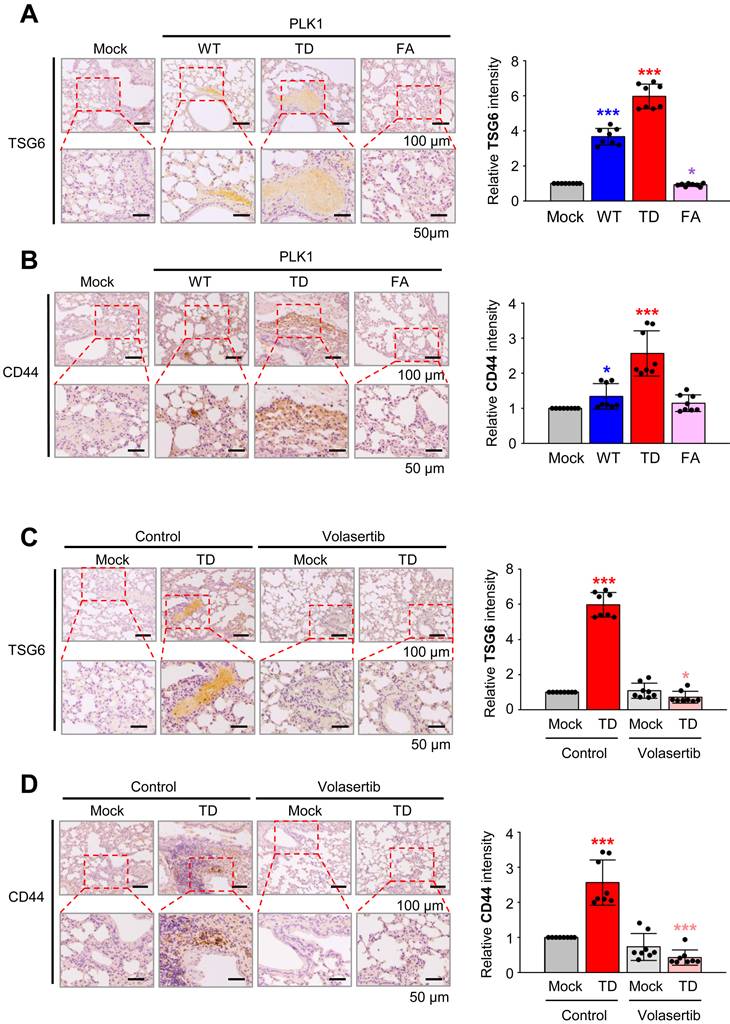
To explore the role of PLK1 in regulating TSG6 and CD44 expression, we conducted immunohistochemistry on lung tissues of BALB/c nude mice that were injected via the tail vein with A549 cells expressing wild-type PLK1 (PLK1-WT), a constitutively active PLK1 (PLK1-TD), or an inactive mutant of PLK1 (PLK1-FA) [12]. The expression of TSG6 and CD44 were higher in tissues of mice injected with A549 cells expressing PLK1-TD compared to those with PLK1-WT or PLK1-FA (Fig. 3A-B). Specifically, the relative intensity of TSG6 (Fig. 3A) or CD44 (Fig. 3B) increased by up to 6.0 or 2.5-fold, respectively, in lung tissues with PLK1-TD. In contrast, TSG6 or CD44 levels decreased by approximately 0.9 or 1.2-fold in tissues from mice injected with A549 cells expressing PLK1-FA compared to that of the mock (Fig. 3A-B). Additionally, treatment with a PLK1 inhibitor volasertib reduced TSG6 or CD44 expression. In this experiment, two weeks after mice were injected in the tail vein with A549 cells expressing PLK1-TD, they were treated intravenously with 20 mg/kg of volasertib weekly for three weeks [12]. After 10 weeks, the lung tissues were stained with antibodies against TSG6 or CD44. As expected, volasertib treatment markedly reduced the relative intensity of TSG6 (Fig. 3C) and CD44 (Fig. 3D), even in mice expressing PLK1-TD (Fig. 3C-D). Thus, the expression of TSG6 and CD44 were regulated by PLK1 activity and can be effectively reduced by PLK1 inhibition.
TSG6 functions as an inducer of EMT by activation of TGF-β/Smad signaling
Given that TSG6 was highly upregulated in TGF-β-induced EMT (Fig. 2) and was the most highly expressed gene in active PLK1-driven EMT of NSCLC cells (Fig. S1A), we investigated whether TSG6 plays a role in inducing EMT in LUAD cells (Fig. 4). To accomplish this, the levels of epithelial and mesenchymal markers were observed by immunoblotting in recombinant human TSG6 (rhTSG6) protein-treated A549 and HCC827 cells (Fig. 4A). The rhTSG6 treatment increased the levels of vimentin and N-cadherin while decreasing the epithelial marker E-cadherin in both cell lines (Fig. 4A). At the same time, the levels of total and its phosphorylated form at T210 of PLK1 were higher than those of control cells (Fig. 4A). In addition, the levels of its receptor CD44 were upregulated in rhTSG6-treated LUAD cells. To understand the signaling pathway for triggering EMT, the levels of Smad2 and p-Smad2 were observed (Fig. 4A). The data showed that the levels of Smad2 and p-Smad2 were higher than those of the control in rhTSG6-treated cells, indicating that TSG6 induces EMT via TGF-β signaling (Fig. 4A). Consistent with these findings, rhTSG6 treatment also increased the mRNA levels of mesenchymal markers (e.g. CDH2 and VIM), PLK1, and CD44 (Fig. 4B).
Co-expression of TSG6 and PLK1 in TGF-β induced EMT of LUAD cells. (A) The gene expression in the GSE114761 dataset in which EMT was induced by treating the LUAD cell line with TGF-β was visualized as a heatmap (left panel), and the ratio of increased expression of each factor in the TGF-β-treated group compared to the control group was plotted (right panel). (B-D) A549 and HCC827 LUAD cells were treated with 2.5 ng/ml of TGF-β for 48 hours. (B) Immunoblotting was performed to measure the protein levels of TSG6, p-PLK1, PLK1, Vimentin, N-cadherin, E-cadherin, CD44, and GAPDH in A549 (left panel) and HCC827 (right panel) cells. The relative band intensities were analyzed and plotted in A549 (right upper panel) and HCC827 (right lower panel) cells. *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 3). (C-D) TNFAIP6, PLK1, HAS2, CD44, CDH2, VIM, and CDH1 expression was measured in in A549 (C) and HCC827 (D) cells by qRT-PCR. *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 3).
Expression of TSG6 in an active PLK1-driven metastatic lung cancer mouse model. (A) Representative TSG6 staining was performed using lung tissue from mice transplanted with wild-type (WT), a constitutively active T210D (TD), and a substrate-nonbinding mutant at W414F/V415A (FA) of the polo-box domain of PLK1 (left panel). The relative density of TSG6 staining was analyzed and plotted (right panel). *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 8). (B) Representative CD44 staining was performed using lung tissue from mice transplanted by various versions of PLK1 (left panel), and the relative density of CD44 staining was analyzed and plotted (right panel). *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 8). (C) Representative TSG6 staining was performed on the lung tissue of mice transplanted with active PLK1-treated volasertib (PLK1 inhibitor) (left panel), and the relative density of TSG6 staining was analyzed and plotted (right panel). *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 8). (D) Representative CD44 staining was performed using lung tissue from mice transplanted with active PLK1-treated volasertib (PLK1 inhibitor) (left panel), and the relative density of CD44 staining was analyzed and plotted (right panel). *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 8).
Next, we investigate the loss of function effects of TSG6 using shRNA targeting TNFAIP6 at positions 530-550 (shTNFAIP6#1) or positions 693-713 (shTNFAIP6#2) in A549 and HCC827 cells (Fig. 4C-D). Depletion of TSG6 reduced the levels of N-cadherin and upregulated the levels of E-cadherin under low levels of TSG6 protein (Fig. 4C, Fig. S2A-B). In addition, the protein levels of CD44, PLK1, Smad2/3, and phosphorylated Smad2 were downregulated by the depletion of TSG6. These changes were similar at the mRNA level. When TSG6 was depleted, the expression of mesenchymal markers, CD44, and PLK1 were downregulated (Fig. 4D). These results indicate that TSG6 is important to induce EMT by activating TGF-β signaling, involving its receptor CD44. To understand the importance of TSG6 in EMT, we performed a rescue experiment in TSG6-depleted cells by treating with rhTSG6 protein (Fig. 4E-F). The levels of TNFAIP6 mRNA reduced by treatment with shRNA (#1 and #2), which was reversed by treatment with rhTSG6 in A549 and HCC827 cells. Under the conditions, the downregulated levels of CDH2, VIM, SMAD2, CD44, and PLK1 caused by TNFAIP6 knockdown were restored by rhTSG6 treatment. These data indicate that TSG6 is crucial for inducing EMT by the activation of TGF-β signaling.
TSG6 induces the polarization of tumor-associated macrophages in LUAD microenvironment
In LUAD with high levels of TNFAIP6 and PLK1, cytokines and chemokines associated with inflammation were markedly upregulated compared to normal tissues (Fig. 1D). Furthermore, microarray analysis of PLK1-TD expressing cells identified that the immune system process plays a key role in the tumor microenvironment (Fig. S3A). To explore whether TSG6 affects the tumor microenvironment, lung fibroblast MRC-5 cells were treated with rhTSG6 (Fig. S3B). Markers for cancer-associated fibroblast (CAF) such as PDGFR, FAP1, and SMA [28] were observed after treatment. However, these markers showed modest changes compared to the control, suggesting that TSG6 does not significantly alter CAF activation in the LUAD TME.
Tumor-secreted factors are known to drive the polarization of M2d-TAM in human monocyte THP-1 cells, particularly through the elevated expression of cytokines in invasive tumor cells [2, 4]. To understand whether TSG6 contributes to macrophage polarization in the TME, we treated human monocyte THP-1 cells with rhTSG6 and assessed marker expression using qRT-PCR (Fig. 5A). Treatment with rhTSG6 led to a slight downregulation of M1 macrophage markers IL12B and iNOS. Conversely, M2 macrophage markers CD163 and CD206 were slightly upregulated with their expression increasing by 2.2 and 2.8-fold, respectively, compared to the control. Furthermore, the mRNA levels of TGF-β and VEGFA, key markers of TAMs in the TME, were also elevated following rhTSG6 treatment (Fig. 5A). These data indicate that TSG6 promotes the TAM polarization of monocytes, contributing to the immune modulation of the LUAD microenvironment.
To clarify the role of TSG6 on TAM polarization within the TME, TSG6 was expressed in A549 cells using a doxycycline-inducible expression system (Fig. 5B). A549 cells expressing TSG6 were cocultured with THP-1 monocytes (Fig. 5B-C). When TNFAIP6 mRNA expression in A549 cells increased by up to 40-fold, the expression of VEGFA or IL6 was also upregulated. In the THP-1 cells cocultured with TSG6-expressing A549 cells, M2 and M2d macrophage markers such as VEGFA, TGFB1, CD206, and CD163 were markedly increased. Conversely, M1 makers IL12B and iNOS were downregulated compared to THP-1 cells cocultured with the mock A549 cells (Fig. 5B-C), indicating that TSG6 secretion from LUAD cells promotes TAM polarization in the surrounding microenvironment.
To confirm this, TSG6 was depleted using shRNA targeting TNFAIP6 in A549, and THP-1 monocytes were cocultured with TSG6-depleted A549 cells (Fig. 5D-E). Depletion of TNFAIP6 in A549 cells led to the downregulation of VEGFA or IL6. In THP-1 cells cocultured with TSG6-depleted A549 cells, M2 and M2d markers, including VEGFA, TGFB1, CD206, and CD163, were markedly reduced (Fig. 5D). In contrast, the levels of M1 marker IL12B in THP-1 cells cocultured with TNFAIP6-depleted A549 cells were higher than those with control shRNA-depleted A549 cells (Fig. 5E). These findings suggest that TSG6 promotes TAM polarization of monocytes, playing a crucial role in modulating the immune microenvironment in LUAD.
TAM polarization was observed in a tail-vein metastasis mouse model driven by active PLK1
To observe TAM polarization in an in vivo model, immunohistochemistry was performed with antibodies against CD163 (a TAM marker) and CD68 (a pan-macrophage marker) on the lung tissue of BALB/c nude mouse injected via the tail vein with A549 cells expressing PLK1-WT, PLK1-TD, or PLK1-FA (Fig. 6A-B), because TSG6 was highly expressed in metastatic tissue nodules expressing PLK-TD (Fig. 3A). The expression of CD68 (Fig. 6A) and CD163 (Fig. 6B) was higher in tissues from mice injected with A549 cells expressing PLK1-TD compared to those injected with PLK1-WT or PLK1-FA. Specifically, the relative intensities of CD68 (Fig. 6A) and CD163 (Fig. 6B) increased approximately 4.5- and 7-fold in lung tissues with PLK1-TD. In contrast, CD68 (Fig. 6A) and CD163 (Fig. 6B) expressions were downregulated approximately 0.5-fold in the tissues of mice injected with A549 cells expressing PLK1-FA compared to those of mock.
Recombinant TSG6 protein induced EMT by activation of TGF-β signaling. (A-B) A549 and HCC827 cells were treated with 200 ng/ml of rhTSG6 for 2 hours. (A) Immunoblotting was performed to measure the protein levels of TSG6, PLK1, p-PLK1T210, E-cadherin, N-cadherin, Vimentin, CD44, p-Smad2S465/S467, Smad2 and GAPDH in A549 (left panel) and HCC827 (right panel) cells. (B) qRT-PCR was used to evaluate CDH1, CDH2, CD44, PLK1, SMAD2, and VIM expression in A549 (right upper panel) and HCC827 (right lower panel) cells. *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 3). (C-F) TSG6 shRNA targeting the positions 530-550 (#1) or 693-713 (#2) was applied to A549 and HCC827 cells. (C) Immunoblotting was performed to measure the protein levels of TSG6, PLK1, CD44, N-cadherin, Smad2/3, p-Smad2S465/S467, and β-actin in A549 (left panel) and HCC827 (right panel) cells. (D) qRT-PCR was used to evaluate TNFAIP6, PLK1, CD44, CDH1, VIM, SMAD2, and CDH2 expression in A549 (upper panel) and HCC827 (lower panel) cells. *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 3). (E-F) qRT-PCR was used to evaluate TNFAIP6, PLK1, CD44, CDH1, VIM, SMAD2, and CDH2 expression in A549 (E) and HCC827 (F) cells after rhTSG6 treatment. *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 3).
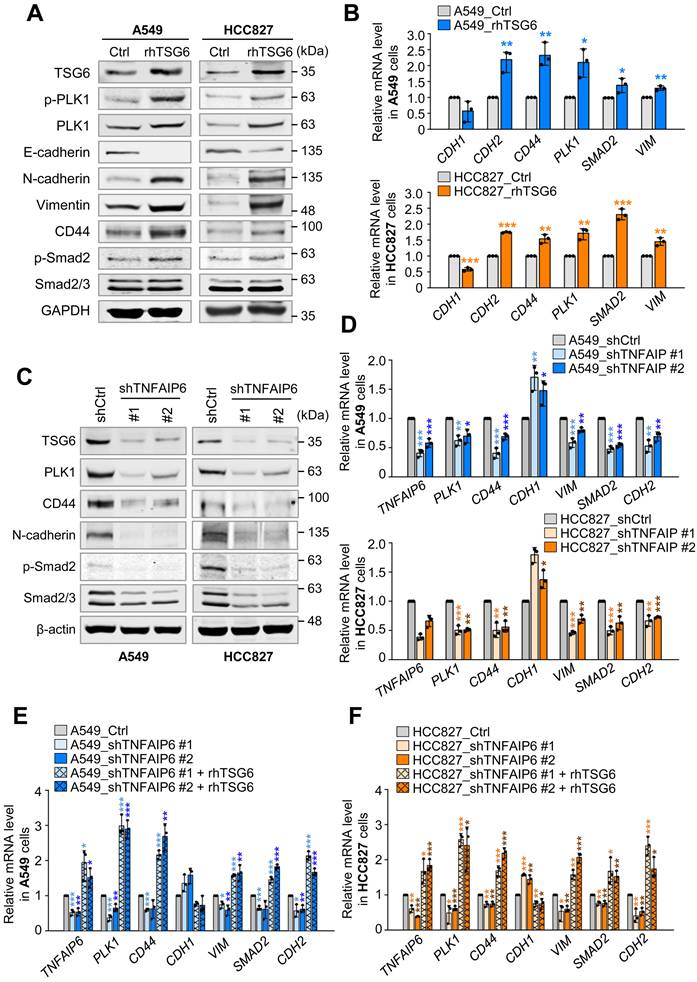
TSG6 induced the polarization of tumor-associated macrophages. (A) Monocyte THP-1 cells were treated with 200 ng/ml of rhTSG6 for 8 hours, and qRT-PCR was used to measure TNFAIP6, TGFB1, VEGFA (M2d macrophage markers), CD206, CD163 (M2 macrophage markers), IL12B, and iNOS (M1 macrophage markers) expression. (B-C) TSG6- overexpressing A549 cells and monocyte THP-1 cells were cocultured. (B) qRT-PCR was used to evaluate TNFAIP6, VEGFA, IL6, IL4 and IL10 expression in A549 cells. *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 3). (C) qRT-PCR was used to evaluate VEGFA, TGFB1 (M2d macrophage markers), CD206, CD163 (M2 macrophage markers), IL12B, and iNOS (M1 macrophage markers) expression in THP-1 cells. *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 3). (D-E) TSG6-depleted A549 cells and monocyte THP-1 cells were cocultured. (D) qRT-PCR was used to evaluate TNFAIP6, VEGFA, IL6, IL4 and IL10 expression in A549 cells. *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 3). (E) qRT-PCR was used to evaluate VEGFA, TGFB1, CD206, CD163, IL12B, and iNOS expression in THP-1 cells. *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 3).
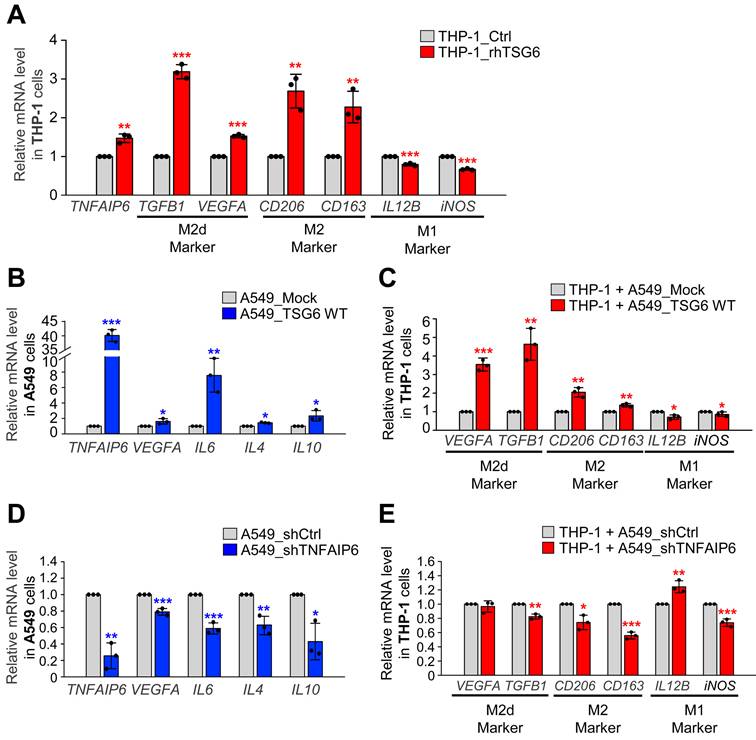
Then the expression of CD68 (Fig. 6C) and CD163 (Fig. 6D) were measured when BALB/c nude mice were treated with volasertib, a PLK1 inhibitor (Fig. 6C-D). Volasertib was injected intravenously (at a dose of 20 mg/kg) weekly for three weeks after A549 cells expressing PLK1-TD were injected into mice [12]. After 10 weeks, lung tissues were stained with antibodies against CD68 (Fig. 6C) and CD163 (Fig. 6D). The relative intensity of CD68 and CD163 were markedly reduced by approximately 6-fold and 14-fold in volasertib-treated PLK1-TD expressing mice, respectively (Fig. 6C-D). Therefore, TAM polarization is promoted in TSG6-upregulated cells driven by active PLK1 within the tumor microenvironment.
TAM polarization was induced in active PLK1-driven metastatic lung cancer of a mouse model. (A) Representative CD68 (pan-macrophage marker) staining was performed on lung tissue of mice transplanted with various versions of PLK1 (left panel), and the relative density of CD68 staining was analyzed and plotted (right panel). *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 8). (B) Representative CD163 (M2 macrophage marker) staining was performed on the lung tissue of mice transplanted with various versions of PLK1 (left panel), and the relative density of CD163 staining was analyzed and plotted (right panel). *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 8). (C) Representative CD68 staining was performed on the lung tissue of mice transplanted with cells containing active PLK1 treated with volasertib (PLK1 inhibitor) (left panel), and the relative density of CD68 staining was analyzed and plotted (right panel). *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 8). (D) Representative CD163 staining was performed using lung tissue from mice transplanted with cells containing active PLK1-treated volasertib (PLK1 inhibitor) (left panel), and the relative density of CD163 staining was analyzed and plotted (right panel). *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 8).
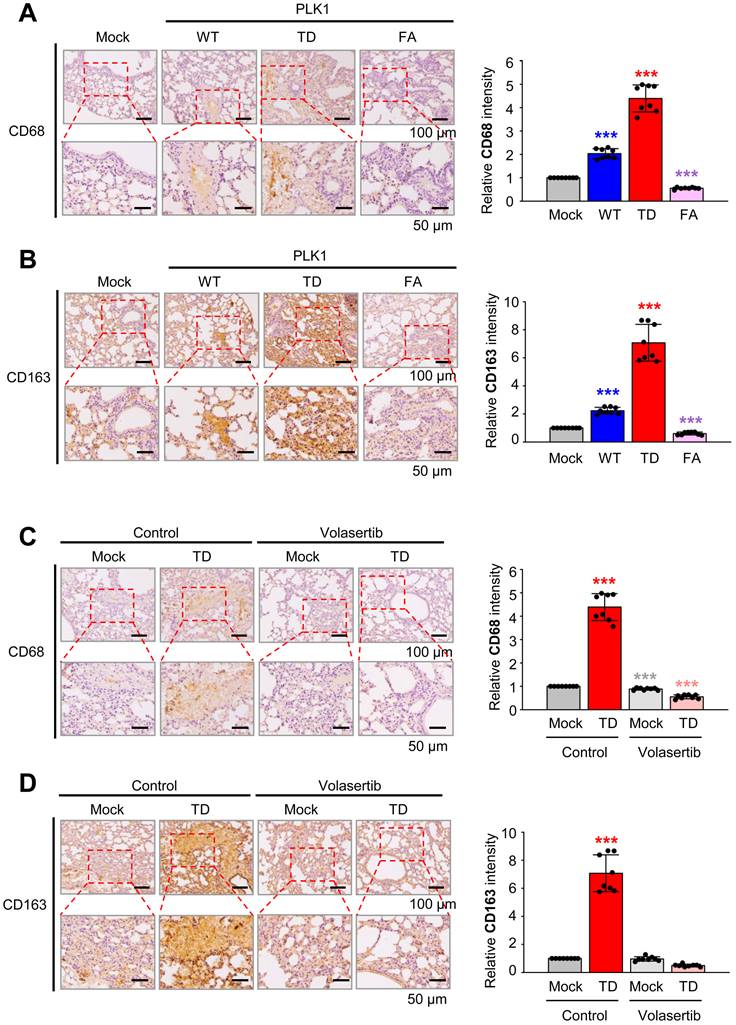
MAPK/ERK and TGFβR1/Smad signaling are the main pathways for TSG6-mediated EMT and TAM polarization. (A) A549 and HCC827 cells were treated with 200 ng/ml of rhTSG6 for 2 hours. Immunoblotting was performed to measure the protein levels of CD44, cleaved CD44 (CD44-ICD), MMP9, p-ERK1/2, ERK1/2, c-Jun, c-Fos, IL-6, and GAPDH in A549 (left panel) and HCC827 (right panel) cells. (B-C) A549 cells were treated with 10 μM trametinib (MEK1/2 inhibitor) for 48 hours and treated with 200 ng/ml of rhTSG6 for 2 hours. (B) Immunoblot was performed to measure the protein levels of TSG6, p-ERK1/2, ERK1/2, vimentin, IL-6, CD44, CD44-ICD, and GAPDH, and the relative band intensity values were analyzed in A549 cells (Sup Fig. S4). *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 3). (C) qRT-PCR was used to evaluate TNFAIP6, VEGFA, IL6, CD44, VIM, CDH1, and CDH2 expression in A549 cells. *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 3). (D-F) A549 cells were treated with 30 μM SB431542 (TGFβR1 inhibitor) for 48 hours and with 200 ng/ml of rhTSG6 for 2 hours. (D) Immunoblot was performed to measure the protein levels of TSG6, p-Smad2, Smad2/3, vimentin, IL-6, CD44, CD44-ICD, and GAPDH (D), and the relative band intensity values were analyzed in A549 cells (E). *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 3). (F) Expression of TNFAIP6, VEGFA, IL6, CD44, VIM, CDH1, and CDH2 in A549 cells was measured by qRT-PCR. *p < 0.05; ** p < 0.01; *** p < 0.001. (n = 3).
TSG6 induces the polarization of TAMs through the activation of MAPK/ERK and TGFβR1/Smad signaling pathways
Next, we aimed to explore which signaling pathway is activated by TSG6 to induce TAM polarization and EMT. Previous studies demonstrated that CD44 interacts with EGFR potentiating MAPK/ERK pathway [13, 29] and TGFβR1 [30] in human cancer cells. In Figure 4, rhTSG6 treatment induces EMT by the activation of TGF-β signaling. To investigate the activation of the MAPK/ERK pathway by TSG6 treatment, we measured the levels of p-ERK1/2 and AP-1, a complex of c-Jun and c-Fos. We discovered that they were upregulated by TSG6. Simultaneously, the levels of MMP9, IL-6, and total CD44 and the cleaved intracellular domain (ICD) of CD44 (CD44-ICD) were upregulated in rhTSG6-treated LUAD cells (Fig. 7A, Fig. S4A-B).
To further dissect the signaling pathway, rhTSG6-treated A549 cells were treated with MEK inhibitor trametinib and TGFβR inhibitor SB431542 (Fig. 7B-F). Trametinib treatment reduced the levels of p-ERK1/2 (normalized to total ERK1/2) by approximately 89% in TSG6-treated cells (vehicle vs trametinib, 3.4 vs 0.4) (Fig. 7B, right panel). Additionally, the levels of total CD44, cleaved CD44-ICD, and TSG6 that were increased by TSG6 treatment were markedly reduced upon trametinib treatment (Fig. 7B, Fig. S4C). Trametinib also suppressed the expression of vimentin and IL-6, indicating that MEK is important for TSG6-mediated EMT and TAM polarization. Furthermore, trametinib downregulated TSG6-induced mRNA levels of TNFAIP6 and CD44 (Fig. 7C). For TAM polarization, the upregulated VEGFA and IL6 mRNAs induced by TSG6 were suppressed in trametinib-treated A549 cells. Similarly, the mesenchymal markers VIM and CDH2 were downregulated following trametinib treatment. Therefore, MAPK/ERK signaling is important for TSG6-mediated EMT and TAM polarization.
To investigate whether TGFβR/Smad signaling is involved in TSG6-mediated EMT, given that TSG6 treatment induced EMT and Smad2 activation (Fig. 4A, C), we examined the effects of TGFβR inhibitor SB431542 on rhTSG6-treated A549 cells (Fig. 7D-F, Fig. S4D). SB431542 treatment reduced the levels of p-Smad2 (normalized to total Smad2) by approximately 83% (vehicle vs SB431542: 2.8 vs 0.5) in rhTSG6-treated cells (Fig. S4D). Additionally, SB431542 treatment decreased the TSG6-induced upregulation of total CD44, cleaved CD44-ICD, vimentin, and IL-6 at both the protein and mRNA levels (Fig. 7D-F). These findings indicate that TGFβR/Smad signaling is critical for TSG6-mediated EMT and TAM polarization. Collectively, both MAPK/ERK and TGFβR/Smad signaling play pivotal roles in TSG6-mediated EMT and TAM polarization.
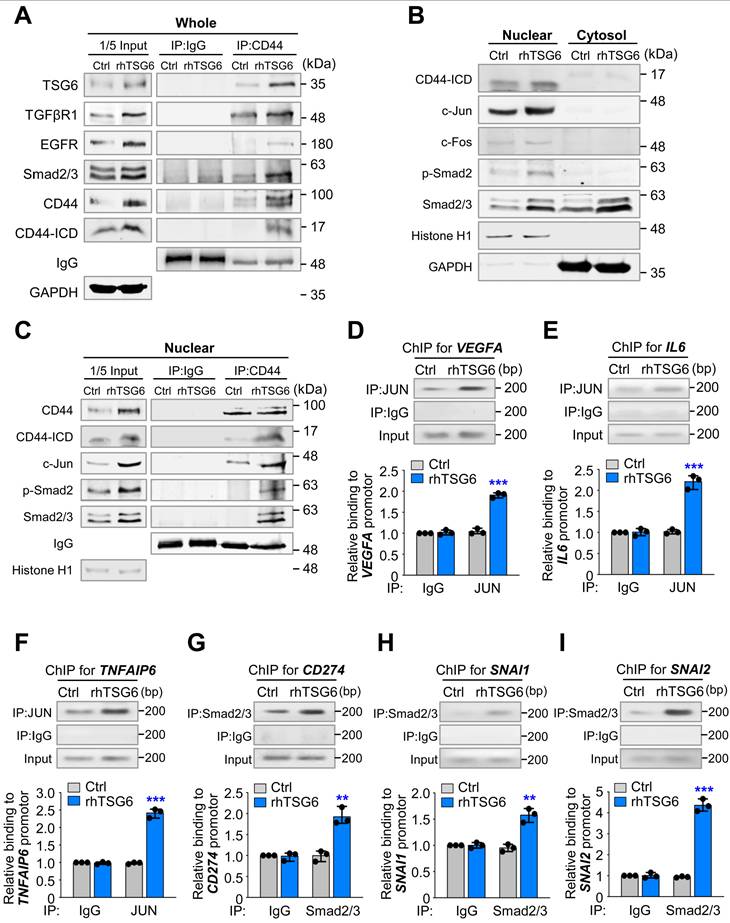
TSG6 promotes the interaction between CD44 and TGFβR or EGFR, driving the expression of genes involved in EMT and TAM polarization
Next, we aimed to investigate whether TSG6 influences the direct interaction between CD44 and TGFβR or EGFR, as MAPK/ERK and TGFβR/Smad signaling were activated in the presence of TSG6 (Fig. 7). Previous studies have reported that CD44 interacts with TGFβR1 [30] or EGFR [29] in cancer. To examine this, we conducted an immunoprecipitation assay using anti-CD44 antibody in rhTSG6-treated A549 cells (Fig. 8A). Our results indicated that CD44 interacted with TSG6 abundantly in rhTSG6-treated A549 cells (Fig. 8A). Additionally, TGFβR1 and EGFR were also detected, although the levels of EGFR were lower than those of TGFβR1 in these cells. Given that cleaved CD44-ICD translocates to the nucleus and regulates transcriptional activity [31], we examined its subcellular localization in rhTSG6-treated A549 cells (Fig. 8B). The levels of CD44-ICD were higher in the nucleus compared to the cytosol and TSG6 treatment upregulates its nuclear location. In rhTSG6-treated cells, c-Jun and c-Fos, components of AP-1, which are transcriptional factors downstream of MAPK/ERK1/2 signaling, were upregulated (Fig. 8B). In addition, p-Smad2 was upregulated in the nucleus after TSG6 treatment, indicating that TGFβR1-mediated signaling is activated (Fig. 8B). Furthermore, CD44-ICD was found to interact with c-Jun, in the nuclear fraction of rhTSG6-treated A549 cells (Fig. 8C). These findings suggest that the nuclear translocation of CD44-ICD enables its interaction with AP-1, contributing to transcriptional regulation in the presence of TSG6. Then to determine whether AP-1 transcriptionally activates genes related to TAM polarization, such as IL-6 and VEGFA, a ChIP assay was conducted in rhTSG6-treated cells using anti-c-Jun antibody (Fig. 8D-E). In rhTSG6-treated cells, the expression of IL6, VEGFA, and TNFAIP6 increased approximately a 2.5-fold compared to controls (Fig. 8D-F), suggesting that AP-1 directly bound to the promoters of IL6, VEGFA, and TNFAIP6. Then to determine whether TGFβR1-mediated signaling is activated for EMT and immune escape by TSG6 treatment, a ChIP assay was performed with anti-Smad2/3 antibody. In rhTSG6-treated A549 cells, the expression of CD274, SNAI1, and SNAI2 increased approximately a 2-, 1.7-, and 4.3-fold increase, respectively, compared to controls, indicating that Smad2/3 directly bound to the promoters of CD274, SNAI1, and SNAI2 for immune escape and EMT (Fig. 8G-I).
In summary, TSG6 treatment promotes interaction between CD44 and TGFβR or EGFR, leading to the activation of TGFβR/Smad or MAPK/ERK signaling pathways and triggering the cleavage of CD44-ICD, which accordingly translocates to the nucleus. In the nucleus, CD44-ICD interacts with transcriptional factors Smad2/3 or AP-1 to induce the expression of genes related to EMT, such as SNAI1 and SNAI2, and TAM polarization, such as VEGFA and IL6.
Discussion
Previously, we observed high expression of TNFAIP6 ranked with top in active PLK1-driven EMT [12]. In this study, we addressed the effects of TSG6 on EMT and the tumor microenvironment, especially monocyte polarization. Microarray analysis of invasive NSCLC cells in active PLK1-induced EMT recognized TSG6 as the most highly expressed factor [12], with the immune processes emerging as a prominent biological process in KEGG pathway analysis. In LUAD patients, highly co-expressed TSG6 and PLK1 correlated with lower OS rates and a higher hazard ratio than 1, as evaluated by KM plot analysis. We further found that the expression of TSG6, CD44, CD68, and CD163 (marker of TAMs) is elevated in metastatic lung cancer tissues of mice driven by active PLK1. The upregulations of TSG6, CD44, CD68, and CD163 were suppressed by the treatment with PLK1 inhibitor volasertib (Fig. 3 and 6), indicating that PLK1 activity is important to regulate the expression of TSG6, CD44, CD68, and CD163 for modulating TME. In our previous study, we also found that β-catenin, a transcriptional factor of TSG6 and CD44, is phosphorylated by PLK1, which enhances its transcriptional activity and extracellular remodeling in advanced NSCLC [20]. Based on this study, the upregulations of TSG6 and CD44 during EMT would be explained by activation of β-catenin by PLK1-mediated phosphorylation in LUAD. The PLK1 inhibitor volasertib suppresses PLK kinase activity, blocking the phosphorylation of β-catenin and reducing its stability. As a result of β-catenin degradation, the expression of its downstream targets, including TSG6 and CD44, would be decreased in LUAD. Notably, TSG6 also showed as an inducer of EMT in LUAD through TGF-β/Smad signaling in Figure 4. In A549 and HCC827 cells treated with TSG6, mesenchymal markers including vimentin and N-cadherin were upregulated (Fig. 4A-B). TSG6-induced EMT was activated by TGF-β signaling, as evidenced by increased phosphorylation of Smad2 in TSG6-treated LUAD cells. Depletion of TSG6 resulted in the downregulation of mesenchymal markers and p-Smad2 (Fig. 4C-D), which was restored by treatment with rhTSG6 (Fig. 4E-F). These data indicate that TSG6 would be a key driver of EMT in LUAD. Because PLK1 is upregulated and activated during the EMT in NSCLC [12], it is possible that TSG6 and PLK1 are indirectly linked through reciprocal regulation.
The function of TSG6 would be associated with HA and CD44, a receptor of both HA or TSG6 [27]. As CD44 serves as a receptor for TSG6, we hypothesized that binding TSG6 to CD44 triggers TGF-β/Smad signaling or MAPK kinase signaling through the formation of a complex with TGFβR1 or EGFR. In the results, inhibition of TGFβR1 or MEK1/2 significantly reduced TSG6-induced EMT and cytokine expression related to TAM polarization (Fig. 7). Notably, the direct interaction between CD44 and TGFβR1 or EGFR was increased following rhTSG6 treatment (Fig. 8A). Therefore, TSG6 facilitates the interaction between CD44 and TGFβR1 or EGFR. The results are supported by the previous studies [30] showing that hyaluronan, another ligand of CD44 and an interacting partner with TSG6, promotes signaling interaction between CD44 and TGFβR1 in metastatic breast tumors [30]. Additionally, the interaction between CD44 and EGFR has been shown to initiate and drive the progression of head and neck squamous cell carcinoma [13, 29]. The cleavage of CD44 may be influenced by its interaction with TGFβR1 or EGFR in the presence of TSG6 by upregulated MMP9. In addition, the upregulation of CD44-ICD was suppressed by treatment with the TGFβR1 inhibitor SB431542 or the MEK inhibitor trametinib. Thus, the increased CD44-ICD, driven by TSG6-CD44-TGFβR1 or EGFR-mediated signaling, translocated into the nucleus, where it contributes to transcriptional regulation via Smad2/3 or AP-1.
TSG6 secretion drives the polarization toward M2d-TAMs from monocytes. In patients with advanced LUAD, the expression levels of mesenchymal markers and cytokines associated with M2d-TAM polarization were significantly higher in LUAD tissues compared to normal tissues (Fig. 1). When rhTSG6 was treated to monocyte cells, the expression of M2d markers increased in THP-1 cells (Fig. 5). Similar increases of M2d inducers were also detected in A549 cells treated with TSG6. These findings demonstrate that TSG6 induces monocyte polarization toward M2d-TAM in the TME. A recent study showed that TSG6 promotes tumor migration and invasion in a colorectal cancer model by reprogramming normal fibroblasts into CAFs [13]. In our study, the reprogramming effects of TSG6 on normal lung fibroblasts into CAFs (Fig. S3B) is potentially due to tissue-specific differences. However, we found that TSG6 strongly promotes M2d-TAM polarization in the LUAD tumor microenvironment (Fig. 5-6). This finding is supported by previous studies showing TSG6-mediated M2 macrophage polarization in alcoholic hepatitis in vivo [32] and in DSS-induced colitis of mice [33]. Thus, the evidence supports our study that TSG6 secreted by LUAD cells drives monocyte polarization into M2d-TAMs within the TME. However, it remains to be determined if TSG6 promotes M2d-TAMs polarization in vivo as a further study.
TSG6 facilitates the interaction between CD44 and TGFβR or EGFR for the expression of genes involved in EMT and TAM polarization. (A) A549 cells were treated with 200 ng/ml of rhTSG6 for 2 hours. Immunoprecipitation and immunoblotting were used to measure the protein levels of CD44, CD44-ICD, TGFβR1, EGFR, and GAPDH in A549 cells. (B) Fractionation of A549 cells treated with rhTSG6 was used to observe the levels of nuclear and cytosolic fraction. Immunoblotting was used to measure the protein levels of CD44, CD44-ICD, c-Jun, c-Fos, Smad2/3, Histone H1 (nuclear loading control), and GAPDH (cytosolic loading control). (C) Nuclear fraction of A549 cells treated with 200 ng/ml of rhTSG6 for 2 hours was used for immunoprecipitation and immunoblotting analysis. Immunoblot analyses were performed using anti-CD44, anti-c-Fos, and anti-c-Jun antibodies. (D-F) ChIP assays for AP-1 (complex of c-Jun and c-Fos) binding to the promoters of VEGFA (D), IL6 (E), and TNFAIP6 (F) in rhTSG6-treated A549 cells. (G-I) ChIP assays for Smad2/3 binding to the promoters of CD274 (G), SNAI1 (H), and SNAI2 (I) in rhTSG6-treated A549 cells. Assays were performed on chromatin fragments using antibody to c-Jun (D-F) or Smad2/3 (G-I) and normalized to pre-immune normal IgG. Immunoprecipitated fractions were assayed by qRT-PCR to determine the binding to each promoter. Data are presented as mean ± SD of three independent experiments (significantly different from the experimental control). *p < 0.05; **p < 0.01; ***p < 0.001; (n = 3).
In conclusion, we suggest that TSG6 would be a crucial factor that contributes to tumor malignancy by inducing TAM polarization in TME. TSG6 treatment facilitates the interaction between CD44 and TGFβR or EGFR, which leads to the activation of TGFβR/Smad or MAPK/ERK signaling pathways and cleavage of CD44 for the production of CD44-ICD. Subsequently, CD44-ICD translocates to the nucleus and induces the expression of genes associated with EMT and TAM polarization through the activation of transcriptional factors Smad2/3 and AP-1. Therefore, targeting TSG6 would provide a promising therapeutic strategy for LUAD treatment by modulating the tumor microenvironment.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We express our sincere gratitude to Hay-Ran Jang, Chang-Hyeon Kim, Rong Xu, Jung-Won Park, Yeo-Bin Kim, Hyun-Jin Kim, and the other members of the Yim lab for their invaluable guidance and assistance in completing this project.
Funding
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (RS-2023-00217123, RS-2025-00515374) to H.Y.
Availability data and materials
The authors declare that all the data supporting the findings of this study are available within the paper and its Supplementary Information files. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Ethical approval and consent to participate
All animal experiments were approved (HY-IACUC-2017-0115A) and managed by the guidelines of the Institutional Animal Care and Use Committee, Hanyang University.
Consent for publication
All authors approved the publication.
Author contributions
H-J. O., G-H. M., D-E. K., - Investigation, Validation, Visualization, and Writing: review and editing; S-B. S., - Validation and Writing: review and editing; H.Y.- Conceptualization, Project administration, Funding acquisition, Investigation, Supervision, and Writing: original draft, review, and editing.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Anderson NM, Simon MC. The tumor microenvironment. Curr Biol. 2020;30:R921-R5
2. Gao J, Liang Y, Wang L. Shaping Polarization Of Tumor-Associated Macrophages in Cancer Immunotherapy. Front Immunol. 2022;13:888713
3. Kim DH, Song NY, Yim H. Targeting dysregulated lipid metabolism in the tumor microenvironment. Arch Pharm Res. 2023;46:855-81
4. Xu R, Lee YJ, Kim CH, Min GH, Kim YB, Park JW. et al. Invasive FoxM1 phosphorylated by PLK1 induces the polarization of tumor-associated macrophages to promote immune escape and metastasis, amplified by IFITM1. J Exp Clin Cancer Res. 2023;42:302
5. Larionova I, Cherdyntseva N, Liu T, Patysheva M, Rakina M, Kzhyshkowska J. Interaction of tumor-associated macrophages and cancer chemotherapy. Oncoimmunology. 2019;8:1596004
6. Milner CM, Day AJ. TSG-6: a multifunctional protein associated with inflammation. J Cell Sci. 2003;116:1863-73
7. Day AJ, Prestwich GD. Hyaluronan-binding proteins: tying up the giant. J Biol Chem. 2002;277:4585-8
8. Selbi W, de la Motte CA, Hascall VC, Day AJ, Bowen T, Phillips AO. Characterization of hyaluronan cable structure and function in renal proximal tubular epithelial cells. Kidney Int. 2006;70:1287-95
9. Misra S, Hascall VC, Markwald RR, Ghatak S. Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer. Front Immunol. 2015;6:201
10. Higman VA, Briggs DC, Mahoney DJ, Blundell CD, Sattelle BM, Dyer DP. et al. A refined model for the TSG-6 link module in complex with hyaluronan: use of defined oligosaccharides to probe structure and function. J Biol Chem. 2014;289:5619-34
11. Lesley J, Gal I, Mahoney DJ, Cordell MR, Rugg MS, Hyman R. et al. TSG-6 modulates the interaction between hyaluronan and cell surface CD44. J Biol Chem. 2004;279:25745-54
12. Shin SB, Jang HR, Xu R, Won JY, Yim H. Active PLK1-driven metastasis is amplified by TGF-beta signaling that forms a positive feedback loop in non-small cell lung cancer. Oncogene. 2020;39:767-85
13. Liu B, Liu T, Liu Y, Feng X, Jiang X, Long J. et al. TSG-6 promotes Cancer Cell aggressiveness in a CD44-Dependent Manner and Reprograms Normal Fibroblasts to create a Pro-metastatic Microenvironment in Colorectal Cancer. Int J Biol Sci. 2022;18:1677-94
14. Lan G, Yu X, Sun X, Li W, Zhao Y, Lan J. et al. Comprehensive analysis of the expression and prognosis for TNFAIPs in head and neck cancer. Sci Rep. 2021;11:15696
15. Chan TC, Li CF, Ke HL, Wei YC, Shiue YL, Li CC. et al. High TNFAIP6 level is associated with poor prognosis of urothelial carcinomas. Urol Oncol. 2019;37:293 e11- e24
16. Yim H, Erikson RL. Plk1-targeted therapies in TP53- or RAS-mutated cancer. Mutat Res Rev Mutat Res. 2014;761:31-9
17. Rizki A, Mott JD, Bissell MJ. Polo-like kinase 1 is involved in invasion through extracellular matrix. Cancer Res. 2007;67:11106-10
18. Wu J, Ivanov AI, Fisher PB, Fu Z. Polo-like kinase 1 induces epithelial-to-mesenchymal transition and promotes epithelial cell motility by activating CRAF/ERK signaling. Elife. 2016;5:e10734
19. Cai XP, Chen LD, Song HB, Zhang CX, Yuan ZW, Xiang ZX. PLK1 promotes epithelial-mesenchymal transition and metastasis of gastric carcinoma cells. Am J Transl Res. 2016;8:4172-83
20. Kim DE, Shin SB, Kim CH, Kim YB, Oh HJ, Yim H. PLK1-mediated phosphorylation of beta-catenin enhances its stability and transcriptional activity for extracellular matrix remodeling in metastatic NSCLC. Theranostics. 2023;13:1198-216
21. Jang HR, Shin SB, Kim CH, Won JY, Xu R, Kim DE. et al. PLK1/vimentin signaling facilitates immune escape by recruiting Smad2/3 to PD-L1 promoter in metastatic lung adenocarcinoma. Cell Death Differ. 2021;28:2745-64
22. Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241
23. Ma L, Hua L, Yu W, Ke L, Li LY. TSG-6 inhibits hypertrophic scar fibroblast proliferation by regulating IRE1alpha/TRAF2/NF-kappaB signalling. Int Wound J. 2023;20:1008-19
24. Yang H, Wu L, Deng H, Chen Y, Zhou H, Liu M. et al. Anti-inflammatory protein TSG-6 secreted by bone marrow mesenchymal stem cells attenuates neuropathic pain by inhibiting the TLR2/MyD88/NF-kappaB signaling pathway in spinal microglia. J Neuroinflammation. 2020;17:154
25. Li H, Huang C, Zhang Z, Feng Y, Wang Z, Tang X. et al. MEK Inhibitor Augments Antitumor Activity of B7-H3-Redirected Bispecific Antibody. Front Oncol. 2020;10:1527
26. Park S, Kim D, Jung H, Choi IP, Kwon HJ, Lee Y. Contribution of HSP90 Cleavage to the Cytotoxic Effect of Suberoylanilide Hydroxamic Acid In Vivo and the Involvement of TXNIP in HSP90 Cleavage. Biomol Ther (Seoul). 2024;32:115-22
27. Wang S, Kim J, Lee C, Jung Y. Tumor necrosis factor-inducible gene 6 interacts with CD44, which is involved in fate-change of hepatic stellate cells. BMB Rep. 2020;53:425-30
28. Gao D, Fang L, Liu C, Yang M, Yu X, Wang L. et al. Microenvironmental regulation in tumor progression: Interactions between cancer-associated fibroblasts and immune cells. Biomed Pharmacother. 2023;167:115622
29. Perez A, Neskey DM, Wen J, Pereira L, Reategui EP, Goodwin WJ. et al. CD44 interacts with EGFR and promotes head and neck squamous cell carcinoma initiation and progression. Oral Oncol. 2013;49:306-13
30. Bourguignon LY, Singleton PA, Zhu H, Zhou B. Hyaluronan promotes signaling interaction between CD44 and the transforming growth factor beta receptor I in metastatic breast tumor cells. J Biol Chem. 2002;277:39703-12
31. Miletti-Gonzalez KE, Murphy K, Kumaran MN, Ravindranath AK, Wernyj RP, Kaur S. et al. Identification of function for CD44 intracytoplasmic domain (CD44-ICD): modulation of matrix metalloproteinase 9 (MMP-9) transcription via novel promoter response element. J Biol Chem. 2012;287:18995-9007
32. Wan YM, Wu HM, Li YH, Xu ZY, Yang JH, Liu C. et al. TSG-6 Inhibits Oxidative Stress and Induces M2 Polarization of Hepatic Macrophages in Mice with Alcoholic Hepatitis via Suppression of STAT3 Activation. Front Pharmacol. 2020;11:10
33. Song WJ, Li Q, Ryu MO, Ahn JO, Ha Bhang D, Chan Jung Y. et al. TSG-6 Secreted by Human Adipose Tissue-derived Mesenchymal Stem Cells Ameliorates DSS-induced colitis by Inducing M2 Macrophage Polarization in Mice. Sci Rep. 2017;7:5187
Author contact
![]() Corresponding author: Hyungshin YIM, Department of Pharmacy, College of Pharmacy, Institute of Pharmaceutical Science and Technology, Hanyang University, Ansan, Gyeonggi-do 15588, Republic of Korea, Phone: +82-31-400-5810, Fax: +82-31-400-5958, E-mail: hsyimac.kr.
Corresponding author: Hyungshin YIM, Department of Pharmacy, College of Pharmacy, Institute of Pharmaceutical Science and Technology, Hanyang University, Ansan, Gyeonggi-do 15588, Republic of Korea, Phone: +82-31-400-5810, Fax: +82-31-400-5958, E-mail: hsyimac.kr.

 Global reach, higher impact
Global reach, higher impact