10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2025; 21(12):5378-5392. doi:10.7150/ijbs.119841 This issue Cite
Review
Mitochondrial RNA in Inflammation
1. Department of Dermatology, the First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, China.
2. Key Laboratory of Dermatology (Anhui Medical University), Ministry of Education, Hefei, Anhui, China.
3. Institute of Dermatology, Anhui Medical University, Hefei 230032, Anhui, China.
4. Department of Occupational Health and Environmental Health, School of Public Health, Anhui Medical University, Hefei, Anhui, China.
*These authors contributed equally to this work
Received 2025-6-18; Accepted 2025-8-4; Published 2025-8-22
Abstract
Mitochondria are dynamic organelles integral to cellular energy metabolism and homeostasis. Beyond their traditional roles, a growing body of evidence underscores the importance of mitochondria as pivotal regulators of innate immune signaling pathways. Recently, mitochondrial RNA (mtRNA) has been identified as a novel modulator of inflammatory responses. mtRNA is detected by intracellular pattern recognition receptors (PRRs), which subsequently activate the mitochondrial antiviral-signaling protein (MAVS) and the interferon regulatory factor 3 (IRF3)/nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling axis, as well as inflammasome pathways. This activation leads to the production of type I interferons and pro-inflammatory cytokines. Furthermore, mtRNA facilitates the propagation of inflammatory signals through exosome-mediated intercellular transfer. Among the various forms of mtRNA, mitochondrial double-stranded RNA (mt-dsRNA) is particularly prone to activating inflammatory responses due to its distinctive double-helical structure. The aberrant accumulation of mt-dsRNA is strongly linked autoimmune diseases, degenerative disease, Liver Disease, kidney disease, cancers, cardiovascular diseases, and respiratory ailments. This review proposes innovative therapeutic strategies aimed at degrading pathological mtRNA or interrupting inflammatory pathways by targeting critical regulatory nodes in mtRNA metabolism and its downstream inflammatory processes.
Keywords: Mitochondrial, Inflammation, mtRNA, mt-dsRNA.
1. Introduction
Mitochondria, semiautonomous organelles in eukaryotic cells, are characterized by a double-membrane system, comprising the outer mitochondrial membrane (OMM), inner mito-chondrial membrane (IMM), and mitochondrial matrix. Their dynamic network properties maintained through the fusion/fission balance are crucial for cellular energy homeostasis [1, 2]. Mitochondria are the energy production centers of cells, participating in processes such as oxidative phosphorylation, ATP synthesis, fatty acid oxidation and decomposition, as well as the synthesis of lipids and heme. Moreover, mitochondria are involved in the regulation of the intracellular calcium ion (Ca2+) signaling transduction and modulate caspase activation and cell death through the release of cytochrome c [3-5]. Numerous studies have demonstrated that under stress conditions, mitochondrial DNA (mtDNA) released into the cytoplasm functions as a damage-associated molecular pattern (DAMP) [6]. Furthermore, extensive research has demonstrated the pivotal role of mitochondria in innate immunity, with mitochondrial RNA (mtRNA) emerging as a novel immunomodulatory molecule critically involved in inflammatory responses [7, 8]. During cellular stress or injury, mtRNA can leak into the cytoplasm or extracellular environment through multiple pathways, functioning as DAMPs that activate pattern recognition receptors (PRRs). This interaction triggers inflammatory cascades through innate immune pathways [9, 10].
At disease level, mtRNA-mediated innate immune activation shows significant associations with various inflammatory pathologies. In osteoarthritis, extracellular mtRNA activates protein kinase R (PKR) and toll-like receptor 3 (TLR3), promoting proinflammatory cytokines production and apoptosis, ultimately leading to chondrocyte degeneration and joint deterioration [11]. Systemic lupus erythematosus (SLE) exhibits increased mtRNA release, which activates inflammatory responses through cytoplasmic PRRs including retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated protein 5 (MDA5), and toll-like receptor 7 (TLR7) [12].
Investigating the metabolic regulation of mtRNA and its interaction with inflammatory processes offers significant insights into the pathogenesis of inflammatory disorders. Furthermore, it aids in the development of targeted therapeutic strategies. These advancements are promising for the progression of precision medicine in diseases associated with inflammation.
2. Structure and Metabolism of mtRNA
2.1 Transcription and Processing of mtRNA
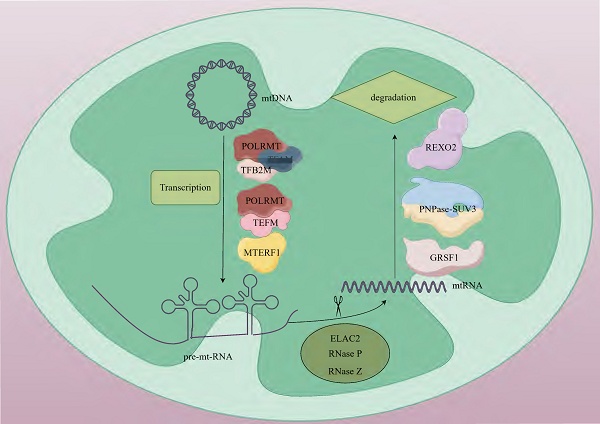
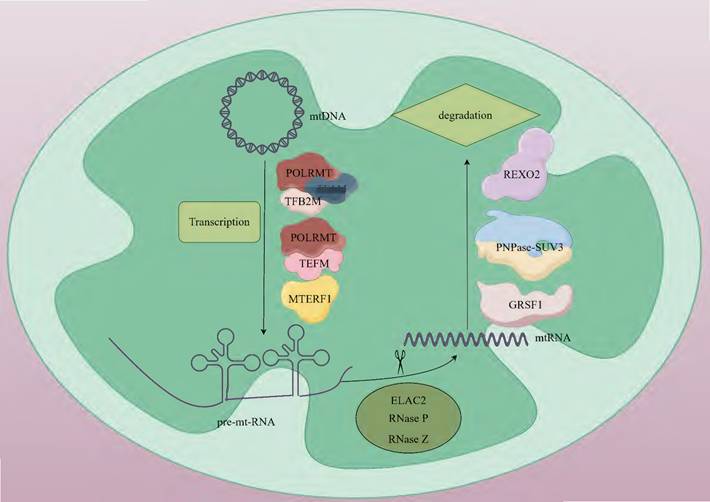
mtRNA transcription operates through a prokaryote-like mechanism, mediated by mtRNA polymerase (POLRMT) with essential support from mitochondrial transcription factor A (TFAM) and mitochondrial transcription factor B2 (TFB2M) [13, 14]. The mitochondrial genome harbors 37 intron-free genes (2 rRNAs, 22 tRNAs, and 13 mRNAs) organized as polycistronic transcription units, which encode 13 critical subunits required for the oxidative phosphorylation (OXPHOS) system [15]. Transcription initiation occurs at the light-strand promoter (LSP) and heavy-strand promoters (HSP) within the control region adjacent to the D-loop structure [16]. During initiation, TFAM binds high-affinity sites 10-15 base pairs upstream of transcription start sites, inducing DNA bending and interacting with POLRMT to recruit it to promoter regions. TFB2M subsequently joins to form a pre-initiation complex that unwinds promoter DNA, enabling transcription commencement [16, 17]. The mitochondrial transcription elongation factor (TEFM) facilitates POLRMT in navigating complex structural regions. The resultant polycistronic RNA undergoes site-specific processing to yield mature mRNAs, tRNAs, and rRNAs. tRNA genes typically flank protein-coding genes, with tRNA maturation triggering release of mRNAs and rRNAs for subsequent maturation steps. Termination is regulated by factors including mitochondrial transcription termination factor 1 (MTERF1) [18] (Fig. 1).
2.2 Types and Functions of mtRNA
mtDNA generates polycistronic transcripts through bidirectional transcription, and these long-chain RNAs need to undergo precise processing. mt-mRNA, mt-tRNA, and mt-rRNA are generated through 5'-end cleavage mediated by the TRMT10C-SDR5C1-PRORP complex and 3'-end cleavage catalyzed by ElaC domain protein 2 (ELAC2) [19, 20] (Table. 1). Among them, mt-mRNA encodes 13 protein subunits of the mitochondrial respiratory chain complex. These subunits, through cooperative assembly with nuclear-encoded subunits, form the core functional units of the electron transport chain [21]. mt-tRNA is responsible for transporting amino acids to the ribosome. It pairs its anticodon with the codon on mRNA to ensure the correct synthesis of the amino acid sequence. There are 22 types of mt-tRNAs. In most cases, each tRNA corresponds to a specific amino acid, but there are also a few tRNAs that can recognize and bind to several structurally similar amino acids [22]. mt-rRNA is a component of the mitochondrial ribosome, including two types, 12S and 16S, which participate in the formation of the small and large subunits, respectively [23]. Ribosomes are the sites of protein synthesis. rRNA, together with ribosomal proteins, forms the structural framework of the ribosome and participates in the catalytic reactions during protein synthesis [24]. During the transcription of mtDNA, long-chain RNA molecules have a relatively high probability of forming double-stranded RNA (dsRNA) structures through complementary base pairing. After the long-chain RNA molecules formed during mitochondrial transcription form mitochondrial dsRNA (mt-dsRNA) structures, they may be recognized and degraded by nucleases within the mitochondria, or they may participate in some regulatory processes, such as regulating mitochondrial gene expression [25]. The formation and degradation of mt-dsRNA are part of the normal operation of mitochondrial functions. The correct processing and degradation of mtRNA are crucial for maintaining mitochondrial functions and cellular energy metabolism [26].
Human mt-RNA Gene Names and Functions.
| RNA Type | Gene Name | Function | References |
|---|---|---|---|
| mt-rRNA | MT-RNR1(12S) | Structural component of the mitochondrial ribosome small subunit (28S), involved in codon-anticodon pairing. | [27] |
| MT-RNR2(16S) | Forms the peptidyl transferase center in the large subunit (39S), catalyzing peptide bond formation. | [27] | |
| mt-mRNA | MT-ND1 | Encodes NADH dehydrogenase (Complex I) subunit ND1, involved in electron transport initiation. | [28] |
| MT-ND2 | Encodes Complex I subunit ND2, participates in the proton transport process across the mitochondrial inner membrane, working in concert with ND1 to maintain the structural integrity and functional activity of Complex I. | [29] | |
| MT-ND3 | Encodes Complex I subunit ND3, Ensures the proper assembly of Complex I and maintains the homeostasis of the mitochondrial respiratory chain. | [30] | |
| MT-ND4 | Encodes Complex I subunit ND4, participates in proton translocation and electron transfer, playing a critical role in the assembly of the membrane arm of Complex I. | [31] | |
| MT-ND4L | Encodes Complex I subunit ND4L (smallest transmembrane subunit), participates in proton translocation in Complex I, assisting in maintaining the transmembrane proton gradient. | [32] | |
| MT-ND5 | Encodes Complex I subunit ND5 (critical for proton pumping), maintains the activity of NADH:ubiquinone oxidoreductase and ensures the proper assembly and stability of the membrane arm of Complex I. | [31] | |
| MT-ND6 | Encodes Complex I subunit ND6 (encoded on the light strand), collaborates with ND4L and ND5 to maintain the structural stability of Complex I. | [33] | |
| MT-CYTB | Encodes cytochrome b (Complex III) (essential for ubiquinol-cytochrome c oxidoreductase activity), participates in the transfer of electrons from coenzyme Q to cytochrome c. | [34] | |
| MT-CO1 | Encodes cytochrome c oxidase (Complex IV) subunit CO1 (largest catalytic subunit), facilitates the transfer of electrons from cytochrome c to molecular oxygen. | [35, 36] | |
| MT-CO2 | Encodes Complex IV subunit CO2, participates in electron transfer and stabilizes the structural integrity of Complex IV. | [35, 37] | |
| MT-CO3 | Encodes Complex IV subunit CO3, assists in proton translocation across the membrane, sustaining the activity of Complex IV. | [35, 38] | |
| MT-ATP6 | Encodes ATP synthase (Complex V) subunit ATP6 (forms the transmembrane proton channel), participates in the formation of proton channels within the mitochondrial inner membrane, driving ATP synthesis. | [39] | |
| MT-ATP8 | Encodes ATP synthase (Complex V) subunit ATP8 (supports proton pumping), forms a bicistronic transcript with ATP6, facilitating the assembly and functionality of the ATP synthase complex. | [40] | |
| mt-tRNA | tRNA-Ala (MT-TA) | Identifying codons GCA, GCC, GCG, GCU, transports alanine. | [41] |
| tRNA-Arg (MT-TR) | Identifying codons CGA, CGC, CGG, CGU, transports arginine. | [42, 43] | |
| tRNA-Asn (MT-TN) | Identifying codons AAC, AAU, transports asparagine. | [44] | |
| tRNA-Asp (MT-TD) | Identifying codons GAC, GAU, transports aspartic acid. | [45] | |
| tRNA-Cys (MT-TC) | Identifying codons UGC, UGU, transports cysteine. | [46] | |
| tRNA-Gln (MT-TQ) | Identifying codons CAA, CAG, transports glutamine. | [47] | |
| tRNA-Glu (MT-TE) | Identifying codons GAA, GAG, transports glutamic acid. | [48] | |
| tRNA-Gly (MT-TG) | Identifying codons GGA, GGC, GGG, GGU, transports glycine. | [49] | |
| tRNA-His (MT-TH) | Identifying codons CAC, CAU, transports histidine. | [50] | |
| tRNA-Ile (MT-TI) | Identifying codons AUA, AUC, AUU, transports isoleucine. | [51] | |
| tRNA-Leu(UUR) (MT-TL1) | Identifying codons UUA, UUG, transports leucine. | [52] | |
| tRNA-Leu(CUN) (MT-TL2) | Identifying codons CUA, CUC, CUG, CUU, transports leucine. | [53] | |
| tRNA-Lys (MT-TK) | Identifying codons AAA, AAG, transports lysine. | [54] | |
| tRNA-Met (MT-TM) | Identifying codons AUA, AUG, transports methionine. | [55] | |
| tRNA-Phe (MT-TF) | Identifying codons UUC, UUU, transports phenylalanine. | [56] | |
| tRNA-Pro (MT-TP) | Identifying codons CCA, CCC, CCG, CCU, transports proline. | [57] | |
| tRNA-Ser(UCN) (MT-TS1) | Identifying codons UCA, UCC, UCG, UCU, transports serine. | [58] | |
| tRNA-Ser(AGY) (MT-TS2) | Identifying codons AGC, AGU, transports serine. | [59] | |
| tRNA-Thr (MT-TT) | Identifying codons ACA, ACC, ACG, ACU, transports threonine. | [60] | |
| tRNA-Trp (MT-TW) | Identifying codons UGG, UGA, transports tryptophan. | [61] | |
| tRNA-Tyr (MT-TY) | Identifying codons UAC, UAU, transports tyrosine. | [62] | |
| tRNA-Val (MT-TV) | Identifying codons GUA, GUC, GUG, GUU, transports valine. | [63] |
2.3 Metabolism of mtRNA
The mtRNA degradation pathway entails the orchestrated activity of various enzymes, which play a crucial role in preserving mtRNA homeostasis by identifying and cleaving RNAs with distinct structures or sequences. This degradation process is primarily executed through the synergistic actions of endonucleases and exonucleases [64]. During the initial processing stage, endonucleases Ribonuclease P (RNase P) and Ribonuclease Z (RNase Z) cleave precursor RNAs to generate mature rRNAs, tRNAs, and mRNAs [20, 65]. The subsequent terminal degradation is mainly mediated by exonuclease complexes. Exonucleases like polynucleotide phosphorylases (PNPase, PNPT1) and suppressor of variegation 3 homolog (SUV3, Suv3p) are responsible for further degrading these RNA molecules [66]. PNPase is a phosphorolytic 3'-5' exoribonuclease that can catalyze the degradation of phosphodiester bonds in RNA and is mainly responsible for the degradation of mitochondrial antisense RNA. In mammals, The localization of PNPase to the mitochondrial inter-membrane space [67] and matrix [68] suggests that it has a dual role in preventing the formation and release of mt-dsRNA into the cytoplasm. PNPase collaborates with SUV3 in the matrix to participate in the degradation of antisense RNA [69]. SUV3 is an RNA helicase. In yeast, it is localized in the mitochondrial matrix and forms the core of the degradosome together with Dss1p, catalyzing the unwinding of RNA duplexes to facilitate Dss1p-mediated degradation [70, 71]. In mammals, SUV3 and PNPase form the mitochondrial degradosome (mtEXO), which maintains the stability of the mitochondrial genome by unwinding RNA and removing abnormal structures such as R-loops [72]. In addition to the core degradation system, other enzymes including MRP1/2, RBP16, LRPPRC, SLIRP, MTPAP, PDE12, and multiple co-factors participate in quality control by regulating RNA stability [73, 74]. In recent years, studies have found that the G-rich sequence factor 1 (GRSF1) plays a specific regulatory role in L-strand RNA metabolism. GRSF1 is an RNA-binding protein that localizes to mitochondria and participates in the post-transcriptional processing of mtRNA and the regulation of gene expression. There are two isoforms of GRSF1 in mitochondria. One is localized in mtRNA granules (MRGs), co-localizing with newly synthesized mtRNA and RNase P. The other is localized in the nucleus and cytoplasm and may be involved in mitochondria-to-nucleus communication [75, 76]. GRSF1 mainly binds to L-strand RNA, and many L-strand RNAs in mitochondria have the potential to form G-quadruplexes. GRSF1 is a key protein that targets these G-quadruplex RNAs and promotes their degradation [76, 77]. GRSF1 acts in concert with the mitochondrial degradosome (the hSuv3-PNPase complex), enhancing the degradation efficiency of G-quadruplex RNAs [76]. RNA exoribonuclease 2 (REXO2), an evolutionarily conserved 3'-5' DEDDh family exonuclease, maintains normal degradation of mt-dsRNA by eliminating nanoRNAs (e.g., ncH2 RNA) generated by the mitochondrial SUV3-PNPase complex, thereby preventing their accumulation and interference with PNPase activity. This process is critical for safeguarding against mitochondrial dysfunction [78] (Fig. 1).
The basic steps of mtRNA metabolism. Human mtDNA is a circular double-stranded molecule. Mitochondrial transcription is orchestrated by POLRMT in coordination with its auxiliary factors, including TFAM, TFB2M, TEFM and MTRES1. This process generates three polycistronic transcripts that undergo subsequent processing. During maturation, precursor RNAs are cleaved by ELAC2, RNase P, and RNase Z to form functional mtRNAs. Terminal RNA degradation requires the coordinated action of GRSF1 and PNPase-SUV3. REXO2 ensures proper degradation of mt-dsRNA by eliminating nanoscale RNA fragments produced during SUV3-PNPase-mediated processing. mtDNA; mitochondrial DNA, POLRMT; mitochondrial RNA polymerase, TFAM; mitochondrial transcription factor A, TFB2M; mitochondrial transcription factor B2, TEFM; mitochondrial transcription elongation factor, MTERF1; mitochondrial transcription termination factor 1, pre-mt-RNA; mitochondrial precursor RNA, ELAC2; ElaC domain protein 2, RNase P; ribonuclease P, RNase Z; ribonuclease Z, mtRNA; mitochondrial RNA, GRSF1; G-rich sequence factor 1, SUV3-PNPase; suppressor of variegation 3-polynucleotide phosphorylase, REXO2; RNA exoribonuclease 2.
2.4 mt-dsRNA and mtRNA
mtDNA is characterized by its circular, double-stranded structure, and its expression results in the production of extensive complementary RNA strands, which have a pronounced tendency to form mt-dsRNA [25]. In instances where mitochondrial function is compromised, the RNA degradation system, such as the PNPase-SUV3 complex, may become dysfunctional. This dysfunction can lead to the inadequate degradation of single-stranded mtRNA, which subsequently folds into stable mt-dsRNA [78]. Consequently, mt-dsRNA represents a distinct subtype of mtRNA, as not all mtRNA molecules exist in a double-stranded form. While mtRNA includes all RNA molecules present within mitochondria, mt-dsRNA specifically refers to those with a double-stranded configuration. Furthermore, mt-dsRNA is more readily recognized by PRRs, which can initiate an inflammatory response.
3. Mechanisms of mtRNA Activating Inflammatory Response
3.1 Activation of Pattern Recognition Receptors
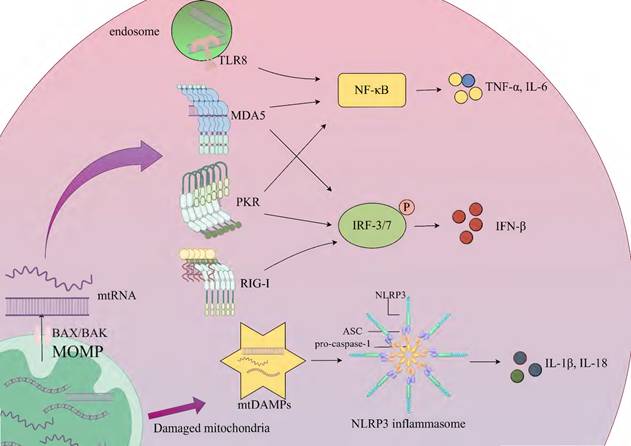
Inflammation, a fundamental innate immune response to noxious stimuli such as infection or tissue injury, serves core functions including pathogen clearance, removal of necrotic cells, and initiation of tissue repair [79]. This response is initiated through the recognition of two key molecular patterns by pattern recognition receptors: pathogen-associated molecular patterns (PAMPs) and DAMPs. PAMPs represent conserved components of pathogens, such as bacterial lipopolysaccharides and viral double-stranded RNA, while DAMPs encompass endogenous molecules released by host cells during injury or stress, including high mobility group box-1 (HMGB1), adenosine triphosphate (ATP), and mitochondrial components (mtDNA and mtRNA) [80]. Upon cellular damage, mtRNA (especially mt-dsRNA) may leak into the cytosol or extracellular space, acting as DAMPs recognized by PRRs. However, certain PRRs (e.g., RIG-I-like receptors), which primarily evolved to detect exogenous PAMPs (e.g., viral RNA), exhibit cross-reactivity toward endogenous mtRNA, potentially leading to aberrant activation of analogous inflammatory pathways [80, 81]. Under physiological conditions, the level of mtRNA in mitochondria is regulated by mtRNA degradation complexes such as PNPase and SUV3. When mitochondrial function is impaired (such as metabolic stress in the tumor microenvironment), stress signals activate the pro-apoptotic proteins BAX (BCL-2-associated X protein) and BAK (BCL-2 antagonist or killer). They oligomerize and insert into the outer mitochondrial membrane to form pores, leading to an increase in outer mitochondrial membrane permeability (MOMP). This process results in the release of mtRNA and mtDNA into the cytoplasm [9]. Double-strand breaks in mtDNA can lead to its linearization and degradation. Meanwhile, mtRNA polymerase generates short-chain mtRNA transcripts at the break sites. These transcripts carry 5'-triphosphate (5'PPP) ends and are partially fold into short-chain mt-dsRNA, which are released into the cytoplasm through BAK/BAX pores. The mtRNA that enters the cytoplasm activates RIG-I-like receptors (RLRs), such as RIG-I and MDA5 [82]. Nucleic acid binding induces conformational changes in RIG-I and MDA5, exposing their N-terminal caspase recruitment domains (CARDs). Subsequently, the CARDs polymerize and interact with the CARD domain of mitochondrial antiviral-signaling protein (MAVS), ultimately triggering a cascade of antiviral signals such as type I interferon [9, 83]. MAVS is located on the outer mitochondrial membrane and serves as a key node in innate immune defense. It can activate downstream signaling pathways by recruiting kinase complexes (such as TBK1/IKKε), including the nuclear factor-κB (NF-κB) and interferon regulatory factor 3/7 (IRF3/7) signaling pathways. The activated NF-κB and IRF3/IRF7 can promote the transcription of related inflammatory factors (such as TNF-α, IL-6) and the expression of type I interferon (such as IFN-β), respectively, thereby triggering an inflammatory response [84]. In addition, fumarase (FH) catalyzes the conversion of fumarate to malate in the mitochondria and cytoplasm of macrophages. When macrophages are treated with the FH inhibitor FHIN1 or a macrophage model with Fh1 gene knockout is constructed by gene editing, intracellular fumarate accumulates, and there is an increase in mtRNA release. In experiments where macrophages stimulated with lipopolysaccharide (LPS) were treated with FHIN1, researchers found that the content of mtRNA in cytoplasmic extracts increased significantly. In tamoxifen-induced Fh1-knockout bone-marrow-derived macrophages (BMDMs), an increase in mtRNA release was also observed. This indicates that FH inhibition is an important cause of increased mtRNA release, and the mtRNA released into the cytoplasm can activate the immune response [85]. Toll-like receptors (TLRs) mediate the recognition of pathogen- and host-derived "danger-associated molecular patterns" (P-/DAMPs), triggering a severe inflammatory response. Human TLR8 can specifically recognize the UR/URR motifs in bacterial and mtRNA. mtRNA activates immune cells through the human TLR8-mediated pathway, inducing the MyD88 (myeloid differentiation primary response 88) adaptor-mediated NF-κB signaling pathway, thus promoting the secretion of pro-inflammatory factors (such as TNF-α, IL-6). This process depends on the endosomal transporter Unc93B1 to deliver TLR8 to the endosome and is independent of TLR7 [86]. Studies have demonstrated that mitochondrial stress conditions, particularly those associated with mitochondrial dysfunction and aging, significantly enhance cytosolic translocation of mt-dsRNAs. These translocated mt-dsRNAs interact with PKR, inducing its dimerization and subsequent autophosphorylation. The phosphorylated PKR (pPKR) functions as an active kinase that phosphorylates eIF2α to suppress global protein synthesis while simultaneously activating NF-κB-mediated pathways. This dual mechanism upregulates pro-inflammatory cytokines (e.g., IL-8, MMP1/13, IFN-β) and enhances expression of interferon-stimulated genes (ISGs) [87, 88]. The NLR family pyrin domain containing 3 (NLRP3) inflammasome is a multi-protein complex composed of NLRP3, apoptosis-associated speck-like protein containing a CARD (ASC), and pro-caspase-1. Mitochondrial damage leads to the release of mitochondrial damage-associated molecular patterns (mtDAMPs), including mtDNA and potentially mtRNA. These mtDAMPs can indirectly activate the NLRP3 inflammasome by binding to the PYD domain of ASC or promoting the formation of ASC specks. After activation, ASC recruits pro-caspase-1 through its CARD domain and promotes its self-cleavage into the active form caspase-1. The latter then cleaves the pro-inflammatory cytokines IL-1β and IL-18 into their active forms [89, 90] (Fig. 2).
3.2 Transcellular Inflammatory Regulatory Effects
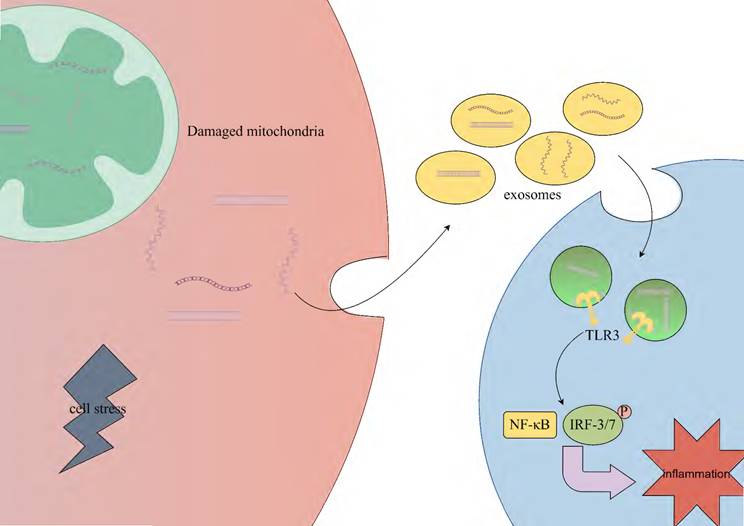
Circulating extracellular vesicles (EVs), including exosomes, microvesicles, and apoptotic bodies, can carry various molecules (such as DNA, RNA) and transmit information between cells [91, 92]. Under conditions of cell stress or injury, mt-dsRNA may be packaged into exosomes. Exosomes are released extracellularly through fusion with the cell membrane and can be taken up by other cells via endocytosis or other mechanisms, thus enabling intercellular communication and information transfer. However, the specific mechanisms remain unclear [91, 93]. The mt-dsRNA released into other cells may activate TLR3, thereby mediating the IRF3/7 and NF-κB signaling pathways, which subsequently promote the transcription of relevant inflammatory cytokines and the expression of type I interferons [94] (Fig. 3).
3.3 Nuclear Regulatory Mechanisms of mtRNA
In the field of diabetes research, under conditions simulating diabetes (high glucose and TNF-α treatment), the association level between mtRNA and chromatin in human umbilical vein endothelial cells (HUVECs) increases. In particular, the association level of SncmtRNA (a mitochondrially encoded long non-coding RNA) significantly increases under stress conditions. Through the combined analysis of single-cell RNA sequencing (scRNA-seq) and single-nucleus RNA sequencing (snRNA-seq), researchers found that the knockdown of SncmtRNA (Snc-KD) significantly inhibited the expression of intercellular adhesion molecule 1 (ICAM1) and vascular cell adhesion molecule 1 (VCAM1). These genes are induced under stress conditions and are closely related to the inflammatory response. Results indicate that mtRNA can act as a retrograde signaling molecule to regulate gene expression through its interaction with nuclear chromatin. In addition, SncmtRNA plays an important role in regulating the expression of genes related to inflammation and immune responses [95].
4. Mechanisms of mtRNA in Various Inflammatory-related Diseases
4.1 Autoimmune Disease
SLE is an autoimmune-mediated connective tissue disorder, characterized by the production of multiple autoantibodies and multisystem damage. The mechanisms by which mt-dsRNA activates the inflammatory response suggest that damage-associated molecular patterns, such as mt-dsRNA released due to mitochondrial dysfunction, may engage the innate immune response via pattern recognition receptors, including RIG-I and MDA5, thereby contributing to the pathogenesis of SLE. mt-dsRNA has been identified as a novel mitochondrial autoantigen target in SLE [96]. In individuals with SLE, autoantibodies against mtRNA (AmtRNA) are present, with research indicating that the serum levels of IgG and IgM subtypes of AmtRNA are significantly elevated compared to healthy controls. The presence of AmtRNA-IgG correlates with anti-mtDNA-IgG, as well as IgG and IgM against anti-β2-glycoprotein I, with even higher levels observed in patients who are positive for anti-double-stranded genomic DNA antibodies. These findings suggest that mt-dsRNA functions as an autoantigen, capable of activating the immune system, inducing the production of autoantibodies, and contributing to the immunopathological processes underlying SLE [96]. Mitochondria-derived mt-dsRNA from erythrocytes plays an important role in SLE. When monocytes in SLE patients phagocytose erythrocytes containing mitochondria (Mito+RBCs), the mt-dsRNA derived from Mito+RBCs can trigger monocytes to release their own mtDNA fragments. These fragments bind to the NLRP3 inflammasome, promoting the production of IL-1β. At the same time, mt-dsRNA also acts in synergy with type I interferon. The MxA protein induced by type I interferon is involved in the unconventional secretion pathway of IL-1β, affecting the inflammatory response [97].
Rheumatoid arthritis (RA) is a complex chronic autoimmune disease. Its main pathological features include progressive joint deformity, synovial inflammation, and cartilage and bone destruction. Its pathogenesis is closely related to mitochondrial dysfunction. Due to the inflammatory-activating effect of mt-dsRNA, when mitochondria are damaged, mt-dsRNA leaks into the cytoplasm, activating the innate immunity and thus releasing inflammatory factors. This process is particularly significant in RA synovial cells, exacerbating joint inflammation [98]. Meanwhile, the release of mt-dsRNA is accompanied by the accumulation of mitochondrial reactive oxygen species (mtROS), which activates the NLRP3 inflammasome and promotes the maturation and secretion of IL-1β and IL-18. Clinical data show that the level of IL-1β in the synovial fluid of RA patients is positively correlated with mtROS [99].
Sjögren's syndrome (SS) is an autoimmune disease primarily characterized by damage to exocrine glands such as salivary glands and lacrimal glands, presenting symptoms like xerostomia and xerophthalmia [88, 100]. Its pathogenesis is closely related to innate immune abnormalities and activation of IFN signaling. The levels of mt-dsRNAs are elevated in the saliva, tears, and salivary glands of SS patients. These mt-dsRNAs can be recognized by pattern recognition receptors such as PKR, MDA5, and TLR3, activating the innate immune response, which in turn activates the Janus Kinase-1 (JAK1) / signal transducer and activator of transcription (STAT) signaling pathway. As a key kinase in this pathway, JAK1 mediates the downstream transmission of IFN signals, promoting the expression of interferon-stimulated genes (ISGs) and forming an inflammatory amplification loop, thereby exacerbating glandular inflammation and dysfunction [88, 100].
Activation of the innate immune response by mtRNA. Key steps of the process are presented on the simplified diagram. Mitochondrial dysfunction triggers stress signals that activate BAX/BAK, which oligomerize and insert into the outer mitochondrial membrane, forming pores that increase MOMP and release mtRNA into the cytoplasm, thereby activating the innate immune response. RIG-I and MDA5 recognize and bind mtRNA, interacting with MAVS to recruit kinase complexes such as TBK1/IKKε, which activate downstream signaling pathways. RIG-I activates IRF3/7 to promote IFN-β expression, while MDA5 activates NF-κB and IRF3/IRF7, inducing the transcription of TNF-α and IL-6, as well as IFN-β expression. mtRNA activates immune cells through the TLR8-mediated pathway, triggering the NF-κB signaling pathway and promoting the secretion of TNF-α and IL-6. This process relies on Unc93B1 to deliver TLR8 to the endosome. mtRNA interacts with PKR, activating NF-κB and IRF3/IRF7-mediated pathways, which upregulate TNF-α, IL-6, and IFN-β. The NLRP3 inflammasome, composed of NLRP3, ASC, and pro-caspase-1, is indirectly activated by mtDAMPs such as mtDNA and mtRNA released into the cytoplasm due to mitochondrial damage. These mtDAMPs bind to the PYD domain of ASC or promote ASC speck formation, ultimately enhancing the expression of IL-1β and IL-18. MOMP; outer mitochondrial membrane permeability, BAX/BAK; BCL-2-associated X protein/BCL-2 antagonist or killer, TLR8; Toll-like receptor 8, MDA5; melanoma differentiation-associated protein 5, PKR; protein kinase R, RIG-I; retinoic acid-inducible gene I, NF-κB; nuclear factor-κB, IRF3/7; interferon regulatory factor 3/7, mtDAMPs; mitochondrial damage-associated molecular patterns, ASC; apoptosis-associated speck-like protein containing a CARD.
Stress-induced mtRNA release triggers inflammation in neighboring cells. Under cellular stress or injury conditions, mt-dsRNA may be encapsulated into exosomes through membrane fusion-mediated extracellular release. These vesicles are subsequently internalized by neighboring cells via endocytic mechanisms. Intracellular recognition of mt-dsRNA by TLR3 triggers the activation of both IRF3/7 and NF-κB signaling pathways, ultimately driving inflammatory responses. TLR3; toll-like receptor 3, NF-κB; nuclear factor-κB, IRF3/7; interferon regulatory factor 3/7.
4.2 Degenerative Disease
Osteoarthritis (OA) is a common degenerative joint disease that mainly affects the synovial joints of adults and is the most common type of arthritis. It often occurs in the knee, hip, and hand joints [101]. The over-activation of PKR is associated with inflammatory degenerative diseases including OA, but the dsRNA molecules that activate it remain largely unclear. Researchers have found that under conditions that induce osteoarthritis, the expression of mt-dsRNA in chondrocytes and its outflow into the cytoplasm are accelerated, leading to innate immune activation [11].
Huntington's disease (HD) is an inherited neurodegenerative disease caused by a mutation in the HTT gene, which encodes the huntingtin protein (Htt). Research has found that in patients with HD and mouse models, the up-regulation of mt-dsRNA expression is related to disease progression. The immune response induced by mt-dsRNA may be involved in the increased vulnerability of the neurons most severely affected in HD patients (striatal projection neurons). In addition, the accumulation of mtRNA in senescent cells promotes neuro-inflammation through the RIG-I/MDA5-MAVS [102, 103].
4.3 Liver Disease
Alcoholic liver disease (ALD) is an inflammatory liver disease caused by long-term and excessive alcohol consumption, which is mediated by multiple inflammatory responses. Research shows that ethanol stress prompts hepatocytes to produce mt-dsRNA. These mt-dsRNA are transmitted to Kupffer cells via exosomes, activating TLR3 in them [104]. Studies have found that treating wild-type Kupffer cells with exosomes directly treated with ethanol (EtOH-Exo) can significantly increase the expression of IL-1β, but this phenomenon does not occur in TLR3-knockout Kupffer cells. This indicates that mt-dsRNA activates TLR3 through exosomes to initiate the immune response. The activated TLR3 makes Kupffer cells express IL-1β, which in turn stimulates γδ T cells to produce IL-17A, participating in the inflammatory response of ALD [104].
Non-alcoholic fatty liver disease (NAFLD) is a type of metabolic stress-induced liver injury disease closely associated with insulin resistance and genetic susceptibility [105]. As a key mitochondrial damage-associated molecular pattern, mt-dsRNA is released into the cytoplasm under certain pathophysiological conditions, triggering innate immune responses. In the alcoholic liver disease model, it can induce the pro-inflammatory activity of Kupffer cells. Studies have shown that a high-fat diet (HFD) can induce the release of mt-dsRNA in wild-type (WT) mice. As a critical DAMP, mt-dsRNA triggers a series of innate immune responses. Specifically, the release of mt-dsRNA activates TLR3, MDA5, and phosphorylated interferon regulatory factor 3 (p-IRF3). This further leads to the upregulated mRNA expression of inflammatory biomarkers such as il-1β and il-6 in the liver, thereby causing hepatic inflammation [106]. In addition, studies have noted that in the alcoholic liver disease model, after being encapsulated by exosomes, mt-dsRNA is transferred from hepatocytes to Kupffer cells in a TLR3-dependent manner, triggering their pro-inflammatory activity. This also supports the role of mt-dsRNA in triggering innate immune responses [106].
4.4 Kidney Disease
Renal tubular atrophy is a key feature of chronic kidney disease (CKD). Studies have found that downregulation of PNPT1 in renal tubular cells leads to leakage of mt-dsRNA into the cytoplasm, activation of PKR, subsequent phosphorylation of eIF2α, and termination of protein translation and apoptosis. Downregulation of PNPT1 and activation of the mt-dsRNA-PKR-eIF2α axis have been observed in human kidney disease patients such as acute tubular necrosis (ATN), diabetic nephropathy (DN), and lupus nephritis (LN), as well as in mouse models of ischemia-reperfusion injury (IRI) and unilateral ureteral obstruction (UUO). Overexpression of PNPT1 or inhibition of PKR activity significantly alleviates renal tubular injury, while renal tubular-specific PNPT1 knockout mice exhibit Fanconi syndrome-like phenotypes and severe mitochondrial damage [107].
DN is a common microvascular complication of diabetes mellitus, characterized by hyperglycemia, persistent increase in urinary albumin excretion, and progressive decline in glomerular filtration rate (GFR). In both type 1 (STZ-induced) and type 2 (db/db) DN mouse models, the expression of mt-dsRNA in tubular cells is significantly increased, with abnormal release from mitochondria to the cytoplasm. After being released into the cytoplasm, mt-dsRNA activates the double-stranded RNA sensor PKR, inducing its phosphorylation (p-PKR), which in turn promotes the phosphorylation of downstream eIF2α (p-eIF2α), ultimately leading to tubular cell apoptosis. In DN mice and HK-2 cells treated with high glucose (HG), elevated mt-dsRNA is accompanied by increased expression of pro-apoptotic proteins (BAX, Cleaved caspase-3) and decreased expression of anti-apoptotic protein (BCL-2); while inhibiting mt-dsRNA release or blocking PKR activation can reduce apoptosis [108].
4.5 Infectious Disease
Viral infections can induce abnormal accumulation of mt-dsRNA in mammalian cells. When these mt-dsRNA are released into the cytoplasm through mitochondrial membrane channels or the mitophagy pathway, they can activate cytoplasmic pattern recognition receptors (such as MDA5/RIG-I), triggering the host antiviral immune response (such as the type I interferon pathway) [109].
4.6 Cancer
The mutational signature of the antiviral DNA deaminase APOBEC has been identified in over 70% of cancers and may represent the predominant component of somatic variations in numerous individual tumors and tumor types [110, 111]. Research has demonstrated that the cytoplasmic leakage of mt-dsRNA is perceived as foreign nucleic acid, thereby activating the interferon response, including the upregulation of APOBEC3A via the RIG-I/MAVS/STAT2-dependent pathway, while concurrently inducing nuclear DNA damage [112]. In B-cell precursor acute lymphoblastic leukemia (B-ALL), studies have indicated that mt-dsRNA serves as a pivotal trigger for the transformation of mesenchymal stromal cells (MSCs) into cancer-associated fibroblasts (CAFs). This transformation is characterized by a robust interferon pathway response, which includes the upregulation of interferon pathway-related genes such as MX1, MX2, and OAS1-3, alongside the secretion of IFN-β. The majority of these genes are implicated in RNA sensing or inhibition processes [113].
4.7 Cardiovascular Disease
Atherosclerosis (AS) is a pathological change in the early stage of cardiovascular diseases, with a high incidence in middle-aged and elderly people. Endothelial cell injury is the initiating factor in the early stage of AS, which can lead to plaque instability and rupture, accelerating the progression of the disease. Endothelial cell injury can be caused by multiple factors, including hyperlipidemia, oxidative stress, and mechanical stress. The release of mtRNA into the cytoplasm, through the activation of PRRs and the NLRP3 inflammasome activation pathway, exacerbates vascular inflammation, disrupts the endothelial barrier function, increases vascular permeability, promotes monocyte infiltration, and lipid deposition. Abnormal mtRNA may promote the development of AS by generating mitochondrial ROS, oxidizing LDL (ox-LDL), and promoting the formation of foam cells [114, 115].
4.8 Respiratory Disease
Asthma is a widely prevalent chronic respiratory disorder that is defined by airway inflammation, heightened bronchial responsiveness, and intermittent airflow obstruction. One of the inflammatory response mechanisms through which mt-dsRNA exerts its effects involves PRRs, particularly TLR3 and the RLRs. Upon detection of mt-dsRNA, these receptors initiate downstream signaling cascades, resulting in the production of pro-inflammatory cytokines, including IFNs, IL-6, and TNF-α [116]. Experimental data indicate a positive correlation between cytoplasmic mt-dsRNA levels and the expression levels of IL-6, IL-5, and IL-13, with a notable elevation of Th2 cytokines in the lungs. Research has demonstrated that epithelial ETS2 is upregulated in asthma and that ETS2 in epithelial cells enhances IL-6, IL-5, and IL-13 levels through ANT2-mediated regulation of mt-dsRNA levels in the cytosol [116].
Mechanisms of mtRNA in Various Inflammatory-related Diseases
| Disease category | Disease name | Possible mechanism | Potential therapeutic strategies | References |
|---|---|---|---|---|
| Autoimmune disease | Systemic lupus erythematosus | 1.mt-dsRNA, functioning as an autoantigen, induces the production of AmtRNA-IgG/IgM by activating cytoplasmic PRRs; 2.After being phagocytosed by monocytes, erythrocyte-derived mt-dsRNA triggers monocytes to release mtDNA fragments. These fragments bind to the NLRP3 inflammasome to promote the production of IL-1β, and synergize with type I interferons to exacerbate inflammation. | 1.Enhance the activity of the PNPase/SUV3 complex 2.Block the activation of the RIG-I/MDA5-MAVS signaling axis or the NLRP3 inflammasome. | [96, 97] |
| Rheumatoid arthritis | 1.Mitochondrial damage leads to the leakage of mt-dsRNA into the cytoplasm, activates innate immune pathways, and induces the release of proinflammatory cytokines; 2.Along with the accumulation of mtROS, it further activates the NLRP3 inflammasome, exacerbating synovial inflammation and joint damage. | 1.Clear cytoplasmic mt-dsRNA through autophagy regulation 2.Inhibit the activity of the NLRP3 inflammasome 3.Reduce mtROS production to decrease mt-dsRNA release | [98, 99] | |
| Sjögren's syndrome | mt-dsRNAs can be recognized by pattern recognition receptors such as PKR, MDA5, and TLR3, activating the innate immune response | 1.JAK1 inhibitors suppress the synthesis or release of mtRNA 2.Ach activates M3 receptors to inhibit mtRNA release and ISG expression | [88, 100] | |
| Degenerative disease | Osteoarthritis | In chondrocytes, abnormal accumulation of mt-dsRNA and its efflux into the cytoplasm activate PKR, induce the production of proinflammatory cytokines and cell apoptosis, and accelerate cartilage degradation. | 1.Inhibit PKR activity via PKR antagonists 2.Enhance the mtRNA degradation system | [11, 101] |
| Huntington disease | Upregulated expression of mt-dsRNA activates neuroinflammation via the RIG-I/MDA5-MAVS signaling pathway, exacerbates damage to striatal projection neurons. | 1.Block the interaction between RIG-I/MDA5 and MAVS 2.Enhance the function of mitochondrial degradation systems to reduce mt-dsRNA accumulation | [102, 103] | |
| Liver disease | Alcoholic liver disease | Ethanol stress induces hepatocytes to produce mt-dsRNA, which is delivered to Kupffer cells via exosomes, activates TLR3, induces IL-1β secretion, thereby stimulating γδ T cells to produce IL-17A, and exacerbates hepatic inflammation. | 1.TLR3 antagonists inhibit TLR3 activity 2.Block exosome-mediated intercellular transfer of mt-dsRNA | [104] |
| Non-alcoholic fatty liver disease | mt-dsRNA released due to mitochondrial dysfunction upregulates the expression of inflammatory factors by activating the TLR3, MDA5, and p-IRF3 pathways, and is transferred via exosomes to amplify inflammation. | 1.Enhance mitophagy to clear damaged mitochondria and abnormal mtRNA 2.Regulate lipid metabolism to improve mitochondrial function | [106, 117] | |
| Kidney disease | Chronic kidney disease | Downregulation of PNPT1 leads to the leakage of mt-dsRNA into the cytoplasm, activates the PKR-eIF2α axis, triggers apoptosis and damage of renal tubular cells, and is associated with acute tubular necrosis, diabetic nephropathy, and other conditions. | 1.Upregulate PNPT1 expression to promote mt-dsRNA degradation 2.Inhibit PKR activity to block eIF2α phosphorylation 3.Repair mitochondrial membrane permeability | [107] |
| Diabetic nephropathy | mt-dsRNA release from mitochondria to the cytoplasm in DN, activating the PKR/eIF2α pathway and inducing tubular cell apoptosis. | 1.inhibiting mt-dsRNA release 2.blocking PKR activation | [108] | |
| Infectious disease | Viral infection | Viruses induce abnormal accumulation of mt-dsRNA in cells, which are released into the cytoplasm via mitochondrial membrane channels or mitophagy pathways, activate MDA5/RIG-I, and trigger type I interferon antiviral immune responses. | 1.Enhance mt-dsRNA degradation to avoid excessive immune activation 2.Regulate PRRs activity to balance antiviral responses | [109] |
| Cancer | Cancer | mtdsRNA that leaks into the cytoplasm is recognized by RIG-I, triggers a type I interferon response through the MAVS signaling axis, upregulates APOBEC3A in a STAT2-dependent manner, and thereby induces nuclear DNA damage. The mutational signature of the antiviral DNA deaminase APOBEC has been identified in over 70% of cancers. | 1.Block RIG-I/MAVS/STAT2 signaling 2.Antagonize IFN-β secretion | [112] |
| B-cell precursor acute lymphoblastic leukemia | mt-dsRNA serves as a pivotal trigger for the transformation of MSCs into cancer-associated fibroblasts CAFs. | 1.Clear cytoplasmic mt-dsRNA through autophagy regulation | [113] | |
| Cardiovascular disease | Atherosclerosis | mtRNA released into the cytoplasm activates PRRs and the NLRP3 inflammasome, exacerbates vascular inflammation, disrupts the endothelial barrier, and promotes monocyte infiltration and foam cell formation. | 1.Clear cytoplasmic mtRNA to inhibit PRRs activation 2.Inhibit the NLRP3 inflammasome to reduce the release of proinflammatory cytokines | [114, 115] |
| Respiratory disease | Asthma | mt-dsRNA is recognized by PRRs (especially TLR3 and RLRs) to activate downstream signaling cascades, inducing the production of pro-inflammatory cytokines. Moreover, ETS2, which is upregulated in epithelial cells in asthma, can regulate the level of cytoplasmic mt-dsRNA through ANT2, thereby increasing cytokines such as IL-6, IL-5, and IL-13, and participating in the inflammatory process of asthma. | 1.Targeted inhibition of PRRs 2. regulation of cytoplasmic mt-dsRNA levels 3.3inhibition of the upstream regulation of the ETS2-ANT2-mt-dsRNA axis. | [116] |
5. Metabolic Regulation of mtRNA and Inflammation
During the inflammatory response, PNPase plays a multi-level immunoregulatory role by regulating mtRNA metabolism, maintaining the stability of the mitochondrial genome, and the homeostasis of energy metabolism, and participating in the regulation of signaling pathways [69]. PNPase and SUV3 form a complex in the mitochondrial matrix, where they synergistically degrade single-stranded mtRNA and unwind dsRNA , effectively preventing the abnormal accumulation of dsRNA. When the function of this complex is impaired, mt-dsRNA accumulates within mitochondria [83]. In addition, The loss of PNPase leads to an accumulation of mt-dsRNA that exceeds the normal retention capacity of mitochondria, which is then released through voltage-dependent anion channels 1/2 (VDAC1/2) and BAK/BAX channels, activating the cytoplasmic pattern recognition receptor MDA5 and triggering excessive type I interferon responses and inflammatory cascades [78, 118, 119]. SUV3 deficiency leads to the accumulation of dsRNA into the mitochondrial matrix, which triggers MDA5-mediated antiviral signaling pathways and type I interferon responses. However, unlike PNPase, SUV3 deficiency does not directly result in the release of dsRNA into the cytoplasm [68, 118]. This functional difference stems from their division of labor in the complex: SUV3 disrupts RNA secondary structures through its helicase activity, while PNPase degrades linearized RNA with its 3'-5' exonuclease activity [120, 121].
6. mtRNA as an Inflammatory Biomarker and Therapeutic Target
Under various disease conditions, changes in mtRNA are closely associated with the inflammatory response. Its stability in peripheral body fluids (such as serum) indicates its potential as an inflammatory biomarker. In the autoimmune disease SS, stimulation with polyinosinic-polycytidylic acid (poly I:C) increases the levels of total mt-dsRNA and cytosolic mt-dsRNA in salivary gland acinar cells, followed by an increase in PKR phosphorylation and enhanced induction of ISGs. Downregulating mt-dsRNAs with 2-CO-methyladenosine (2-CM) attenuates the IFN signaling signature [88].
Studies have confirmed that mt-dsRNA is closely associated with Sjögren's syndrome in terms of expression levels, disease activity, and differential diagnosis. In terms of expression levels, there are significant differences in mtRNA levels in saliva and plasma between SS patients and healthy individuals. For example, plasma ND1 levels decrease and ND4 levels increase in Sjögren's syndrome patients; saliva ND1 levels increase, while ND5 and others decrease in SS patients. Regarding disease activity, saliva mtRNA scores are positively correlated with objective disease activity indicators (such as ESSDAI and ClinESSDAI) and Raynaud phenomenon, and some plasma mtRNAs are positively correlated with patients' global assessment. In terms of differential diagnosis, the ability of mtRNA composite scores to distinguish patients from healthy individuals is better than that of ISG scores, and the plasma mtRNA scores of patients are significantly higher than those of patients with rheumatoid arthritis and systemic lupus erythematosus, which is conducive to differentiation. This indicates that mtRNA may be a potential biomarker for disease monitoring and stratification in SS [100].
Besides autoimmune diseases, in degenerative diseases like OA, changes in mtRNA are also closely related to the inflammatory response. The expression of mt-dsRNA is significantly increased in the damaged cartilage of OA patients, synovial fluid, and the cartilage of surgically induced OA mice. The level of mt-dsRNA in the cartilage of patients is positively correlated with the senescence-associated secretory phenotype (SASP) and the expression of ISGs, and it increases with the aggravation of cartilage damage [11]. Many studies have shown that activating autophagy can clear mt-dsRNA in the cytoplasm, thereby rescuing the phenotype of mitochondrial dysfunction and alleviating OA-related changes in articular chondrocytes [11].
In liver diseases, research on ALD has also revealed the association between mt-RNA and the inflammatory response. Immunostaining shows that in ethanol-treated mice and hepatocytes of healthy individuals, mt-dsRNA co-localizes with mitochondria, indicating that the accumulation of mt-dsRNA is related to the inflammatory state of hepatocytes under alcohol stimulation and can serve as a potential inflammatory biomarker [104]. From a therapeutic perspective, studies have found that increasing the expression level of PNPase or enhancing its activity can effectively reduce the accumulation of mt-dsRNA. Meanwhile, experiments have demonstrated that when Kupffer cells lack TLR3, the migration of γδ T cells co-cultured with them and the expression of IL-17A are abnormal. This directly indicates that TLR3 is a crucial link for Kupffer cells to receive mt-dsRNA signals and transmit them to γδ T cells, enabling the production of IL-17A and cell migration. It further proves the indispensability of TLR3 in the mt-dsRNA-mediated inflammatory response, suggesting that developing antagonists targeting TLR3 can inhibit the inflame-matory response and providing a potential therapeutic target for ALD [104].
Existing studies have confirmed that cytoplasmic leakage of mt-dsRNA drives the SASP through the RIG-I/MDA5-MAVS pathway, triggering inflame-matory responses, and this process depends on mitochondrial membrane permeability changes mediated by BAX/BAK. BAX/BAK, RIG-I/MDA5, and MAVS may serve as potential targets for inhibiting senescence-related inflammation, provi-ding new insights for intervening in age-related diseases such as tissue fibrosis [122].
7. Current Perspectives and Future Directions
As one of the key regulators of mitochondrial function, the interaction mechanism between mtRNA and the inflammatory response has become a research frontier in the field of immunometabolism. With the role of mtRNA in inflammation regulation being gradually revealed, it is of increasing importance to further explore the specific mechanisms of mtRNA in different inflammation-related diseases, especially its dynamic changes during disease occurrence, development, and outcome, as well as its regulatory mechanisms. Verifying the feasibility and accuracy of using the levels of mtRNA in blood or other body fluids as early diagnostic markers for inflammation-related diseases can provide a reliable basis for clinical diagnosis. For example, in certain autoimmune and infectious diseases, the levels of mtRNA are significantly elevated and are positively correlated with disease activity.
Regarding intervention strategies targeting mtRNA, current research focuses on regulating its metabolism and inflammation-activation pathways. For instance, enhancing the activity of PNPase may promote mtRNA degradation, thereby suppressing mitochondrion-derived inflammation [80]. Addi-tionally, by activating mitophagy (such as using inducers like urolithin A) to remove dysfunctional mitochondria, abnormal mtRNA release can be indirectly reduced (as seen in studies related to NLRP3 inflammasome activation). However, the specificity and safety of these therapies still need further evaluation. For example, the role of mitophagy is context-specific: in chronic or mild stress, it exerts a protective effect by clearing damaged mitochondria and inhibiting inflammation (such as limiting the overactivation of the NLRP3 inflammasome); however, in acute and severe infectious inflammation, its excessive activation may suppress the immune response, hinder pathogen clearance, and produce harmful effects [123]. During systemic administration, PNPase enhancers may non-specifically affect RNA in other tissues, potentially causing side effects and leading to off-target effects.
Mitochondria play a crucial role in aging-related inflammation. Mitochondria regulate the secretion of aging-related cytokines and chemokines through multiple pathways. The release of signaling components such as mtDNA, mtRNA, N-formylated peptides, and ROS can activate the inflammatory response. Future research can further clarify the specific mechanisms of mt-dsRNA release and its interactions with other aging-related signaling pathways [84]. For example, the POLRMT inhibitor IMT1 can reduce mtRNA synthesis, thereby decreasing the cytoplasmic release of mt-dsRNA and effectively alleviating inflammation [124].
Acknowledgements
Funding
This work was supported by the National Natural Science Foundation of China under Grant [82304106 and 82173494].
Author contributions
Jian Chen contributed to conceptualization, writing - original draft. Chen You contributed to writing - original draft. Haibo Xie and Qixing Zhu contributed to conceptualization, project administration, writing - review & editin, and funding acquisition.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Dorn GW. Mitochondrial dynamism and heart disease: changing shape and shaping change. EMBO Molecular Medicine. 2015;7:865-877
2. Sabbir MG, Dar NJ, Bhat SA, Alanazi HH, Perry J. Editorial: Proteins and protein-complexes underlying mitochondrial structure-function and metabolism: implications in diseases. Front Cell Dev Biol. 2024;12:1386787
3. Malpartida AB, Williamson M, Narendra DP, Wade-Martins R, Ryan BJ. Mitochondrial Dysfunction and Mitophagy in Parkinson's Disease: From Mechanism to Therapy. Trends Biochem Sci. 2021;46:329-343
4. Liang H, Ma Z, Zhong W, Liu J, Sugimoto K, Chen H. Regulation of mitophagy and mitochondrial function: Natural compounds as potential therapeutic strategies for Parkinson's disease. Phytother Res. 2024;38:1838-1862
5. Rossmann MP, Dubois SM, Agarwal S, Zon LI. Mitochondrial function in development and disease. Dis Model Mech. 2021;14:dmm048912
6. Yan C, Liu X, Xu H, Wang L. Cytoplasmic mtDNA clearance suppresses inflammatory immune responses. Trends Cell Biol. 2024;34:897-900
7. López-Polo V, Maus M, Zacharioudakis E, Lafarga M, Attolini CS-O, Marques FDM. et al. Release of mitochondrial dsRNA into the cytosol is a key driver of the inflammatory phenotype of senescent cells. Nature Communications. 2024;15:7378
8. Szymanski J, Janikiewicz J, Michalska B, Patalas-Krawczyk P, Perrone M, Ziolkowski W. et al. Interaction of Mitochondria with the Endoplasmic Reticulum and Plasma Membrane in Calcium Homeostasis, Lipid Trafficking and Mitochondrial Structure. Int J Mol Sci. 2017;18:1576
9. Pandey S, Anang V, Schumacher MM. Mitochondria driven innate immune signaling and inflammation in cancer growth, immune evasion, and therapeutic resistance. Int Rev Cell Mol Biol. 2024;386:223-247
10. Jorgensen AN, Rashdan NA, Rao KNS, Delgadillo LF, Kolluru GK, Krzywanski DM. et al. Neurogranin expression regulates mitochondrial function and redox balance in endothelial cells. Redox Biol. 2024;70:103085
11. Kim S, Lee K, Choi YS, Ku J, Kim H, Kharbash R. et al. Mitochondrial double-stranded RNAs govern the stress response in chondrocytes to promote osteoarthritis development. Cell Rep. 2022;40:111178
12. Min Y, O'Neill LAJ. Targeting mitochondrial metabolites and nucleic acids as an anti-inflammatory strategy. Frontiers in Drug Discovery. 2023 3
13. Basu U, Bostwick AM, Das K, Dittenhafer-Reed KE, Patel SS. Structure, mechanism, and regulation of mitochondrial DNA transcription initiation. J Biol Chem. 2020;295:18406-18425
14. Barchiesi A, Vascotto C. Transcription, Processing, and Decay of Mitochondrial RNA in Health and Disease. Int J Mol Sci. 2019;20:2221
15. Falkenberg M, Larsson NG, Gustafsson CM. Replication and Transcription of Human Mitochondrial DNA. Annu Rev Biochem. 2024;93:47-77
16. Gustafsson CM, Falkenberg M, Larsson NG. Maintenance and Expression of Mammalian Mitochondrial DNA. Annu Rev Biochem. 2016;85:133-160
17. Lai HC, Ho UY, James A, De Souza P, Roberts TL. RNA metabolism and links to inflammatory regulation and disease. Cell Mol Life Sci. 2021;79:21
18. Santonoceto G, Jurkiewicz A, Szczesny RJ. RNA degradation in human mitochondria: the journey is not finished. Human Molecular Genetics. 2024;33:R26-R33
19. Shao Z, Yan W, Peng J, Zuo X, Zou Y, Li F. et al. Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate. Nucleic Acids Research. 2014;42:509-525
20. Kotrys AV, Szczesny RJ. Mitochondrial Gene Expression and Beyond-Novel Aspects of Cellular Physiology. Cells. 2019;9:17
21. Li Y, Giorgi EE, Beckman KB, Caberto C, Kazma R, Lum-Jones A. et al. Association between mitochondrial genetic variation and breast cancer risk: The Multiethnic Cohort. PLoS One. 2019;14:e0222284
22. Carbajosa G, Ali AT, Hodgkinson A. Identification of human mitochondrial RNA cleavage sites and candidate RNA processing factors. BMC Biol. 2022;20:168
23. Sanchez MI, Mercer TR, Davies SM, Shearwood AM, Nygard KK, Richman TR. et al. RNA processing in human mitochondria. Cell Cycle. 2011;10:2904-2916
24. Ramaswamy P, Woodson SA. Global stabilization of rRNA structure by ribosomal proteins S4, S17, and S20. J Mol Biol. 2009;392:666-677
25. Recazens E, Jourdain AA. Cytosine methylation flags mitochondrial RNA for degradation. Trends Biochem Sci. 2024;49:843-845
26. Chen YG, Hur S. Cellular origins of dsRNA, their recognition and consequences. Nat Rev Mol Cell Biol. 2022;23:286-301
27. Bruni F. Human mtDNA-Encoded Long ncRNAs: Knotty Molecules and Complex Functions. International Journal of Molecular Sciences. 2024;25:1502
28. Xu Y-c, Su J, Zhou J-j, Yuan Q, Han J-s. Roles of MT-ND1 in Cancer. Current Medical Science. 2023;43:869-878
29. Gusdon AM, Votyakova TV, Reynolds IJ, Mathews CE. Nuclear and Mitochondrial Interaction Involving mt-Nd2 Leads to Increased Mitochondrial Reactive Oxygen Species Production. Journal of Biological Chemistry. 2007;282:5171-5179
30. Xie M, Gu S, Liu Y, Yang H, Wang Y, Yin W. et al. 2-Hydroxyisobutyric acid targeted binding to MT-ND3 boosts mitochondrial respiratory chain homeostasis in hippocampus to rescue diabetic cognitive impairment. Redox Biology. 2025;79:103446
31. Bourges I, Ramus C, Mousson de Camaret B, Beugnot R, Remacle C, Cardol P. et al. Structural organization of mitochondrial human complex I: role of the ND4 and ND5 mitochondria-encoded subunits and interaction with prohibitin. Biochemical Journal. 2004;383:491-499
32. Destiarani W, Mulyani R, Yusuf M, Maksum IP. Molecular Dynamics Simulation of T10609C and C10676G Mutations of MitochondrialND4LGene Associated With Proton Translocation in Type 2 Diabetes Mellitus and Cataract Patients. Bioinformatics and Biology Insights. 2020;14:1177932220978672
33. Bai Y. The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. The EMBO Journal. 1998;17:4848-4858
34. Dirican E, Savrun ŞT, Aydın İE, Gülbay G, Karaman Ü. Analysis of mitochondrial DNA cytochrome-b (CYB) and ATPase-6 gene mutations in COVID-19 patients. Journal of Medical Virology. 2022;94:3138-3146
35. Das S, Ferlito M, Kent OA, Fox-Talbot K, Wang R, Liu D. et al. Nuclear miRNA Regulates the Mitochondrial Genome in the Heart. Circulation Research. 2012;110:1596-1603
36. Wang Y, Das S, Bedja D, Campbell N, Dunkerly B, Chenna V. et al. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. PLoS ONE. 2014;9:e96820
37. Das S, Bedja D, Campbell N, Dunkerly B, Chenna V, Maitra A. et al. Mutation on MT-CO2 gene induces mitochondrial disease associated with neurodegeneration and intracerebral iron accumulation (NBIA). Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2024;1870:166856
38. Smith KK, Moreira JD, Wilson CR, Padera JO, Lamason AN, Xue L. et al. A systematic review on the biochemical threshold of mitochondrial genetic variants. Genome Research. 2024;34:341-365
39. Capiau S, Smet J, De Paepe B, Yildiz Y, Arslan M, Stevens O. et al. Clinical Heterogeneity in MT-ATP6 Pathogenic Variants: Same Genotype—Different Onset. Cells. 2022;11:489
40. Del Dotto V, Musiani F, Baracca A, Solaini G. Variants in Human ATP Synthase Mitochondrial Genes: Biochemical Dysfunctions, Associated Diseases, and Therapies. International Journal of Molecular Sciences. 2024;25:2239
41. Xu M, Lewis RV. Structure of a protein superfiber: spider dragline silk. Proc Natl Acad Sci U S A. 1990;87:7120-7124
42. Suzuki T, Yashiro Y, Kikuchi I, Ishigami Y, Saito H, Matsuzawa I. et al. Complete chemical structures of human mitochondrial tRNAs. Nature Communications. 2020;11:4269
43. Ding Y, Gao B, Huang J. Mitochondrial Cardiomyopathy: The Roles of mt-tRNA Mutations. Journal of Clinical Medicine. 2022;11:6431
44. Osawa S, Jukes TH, Watanabe K, Muto A. Recent evidence for evolution of the genetic code. Microbiol Rev. 1992;56:229-264
45. Moulinier L, Eiler S, Eriani G, Gangloff J, Thierry JC, Gabriel K. et al. The structure of an AspRS-tRNA(Asp) complex reveals a tRNA-dependent control mechanism. EMBO J. 2001;20:5290-5301
46. Santorelli FM, Siciliano G, Casali C, Basirico MG, Carrozzo R, Calvosa F. et al. Mitochondrial tRNA(Cys) gene mutation (A5814G): a second family with mitochondrial encephalopathy. Neuromuscul Disord. 1997;7:156-159
47. Rinehart J, Krett B, Rubio MA, Alfonzo JD, Soll D. Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion. Genes Dev. 2005;19:583-592
48. Van Hove JL, Freehauf C, Miyamoto S, Vladutiu GD, Pancrudo J, Bonilla E. et al. Infantile cardiomyopathy caused by the T14709C mutation in the mitochondrial tRNA glutamic acid gene. Eur J Pediatr. 2008;167:771-776
49. Yang H, Zhang VW, Ai L, Gan S, Wu L. Multisystem Mitochondrial Disease Associated With a Mare m.10000G>A Mitochondrial tRNA (Gly) (MT-TG) Variant. Front Neurol. 2022;13:795060
50. Mimaki M, Ikota A, Sato A, Komaki H, Akanuma J, Nonaka I. et al. A double mutation (G11778A and G12192A) in mitochondrial DNA associated with Leber's hereditary optic neuropathy and cardiomyopathy. J Hum Genet. 2003;48:47-50
51. Casali C, d'Amati G, Bernucci P, DeBiase L, Autore C, Santorelli FM. et al. Maternally inherited cardiomyopathy: clinical and molecular characterization of a large kindred harboring the A4300G point mutation in mitochondrial deoxyribonucleic acid. J Am Coll Cardiol. 1999;33:1584-1589
52. Mariotti C, Tiranti V, Carrara F, Dallapiccola B, DiDonato S, Zeviani M. Defective respiratory capacity and mitochondrial protein synthesis in transformant cybrids harboring the tRNA(Leu(UUR)) mutation associated with maternally inherited myopathy and cardiomyopathy. J Clin Invest. 1994;93:1102-1107
53. Grasso M, Diegoli M, Brega A, Campana C, Tavazzi L, Arbustini E. The mitochondrial DNA mutation T12297C affects a highly conserved nucleotide of tRNA(Leu(CUN)) and is associated with dilated cardiomyopathy. Eur J Hum Genet. 2001;9:311-315
54. Virgilio R, Ronchi D, Bordoni A, Fassone E, Bonato S, Donadoni C. et al. Mitochondrial DNA G8363A mutation in the tRNA Lys gene: clinical, biochemical and pathological study. J Neurol Sci. 2009;281:85-92
55. Bilbille Y, Gustilo EM, Harris KA, Jones CN, Lusic H, Kaiser RJ. et al. The human mitochondrial tRNAMet: structure/function relationship of a unique modification in the decoding of unconventional codons. J Mol Biol. 2011;406:257-274
56. Yang M, Zhang X, Liu G, Yin Y, Chen K, Yun Q. et al. The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.). PLoS One. 2010;5:e12762
57. Kubyshkin V, Davis R, Budisa N. Biochemistry of fluoroprolines: the prospect of making fluorine a bioelement. Beilstein J Org Chem. 2021;17:439-460
58. Lin Y, Xu X, Wang W, Liu F, Zhao D, Li D. et al. A mitochondrial myopathy-associated tRNA(Ser(UCN)) 7453G>A mutation alters tRNA metabolism and mitochondrial function. Mitochondrion. 2021;57:1-8
59. Shimada N, Suzuki T, Watanabe K. Dual mode recognition of two isoacceptor tRNAs by mammalian mitochondrial seryl-tRNA synthetase. J Biol Chem. 2001;276:46770-46778
60. Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J. et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457-465
61. Baric I, Fumic K, Petkovic Ramadza D, Sperl W, Zimmermann FA, Muacevic-Katanec D. et al. Mitochondrial myopathy associated with a novel 5522G>A mutation in the mitochondrial tRNA(Trp) gene. Eur J Hum Genet. 2013;21:871-875
62. Lamichhane TN, Arimbasseri AG, Rijal K, Iben JR, Wei FY, Tomizawa K. et al. Lack of tRNA-i6A modification causes mitochondrial-like metabolic deficiency in S. pombe by limiting activity of cytosolic tRNATyr, not mito-tRNA. RNA. 2016;22:583-596
63. Arredondo JJ, Gallardo ME, Garcia-Pavia P, Domingo V, Breton B, Garcia-Silva MT. et al. Mitochondrial tRNA valine as a recurrent target for mutations involved in mitochondrial cardiomyopathies. Mitochondrion. 2012;12:357-362
64. Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation and degradation of human mitochondrial RNA: the prokaryotic past leaves its mark. Mol Cell Biol. 2005;25:6427-6435
65. Siira SJ, Shearwood AJ, Bracken CP, Rackham O, Filipovska A. Defects in RNA metabolism in mitochondrial disease. Int J Biochem Cell Biol. 2017;85:106-113
66. Boughanem H, Bottcher Y, Tome-Carneiro J, Lopez de Las Hazas MC, Davalos A, Cayir A. et al. The emergent role of mitochondrial RNA modifications in metabolic alterations. Wiley Interdiscip Rev RNA. 2023;14:e1753
67. Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM. et al. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456-467
68. Dhir A, Dhir S, Borowski LS, Jimenez L, Teitell M, Rotig A. et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature. 2018;560:238-242
69. Wang G, Shimada E, Koehler CM, Teitell MA. PNPASE and RNA trafficking into mitochondria. Biochim Biophys Acta. 2012;1819:998-1007
70. Uchiumi T, Kang D. The role of TFAM-associated proteins in mitochondrial RNA metabolism. Biochim Biophys Acta. 2012;1820:565-570
71. Dziembowski A, Piwowarski J, Hoser R, Minczuk M, Dmochowska A, Siep M. et al. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J Biol Chem. 2003;278:1603-1611
72. Silva S, Camino LP, Aguilera A. Human mitochondrial degradosome prevents harmful mitochondrial R loops and mitochondrial genome instability. Proceedings of the National Academy of Sciences. 2018;115:11024-11029
73. Nicholls TJ, Rorbach J, Minczuk M. Mitochondria: mitochondrial RNA metabolism and human disease. Int J Biochem Cell Biol. 2013;45:845-849
74. Fisk JC, Presnyak V, Ammerman ML, Read LK. Distinct and overlapping functions of MRP1/2 and RBP16 in mitochondrial RNA metabolism. Mol Cell Biol. 2009;29:5214-5225
75. Jedynak-Slyvka M, Jabczynska A, Szczesny RJ. Human Mitochondrial RNA Processing and Modifications: Overview. Int J Mol Sci. 2021;22:7999
76. Pietras Z, Wojcik MA, Borowski LS, Szewczyk M, Kulinski TM, Cysewski D. et al. Dedicated surveillance mechanism controls G-quadruplex forming non-coding RNAs in human mitochondria. Nat Commun. 2018;9:2558
77. Kim SJ, Chun M, Wan J, Lee C, Yen K, Cohen P. GRSF1 is an age-related regulator of senescence. Sci Rep. 2019;9:5546
78. Xavier VJ, Martinou JC. RNA Granules in the Mitochondria and Their Organization under Mitochondrial Stresses. Int J Mol Sci. 2021;22:9502
79. Meizlish ML, Franklin RA, Zhou X, Medzhitov R. Tissue Homeostasis and Inflammation. Annu Rev Immunol. 2021;39:557-581
80. Grochowska J, Czerwinska J, Borowski LS, Szczesny RJ. Mitochondrial RNA, a new trigger of the innate immune system. Wiley Interdiscip Rev RNA. 2022;13:e1690
81. Kim S, Ku Y, Ku J, Kim Y. Evidence of Aberrant Immune Response by Endogenous Double-Stranded RNAs: Attack from Within. Bioessays. 2019;41:e1900023
82. Tadepalle N, Shadel GS. RNA reports breaking news from mitochondria. Mol Cell. 2021;81:1863-1865
83. Borowski LS, Dziembowski A, Hejnowicz MS, Stepien PP, Szczesny RJ. Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res. 2013;41:1223-1240
84. Kalykaki M, Rubio-Tomas T, Tavernarakis N. The role of mitochondria in cytokine and chemokine signalling during ageing. Mech Ageing Dev. 2024;222:111993
85. Hooftman A, Peace CG, Ryan DG, Day EA, Yang M, McGettrick AF. et al. Macrophage fumarate hydratase restrains mtRNA-mediated interferon production. Nature. 2023;615:490-498
86. Kruger A, Oldenburg M, Chebrolu C, Beisser D, Kolter J, Sigmund AM. et al. Human TLR8 senses UR/URR motifs in bacterial and mitochondrial RNA. EMBO Rep. 2015;16:1656-1663
87. Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2α kinases: their structures and functions. Cellular and Molecular Life Sciences. 2013;70:3493-3511
88. Yoon J, Lee M, Ali AA, Oh YR, Choi YS, Kim S. et al. Mitochondrial double-stranded RNAs as a pivotal mediator in the pathogenesis of Sjӧgren's syndrome. Mol Ther Nucleic Acids. 2022;30:257-269
89. Saxena AR, Gao LY, Srivatsa S, Bobersky EZ, Periasamy S, Hunt DT. et al. Oxidized and degraded mitochondrial polynucleotides (DeMPs), especially RNA, are potent immunogenic regulators in primary mouse macrophages. Free Radical Biology and Medicine. 2017;104:371-379
90. Liao Y, Liu K, Zhu L. Emerging Roles of Inflammasomes in Cardiovascular Diseases. Frontiers in Immunology. 2022;13:834289
91. Kim KM, Meng Q, Perez de Acha O, Mustapic M, Cheng A, Eren E. et al. Mitochondrial RNA in Alzheimer's Disease Circulating Extracellular Vesicles. Front Cell Dev Biol. 2020;8:581882
92. Fabbiano F, Corsi J, Gurrieri E, Trevisan C, Notarangelo M, D'Agostino VG. RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins? J Extracell Vesicles. 2020;10:e12043
93. Lekva T, Sundaram AYF, Roland MCP, Asheim J, Michelsen AE, Norwitz ER. et al. Platelet and mitochondrial RNA is decreased in plasma-derived extracellular vesicles in women with preeclampsia-an exploratory study. BMC Med. 2023;21:458
94. Dela Justina V, Giachini FR, Priviero F, Webb RC. Double-stranded RNA and Toll-like receptor activation: a novel mechanism for blood pressure regulation. Clin Sci (Lond). 2020;134:303-313
95. Sriram K, Qi Z, Yuan D, Malhi NK, Liu X, Calandrelli R. et al. Regulation of nuclear transcription by mitochondrial RNA in endothelial cells. Elife. 2024;13:e86204
96. Becker Y, Marcoux G, Allaeys I, Julien AS, Loignon RC, Benk-Fortin H. et al. Autoantibodies in Systemic Lupus Erythematosus Target Mitochondrial RNA. Front Immunol. 2019;10:1026
97. Caielli S, Balasubramanian P, Rodriguez-Alcazar J, Balaji U, Wan Z, Baisch J. et al. An unconventional mechanism of IL-1beta secretion that requires Type I IFN in lupus monocytes. bioRxiv. 2023
98. Jing W, Liu C, Su C, Liu L, Chen P, Li X. et al. Role of reactive oxygen species and mitochondrial damage in rheumatoid arthritis and targeted drugs. Front Immunol. 2023;14:1107670
99. Chen Y, Zhou Z, Min W. Mitochondria, Oxidative Stress and Innate Immunity. Front Physiol. 2018;9:1487
100. Ha YJ, Choi YS, Choi SR, Yoon J, Ku D, Kim Y. et al. Association of mitochondrial RNA expression levels in saliva and plasma with interferon signature gene expression and disease activity in patients with Sjogren disease. RMD Open. 2025;11:e005166
101. O'Neill TW, Felson DT. Mechanisms of Osteoarthritis (OA) Pain. Curr Osteoporos Rep. 2018;16:611-616
102. Luna-Sanchez M, Bianchi P, Quintana A. Mitochondria-Induced Immune Response as a Trigger for Neurodegeneration: A Pathogen from Within. Int J Mol Sci. 2021;22:8523
103. Lee H, Fenster RJ, Pineda SS, Gibbs WS, Mohammadi S, Davila-Velderrain J. et al. Cell Type-Specific Transcriptomics Reveals that Mutant Huntingtin Leads to Mitochondrial RNA Release and Neuronal Innate Immune Activation. Neuron. 2020;107:891-908 e8
104. Lee JH, Shim YR, Seo W, Kim MH, Choi WM, Kim HH. et al. Mitochondrial Double-Stranded RNA in Exosome Promotes Interleukin-17 Production Through Toll-Like Receptor 3 in Alcohol-associated Liver Injury. Hepatology. 2020;72:609-625
105. Zheng Y, Wang S, Wu J, Wang Y. Mitochondrial metabolic dysfunction and non-alcoholic fatty liver disease: new insights from pathogenic mechanisms to clinically targeted therapy. J Transl Med. 2023;21:510
106. Huang YH, Wang FS, Wang PW, Lin HY, Luo SD, Yang YL. Heat Shock Protein 60 Restricts Release of Mitochondrial dsRNA to Suppress Hepatic Inflammation and Ameliorate Non-Alcoholic Fatty Liver Disease in Mice. Int J Mol Sci. 2022;23:577
107. Zhu Y, Zhang M, Wang W, Qu S, Liu M, Rong W. et al. Polynucleotide phosphorylase protects against renal tubular injury via blocking mt-dsRNA-PKR-eIF2alpha axis. Nat Commun. 2023;14:1223
108. Duan T, Sun L, Xi Y, Li C, Zhao Q, Xu L. et al. Mito-tempo ameliorates tubular injury of diabetic nephropathy via inhibiting mt-dsRNA release and PKR/eIF2alpha pathway activation. Free Radic Biol Med. 2025;237:147-159
109. Fu C, Cao N, Liu W, Zhang Z, Yang Z, Zhu W. et al. Crosstalk between mitophagy and innate immunity in viral infection. Front Microbiol. 2022;13:1064045
110. Bergstrom EN, Luebeck J, Petljak M, Khandekar A, Barnes M, Zhang T. et al. Mapping clustered mutations in cancer reveals APOBEC3 mutagenesis of ecDNA. Nature. 2022;602:510-517
111. Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y. et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94-101
112. Wick C, Moghadasi SA, Becker JT, Fanunza E, Oh S, Bournique E. et al. Mitochondrial double-stranded RNA triggers induction of the antiviral DNA deaminase APOBEC3A and nuclear DNA damage. J Biol Chem. 2023;299:105073
113. Burt RJ, Dey A, Akarca A, Allen H, Amerikanou R, Atkinson S. et al. Mitochondrial dsRNA from B-ALL cells stimulates mesenchymal stromal cells to become cancer-associated fibroblasts. Blood Adv. 2024;8:5696-5709
114. Wojtasinska A, Frak W, Lisinska W, Sapeda N, Mlynarska E, Rysz J. et al. Novel Insights into the Molecular Mechanisms of Atherosclerosis. Int J Mol Sci. 2023;24:13434
115. Wang R, Wang M, Ye J, Sun G, Sun X. Mechanism overview and target mining of atherosclerosis: Endothelial cell injury in atherosclerosis is regulated by glycolysis (Review). Int J Mol Med. 2021;47:65-76
116. Jiang H, Jiang Y, Dong R, Fu CY. ETS2 aggravate allergic airway inflammation by regulating ANT2-mediated cytosolic mitochondrial DsRNA levels. Respir Res. 2025;26:159
117. Ajith TA. Role of mitochondria and mitochondria-targeted agents in non-alcoholic fatty liver disease. Clin Exp Pharmacol Physiol. 2018;45:413-421
118. Krieger MR, Abrahamian M, He KL, Atamdede S, Hakimjavadi H, Momcilovic M. et al. Trafficking of mitochondrial double-stranded RNA from mitochondria to the cytosol. Life Sci Alliance. 2024;7:e202302396
119. Yoon J, Kim S, Lee M, Kim Y. Mitochondrial nucleic acids in innate immunity and beyond. Exp Mol Med. 2023;55:2508-2518
120. Wang DD, Shu Z, Lieser SA, Chen PL, Lee WH. Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3'-to-5' directionality. J Biol Chem. 2009;284:20812-20821
121. Begeman A, Smolka JA, Shami A, Waingankar TP, Lewis SC. A spatial atlas of mitochondrial gene expression reveals dynamic translation hubs and remodeling in stress. bioRxiv. 2024
122. Victorelli S, Eppard M, Woo SH, Everts SPA, Martini H, Pirius N. et al. Mitochondrial RNA cytosolic leakage drives the SASP. Res Sq. 2024 rs.3.rs-4876596
123. Lima T, Li TY, Mottis A, Auwerx J. Pleiotropic effects of mitochondria in aging. Nat Aging. 2022;2:199-213
124. Bonekamp NA, Peter B, Hillen HS, Felser A, Bergbrede T, Choidas A. et al. Small-molecule inhibitors of human mitochondrial DNA transcription. Nature. 2020;588:712-716
Author contact
![]() Corresponding authors: Department of Dermatology, the First Affiliated Hospital of Anhui Medical University, Hefei 230032, Anhui, China. Phone: +86-6292-3009. E-mail address: xiehaibo666com (Haibo Xie); Department of Dermatology, the First Affiliated Hospital of Anhui Medical University, Hefei 230032, Anhui, China. Phone: +86-6292-3009. E-mail address: zqxingedu.cn (Qixing Zhu).
Corresponding authors: Department of Dermatology, the First Affiliated Hospital of Anhui Medical University, Hefei 230032, Anhui, China. Phone: +86-6292-3009. E-mail address: xiehaibo666com (Haibo Xie); Department of Dermatology, the First Affiliated Hospital of Anhui Medical University, Hefei 230032, Anhui, China. Phone: +86-6292-3009. E-mail address: zqxingedu.cn (Qixing Zhu).

 Global reach, higher impact
Global reach, higher impact