10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2025; 21(14):6081-6112. doi:10.7150/ijbs.115401 This issue Cite
Review
Role of Ubiquitin-regulated EMT in Cancer Metastasis and Chemoresistance
1. National “111” Center for Cellular Regulation and Molecular Pharmaceutics, Hubei University of Technology, Wuhan 430068, China.
2. Hubei Key Laboratory of Industrial Microbiology, Hubei University of Technology, Wuhan 430068, China.
3. School of Life Sciences, Zhengzhou University, Zhengzhou 450001, China.
4. Membrane Protein Disease Research Group, Department of Physiology, Faculty of Medicine and Dentistry, University of Alberta, Edmonton, AB T6G 2R3, Canada.
† Co-first authors: Shuai Xiao and Lingli Tian.
Received 2025-4-9; Accepted 2025-9-11; Published 2025-9-29
Abstract
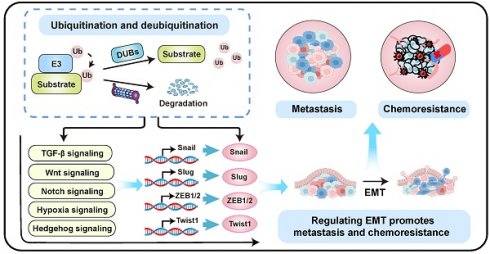
Epithelial-mesenchymal transition (EMT) is a fundamental biological process that promotes cancer metastasis and chemoresistance. However, the therapeutic efficacy of EMT inhibitors remains limited. Ubiquitination, a critical post-translational modification, involves attaching ubiquitin molecules to proteins to regulate their function and stability. It modulates EMT by controlling key EMT transcription factors (EMT-TFs) and associated signaling pathways. Evidence indicates that ubiquitination-dependent regulation of EMT serves as a central mechanism underlying tumor metastasis and chemoresistance. Targeting specific deubiquitinases (DUBs) or E3 ligases can effectively reverse EMT-induced cancer progression and treatment resistance. These findings highlight the therapeutic potential of E3 ligase and DUB inhibitors in oncology. Collectively, ubiquitination-regulated EMT is pivotal in mediating metastasis and chemoresistance in malignant tumors. This review summarizes the molecular mechanisms of EMT and emphasizes ubiquitination's essential role in regulating EMT to promote tumor metastasis and chemoresistance. Consequently, developing inhibitors against specific E3 ligases and DUBs offers a promising strategy to improve cancer treatment outcomes.
Keywords: Ubiquitination, Epithelial-mesenchymal transition, tumor metastasis, Drug resistance
Introduction
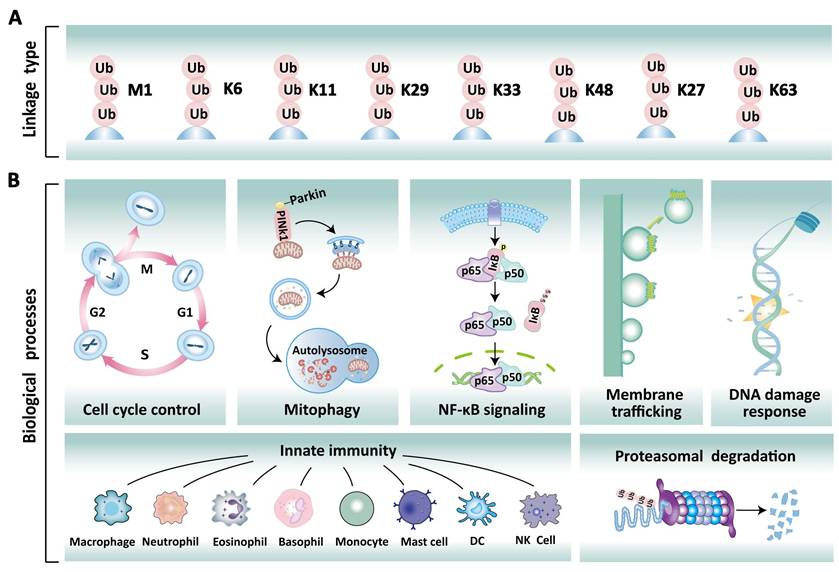
Ubiquitination is a fundamental and reversible post-translational modification (PTM) that plays a pivotal role in eukaryotic cellular homeostasis by dynamically regulating protein stability, activity, localization, and function [1]. This modification process primarily involves ubiquitin molecules, which consist of 76 amino acids and are highly conserved across eukaryotes [2]. In mammalian cells, polyubiquitination typically occurs through the conjugation of ubiquitin via the initial methionine (M1) and seven lysine residues (K6, K11, K27, K29, K33, K48, K63) (Figure 1A) [3]. These distinct ubiquitin modifications exhibit functional diversity. For instance, K48- and K11-linked polyubiquitination predominantly serve as proteolytic signals directing 26S proteasome-mediated substrate recognition and degradation [4, 5]. In contrast, K63-linked polyubiquitination can influence the functions of proteins involved in DNA damage response, signal transduction, and cell cycle control [6]. Notably, the M1-linked linear ubiquitination is formed through the N-terminal methionine residue of ubiquitin, playing a pivotal role in immune regulation and inflammatory responses by activating the NF-κB transcription factor [5]. Furthermore, the remaining linkage types (K6, K27, K29, K33) exhibit pleiotropic roles that span protein activity modulation, intracellular signaling, genomic stability maintenance, cell cycle checkpoint control, and innate immune regulation [6, 7]. Deubiquitination is the reverse reaction catalyzed by deubiquitinases (DUBs), which remove ubiquitin chains to stabilize proteins and modulate biological processes. Collectively, the dynamic balance between ubiquitination and deubiquitination is crucial for maintaining the normal physiological functions of the cell, including protein degradation, DNA damage repair, the cell cycle, and signal transduction (Figure 1B).
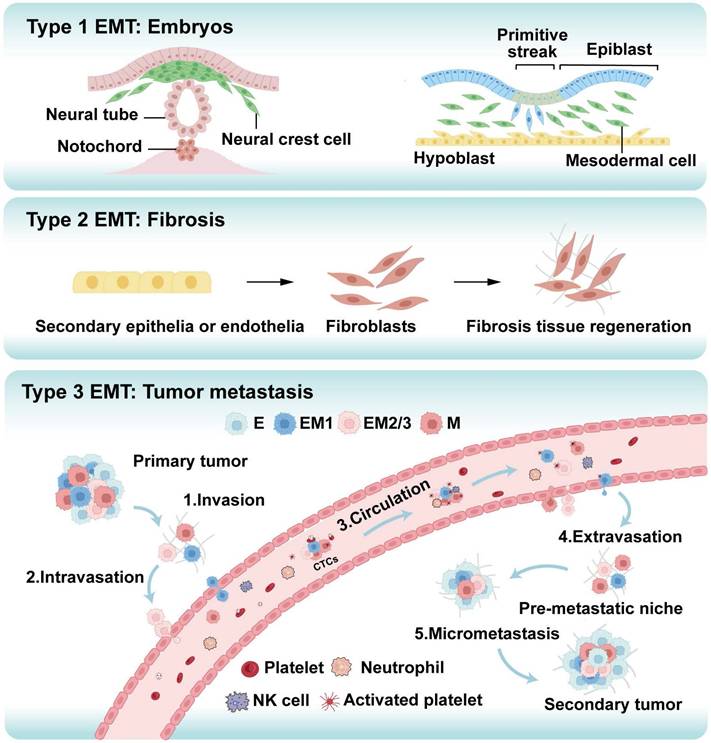
Epithelial-Mesenchymal Transition (EMT) is a cellular reprogramming process in which epithelial cells lose their polarity and cell-cell adhesion properties while acquiring migratory and invasive mesenchymal characteristics [8]. This process, initially characterized in embryonic development and wound healing, is hijacked during cancer progression to drive metastasis, the leading cause of cancer-related mortality [9]. EMT is classified into three distinct types: Type 1 EMT governs normal developmental processes such as embryogenesis; Type 2 EMT facilitates tissue repair and is linked to inflammation and fibrosis; Type 3 EMT, associated with tumor progression, promotes invasion and metastasis (Figure 2) [10-12]. In oncology, EMT enables tumor cells to disseminate from the primary site, enhance invasiveness, and initiate systemic spread [13-15]. This process enhances cellular plasticity, allowing for transitions to hybrid epithelial/mesenchymal states that are highly aggressive and prone to metastasis [16]. Cytoskeletal reorganization during EMT alters cell shape and motility, promoting migration. Moreover, EMT activates cancer stem cell (CSC) properties, upregulating stemness genes that amplify metastatic potential and confer treatment resistance, partly due to acquired mesenchymal resilience to therapies [17, 18]. Overall, EMT promotes metastasis by enabling profound cellular plasticity.
Different types of ubiquitinated chains and various physiological roles. (A) Ubiquitin chain can be classified into eight based on linkage types: Met1, K6, K11, K27, K29, K33, K48, and K63. (B) Different ubiquitination modifications play a specific role in various cellular processes, including cell cycle regulation, mitophagy, NF-κB signaling, membrane trafficking, DNA damage repair, innate immune response, and proteasomal degradation.
The three types of epithelial-mesenchymal transition (EMT) play distinct yet significant roles in various biological processes. Type 1 EMT is crucial for embryonic development, facilitating cell migration and differentiation, which are essential for processes such as gastrulation, neural crest cell migration, and organogenesis. Type 2 EMT is associated with fibrosis, promoting the transformation of epithelial cells into mesenchymal cells, which leads to excessive extracellular matrix (ECM) deposition, tissue remodeling, and ultimately organ dysfunction. Type 3 EMT is particularly critical in cancer metastasis, where primary tumor cells acquire invasive capabilities through EMT. These cells detach from the primary site and undergo five key steps: local invasion, intravasation, survival in the circulation, extravasation, and the formation of metastatic lesions. This process enables the spread of cancer to other parts of the body, resulting in the establishment of secondary tumors or metastatic disease. E: epithelial tumor cell, M: mesenchymal tumor cell, EM1, EM2/3:intermediate cell states, CTCs: circulating tumor cells.
Ubiquitination critically regulates EMT by controlling the degradation and stability of key proteins through the ubiquitin-proteasome system (UPS) [19]. This process is primarily mediated by E3 ligases and DUBs, which regulate the key EMT transcription factors (EMT-TFs) and EMT-associated signaling pathways [19]. Notably, the stability of Snail, a critical EMT transcription factor, is dynamically controlled by ubiquitination; dysregulation of this process enhances EMT and cancer progression [20]. For instance, in colorectal cancer (CRC), mitogen and stress-activated protein kinase 1 (MSK1) recruits USP5 to deubiquitinate and stabilize Snail, facilitating EMT and metastasis [21]. Conversely, in triple-negative breast cancer (TNBC), the E3 ligase Membrane Associated Ring-CH-Type Finger 2 (MARCH2) ubiquitinates Snail, driving its degradation and suppressing tumor growth and metastasis [22]. In non-small cell lung cancer (NSCLC), RNF187 promotes EMT and apoptosis resistance by activating MAPK/PI3K signaling pathways [23]. Dysregulated expression of ubiquitination-related enzymes, such as E3 ligases or DUBs, frequently promotes EMT activation across various cancers, driving enhanced cell migration, invasion, metastasis, and therapy resistance [24, 25]. Therefore, ubiquitination profoundly influences tumor invasion, metastasis, and drug resistance by modulating EMT dynamics. This review summarizes the molecular mechanisms of EMT and highlights its pivotal roles in driving tumor metastasis and chemoresistance. We further elaborate on how EMT is regulated by specific E3 ligases and DUBs, which modulate key EMT inducers like ZEB1 and Snail via ubiquitination-mediated degradation or stabilization. Notably, targeting specific E3 ligases or DUBs can reverse EMT-associated metastasis and chemoresistance. These insights enhance our understanding of ubiquitination in EMT-driven tumor metastasis and drug resistance, supporting the development of E3 ligase or DUB inhibitors as promising antitumor therapies.
Overview of ubiquitination and deubiquitination
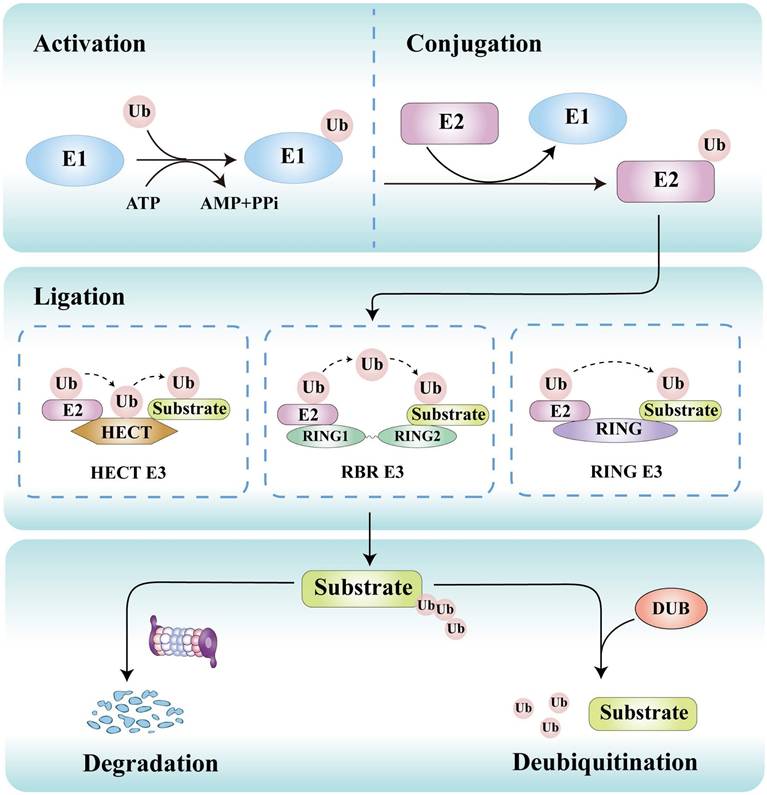
Ubiquitination is initiated by E1 activating enzymes that form a thioester bond with ubiquitin in an ATP-dependent manner. This ubiquitin is then transferred to E2 conjugating enzymes, which collaborate with E3 ligases to attach ubiquitin to lysine residues or other sites on substrate proteins (Figure 3) [26]. This process can lead to proteasomal degradation or functional alterations of target proteins. The primary function of E1 activating enzymes is to initiate the activation process of ubiquitin molecules. Currently, eight distinct E1 enzymes have been identified, including conventional enzymes such as UBA1, UBA6, UBA7, SAE, and NAE, as well as non-conventional enzymes like UBA4, UBA5, and ATG7 [27]. Around 40 E2 enzymes have been identified, which are responsible for selecting the specific lysine residues located on the target protein that will undergo covalent binding with the ubiquitin molecule [28]. E3 ligases play a critical role in determining substrate specificity by directly recognizing the target protein [29]. More than 800 E3 ligases have been discovered, which are classified into three distinct families: the really interesting new gene (RING), homologous to E6AP carboxyl terminus (HECT), and RING-between-RING (RBR) E3 ligases [30]. They employ unique catalytic strategies to regulate protein fate.
Deubiquitination counterbalances ubiquitination through removing ubiquitin from substrate proteins by a family of over 100 DUBs, which catalyze the hydrolysis of ubiquitin chains from substrates [31-33]. DUBs are implicated in a wide range of cellular processes, such as protein stability regulation, signal transduction, cell cycle control, DNA repair, and tumorigenesis [34]. These enzymes can be categorized into seven primary families: ubiquitin-specific proteases (USPs), ubiquitin carboxy-terminal hydrolases (UCHs), motif interacting with ubiquitin-containing novel DUB (MINDYs), JAMM/MPN domain-associated metallopeptidases (JAMMs), ovarian tumor-related proteases (OTUs), Machado-Joseph domain proteases (MJDs), and Zinc finger and UFSP domain protein (ZUFSP) [35]. Dysregulation of DUB activity is implicated in various diseases, particularly tumors [36] and neurodegenerative diseases [37]. DUBs have been shown to influence tumor progression and metastasis, with some DUBs acting as inhibitors while others promote tumor development. For instance, USP10 stabilizes p53 by deubiquitinating, thus counteracting Murine double minute 2 (MDM2)-mediated ubiquitination and inhibiting the growth of renal cell carcinoma (RCC) cells [38]. Furthermore, USP14 stabilizes the oncogene protein B-cell lymphoma 6 through deubiquitination, thereby promoting the proliferation of ovarian cancer (OC) cells [39]. Collectively, dysregulation of ubiquitination or deubiquitination disrupts this homeostasis, contributing to pathological conditions such as cancer, neurodegenerative disorders, cardiovascular diseases, and metabolic syndromes.
EMT: Mechanisms and roles in cancer
The EMT process involves key molecular alterations, including the downregulation of epithelial markers and the upregulation of mesenchymal markers, which collectively modify cell adhesion properties [40]. These molecular shifts are intrinsically coupled with cytoskeletal remodeling, directly impacting cellular morphology and motility. The EMT is regulated by key EMT-TFs, signaling pathways, and epigenetic modifications that influence gene expression changes [41, 42]. Among these EMT-TFs, zinc-finger proteins (Snail and Slug), zinc-finger E-box binding homeobox factors (ZEB1 and ZEB2), and basic helix-loop-helix proteins (Twist1 and Twist2) have been studied the most extensively [43, 44]. Specifically, Snail and Slug bind to the E-box motif in the CDH1 promoter region to repress E-cadherin expression while activating mesenchymal gene transcription, thereby driving EMT progression [45, 46]. Similarly, Twist1 and Twist2 suppress epithelial genes and promote the expression of mesenchymal genes, contributing to EMT-associated metastasis [47]. ZEB1 and ZEB2 further inhibit CDH1 transcription via E-box binding, accelerating the transition to a mesenchymal phenotype [48, 49]. Collectively, these TFs orchestrate the expression of mesenchymal phenotypic markers and underpin the molecular dynamics of EMT.
Ubiquitination is a dynamic and reversible process. The process of ubiquitin transfer necessitates the coordinated action of three types of ubiquitinating enzymes. Initially, the E1 ubiquitin-activating enzyme activates ubiquitin in an ATP-dependent manner. Subsequently, the activated ubiquitin molecule is transferred from the E1 enzyme to E2 ubiquitin-conjugating enzymes. Finally, E3 ubiquitin ligases facilitate the transfer of ubiquitin from E2 to target substrates, which may occur either directly or indirectly, depending on their structural characteristics and functional roles. Conversely, deubiquitinases (DUBs) can remove ubiquitin from substrates, thereby regulating the level of ubiquitination and the stability of proteins.
Furthermore, the regulation of EMT involves multiple signaling pathways, including the TGF-β, Wnt/β-catenin, Notch, Hedgehog (Hh), and Hypoxia signaling [50]. These extracellular signals promote the transcription of EMT-TF, thereby regulating the EMT process [50]. Specifically, TGF-β signaling induces morphological and functional alterations in cells through both SMAD-dependent and non-SMAD-dependent mechanisms [51]. In the Smad-dependent pathway, TGF-β activation results in the transcriptional regulation of EMT-TFs, facilitating the downregulation of epithelial markers and upregulation of mesenchymal markers [52]. Additionally, in non-SMAD pathways, TGF-β engages MAPK (including ERK, JNK, and p38), Rho-like GTPase, and PI3K/AKT signaling to modulate EMT [53-55]. The Wnt/β-catenin signaling pathway plays a critical role in stabilizing EMT-TFs and enhancing the transcription of EMT-related genes, contributing to metastasis [56]. Under hypoxic conditions, Notch signaling amplifies hypoxia-inducible factor-1α (HIF-1α)-mediated activation of the lysyl oxidase (LOX) gene, which facilitates Snail expression and EMT progression [57]. Furthermore, Notch signaling interacts with Wnt and TGF-β signaling, jointly regulating the expression of EMT-TF and enhancing the EMT in tumor contexts [58]. Hh signaling activation occurs via ligand binding, leading to Gli transcription factor nuclear translocation and subsequent regulation of EMT-TFs, including Snail and Twist family members [59, 60].
EMT involves the transformation of epithelial cells into mesenchymal phenotypes, enhancing cell motility, invasiveness, and stemness [61]. This process is reactivated in cancers and directly contributes to tumor metastasis and treatment resistance [62, 63]. Mechanistically, key signaling pathways (TGF-β, Wnt/β-catenin, Notch, Hh, and Hypoxia) promote the expression of EMT-TFs (Snail, Twist, ZEB), which collectively repress E-cadherin while upregulating mesenchymal markers like vimentin and N-cadherin [62, 64-66]. Matrix metalloproteinases (MMPs) facilitate invasion by degrading extracellular matrix components and activating EMT-associated signals [67]. EMT also enhances cellular adaptability within hypoxic tumor microenvironments, contributing to survival under metabolic stress [68]. Beyond promoting motility and invasiveness, EMT is linked to increased stemness and the activation of anti-apoptotic mechanisms and multidrug resistance efflux pumps, which together heighten tumor heterogeneity and treatment resistance [69]. Moreover, EMT cooperates with immunosuppressive elements in the tumor microenvironment, reducing sensitivity to immunotherapies. In NSCLC, EMT-induced immunosuppression correlates with poor patient outcomes [70]. Furthermore, in mesenchymal tumors such as osteosarcoma (OS), the EMT phenotype is also associated with chemotherapy resistance [69, 71]. Furthermore, the high expression of EMT-TFs such as Snail and Slug significantly enhanced the cisplatin resistance in ovarian carcinoma (OC) [72]. Collectively, the EMT process is closely related to tumor cell invasion, metastasis, and treatment resistance.
Ubiquitination in regulating EMT
Ubiquitination plays a key role in regulating EMT [19]. Several E3 ligases (ubiquitination) and DUBs (deubiquitination) critically regulate EMT by modulating core EMT-TFs such as Snail/Slug, ZEB1/2, and Twist1 (Figure 4, Table 1, and Table 2), as well as key EMT-associated signaling networks such as TGF-β, Wnt/β-catenin signaling, Notch, Hh, and hypoxia signaling (Figure 5, Table 1, and Table 2).
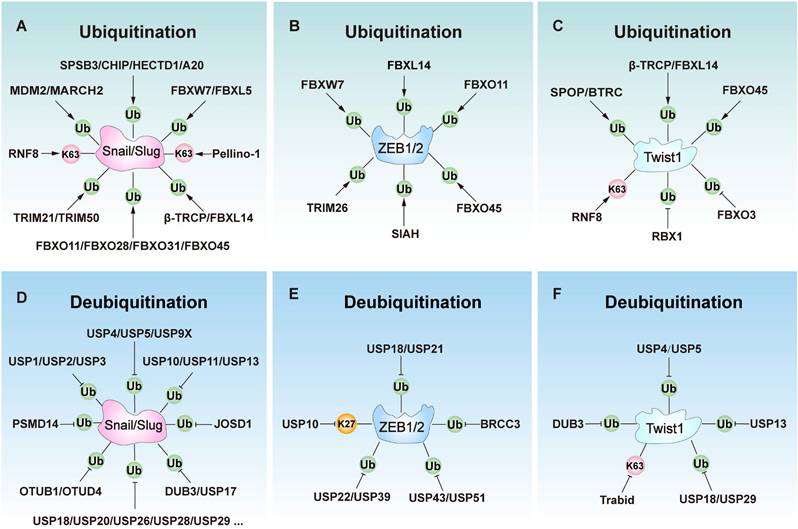
The role of E3s and deubiquitinases (DUBs) involves key transcript factors in Epithelial-mesenchymal transition (EMT) regulation, including Snail, Slug, ZEB1, ZEB2, and Twist1. (A) E3 ligases ubiquitinate Slug or Snail to regulate EMT. (B) E3 ligases ubiquitinate ZEB1 or ZEB2 to regulate EMT. (C) DUBs ubiquitinate Twist1 to regulate EMT. (D) DUBs ubiquitinate Slug or Snail to regulate EMT. (E) E3 ligases ubiquitinate ZEB1 or ZEB2 to regulate EMT. (F) DUBs ubiquitinate Twist1 to regulate EMT.
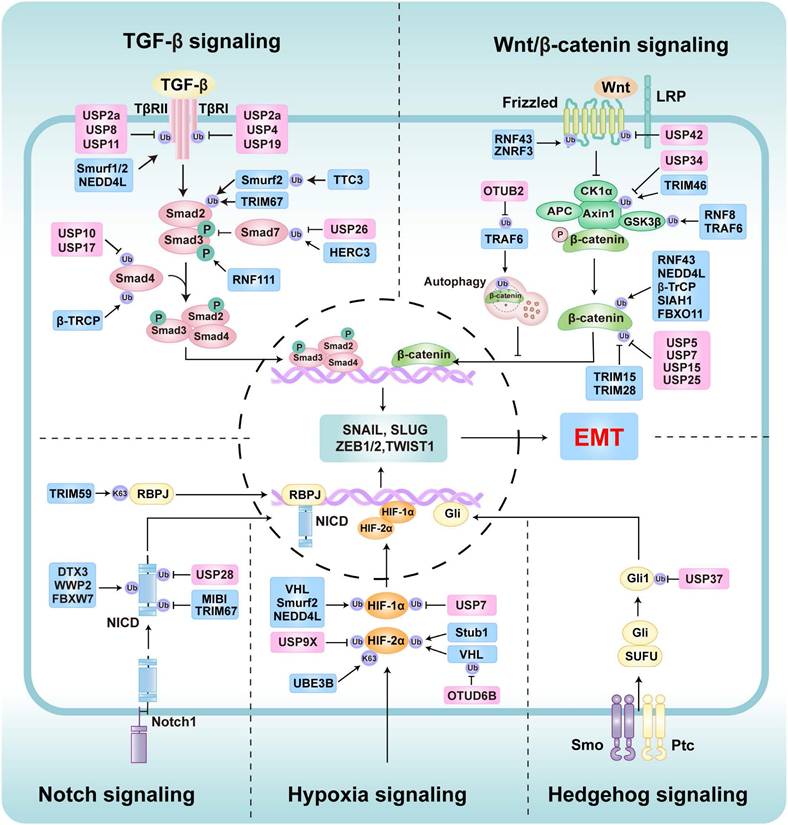
The role of E3s and DUBs involves key signaling pathways in EMT regulation, including TGF-β signaling, Wnt/β-catenin signaling, Hypoxia signaling, Hedgehog signaling, and Notch signaling. These pathways modulate the activity of EMT transcription factors through various mechanisms, thereby promoting or inhibiting the EMT process. The E3s are marked with blue icons, and DUBs are marked with pink icons.
E3 ligases in EMT regulation
| Protein | Substrates | Effect on EMT | Ref. |
|---|---|---|---|
| SPSB3 | Snail | Negatively regulates EMT by degrading Snail | [74] |
| CHIP | Snail, Slug | Negatively regulates EMT by degrading Snail and Slug | [75, 76] |
| MARCH2 | Snail | Negatively regulates EMT by promoting the degradation of Snail. | [22] |
| HECTD1 | Snail | Negatively regulates EMT by degrading Snail. | [92] |
| Pellino-1 | Snail, Slug | Positively regulates EMT by stabilizing Slug and Snail. | [93, 94] |
| MDM2 | Snail, Slug, SMAD2/3 | Positively regulates EMT by stabilizing Slug and Snail; activating the TGF-β signaling pathway. | [77, 78, 163] |
| A20 (TNFAIP3) | Snail | Positively regulates EMT by stabilizing Snail through monoubiquitination | [96] |
| β-TRCP1 (FBXW1) | Snail, Slug, Twist1, SMAD4, β-catenin | Negatively regulates EMT by degrading Slug, Snail, Twist1, and inhibiting TGF-β and Wnt/β-catenin signaling. | [79, 80, 144, 158, 184] |
| FBXW7 (FBW7) | Snail, ZEB1, ZEB2, Notch1 | Negatively regulates EMT by regulating Snail, ZEB1, and ZEB2; inhibiting Notch signaling. | [81, 131, 132] |
| FBXL5 | Snail | Negatively regulates EMT by degrading Snail. | [82, 83] |
| FBXL14 (Ppa) | Snail, Slug, Twist, ZEB2 | Negatively regulates EMT by degrading Snail, Slug, ZEB2, and Twist. | [84, 85, 145] |
| FBXO3 | Twist1 | Positively regulates EMT by enhancing USP4-induced Twist1 stabilization. | [148] |
| FBXO11 | Snail, ZEB1, β-catenin | Negatively regulates EMT by degrading Snail, ZEB1, and inhibiting Wnt/β-catenin signaling. | [86, 128] |
| FBXO28 | Snail | Negatively regulates EMT by degrading Snail. | [87] |
| FBXO31 | Snail | Negatively regulates EMT by degrading Snail. | [88] |
| FBXO45 | Twist, Snail, Slug, and ZEB2 | Negatively regulates EMT by degrading Twist, Snail, Slug, and ZEB2. | [89] |
| TRIM15 | / | Positively regulates EMT by activating the Wnt/β-catenin signaling. | [191] |
| TRIM28 | / | Positively regulates EMT by activating the Wnt/β-catenin signaling. | [192] |
| TRIM21 | Snail | Negatively regulates EMT by degrading Snail. | [90] |
| TRIM26 | ZEB1 | Negatively regulates EMT by degrading ZEB1. | [91, 129] |
| TRIM46 | Axin1 | Positively regulates EMT by degrading Axin1 and activating the Wnt/β-catenin signaling | [190] |
| TRIM50 | Snail | Negatively regulates EMT by degrading Snail. | [91] |
| TRIM59 | RBPJ | Positively regulates EMT by stabilizing RBPJ and activating Notch signaling. | [213] |
| TRIM67 | SMAD3 | -Negatively regulates EMT by degrading SMAD3 and inhibiting TGF-β signaling. -Positively regulates EMT by activating Notch signaling. | [159, 210] |
| TRAF6 | β-catenin | -Positively regulates EMT by degrading GSK3β and activating the Wnt/β-catenin signaling. -Negatively regulates EMT by inhibiting Wnt/β-catenin signaling. | [188, 189] |
| HERC3 | SMAD7, EIF5A2 | Positively regulates EMT by degrading Smad7 and activating TGF-β signaling Negatively regulates EMT by degrading EIF5A2. | [161, 162] |
| SIAH | ZEB1 | Negatively regulates EMT by degrading ZEB1. | [130] |
| NEDD4L | TGF-β, TβRII, β-catenin, HIF1α | Negatively regulates EMT by inhibiting TGF-β, Wnt/β-catenin, and Hypoxia signaling. | [160, 187] |
| RBX1 | Twist1 | Positively regulates EMT by inhibiting FBXO45-induced Twist1 degradation. | [147] |
| RNF8 | Slug, Twist1, GSK3β/β-catenin | Positively regulates EMT by stabilizing Slug and Twist1; activating Wnt/β-catenin signaling | [95, 146, 193] |
| RNF43 | β-catenin | Negatively regulates EMT by inhibiting Wnt/β-catenin signaling. | [183] |
| RNF61 (MKRN1) | SNIP1 | Positively regulates EMT by degrading SNIP1 and activating TGF-β signaling. | [164] |
| RNF111 | SMAD3 | Positively regulates EMT by activating TGF-β/SMAD3 signaling. | [165, 166] |
| TTC3 | Smurf2 | Positively regulates EMT by inhibiting Smurf2-induced TGFR and SMAD degradation and activating TGF-β signaling. | [167] |
| SPOP | Twist1 | Negatively regulates EMT by degrading Twist1. | [142] |
| Smurf1 | TGF-βRII | Negatively regulates EMT by degrading TGF-βRII and inhibiting TGF-β signaling. | [154] |
| Smurf2 | SMAD1/2, TGF-βRI, HIF1α | Negatively regulates EMT by inhibiting TGF-β and Hypoxia signaling. | [155-157, 227] |
| Siah1-SIP-Skp1 | β-catenin | Negatively regulates EMT by degrading β-catenin and inhibiting Wnt/β-catenin signaling. | [186] |
| VHL | HIF1α, HIF2α | Negatively regulates EMT by degrading HIF1α/ HIF2α and inhibiting Hypoxia signaling. | [222, 223] |
| Stub1 | HIF2α | Negatively regulates EMT-associated vascular remodeling by degrading HIF2α and inhibiting Hypoxia signaling under acute hypoxia conditions. | [229] |
| DTX3 | NICD | Negatively regulates EMT by degrading NICD and inhibiting Notch signaling. | [206] |
| WWP2 | NICD | Negatively regulates EMT by downregulating Notch activity. | [207] |
| MIB1 | / | Positively regulates EMT by activating Notch signaling. | [211] |
| BTRC | Twist1 | Negatively regulates EMT by ubiquitination-mediated degradation of Twist1. | [143] |
DUBs in EMT regulation
| Protein | Substrates | Effect on EMT | Ref. |
|---|---|---|---|
| USP1 | Snail, TAK1 | Positively regulates EMT by stabilizing Snail; activating TGF-β signaling by stabilizing TAK1. | [97, 168] |
| USP2 | Snail | Positively regulates EMT by stabilizing Snail. | [99] |
| USP2a | TβRI/II | Positively regulates EMT by stabilizing TβRI/II and activating TGF-β signaling. | [174] |
| USP3 | Snail, SUZ12 | Positively regulates EMT by stabilizing Snail and SUZ12. | [100, 175] |
| USP4 | Snail, Twist1, TβRI | Positively regulates EMT by stabilizing Snail and Twist1; activating TGF-β signaling. | [101, 148, 169, 170] |
| USP5 | Snail, Slug, Twist1, β-catenin | Positively regulates EMT by stabilizing Snail, Slug, and Twist1; activating β-catenin signaling. | [21, 122, 149, 196] |
| USP7 | β-catenin, HIF-1α | Positively regulates EMT by activating β-catenin and Hypoxia signaling. | [197-199, 230] |
| USP8 | TβRII | Positively regulates EMT by stabilizing TβRII and activating TGF-β signaling. | [176] |
| USP9X | Snail, HIF-2α | Positively regulates EMT by stabilizing Snail; activating Hypoxia signaling. | [102, 231] |
| USP10 | Snail, Slug, ZEB1, SMAD4 | -Positively regulates EMT by stabilizing Snail and Slug; activating TGF-β signaling. -Negatively regulates EMT by degrading ZEB1. | [103, 123, 140, 172] |
| USP11 | Snail, TβRII | Positively regulates EMT by stabilizing Snail; activating TGF-β signaling by stabilizing TβRII. | [104, 177] |
| USP13 | Snail, Twist1, WISP1 | Positively regulates EMT by stabilizing Snail and Twist1; activating Wnt/β-catenin signaling by stabilizing WISP1. | [105, 150, 200] |
| USP15 | β-catenin | Positively regulates EMT by stabilizing β-catenin and activating Wnt/β-catenin signaling. | [201] |
| USP17 | Snail, SMAD4 | Positively regulates EMT by stabilizing Snail; activating TGF-β signaling. | [106, 171] |
| USP18 | Snail, ZEB1, Twist1 | Positively regulates EMT by stabilizing Snail, Twist1, and ZEB1. | [107, 136, 151] |
| USP19 | TβRI | -Positively regulates EMT by USP19-CY, which stabilizes TβRI and TβRII to activate TGF-β signaling. -Negatively regulates EMT by USP19-ER, which inhibits EMT by reducing TβRI surface expression. | [173] |
| USP20 | Slug | Positively regulates EMT by stabilizing Slug. | [124] |
| USP21 | ZEB1 | Positively regulates EMT by stabilizing ZEB1. | [137] |
| USP22 | ZEB1 | Positively regulates EMT by stabilizing ZEB1. | [133, 195] |
| ADAM9 | Negatively regulates EMT by stabilizing ADAM9 and Wnt/β-catenin signaling. | ||
| USP25 | β-catenin | Positively regulates EMT by stabilizing β-catenin and activating Wnt/β-catenin signaling. | [202] |
| USP26 | Snail | -Positively regulates EMT by stabilizing Snail. -Negatively regulates EMT by stabilizing SMAD7 and inhibiting TGF-β signaling. | [108, 178] |
| USP27X | Snail | Positively regulates EMT by stabilizing Snail. | [109] |
| USP28 | Snail, NICD | Positively regulates EMT by stabilizing Snail and NICD, activating Notch-induced EMT | [110, 208] |
| USP29 | Snail, Twist1 | Positively regulates EMT by stabilizing Snail and Twist1. | [111, 152] |
| USP30 | Snail | Positively regulates EMT by stabilizing Snail. | [112] |
| USP34 | Axin1 | Negatively regulates EMT by stabilizing Axin1 and Wnt/β-catenin signaling. | [194] |
| USP35 | Snail | Positively regulates EMT by stabilizing Snail. | [113] |
| USP36 | DOCK4 | Positively regulates EMT by stabilizing DOCK4 and activating Wnt/β-catenin signaling. | [203] |
| USP37 | Snail, Gli-1 | Positively regulates EMT by stabilizing Snail; activating Hh signaling. | [114, 238] |
| USP39 | ZEB1 | Positively regulates EMT by stabilizing ZEB1. | [129] |
| USP41 | Snail | Positively regulates EMT by stabilizing Snail. | [115] |
| USP42 | FZD | Negatively regulates EMT by stabilizing FZD and suppressing the Wnt signaling pathway. | [182] |
| USP43 | ZEB1 | Positively regulates EMT by stabilizing ZEB1. | [138] |
| USP47 | Snail | Positively regulates EMT by stabilizing Snail. | [116] |
| USP51 | ZEB1 | Positively regulates EMT by stabilizing ZEB1. | [134, 135] |
| BRCC3 | ZEB1 | Positively regulates EMT by stabilizing ZEB1. | [139] |
| DUB3 | Snail, Slug, Twist | Promotes EMT by stabilizing Snail, Slug, and Twist. | [98, 125] |
| OTUB1 | Snail | Positively regulates EMT by stabilizing Snail. | [117] |
| OTUB2 | β-catenin | Positively regulates EMT by stabilizing β-catenin and activating Wnt/β-catenin. | [204] |
| OTUD4 | Snail | Positively regulates EMT by stabilizing Snail. | [118] |
| OTUD6B | VHL | Negatively regulates EMT by stabilizing VHL or mutated VHL, inhibiting Hypoxia signaling through downregulating HIF-1α or HIF-2α. | [225, 226] |
| Trabid | Twist1 | Negatively regulates EMT by degrading Twist1 | [153] |
| EIF3H | Snail | Positively regulates EMT by stabilizing Snail. | [119] |
| JOSD1 | Snail | Positively regulates EMT by stabilizing Snail. | [120] |
| PSMD14 | Snail | Positively regulates EMT by stabilizing Snail. | [121] |
Ubiquitination regulation of EMT-TFs
Snail/Slug regulation
Snail and Slug are key EMT-TFs involved in the regulation of EMT by suppressing E-cadherin expression [73]. The stability and activity of Snail/Slug are primarily governed by E3 ligases and DUBs. Generally, E3 ligases inhibit EMT by facilitating the ubiquitination and proteasomal degradation of these proteins (Table 1). For instance, SPRY Domain-Containing SOCS Box Protein 3 (SPSB3) promotes Snail degradation in a GSK-3β phosphorylation-dependent manner to limit EMT [74]. C-terminus of HSC70-interacting protein (CHIP) ubiquitinates Snail by K48-linked ubiquitin chains, leading to its degradation and inhibiting the EMT process [75, 76]. Similarly, MARCH2 directly interacts with Snail to induce its ubiquitination and subsequent proteasomal degradation, thereby suppressing EMT [22]. Other E3 ligases, such as MDM2 [77, 78], FBXW1/β-TRCP1 [79, 80], FBXW7/FBW7 [81], FBXL5 [82, 83], FBXL14/Ppa [84, 85], FBXO11[86], FBXO28 [87], FBXO31 [88], FBXO45 [89], TRIM21 [90], TRIM50 [91], and HECTD1 [92] can degrade Snail or Slug to inhibit EMT. F-box proteins play a major role in regulating the functions of Snail and Slug proteins, and are also closely related to tumor metastasis. These E3 ligases are usually downregulated in aggressive cancers, enabling Snail accumulation to promote EMT. However, a few E3 ligases like Pellino-1 [93, 94] and RNF8 [95] can stabilize Snail or Slug by K63-linked ubiquitin chains to facilitate EMT. Additionally, A20 (TNFAIP3) stabilizes Snail through monoubiquitination, thereby promoting EMT in response to TGF-β1 [96].
Conversely, DUBs stabilize Snail or Slug by removing ubiquitin modifications, thereby driving the EMT process (Table 2). For instance, USP1 stabilizes Snail by removing K48-linked polyubiquitin chains, increasing its stability and promoting EMT progression [97]. Similarly, DUB3 can also stabilize Snail through deubiquitination, thereby promoting the EMT process [98]. Other DUBs such as USP2 [99], USP3 [100], USP4 [101], USP5 [21], USP9X [102], USP10 [103], USP11 [104], USP13 [105], USP17 [106], USP18 [107], USP26 [108], USP27X [109], USP28 [110], USP29 [111], USP30 [112], USP35 [113], USP37 [114], USP41 [115], USP47 [116], OTUB1 [117], OTUD4 [118], Eukaryotic translation initiation factor 3 subunit H (EIF3H) [119], Josephin domain-containing 1 (JOSD1) [120], PSMD14 [121] enhance the stability of Snail by removing ubiquitin chains to stabilize it, facilitating EMT progression. Furthermore, only a few DUBs like USP5 [122], USP10 [123], USP20 [124], and DUB3 [125] can stabilize Slug by deubiquitinating to promote the EMT. Thus, DUBs mainly promote EMT by stabilizing Snail.
ZEB1/2 regulation
ZEB1 and ZEB2 act as master transcriptional repressors of EMT, primarily inhibiting E-cadherin expression to disrupt intercellular junctions and initiate EMT [126]. They also induce the expression of mesenchymal markers, including vimentin, thereby enhancing cellular migration and invasion capabilities [127]. The stability of ZEB1 and ZEB2 is dynamically regulated by ubiquitination and deubiquitination (Table 1 and Table 2). Specific E3 ligases, such as FBXO11, directly ubiquitinate and degrade ZEB1 to inhibit EMT [128]. Additional E3 ligases such as TRIM26 [129], SIAH [130], FBXO45 [89], FBXW7 [131, 132], and FBXL14 [85], which can also negatively regulate EMT by targeting ZEB1 or ZEB2 for degradation.
DUBs critically regulate ZEB1 stability through deubiquitination to regulate EMT and metastasis. For instance, USP22 stabilizes ZEB1 to activate ZEB1-mediated transcriptional activation and drive EMT [133]. Similarly, USP51 stabilizes ZEB1 through deubiquitination, thereby promoting mesenchymal activation and stromal recruitment in gastric cancer (GC) and lung adenocarcinoma (LUAD) [134, 135]. Moreover, CDK4/6 further amplifies this process in LUAD by phosphorylating USP51 [134]. Other DUBs such as USP18 [136], USP21 [137], USP39 [129]. USP43 [138], and BRCA1-BRCA2-containing complex subunit 3 (BRCC3) [139] enhance the stability of ZEB1 by removing ubiquitin chains, facilitating cell migration and EMT progression. Conversely, USP10 promotes ZEB1 degradation in CRC by removing K27-linked ubiquitin chains, thereby inhibiting EMT [140]. These findings underscore the critical dynamic balance between ubiquitination by E3 ligases and deubiquitination by DUBs in controlling ZEB1 stability, thereby regulating the EMT process.
Twist1 regulation
Twist1 is another critical transcription factor in regulating EMT by directly binding to the E-box motif to repress E-cadherin expression and activate mesenchymal genes [141]. Its stability is mainly regulated through the ubiquitination and deubiquitination processes to control the progression of epithelial-mesenchymal transition (Table 1 and Table 2). For instance, the E3 ligase SPOP (speckle-type POZ protein) ubiquitinates and degrades Twist1 to suppress EMT progression [142]. Other E3 ligases like BTRC [143], β-TRCP [144], FBXL14 [145], and FBXO45 [89] promote the degradation of Twist1 to inhibit EMT. Conversely, E3 ligase RNF8 [146], RBX1[147], and FBXO3 [148] stabilize Twist1 to activate EMT and cancer progression. Furthermore, DUBs can stabilize Twist1 to promote EMT. For instance, USP5 stabilizes Twist1 through deubiquitination, thereby activating EMT in bladder cancer [149]. Moreover, USP13 similarly stabilizes Twist1 to facilitate EMT [150]. Other DUBs like DUB3 [125], USP4 [148], USP18 [151], and USP29 [152] can also stabilize Twist1 through deubiquitination, thereby facilitating EMT progression. Conversely, DUB TRAF-binding domain (Trabid) can promote the degradation of Twist1 by removing K63-linked ubiquitin chains, leading to its degradation and EMT inhibition in hepatocellular carcinoma (HCC) [153]. Together, E3 ligases and DUBs regulate Twist stability to control EMT progression.
Ubiquitination Regulation in EMT-Related Signaling Pathways
TGF-β signaling regulation
TGF-β signaling is a prominent pathway for the induction of EMT [8]. The canonical SMAD pathway involves TGF-β ligands binding to TGF-β type II receptor (TβRII), which then recruits and activates TGF-β type I receptor (TβRI) [52]. Activated TβRI phosphorylates SMAD2 and SMAD3, which form complexes with SMAD4 that translocate to the nucleus to regulate EMT-related gene expression [52]. However, SMAD7 inhibits this pathway by directly binding TβRI or disrupting SMAD complex formation [52]. Ubiquitination critically regulates TGF-β/SMAD-induced EMT by targeting pathway components (Table 1). For instance, Smurf1 directly ubiquitinates TβRII by K48-linked polyubiquitin chains to degrade it, suppressing TGF-β-induced EMT [154]. Similarly, Smurf2 promotes proteasomal degradation of SMAD2 and TGFβ receptor, thereby suppressing TGF-β-induced EMT [155-157]. Furthermore, other E3 ligases like β-TrCP [158], TRIM67 [159], and NEDD4L [160] also negatively regulate TGF-β-induced EMT by SMAD proteins or TGF-β receptors. Conversely, HERC3 promotes the autophagic degradation of Smad7 through K63-linked polyubiquitin chains, thereby enhancing TGF-β/SMAD-induced EMT [161]. Intriguingly, another research showed that HERC3 ubiquitinates and degrades EIF5A2, thereby inhibiting the EMT induced by the EIF5A2/TGF-β/Smad2/3 signaling pathway in CRC [162]. These two opposite results further demonstrate the complexity and heterogeneity of tumor cell signal regulation. Other E3 ligases, including MDM2 [163], RNF61 [164], RNF111 [165, 166], and TTC3 [167], enhance TGF-β signaling and EMT by stabilizing receptors or facilitating signal transduction.
The regulatory balance is further influenced by DUBs, which remove ubiquitination levels to modulate TGF-β/SMAD signaling pathways (Table 2). For example, USP1 stabilizes AK1 to promote TGF-β-induced EMT in TNBC cells [168]. USP4 stabilizes TβRI by removing the ubiquitination, thereby accelerating TGF-β1-induced EMT and contributing to renal interstitial fibrosis and HCC [169, 170]. Moreover, USP10 and USP17 stabilized SMAD4 through their deubiquitinase activity, thereby enhancing TGF-β SMAD-dependent signaling and promoting EMT in OS and HCC [171, 172]. Specifically, USP19 exhibits isoform-dependent functions in regulating TGF-β signaling and EMT. In breast cancer (BC) models, the cytoplasmic isoform USP19 stabilizes both the TβRI and TβRII to enhance TGF-β-induced EMT and cell migration, whereas the endoplasmic reticulum-localized isoform USP19 inhibits EMT by reducing TβRI surface expression [173]. Other DUBs, such as USP2a [174], USP3 [175], USP8 [176], and USP11[177], drive TGF-β-induced EMT by stabilizing key TGF-β receptors. Conversely, USP26 inhibits this pathway by deubiquitinating and stabilizing SMAD7, preventing formation of the SMAD2/3-TβRI complex and suppressing TGF-β-induced migration and invasion in glioblastoma (GBM) [178]. Collectively, this intricate interplay of ubiquitination and deubiquitination mechanisms precisely regulates TGF-β-mediated EMT.
Wnt/β-catenin signaling regulation
The Wnt/β-catenin signaling pathway plays a pivotal role in regulating EMT, facilitating the shift from epithelial to mesenchymal characteristics during processes such as cancer metastasis [179, 180]. Ubiquitination modulates the Wnt/β-catenin signaling pathway, thereby regulating EMT (Table 1). Signal initiation occurs through the binding of Wnt ligands to Frizzled (FZD) receptors and LRP5/6 co-receptors at the cell surface, which stabilizes β-catenin by inhibiting its degradation complex and preventing ubiquitination-mediated proteasomal targeting [181]. Specific E3 ubiquitin ligases such as RNF43/ZNRF3 negatively regulate Wnt signaling by promoting FZD receptor ubiquitination and degradation, thereby suppressing EMT [182]. Furthermore, RNF43 inhibits Wnt/β-catenin signaling by downregulating β-catenin, thereby enhancing EMT in TNBC cells [183]. Other E3s like β-TrCP [184], FBXO11 [185], SIAH1-SIP-Skp1 complex [186], and NEDD4L [187] can also induce ubiquitination and degradation of β-catenin to inhibit Wnt/β-catenin signaling and negatively regulate EMT. Conversely, TNF receptor-associated factor 6 (TRAF6) activates β-catenin signaling and EMT by mediating ubiquitination and degradation of GSK3β [188]. However, another study showed that TRAF6 paradoxically inhibits the Wnt pathway by promoting the autophagic degradation of β-catenin, thereby suppressing EMT in CRC [189]. Other E3s like TRIM46 promote Wnt/β-catenin signaling by degrading Axin1 to promote hypoxia-induced EMT in HK2 cells [190]. Similarly, TRIM15 [191], TRIM28 [192], and RNF8 [193] can also activate Wnt/β-catenin signaling and EMT by stabilizing β-catenin.
Furthermore, DUBs regulate EMT by removing ubiquitin chains to stabilize key components of Wnt/β-catenin signaling (Table 2). For instance, USP34 stabilizes Axin1 through deubiquitination to maintain the integrity of the β-catenin destruction complex, thereby reducing β-catenin levels and inhibiting EMT [194]. USP22 deubiquitinates and stabilizes ADAM9 to inhibit Wnt/β-catenin signaling and EMT in trophoblast cells [195]. Moreover, USP42 suppresses EMT by stabilizing ZNRF3/RNF43, which promotes FZD receptor ubiquitination and degradation [182]. Conversely, USP5 stabilizes β-catenin by removing ubiquitin, driving Wnt/β-catenin signaling-induced EMT [196]. Additionally, other DUBs like USP7 [197-199], USP13 [200], USP15 [201], USP25 [202], USP36 [203], and OTUB2 [204] similarly promote the stabilization of β-catenin or other elements, enhancing Wnt/β-catenin signaling to facilitate EMT. Collectively, ubiquitination dynamics serve as a critical molecular switch governing the activity of the Wnt/β-catenin pathway and thereby modulating EMT.
Notch signaling regulation
The Notch signaling pathway, an evolutionarily conserved system of intercellular communication, governs diverse cellular processes including differentiation, proliferation, apoptosis, and stem cell self-renewal [205]. Substantial evidence demonstrates that Notch signaling modulates EMT through direct transcriptional regulation and intricate crosstalk with pathways such as TGF-β and Wnt/β-catenin [52]. E3 ligases critically regulate this process by substrate-specific ubiquitination of Notch signaling components, thereby determining their stability, subcellular localization, and signaling output (Table 1 and Table 2). For instance, Deltex E3 ubiquitin ligase 3 (DTX3) and WW domain containing E3 ubiquitin protein ligase 2 (WWP2) bind to Notch intracellular domain (NICD), promoting its ubiquitination and degradation to suppress Notch-induced EMT [206, 207]. Conversely, USP28 stabilizes NICD to activate Notch-induced EMT [208]. Furthermore, FBXW7 ubiquitinates and degrades Notch1 to suppress Notch signaling-induced EMT [209], while E3 ligase MIB1 and TRIM67 promote EMT and cell invasion in NSCLC by positively regulating the Notch signaling [210, 211]. Under Hypoxia, NICD overexpression causes degradation of ataxin-1 (ATXN1) by MDM2, thereby enhancing Snail expression to induce EMT in cervical cancer cells [212]. Other mechanisms involve E3 ligases such as TRIM59 that stabilize recombination signal binding protein for immunoglobulin kappa J region (RBPJ) to activate Notch signaling to induce EMT and metastasis [213].
Hypoxia-induced signaling regulation
Hypoxia, a hallmark of solid tumors, critically promotes EMT to enhance cancer cell migration and invasion [214, 215]. This induction is primarily mediated by hypoxia-inducible factors (HIFs), a member of key TFs activated under low oxygen conditions [215]. HIFs, particularly HIF-1α, directly regulate the expression of EMT-related genes, including Snail, Slug, ZEB1/2, and Twist1/2 [216, 217]. This regulation occurs through HIF-1α binding to hypoxia response elements in the promoter regions of these genes, driving their expression and promoting EMT. Furthermore, Multiple signaling pathways, including TGF-β, Wnt/β-catenin, PI3K/AKT/mTOR, and Notch, are modulated by hypoxia to further induce EMT [218-221]. The E3 ligase von Hippel-Lindau (VHL) is a crucial tumor inhibition factor, which plays a key role in Hypoxia-induced EMT by ubiquitinating and degrading HIFs [222, 223]. VHL knockdown in ccRCC leads to HIF-1α/2α accumulation, which upregulates N-cadherin and vimentin while suppressing E-cadherin, ultimately facilitating tumor invasion [224]. Moreover, deubiquitylase ovarian tumor domain-containing 6B (OTUD6B) stabilizes VHL to suppress HIF-1α/2α-mediated EMT and metastasis in HCC and ccRCC [225, 226]. Other E3 ligases like Smurf2 and NEDD4L also regulate HIF-1α stability to constrain EMT [187, 227]. In contrast, E3 ligase UBE3B stabilizes HIF-2α by K63-linked polyubiquitin chains to promote lung metastasis [228]. Acute hypoxia induces acetylation of STIP1 homology and U-box containing protein 1 (Stub1), which promotes deubiquitinate of HIF-2α and inhibits EMT-associated vascular remodeling [229]. Additional DUBs like USP7 [230] and USP9X [231] enhance HIF-1α or HIF-2α signaling by counteracting ubiquitination, thereby indirectly influencing the expression of EMT-TFs (Table 1 and Table 2).
Hh signaling regulation
Hh signaling pathway is an evolutionarily conserved mechanism involving key components such as Patched (Ptc), Smoothened (Smo), and Glioblastoma-associated oncogene homolog (Gli) [232]. It plays a significant role in EMT by upregulating EMT-TFs, which are critical for cancer metastasis and chemoresistance [233]. Gli1 acts as a primary effector with its aberrant activity linked to the Hh-dependent induction of key EMT regulators, including Snail, Slug, and Twist [234]. The stability of Gli, Ptc, and Smo is regulated by ubiquitination, which becomes a crucial regulatory mechanism in the Hh signaling pathway [235]. For instance, novel E3 ligases like Btbd9 and Kctd3 positively regulate Hh signaling [236], while Smurf1 and Smurf2 suppress the Hh/Gli signaling pathway by ubiquitinating and degrading Gli1 via K48-linked polyubiquitination [237]. Despite numerous studies showing that E3 ligases can regulate the Hh signaling by regulating Gli, E3 ligases in the Hh-mediated EMT process remain limited. DUBs like USP37 enhance Gli1 stability and activate Hh signaling by deubiquitinating, facilitating EMT-induced metastasis and chemoresistance in BC cells [238]. Similarly, USP5 and USP7 promote Hh signaling by stabilizing Gli1 through deubiquitination [239, 240], and UCHL5/UCH37 stabilizes Smo to enhance Hh signaling [241]. However, whether they are involved in regulating EMT in tumors requires further investigation. Consequently, the dysregulation of ubiquitination in the Hh pathway is closely associated with EMT and cancer progression. Targeting ubiquitination-related enzymes may offer new therapeutic strategies for Hh signal-driven EMT and metastasis.
Ubiquitination-regulated EMT in cancer metastasis
EMT functions as a key driver of tumor metastasis, a complex biological process finely controlled by ubiquitination and deubiquitination [50]. Dysregulation of EMT boosts cancer spread by enhancing cell migration, invasion, and resistance to apoptosis [242]. This section aims to explore the role of E3 ligases or DUBs-regulated EMT in tumor metastasis and analyze their potential promoting mechanisms (Table 3).
Stabilizing Snail/Slug and promoting metastasis
Snail and Slug critically regulate tumor metastasis by inducing EMT, a process governed by their modulation of genes involved in cell adhesion, polarity, and cytoskeletal dynamics [243]. Under normal conditions, the levels of Snail or Slug are maintained at low steady-state concentrations through proteasomal degradation, ensuring precise control of EMT progression [243]. However, dysregulation of ubiquitination often leads to the stabilization of the Snail/Slug proteins, thereby facilitating EMT and metastatic dissemination (Table 3) [244]. For instance, the E3 ligase Pellino-1 is abnormally highly expressed in LUAD and TNBC, which often leads to metastasis and a lower survival rate for patients. Mechanistically, the highly expressed Pellino-1 stabilizes Snail or Slug by K63 ubiquitination to promote EMT and metastasis in vitro and in vivo [93, 94]. Similarly, RNF8 also stabilizes Slug via K63 ubiquitination, thereby driving EMT and metastasis in lung cancer cells in vivo [95]. Furthermore, A20 promotes the monoubiquitination of Snail, thereby facilitating TGF-β1-induced EMT and metastasis in BC cells in vivo [96]. Since most E3 ligases degrade Snail or Slug through the K48-linked ubiquitin chain, they inhibit the EMT process and tumor metastasis. Therefore, multiple E3 ligases are expressed at low levels in tumors, which facilitates the EMT-induced metastasis. Conversely, DUBs play a crucial role in promoting EMT-induced cancer metastasis.
E3 and DUBs promote metastasis by regulating EMT
| Target | E3/DUBs | Regulatory mechanism | Ref. | |
|---|---|---|---|---|
| Snail/Slug | Pellino-1 | E3 | Pellino-1 stabilizes Snail and Slug by K63-linked ubiquitin chains, facilitating lung tumorigenesis and metastasis in vitro and in vivo. | [93, 94] |
| Slug | RNF8 | E3 | RNF8 stabilizes Slug by K63-linked ubiquitin chains, promoting EMT and migration in LC cells in vivo. | [95] |
| Snail | A20 | E3 | A20 stabilizes Snail by monoubiquitination, thereby facilitating TGF-β1-induced EMT and metastasis in BC cells in vivo. | [96] |
| Snail | USP1 | DUB | USP1 promotes the metastasis of OC cells by stabilizing Snail in vitro and in vivo. | [97] |
| Snail | USP2 | DUB | USP2 promotes the proliferation and metastasis of choroidal melanoma cells by stabilizing the Snail protein. | [99] |
| Snail | USP4 | DUB | USP4 stabilizes Snail via deubiquitination, driving EMT and metastasis in HCC in vitro and in vivo. | [101] |
| Snail/Slug | USP5 | DUB | USP5 stabilizes Slug, promoting EMT and metastasis in bladder cancer and HCC in vitro and in vivo. USP5 stabilizes Snail, promoting EMT and metastasis in CRC cells in vitro and in vivo. | [21, 122, 245] |
| Snail | USP9X | DUB | USP9X stabilizes Snail, promoting the migration, invasion, and metastasis of TNBC cells in vitro and in vivo. | [102] |
| Snail/Slug | USP10 | DUB | USP10 promotes EMT and the metastasis of BC cells by stabilizing Snail and Slug in vitro and in vivo. | [103, 123] |
| Snail | USP11 | DUB | USP11 is significantly upregulated in OC tissues and promotes invasion and metastasis by deubiquitinating Snail in vitro and in vivo. | [104] |
| Snail | USP13 | DUB | USP13 is highly expressed in GC and promotes EMT and metastasis by stabilizing Snail in vitro and in vivo. | [105] |
| Snail | USP17 | DUB | USP17 promotes the migration and invasion of OSCC cells by stabilizing Snail in vitro. | [106] |
| Snail | USP18 | DUB | USP18 is highly expressed in CRC tissues and promotes proliferation, migration and invasion by stabilizing Snail in vitro. | [107] |
| Slug | USP20 | DUB | USP20 positively regulates Slug to promote BC metastasis and invasion in vitro and in vivo. | [124] |
| Snail | USP26 | DUB | USP26 is highly expressed in ESCC and stabilizes Snail to promote the migration and invasion of ESCC cells in vitro. | [108] |
| Snail | USP27X | DUB | USP27X promotes the migration, invasion of BC cells by stabilizing Snail in vitro. | [109] |
| Snail | USP28 | DUB | USP28 promotes BC metastasis by stabilizing Snail through deubiquitination in vitro and in vivo. | [110] |
| Snail | USP29 | DUB | USP29 enhances the interaction between Snail and SCP1, stabilizing Snail and promoting the migration of GC cells in vitro and in vivo. | [111] |
| Snail | USP30 | DUB | USP30 stabilizes Snail to facilitate the EMT and metastasis of BC cells in vitro and in vivo. | [112] |
| Snail | USP35 | DUB | USP35 stabilizes Snail to facilitate the EMT and metastasis of GC tissues in vitro and in vivo. | [113] |
| Snail | USP37 | DUB | USP37 stabilizes Snail via deubiquitination, promoting GC and LUAD cells metastasis in vitro and in vivo. | [114, 249] |
| Snail | USP41 | DUB | USP41 increases the migration of breast cancer cells by stabilizing Snail, associated with poor prognosis in BC patients in vitro and in vivo. | [115] |
| Snail | USP47 | DUB | USP47 stabilizes Snail to facilitate EMT and in CRC and BC cells in vitro and in vivo. | [116, 246] |
| Snail | DUB3 | DUB | DUB3 stabilizes Snail to promote the EMT and metastasis of HCC and BC cells in vitro and in vivo. | [248] |
| Snail | OTUB1 | DUB | OTUB1 is highly expressed in ESCC, which stabilizes Snail to promote ESCC metastasis in vitro and in vivo. | [117] |
| Snail | OTUD4 | DUB | OTUD4 stabilizes Snail, which is identified as a novel therapeutic target for melanoma and BC metastasis in vitro and in vivo. | [118, 247] |
| Snail | JOSD1 | DUB | JOSD1 is significantly overexpressed in LUAD, stabilizing Snail to promote EMT and metastasis in vitro. | [120] |
| Snail | PSMD14 | DUB | PSMD14 stabilizes Snail to promote metastasis in ESCC in vitro and in vivo. | [121] |
| Snail | EIF3H | DUB | EIF3H interacts with and stabilizes Snail through deubiquitination, promoting EMT in ESCC in vitro and in vivo. | [119] |
| ZEB1 | USP18 | DUB | USP18 stabilizes ZEB1 through deubiquitination and enhances EMT and metastasis in ESCC in vitro and in vivo. | [136] |
| ZEB1 | USP21 | DUB | USP21 stabilizes ZEB1 to promote the EMT and metastasis of CRC in vitro, thereby contributing to poor prognosis. | [137] |
| ZEB1 | USP22 | DUB | USP22 stabilizes ZEB1 to enhance angiogenesis and EMT, thereby promoting the progression of OC in vitro. | [133] |
| ZEB1 | USP39 | DUB | USP39 stabilizes ZEB1 through deubiquitination, enhancing the proliferation and migration abilities of MM and HCC in vitro and in vivo. | [129, 251]. |
| ZEB1 | USP43 | DUB | USP43 is highly expressed in colorectal cancer tissues. It stabilizes ZEB1 to promote the migration and invasion of CRC in vitro. | [138] |
| ZEB1 | USP51 | DUB | USP51 plays an important role in the metastasis of GC, BC, and LUAD by stabilizing ZEB1 in vitro and in vivo. | [134, 252] |
| ZEB1 | BRCC3 | DUB | BRCC3 stabilizes ZEB1to promote EMT and metastasis in TNBC in vitro and in vivo. | [139] |
| Twist1 | USP4 | DUB | USP4 stabilizes Twist1 to drive EMT-associated stemness and malignancy in LC in vitro and TNBC invasion, migration, and tumor metastasis in vitro and in vivo. | [148, 254] |
| Twist1 | USP13 | DUB | USP13 stabilizes Twist1 to drive EMT, which promotes BC metastasis to the lung in vitro and in vivo. | [150] |
| Twist1 | USP18 | DUB | USP18 is highly expressed in GBM and promotes the migration and invasion of GBM cells by stabilizing Twist1 in vitro and in vivo. | [151] |
| Twist1 | USP29 | DUB | USP29 promotes the malignant phenotypes in TNBC cells by deubiquitinating Twist1 in vitro and in vivo. | [152] |
| Twist1 | USP51 | DUB | USP51 stabilizes Twist1 to drive EMT-associated stemness and malignancy in TNBC cells in vitro. | [255] |
| Twist1 | RNF8 | E3 | RNF8 stabilizes Twist by K63-linked ubiquitin chains, promoting EMT, migration, and invasion in BC cells in vitro and in vivo. | [146] |
| Twist1 | FBXO3 | E3 | FBXO3 stabilizes Twist1 by stabilizing USP4, enhancing BC cell migration and tumor metastasis in vitro and in vivo. | [148] |
| Twist1 | RBX1 | E3 | RBX1 degrades FBXO45 to stabilize Twist1 and promotes the migration and invasion of TNBC both in vitro and in vivo. | [147] |
| TGF-β signaling | ||||
| SNIP1 | RNF61 (MKRN1) | E3 | RNF61 promotes TGF-β signaling by degrading SNIP1, facilitating EMT and metastasis in CRC in vitro and in vivo. | [164] |
| SMAD3 | RNF111 | E3 | RNF111 activates TGF-β signaling by targeting SMAD3 to enhance Snail expression and promote NSCLC invasion and migration in vitro. | [166] |
| SMAD7 | HERC3 | E3 | HERC3 induces SMAD7 degradation in an autolysosome-dependent manner, activating the TGF-β signaling and GBM cell invasion and tumor metastasis in vitro and in vivo. | [161] |
| SMAD2/3 | MDM2 | E3 | MDM2 activates the Smad pathway to promote EMT during OC cell invasion and migration in vitro. | [163] |
| TAK1 | USP1 | DUB | USP1/WDR48 enhances TGF-β-mediated EMT and TNBC cell migration by stabilizing TAK1 in vitro. | [168] |
| TβRI | USP2a | DUB | USP2a activates TGF-β signaling by deubiquitinating TβRI, promoting metastasis in LC in vivo. | [174] |
| SUZ12 | USP3 | DUB | USP3 enhances TGF-β1-induced EMT and metastasis of GC cells by destabilizing SUZ12 in vitro. | [175] |
| TβRI | USP4 | DUB | USP4 interacts with and deubiquitinates βRI, promoting TGF-β signaling-induced EMT and metastasis in HCC in vitro and in vivo. | [170] |
| TβRII | USP8 | DUB | USP8 stabilizes TβRII to promote TGF-β/SMAD-induced EMT, invasion, and metastasis in BC cells in vitro and in vivo. | [176] |
| SMAD4 | USP10 | DUB | USP10 interacts with and stabilizes SMAD4 by removing Lys-48-linked ubiquitin chains, thereby promoting HCC metastasis in vitro and in vivo. | [172] |
| TβRII | USP11 | DUB | USP11 stabilizes TβRII to activate the TGF-β signaling, promoting EMT and metastasis in BC in vitro and in vivo. | [177] |
| SMAD4 | USP17 | DUB | USP17 stabilizes SMAD4 to activate the TGF-β signaling, thereby promoting EMT and OS cell migration and invasion in vitro. | [171] |
| TβRI | USP19 | DUB | USP19-CY (cytoplasmic isoform) stabilizes TβRI and TβRII to enhance TGF-β-induced EMT and migration in BC in vitro. | [173] |
| Wnt/β-catenin signaling | ||||
| GSK3β/ β-catenin | RNF8 | E3 | RNF8 inhibits GSK-3β and subsequently activates β-catenin signaling, promoting EMT and BC cell migration and tumor metastasis in vitro and in vivo | [193] |
| β-catenin | TRIM15 | E3 | TRIM15 activates the Wnt/β-catenin signaling to promote EMT, driving ESCC cell migration, invasion, and tumor metastasis in vitro and in vivo. | [191] |
| β-catenin | TRIM28 | E3 | TRIM28 activates the Wnt/β-catenin signaling to promote EMT, migration, and invasion of OC cells in vitro. | [192] |
| β-catenin | USP5 | DUB | USP5 stabilizes β-catenin to activate the Wnt/β-catenin signaling, enhancing EMT and migration in NSCLC cells in vitro. | [196] |
| DDX3X, β-catenin | USP7 | DUB | USP7 stabilizes β-catenin and DDX3X to activate Wnt/β-catenin signaling, promoting the metastasis of CRC and OS cells in vitro. | [197, 198] |
| WISP1 | USP13 | DUB | USP13 stabilizes WISP1 to activate the Wnt/β-catenin signaling, promoting ESCC cell migration and tumor metastasis in vitro and in vivo. | [200] |
| β-catenin | USP15 | DUB | USP15 promotes the nuclear translocation of β-catenin and activates the Wnt/β-catenin signaling, enhancing the EMT and invasion in GC cells in vitro. | [201] |
| β-catenin | USP25 | DUB | USP25 stabilizes β-catenin through interaction with TRIM21, activating β-catenin signaling-induced EMT and driving cell migration, invasion, and tumor metastasis in vitro and in vivo. | [202] |
| β-catenin | OTUB2 | DUB | OTUB2 stabilizes β-catenin by suppressing TRAF6-mediated autophagy-dependent degradation, promoting Wnt/β-catenin signaling and driving EMT cell invasion and tumor metastasis in vitro and in vivo. | [204] |
DUBs inhibit the degradation of the Sanil protein mediated by the K48 ubiquitin chain, thereby promoting EMT and driving tumor metastasis (Table 3). For instance, USP1 [97] and USP11 [104] stabilize Snail to promote the invasion and metastasis of OC cells in vitro and in vivo. In choroidal melanoma, USP2 overexpression facilitates EMT migration and invasion by stabilizing Snail in vitro [99]. Furthermore, USP4 [101] and DUB3 [98] stabilize Snail and induce EMT, thereby promoting HCC cell migration, invasion, and tumor metastasis in vitro and in vivo. In bladder cancer and HCC, USP5 deubiquitinates and stabilizes Slug to induce EMT, facilitating cancer cell migration, invasion, and tumor metastasis in vitro and in vivo [122, 245]. Moreover, USP5 [21], USP18[107], and USP47 [116] suppress Snail degradation to induce EMT, thereby promoting CRC cell migration and tumor metastasis in vitro and in vivo. In BC, USP9X [102], USP10 [103], USP20 [124], USP27X [109], USP28 [110], USP30 [112], USP41[115], USP47 [246], OTUD4 [247], and DUB3 [248] are often abnormally highly expressed. Those DUBs stabilize Snail or Slug by removing ubiquitination, thereby increasing EMT and BC cell migration and tumor metastasis in vitro and in vivo. Moreover, OTUD4 also directly deubiquitinates and stabilizes Snail to promote melanoma cell migration and tumor metastasis in vitro and in vivo [118]. Similarly, USP13 [105], USP29 [111], USP35 [113], and USP37 [249] stabilize Snail to repress E-cadherin, enhancing epithelial-mesenchymal plasticity and driving GC cell migration and tumor metastasis in vitro and in vivo. In oral squamous cell carcinoma cells (OSCC), USP17 promotes the stability of Snail, leading to migration and invasion involving EMT in vitro [106]. Furthermore, USP26 [108], PSMD14 [121], OTUB1 [117], and EIF3H [119] deubiquitinate and stabilize Snail, enhancing esophageal squamous cell carcinoma (ESCC) cell migration and tumor metastasis in vivo and in vitro. In LUAD, USP37 [114] and JOSD1 [120] deubiquitinate Snai1 to activate EMT, thereby promoting cell invasion and migration in vitro. Therefore, the abnormal expression of these DUBs in tumors is a key factor that promotes the stability of Snail and tumor metastasis.
Stabilizing ZEB1 and promoting metastasis
ZEB1 promotes cell migration and invasion by regulating cytoskeletal remodeling, cell-cell adhesion, and increasing the expression of vimentin [250]. Several DUBs stabilize ZEB proteins through deubiquitination, thereby enhancing EMT and tumor metastasis (Table 3). For instance, USP21 and USP43 deubiquitinate and stabilize ZEB1 to induce EMT, enhancing cell migration and stemness in CRC in vitro [137, 138]. Similarly, USP18 stabilizes ZEB1 by deubiquitinating to facilitate ESCC cell migration and tumor metastasis in vitro and in vivo [136]. USP22 stabilizes ZEB1 to induce angiogenesis and EMT, thereby promoting the invasion and migration of OC in vitro [133]. In HCC and multiple myeloma (MM) cells, USP39 stabilizes ZEB1 by deubiquitination to induce EMT, thereby promoting cell migration and tumor metastasis in vitro and in vivo zebrafish experiments [129, 251]. Similarly, USP51 stabilizes ZEB1 through deubiquitination, promoting metastatic dissemination in GC in vitro and in vivo [135]. Further research showed that CDK4/6 phosphorylation of USP51 is required for ZEB1-mediated metastasis in LUAD and BC in vitro and in vivo [134, 252]. Concurrently, BRCC3 stabilizes ZEB1 by deubiquitination to induce EMT, thereby promoting TNBC cell migration, invasion, and tumor metastasis in vitro and in vivo [139]. However, USP10 degrades ZEB1 by removing K27-linked ubiquitin chains, thereby suppressing CRC cell migration mediated by ZEB1 in vitro [140]. Conversely, ERK-mediated phosphorylation of USP10 at Ser236 impairs its interaction with ZEB1, thereby stabilizing ZEB1 and promoting CRC metastasis in vivo [140]. Together, these mechanisms illustrate how the interplay between E3 ligases and DUBs regulates ZEB stability to drive cancer progression.
Stabilizing Twist1 and promoting metastasis
Twist1 represses E-cadherin transcription, promoting tumor cell migration, invasion, and metastasis [243]. Clinically, elevated Twist1 correlates with increased lymph node metastasis, distant metastasis, and advanced tumor stage across multiple cancers [253]. The protein stability of Twist1 is dynamically regulated by E3s and DUBs, which promote EMT and tumor metastasis by stabilizing Twist1(Table 3). For instance, USP13 directly interacts with Twist1 and cleaves FBXL14-induced K48-linked polyubiquitin chains, increasing Twist1 protein levels and facilitating BC cell migration and tumor metastasis in vitro and in vivo [150]. In GBM, USP18 deubiquitinates and stabilizes Twist1, thereby inducing EMT and promoting cell migration and tumor metastasis in vitro and in vivo [151]. Similarly, USP29 stabilizes Twist1 through deubiquitination, thereby driving malignant phenotypes in TNBC in vitro and in vivo [152]. Furthermore, specific E3 ligases can also stabilize Twist1 to facilitate metastasis. For instance, RNF8 can stabilize Twist1 to induce EMT through K63-linked ubiquitin chains, thereby enhancing BC cell migration, invasion, and tumor metastasis in vitro and in vivo [146]. FBXO3 disrupts the DNPEP-mediated degradation of USP4, stabilizing Twist1 and promoting BC cell migration and tumor metastasis in vitro and in vivo [148]. RBX1 ubiquitinates and degrades FBXO45, consequently stabilizing Twist1to drive EMT and facilitating TNBC cell invasion, migration, and tumor metastasis in vitro and in vivo [147]. Additionally, USP4 [254] and USP51 [255] stabilize Twist1 polyubiquitination, enhancing EMT-associated stemness and malignancy in lung cancer in vitro. This regulatory network highlights Twist1 as a critical convergence point for post-translational modifications that orchestrate EMT and metastatic progression.
Activating TGF-β signaling to drive EMT-induced metastasis
TGF-β-induced EMT is a pivotal factor for tumor metastasis, involving multiple EMT-TFs and EMT signaling pathways [256]. E3 ligases and DUBs act as key regulators by modulating the stability and activity of TGF-β pathway components, thereby influencing EMT and tumor metastasis (Table 3). For instance, E3 ligase RNF61 degrades Smad nuclear-interacting protein 1 (SNIP1) to activate TGF-β-mediated EMT, promoting CRC cell invasion and tumor metastasis in vitro and in vivo [164]. Furthermore, HERC3 promotes the autophagic degradation of Smad7 through ubiquitination, thereby activating the TGF-β signaling and driving EMT and GBM cell invasion and tumor metastasis in vitro and in vivo [161]. In OC, MDM2 promotes EMT by activating the TGF-β-Smads-Snail/Slug pathway, enhancing OC cell invasion and migration in vitro [163]. Similarly, RNF111 is highly expressed in the high-metastatic NSCLC cell line 95D, which activates TGF-β signaling-induced EMT to enhance NSCLC cell invasion and migration in vitro [166]. Furthermore, DUBs stabilize core TGF-β signaling proteins by removing ubiquitin chains, thereby promoting EMT and tumor metastasis. For instance, the USP1/WDR48 complex stabilizes TAK1 through deubiquitination to enhance EMT and cell migration in TNBC in vitro [168]. In lung cancer, USP2a stabilizes TβRI by removing K33-linked polyubiquitin chains to promote nuclear translocation of SMAD2/3, activating TGF-β-induced EMT and metastasis in vivo [174]. Similarly, USP4 and USP10 activate the TGF-β signaling through deubiquitinating TβRI and Smad4, thereby promoting EMT-induced cell migration, invasion, and metastasis of HCC in vitro and in vivo [170, 172]. Other mechanisms involve USP3 interacts with and stabilizes SUZ12 by deubiquitination, promoting TGF-β1-induced EMT and cell migration and invasion in GC in vitro [175]. Furthermore, USP8 [176] and USP11 [177] enhance TGF-β/SMAD signaling by deubiquitinating and stabilizing TβRII, increasing plasma membrane expression and promoting EMT, invasion, and metastasis in BC cells in vitro and in vivo. USP17 promotes TGF-β-induced EMT by stabilizing SMAD4, thereby promoting OS cell migration and invasion in vitro [171]. Specifically, the cytoplasmic isoform USP19 expression is higher in BC tissues and is correlated with poor prognosis. Mechanistically, cytoplasmic isoform USP19 stabilizes TβRI and TβRII to enhance TGF-β-induced EMT and BC cell migration and extravasation in vitro [173]. Conversely, endoplasmic reticulum-localized isoform USP19 inhibits BC cell migration [173]. Therefore, the ubiquitination-related factors play a significant role in promoting tumor metastasis through TGF-β-mediated EMT.
Activating Wnt/β-catenin signaling to drive EMT-induced metastasis
The dysregulation of Wnt/β-catenin signaling promotes EMT and tumor metastasis across various malignancies through ubiquitination and deubiquitination events that regulate β-catenin stability (Table 3) [257, 258]. Emerging evidence showed that RNF8 is overexpressed in highly metastatic BC cell lines. It activates β-catenin-induced EMT by inactivating GSK-3β, thereby promoting BC cell migration and tumor metastasis in vitro and in vivo [193]. Similarly, the non-SMC concentrate I complex subunit (NCAPG) stabilizes β-catenin through competitive binding with SIP to inhibit SIAH1 activity, promoting β-catenin-induced EMT and HCC cell migration and tumor metastasis in vitro and in vivo [186]. Furthermore, elevated expression of TRIM15 in ESCC tissues and cell lines activates Wnt/β-catenin-induced EMT, leading to cell migration, invasion, and tumor metastasis in vitro and in vivo [191]. Additionally, TRIM28 has been implicated in OC cell metastasis in vitro, as its knockdown significantly attenuates Wnt/β-catenin signaling and suppresses EMT processes [192].
DUBs can stabilize these key components through deubiquitination, thereby enhancing Wnt/β-catenin signaling and tumor metastasis. For instance, USP5 stabilizes β-catenin to activate the Wnt/β‑catenin and EMT, thereby promoting NSCLC cell migration and invasion in vitro [196]. Similarly, USP7 activates Wnt/β‑catenin-induced EMT by stabilizing β-catenin, thereby enhancing OS cell migration and invasion in vitro [198]. Furthermore, in CRC, USP7 augments Wnt/β-catenin signaling by stabilizing DDX3X, promoting EMT and cell migration in vitro [197]. Similarly, USP13 stabilizes WISP1 to promote the Wnt/β-catenin-induced EMT, driving ESCC cell migration and tumor metastasis in vitro and in vivo [200]. USP15-mediated β-catenin stabilization facilitates EMT and GC cell invasion in vitro [201]. In HCC, the USP25-TRIM21 axis activates β-catenin signaling-induced EMT and drives cell migration, invasion, and tumor metastasis in vitro and in vivo [202]. Furthermore, OTUB2 stabilizes β-catenin by suppressing TRAF6-mediated autophagy-dependent degradation and activating Wnt/β-catenin-induced EMT, thereby driving intrahepatic cholangiocarcinoma (iCCA) cell invasion and tumor metastasis in vitro and in vivo [204]. These findings demonstrate that ubiquitination-mediated regulation of Wnt/β-catenin signaling constitutes a critical mechanistic node controlling EMT and metastasis across diverse malignancies.
Ubiquitination-regulated EMT in chemoresistance and strategies
Ubiquitination-regulated EMT in chemoresistance
Chemotherapy resistance is a major challenge in cancer treatment [259]. The resistance mechanism often involves EMT activation, which causes cancer cells to gain stem cell-like qualities, increased migration ability, and decreased sensitivity to chemotherapy [260]. Emerging evidence shows that the development of chemoresistance in cancer therapy is frequently connected to ubiquitination-regulated EMT [50]. In this section, we aim to explore the role of E3 ligases or DUBs-regulated EMT in tumor chemoresistance and highlight that E3 ligases or DUBs inhibitors are crucial for overcoming tumor metastasis and chemoresistance (Table 4) [232-234].
Cisplatin
Platinum-based chemotherapy is a primary treatment for solid tumors, but its efficacy is frequently undermined by the development of drug resistance [261]. Cisplatin, a DNA-damaging agent and widely utilized in NSCLC, often encounters resistance mediated by ubiquitin-regulated EMT (Table 4) [262]. For instance, the lipid metabolism enzyme carnitine palmitoyltransferase 1C (CPT1C), highly expressed in NSCLC cells, contributes to cisplatin resistance by inducing EMT in vitro [263]. Mechanistically, cisplatin treatment induces the degradation of the E3 ligase NEDD4L, leading to enhanced CPT1C stability and subsequent EMT-driven cisplatin resistance [263]. Intriguingly, NEDD4 exhibits context-dependent roles. In cisplatin-resistant nasopharyngeal carcinoma (NPC) cells, NEDD4 expression contributes to EMT, and its downregulation reverses resistance in vitro [264]. Similarly, the E3 ligase FBXW7 suppresses EMT and chemoresistance in NSCLC by degrading Snail, whereas reduced FBXW7 expression in patient tissues correlates with poorer treatment response in vitro [265]. Conversely, the downregulation of cyclin D3 in cisplatin-resistant LUAD cells impairs PARK2-mediated vimentin degradation, stabilizing vimentin and promoting EMT and chemoresistance in vitro and in vivo [266]. Other E3 ligases contribute to EMT-induced resistance: TRAF6 mediates the ubiquitination and degradation of GSK3β, activating β-catenin signaling to promote EMT and cisplatin resistance in vitro and in vivo [188]. Furthermore, Hakai stabilizes phosphorylated AKT to enhance EMT, leading to cisplatin resistance in NSCLC in vitro [267].
DUBs significantly facilitate EMT to promote cisplatin resistance in various cancers. For instance, USP1 stabilizes Snail through deubiquitination after platinum-based treatments, thereby inducing EMT and conferring resistance in OC cells in vitro [97]. Similarly, USP9X enhances Snail stability by removing K48-linked ubiquitin chains, contributing to cisplatin and doxorubicin (Dox) resistance in TNBC in vitro and in vivo [102], while TGF-β-induced USP27X stabilizes Snail and activates cancer-associated fibroblasts, reducing cisplatin sensitivity in TNBC in vitro and in vivo [109]. Additionally, PSMD14 stabilizes Snail by deubiquitination to drive EMT and diminish cisplatin efficacy in ESCC cells in vitro and in vivo [121, 268]. Notably, key EMT-TFs like ZEB1 and Twist directly promote cisplatin resistance by enabling EMT-linked chemoresistance. For instance, USP51 stabilizes ZEB1 to promote A549 cells' cisplatin resistance in vitro [269], and USP29 stabilizes Twist1 to enhance EMT, metastasis, and cisplatin resistance in TNBC in vitro and in vivo [152]. DUBs also regulate TGF-β signaling components, such as USP32 stabilizes SMAD2 to activate TGF-β-mediated proliferation and migration, augmenting cisplatin resistance in GC in vitro and in vivo [270]. In LUAD, USP7 suppresses c-Myc degradation to promote EMT and cisplatin resistance in vitro [271], and USP22 stabilizes c-Myc and ALDH1A3 to facilitate EMT and resistance in TNBC and lung cancer in vitro and in vivo [272, 273]. Furthermore, USP37 stabilizes Gli-1 to activate Hh signaling, driving EMT and cisplatin resistance in BC cells in vitro and in vivo [238]. Collectively, DUBs and E3 ligases converge on EMT regulation, establishing them as key mediators of cisplatin resistance and promising therapeutic targets.
E3 and DUBs promote resistance by regulating EMT
| Drugs | Targeting | Drug resistance regulatory mechanism | Cancers | Ref |
|---|---|---|---|---|
| Cisplatin | NEDD4L | Cisplatin induces a decrease in NEDD4, stabilizing CPT1CAK to drive EMT and cisplatin resistance in vitro. | NSCLC | [263] |
| FBXW7 | Lower expression of FBXW7 stabilizes Snail, driving EMT and cisplatin resistance in vitro. | NSCLC | [265] | |
| PARK2 | Cisplatin inhibits PARK2-mediated vimentin, promoting EMT and cisplatin resistance in vitro and in vivo. | LUAD | [266] | |
| TRAF6 | TRAF6 degrades GSK3β, thereby activating β-catenin signaling and promoting EMT and cisplatin resistance in vitro and in vivo. | NPC | [188] | |
| Hakai | Stabilizes p-AKT to enhance EMT and cisplatin resistance in vitro. | NSCLC | [267] | |
| USP1 | USP1 stabilizes Snail to enhance EMT and cisplatin resistance in vitro. | OC | [97] | |
| USP7 | USP7 stabilizes c-Myc to enhance EMT and cisplatin resistance in vitro. | LUAD | [271] | |
| USP9X | USP9X stabilizes Snail to enhance EMT and cisplatin resistance in vitro and in vivo. | TNBC | [102] | |
| USP22 | USP22 stabilizes c-Myc and ALDH1A3 to enhance EMT and cisplatin resistance in vitro and in vivo. | TNBC, LUAD | [272, 273] | |
| USP27X | USP27X stabilizes Snail to enhance EMT and cisplatin resistance in vitro and in vivo. | TNBC | [109] | |
| USP29 | USP29 stabilizes Twist1 to enhance EMT and cisplatin resistance in vitro and in vivo. | TNBC | [152] | |
| USP32 | USP32 stabilizes SMAD2 to enhance EMT and cisplatin resistance in vitro and in vivo. | GC | [270] | |
| USP37 | USP37 stabilizes Gli-1 to enhance EMT and cisplatin resistance in vitro and in vivo. | BC | [238] | |
| USP51 | USP51 stabilizes ZEB1 to enhance EMT and cisplatin resistance in vitro. | LC | [269] | |
| PSMD14 | PSMD14 stabilizes Snail to enhance EMT and cisplatin resistance in vitro and in vivo. | ESCC | [121, 268] | |
| Oxaliplatin | OTUB2 | OTUB2 stabilizes SP1 and GINS1 to enhance EMT and Oxaliplatin resistance in vitro and in vivo. | CRC | [275] |
| FBXW7 | Lower expression of FBXW7 stabilizes ZEB2, driving EMT and Oxaliplatin resistance in vitro and in vivo. | CRC | [132] | |
| Doxorubicin | RNF8 | RNF8 stabilizes Twist via K63-linked polyubiquitin chains, driving EMT and Dox resistance in vivo. | TNBC | [146 |
| SIAH1 | Dox induces SIAH1 decrease, stabilizing ZEB1 to drive EMT and Dox resistance in vitro. | OS, HCC | [278, 279] | |
| USP9X | USP9X stabilizes Snail to enhance EMT and Dox resistance. | TNBC | [109] | |
| USP14 | USP14 modulates Wnt signaling to enhance Dox resistance in vitro. | MM | [282] | |
| USP29 | USP29 stabilizes Snail to enhance EMT and Dox resistance in vitro and in vivo. | NSCLC | [283] | |
| USP45 | USP29 stabilizes MYC to enhance EMT and Dox resistance in vivo. | CC | [284] | |
| Gemcitabine | UBR5 | UBR5 degrades O-GlcNAcase (OGA), inducing EMT and GEM resistance in vitro and in vivo. | PC | [286] |
| TRIM59 | TRIM59 stabilizes RBPJ, activating Notch signaling to induce EMT and drive GEM in vitro and in vivo. | PC | [213] | |
| RNF126 | RNF126 ubiquitinates and degrades PTEN, activating the AKT/GSK-3β/β-catenin pathway to induce EMT and drive GEM resistance in vitro and in vivo. | PC | [287] | |
| FBW7 | MiR-223 downregulates FBW7 to activate Notch-1-induced EMT and GEM resistance in vitro. | PC | [209] | |
| Smurf2 | miR-15b downregulates Smurf2 to stabilize Smad2/3, activating the TGF-β-induced EMT and driving resistance in vitro. | PC | [289] | |
| Paclitaxel | USP29 | USP29 stabilizes Snail to enhance EMT and paclitaxel resistance in vitro | NSCLC | [283] |
| USP30 | USP30 stabilizes Snail to enhance EMT and paclitaxel resistance in vitro and in vivo. | BC | [112] | |
| 5-Fu | FBXW7 | Lower expression of FBXW7 stabilizes ZEB2, driving EMT and Oxaliplatin resistance in vitro and in vivo. | CRC | [132] |
| Sunitinib | TRIM21 | TRIM21 stabilizes AXL to enhance sunitinib resistance in vitro and in vivo. | KIRC | [295] |
| Sorafenib | RNF8 | RNF8 upregulates N-cadherin and Snail to enhance sorafenib resistance in vitro. | HCC | [294] |
| Lenvatinib | RNF8 | RNF8 upregulates N-cadherin and Snail to enhance lenvatinib resistance in vitro. | HCC | [294] |
| Temozolomide | HERC3 | HERC3 degrades SMAD7 to activate TGF-β signaling, inducing EMT and autophagy, and TMZ resistance in vitro and in vivo | GBM | [161] |
| Vemurafenib | OTUD4 | OTUD4 stabilizes Snail to enhance EMT and Vemurafenib resistance in vitro. | MEL | [118] |
| PLX-4720 | OTUD4 | OTUD4 stabilizes Snail to enhance EMT and PLX-4720 resistance in vitro. | MEL | [118] |
Oxaliplatin
Oxaliplatin is a standard chemotherapeutic agent primarily employed in the treatment of CRC [274]. The resistance of Oxaliplatin is largely through dysregulation of ubiquitin-mediated pathways involving EMT, which play a critical role in tumor metastasis and chemoresistance (Table 4). For instance, OTUB2 stabilizes transcription factor SP1 by removing K48-linked ubiquitin ligases, enhancing EMT and oxaliplatin resistance in CRC in vitro and in vivo [275]. Furthermore, FBXW7 suppresses EMT and chemoresistance in NSCLC by degrading ZEB2, while FBXW7 deletion promotes EMT by enhancing the stabilization of ZEB2, thereby promoting oxaliplatin and 5-fluorouracil (5-FU) resistance in CRC in vitro, ex vivo, and in vivo [132]. Therefore, the dysregulation of ubiquitination leads to EMT, which significantly promotes the occurrence of Oxaliplatin resistance.
Doxorubicin
Dox, an anthracycline antibiotic, is widely used in cancer chemotherapy for its broad-spectrum activity against various malignancies [276]. However, acquired resistance often develops, with ubiquitination pathways playing a crucial role by regulating EMT to confer resistance in different types of cancers (Table 4) [277]. In Dox-resistant HCC (HCC/Dox) cells, elevated ZEB1 expression drives EMT-mediated resistance [278]. Mechanistically, SIAH1 downregulation inhibits ubiquitination-mediated degradation of ZEB1, thereby enhancing ZEB1 stability to promote chemoresistance in HCC in vitro [278]. Similarly, the Dox resistance of OS cells is attributed to SIAH1 downregulation, which reduces ZEB1 ubiquitination and stabilizes ZEB1, thereby facilitating EMT and Dox resistance in vitro [279]. Furthermore, RNF8 stabilizes Twist via K63-linked polyubiquitin chains, promoting nuclear translocation and Dox resistance in TNBC in vitro and in vivo [146]. Conversely, FBXW7 suppresses EMT in HCC cells to enhance Dox sensitivity in vitro [280]. Further study showed that MiR-223 targets FBXW7 to enhance Dox resistance in CRC in vivo [281]. Additionally, DUBs augment Dox resistance in various tumors. For instance, USP14 modulates Wnt signaling to enhance resistance in MM in vitro [282], and USP29 stabilizes Snail through deubiquitination to boost resistance to Dox and paclitaxel in NSCLC cells in vitro and in vivo [283]. Moreover, USP45 stabilizes MYC in cervical cancer, upregulating vimentin, N-cadherin, and cancer stem cell protein expression to confer Dox resistance in vivo [284]. These findings collectively showed that ubiquitin-mediated regulation of EMT regulators constitutes a fundamental molecular mechanism for acquiring Dox resistance.
Gemcitabine
Gemcitabine (GEM) is the first-line chemotherapy for pancreatic cancer (PC), but its efficacy is frequently compromised by acquired resistance [285]. Ubiquitin-mediated EMT is emerging as a critical mechanism underpinning this phenomenon [286]. Key ubiquitin-related regulators contribute to this resistance (Table 4). The ubiquitin-protein ligase E3 module N-recognition 5 (UBR5) is markedly upregulated in GEM-resistant PC cells and clinical samples, where it promotes resistance through EMT activation [286]. Mechanistically, UBR5 degrades O-GlcNAcase (OGA) by K48-linked ubiquitin chains, thereby inducing EMT and GEM resistance in vitro and in vivo [286]. Concurrently, TRIM59 stabilizes RBPJ by K63-linked ubiquitin chains, activating Notch signaling to induce EMT and drive GEM resistance in PC in vitro and in vivo [213]. RNF126 ubiquitinates and degrades PTEN, thereby activating the AKT/GSK-3β/β-catenin pathway to induce EMT and drive GEM resistance in PC in vitro and in vivo [287]. Moreover, the ubiquitin-like protein FAT10 stabilizes forkhead box protein M1 (FOXM1) by inhibiting ubiquitin-mediated degradation, thereby facilitating EMT and GEM resistance in PC in vitro [288]. Additionally, microRNA participates in this resistance network, such as miR-15b downregulates Smurf2 to inhibit Smad2/3 ubiquitination and degradation, thereby activating the TGF-β-induced EMT and driving resistance in PC in vitro [289], while MiR-223 downregulates FBW7 to exacerbate EMT and resistance in GEM-resistant PC in vitro [209]. Collectively, these mechanisms highlight ubiquitin-regulated EMT as a pivotal driver of GEM resistance in PC.
Others
Beyond mentioned above, multiple therapeutic agents exhibit resistance mechanisms linked to ubiquitin-mediated EMT processes, including temozolomide (TMZ), sorafenib, and Lenvatinib (Table 4) [290, 291]. TMZ, a chemotherapeutic alkylating agent commonly used for GBM treatment, is limited in efficacy by resistance mechanisms and the blood-brain barrier despite its first-line status [292, 293]. Dysregulation of ubiquitination enhances TGF-β signaling, which contributes to tumor progression and TMZ resistance. For instance, the E3 ligase HERC3 degrades SMAD7 by K48-linked ubiquitin ligase, thereby activating TGF-β signaling, inducing EMT and autophagy, and conferring TMZ resistance in GBM in vitro and in vivo [161]. In HCC, RNF8 promotes EMT and increases sorafenib and Lenvatinib resistance; its silencing suppresses EMT and enhances drug sensitivity by downregulating N-cadherin and Snail in vitro [294]. Similarly, STAM Binding Protein Like 1 (STAMBPL1) mitigates TRIM21-mediated lysosomal degradation of AXL, reinforcing the mesenchymal phenotype and sunitinib resistance in Kidney Renal Clear cell carcinoma (KIRC) in vitro and in vivo [295]. In BC, USP30 stabilizes Snail through removing K48-linked polyubiquitin chains, accelerating EMT and promoting paclitaxel chemosensitivity in vitro and in vivo [112]. Similarly, OTUD4 stabilizes Snail via ubiquitination to promote malignant phenotypes, driving resistance to BRAF inhibitors like vemurafenib and PLX4720 in melanoma in vitro [118]. Collectively, ubiquitination and deubiquitination are critical regulators of EMT-associated therapeutic resistance in human malignancies.
Targeting ubiquitination reverses EMT-induced metastasis and chemoresistance
Targeting ubiquitination-related factors is a prominent research direction in cancer therapy, particularly for countering EMT-mediated metastasis and drug resistance [296]. Multiple inhibitors specific to E3 ligases or DUBs effectively reverse these processes [297]. For instance, PSMD14 inhibitor Thiolutin reduces Snail stability to inhibit EMT, suppressing motility and stemness while enhancing cisplatin sensitivity in ESCC in vitro and in vivo [268]. Similarly, the Hakai inhibitor Hakin-1 impedes E-cadherin ubiquitination to hinder EMT and tumor progression in CRC in vitro and in vivo [298]. The MDM2 antagonist Nutlin-3 suppresses TGF-β-Smads-mediated EMT and metastasis in OC in vitro [163]. DUB inhibitors like WP1130 promote Snail degradation by targeting Dub3 or USP9X, inhibiting EMT and metastasis in BC while sensitizing TNBC to cisplatin and paclitaxel in vitro and in vivo [102, 248]. Other examples include Nucleoredoxin interacts with DUB3 to promote Snail degradation via the ubiquitin-proteasome system and suppress HCC progression in vitro and in vivo [98]. Natural compounds such as Erianin from Dendrobium chrysotoxum induce Snail degradation via OTUB1 targeting to suppress metastasis in ESCC models in vitro [299]. Biochanin A facilitates ZEB1 ubiquitination and degradation, reversing EMT-associated cisplatin resistance in LUAD in vitro and in vivo [300]. Cinobufotalin inhibits USP7-mediated MYC deubiquitination to suppress EMT and increase cisplatin sensitivity in LUAD [301], while P5091 reduces USP47-induced EMT in breast epithelial cells [246]. Other small molecules like compound 3d disrupt PELI1-Snail/Slug interactions to inhibit EMT and metastasis in TNBC in vitro and in vivo [302]. Similarly, the USP2 inhibitor ML364 induces Snail degradation to inhibit proliferation and metastasis in choroidal melanoma in vitro and in vivo [99]. The USP4 inhibitor U4-I05 degrades β-catenin and Twist1, inhibiting metastasis and enhancing sensitivity to oxaliplatin and 5-FU in CRC in vitro and in vivo [303]. Additionally, the USP28 inhibitor compound 19, a [1,2,3] triazolo [4,5-d] pyrimidine derivative, blocks proliferation and EMT in GC [304]. Collectively, these research results indicate that targeting ubiquitination-related factors possesses great potential in addressing EMT-driven tumor metastasis and chemoresistance.
Drug development targeting E3 ligases and DUBs
Drug development targeting E3 ligases
E3 ligases act as key regulators by mediating the ubiquitination of specific substrates, ultimately degrading the substrates or enabling signal transmission [305]. The dysregulation of E3 ligases is associated with several human diseases, particularly cancers, making them attractive targets for the development of new drugs [305]. In this section, we systematically summarize the current situation of drug development targeting E3 ligases and DUBs and their potential mechanisms (Table 5).
Summary of pharmacological strategies directly targeting the E3 ligase or DUBs for cancer therapy in clinical trials
| Target | Drug | Mechanism | Cancer types | Phase | Identifier |
|---|---|---|---|---|---|
| Drug targeting E3 ligases | |||||
| CRBN | ARV-110 | An inhibitor of PROTACs with E3 CRBN, targeting the androgen receptor. | Prostate cancer | Phase I Phase II | NCT05177042, NCT03888612 |
| ARV-471 | An inhibitor of PROTACs with E3 CRBN, targeting the estrogen receptor. | Breast cancer | Phase I Phase II Phase III | NCT04072952, NCT05548127, NCT05732428, NCT05573555, NCT05654623, NCT05501769, NCT06125522, NCT05463952 | |
| CC92480 (Mezigdomide) | A novel E3 ligase CRBN modulator, targeting IKZF1 and ZFP91. | Multiple myeloma | Phase I Phase II | NCT05707390, NCT06645678, NCT07032714, NCT05552976, NCT06163898, NCT06988488, NCT05519085, NCT03989414, NCT03374085 | |
| VHL | HP518 | Recruits VHL to ubiquitinate and degrade the AR protein. | Prostate cancer | Phase I Phase II | NCT05252364, NCT06155084 |
| CRL4-CRBN | CC-90009 | Recruits the CRL4-CRBN E3 complex to ubiquitinate and degrade GSPT1. | Acute myeloid leukemia | Phase I Phase II | NCT04336982, NCT02848001 |
| KPG-818 | Specifically binds to CRBN, modulating the activity of the CRL4-CRBN complex. | Hematological malignancies | Phase I | NCT04283097 | |
| Cbl-b | NX-1607 | Binds to Cbl-b, preventing activation and inhibiting function. | Advanced malignancies | Phase 1 | NCT05107674 |
| MDM2 | RG7112 | Binds to MDM2 and disrupts MDM2-p53 interaction, stabilizing p53. | Myelogenous leukemia, Neoplasms, Sarcoma Hematologic neoplasms, | Phase I | NCT01677780, NCT00559533, NCT00623870, NCT01143740, NCT01164033, NCT01605526 |
| APG115 | Disrupts MDM2-p53 interaction, inducing cell-cycle arrest and apoptosis. | Solid tumor or lymphoma | Phase I | NCT02935907, CTR20170975 | |
| JNJ-26854165 (Serdemetan) | Inhibits HDM2-P53 interaction, stabilizing p53. | Neoplasms | Phase I | NCT00676910 | |
| ALRN-6924 | Binds to MDM2 and MDMX to disrupt MDM2-p53 interaction, stabilizing p53. | Leukemia, Solid Tumors | Phase I | NCT03654716, NCT02909972, NCT02264613, NCT03725436, NCT05622058 | |
| RG7388 (Idasanutlin) | Binds to MDM2 to disrupt MDM2-p53 interaction, stabilizing p53. | Acute myeloid leukemia, Solid tumors | Phase I | NCT02670044, NCT03362723, NCT02828930, NCT01773408, NCT01462175, NCT01901172 | |
| MK-8242 | Prevents HDM2-P53 interaction, stabilizing p53. | Solid tumors, Acute myeloid leukemia | Phase I | NCT01463696, NCT01451437 | |
| SAR405838 | High-affinity binds to MDM2 to disrupt MDM2-p53 interaction, stabilizing p53. | Neoplasm malignant | Phase I | NCT01636479, NCT01985191 | |
| CGM097 | Binds to MDM2 to disrupt MDM2-p53 interaction, stabilizing p53. | Solid tumors | Phase I | NCT01760525 | |
| Milademetan (DS-3032b) | Specifically binds to MDM2 to disrupt MDM2-p53 interaction, stabilizing p53. | Advanced solid tumor, Myeloid leukemia, Myeloma, Dedifferentiated liposarcoma | Phase I | NCT01877382, NCT03671564, NCT03614455, NCT02579824, NCT04979442 | |
| Siremadlin (HDM-201) | Binds to the binding pocket of p53 in MDM2, competitively inhibiting MDM2-p53 interaction to stabilize p53. | Colorectal cancer, Solid and hematological tumors, Liposarcoma | Phase I | NCT02890069, NCT02143635, NCT02343172 | |
| AMG232 | Binds to MDM2 to disrupt MDM2-p53 interaction, stabilizing p53. | Malignancy | Phase I | NCT01723020, NCT02110355, NCT02016729 | |
| RO6839921 | An inactive pegylated prodrug of the oral MDM2 antagonist to disrupt MDM2-p53 interaction, stabilizing p53. | Acute myeloid leukemia | Phase I | NCT02098967 | |
| BI907828 | Binds to MDM2 to disrupt MDM2-p53 interaction, stabilizing p53. | Different types of advanced cancer, Glioblastoma | Phase I | NCT03449381, NCT05376800, NCT03964233 | |
| IAPs | GDC-0152 | Binds to BIR domains of ML-IAP, XIAP, cIAP1, and cIAP2 to degrade cIAP1, promoting cell apoptosis. | Advanced or metastatic malignancies | Phase I | NCT00977067 |
| LCL161 | Binds to the BIR3 domain of cIAPs, inducing their autoubiquitination and degradation. | Colorectal cancer, Multiple myeloma, Solid tumors, Neoplasms, Small cell lung cancer, Breast cancer | Phase I Phase II | NCT02890069, NCT03111992, NCT01240655, NCT01968915, NCT01617668, NCT01098838, NCT02098161, NCT02649673 | |
| AT-406 | Binds to XIAP and cIAPs, inducing cIAP1 degradation and caspase activation. | Adenocarcinoma of the pancreas, Squamous cell carcinoma, Solid tumors | Phase I Phase II | NCT04122625, NCT03871959, NCT02022098, NCT03270176, NCT01078649 | |
| TL-32711 | Binds to the BIR3 domain of cIAPs, inducing their autoubiquitination and degradation | Ovarian cancer | Phase I | NCT01940172 | |
| APG-1387 | A next-generation IAP inhibitor mimics endogenous SMAC to degrade IAPs. | Solid tumors | Phase I | NCT03386526 | |
| AEG-40826 | A selectively inhibits IAP biological activity, restores apoptotic signaling. | Advanced solid tumors | Phase I | NCT00708006 | |
| BI-891065 | Binds to CIAPs and promotes their degradation, inducing tumor cell apoptosis. | Neoplasm, Non-small-cell lung carcinoma | Phase I | NCT04138823, NCT03166631 | |
| CRLs | MLN4924 (Pevonedistat) | Blocks the activation of NEDD8 by competitively binding to the adenosineylation site of NAE, thereby inhibiting cullin ring neddylation. | Multiple myeloma, Myeloid leukemia, Mesothelioma, Solid neoplasm, Lymphoblastic leukemia, Metastatic melanoma | Phase I Phase II | NCT03770260, NCT04712942, NCT03319537, NCT03330106, NCT03814005, NCT03349281, NCT02610777, NCT02782468, NCT03486314, NCT03459859, NCT01862328, NCT01814826, NCT03057366, NCT02122770, NCT00911066, NCT00722488, NCT00677170, NCT01011530 |
| Drug targeting DUBs | |||||
| USP14 UCHL5 | VLX1570 | Binds to USP14 and UCHL5 to inhibit their function. | Multiple myeloma | Phase I Phase II | NCT02372240 |
| USP1 | KSQ-4279 | A selective small molecule inhibitor of USP1 with anti-proliferative activity in tumors with HRR mutations. | Advanced solid tumors | Phase I | NCT05240898 |
| TNG348 | A reversible allosteric inhibitor, inhibiting its deubiquitinase activity by binding to the allosteric site of USP1 | BRCA1/2 mutant tumors or HRD+ solid tumors | Phase I Phase II | NCT06065059 | |
| XL309 | A selective USP1 inhibitor with an unknown binding mechanism | Advanced solid tumors | Phase I | NCT05932862 | |
| SIM0501 | A selective USP1 inhibitor with an unknown binding mechanism | Advanced solid tumors | Phase I | NCT06331559 | |
| HSK39775 | A selective USP1 inhibitor with an unknown binding mechanism | Advanced solid tumors | Phase I Phase II | NCT06314373 | |
| UCHL3 | Perifosine | A selective UCHL3 inhibitor with an unknown binding mechanism | Pediatric solid tumors, Refractory tumors, Leukemia, Breast cancer, Colorectal cancer | Phase I Phase II Phase III | NCT01049841, NCT00873457, NCT00391560, NCT00054145, NCT01097018 |
PROTACs and Molecular Glue Degraders
E3 ligases play a crucial role in the ubiquitination process and are therefore essential components of PROTACs and molecular glues [306, 307]. Proteolysis-targeting chimera (PROTAC) technology plays a pivotal role in the development of small-molecule drugs, leveraging the ubiquitin-proteasome system to induce protein degradation [308]. PROTACs are heterobifunctional molecules that facilitate the proximity between a target protein and an E3 ligase, leading to ubiquitination and subsequent proteasomal degradation of the target protein [309]. Several PROTACs are currently undergoing clinical trials for cancer therapy. For instance, ARV-110, also known as bavdegalutamide, is an investigational drug being evaluated in phase 1/2 clinical trials for the treatment of metastatic castration-resistant prostate cancer (mCRPC) [310]. A phase 1b study is also evaluating the combination of ARV-110 with abiraterone in patients with metastatic prostate cancer [311]. Furthermore, preclinical studies have demonstrated that ARV-110 can degrade AR by ≥95% and exhibits antitumor activity in enzalutamide-naive and resistant prostate cancer xenograft models [312]. ARV-471 is an orally bioavailable PROTAC degrader, which has potential antitumor activity in BC treatment by targeting estrogen receptor (ER) [313]. For instance, ARV-471 therapy was well-tolerated and showed antitumor activity in patients with ER+/HER2- locally advanced or metastatic BC in a phase I clinical trial [313]; current phase III trials are evaluating its utility in locally advanced and metastatic BC [314]. Despite PROTACs having shown great potential, challenges remain in their clinical development, including poor oral bioavailability, large molecular weights, and dependencies on specific E3 ligase receptors [315].
Molecular glues offer an alternative approach to targeted protein degradation [307]. These small molecules induce interactions between a target protein and an E3 ligase, leading to the degradation of the target protein [307]. Compared to PROTAC, molecular glues have several advantages, including smaller molecules, improved cell permeability, and tissue specificity, enabling more efficient promotion of E3 ligase-target protein interactions and ubiquitination [308, 316]. Several molecular glue degraders (MGDs) targeting E3 ligases are currently being evaluated in preclinical and clinical studies for cancer therapy [317]. For instance, CC-90009 recruits the cullin4-RING E3 ubiquitin ligase (CRL4)-cereblon (CRBN) complex to induce proteasomal degradation of G1-to-S phase transition 1 (GSPT1), leading to potent suppression of acute myeloid leukemia (AML) in preclinical studies [318]. Furthermore, the clinical trial of CC-90009 to treat leukemia is under investigation [318]. Overall, targeted protein degradation using PROTACs and molecular glues is a promising therapeutic strategy for treating various diseases, including cancer [319]. While challenges remain in their clinical development, ongoing research is focused on improving their selectivity, bioavailability, and efficacy.
MDM2 inhibitors
MDM2 inhibitors represent a significant therapeutic approach targeting MDM2, an E3 ligase that negatively regulates p53 activity [320]. These inhibitors aim to restore p53 function in tumors harboring wild-type TP53 by disrupting he interaction between MDM2 and p53 [321]. Several MDM2 inhibitors have progressed to clinical trials, evaluating their safety, efficacy, and optimal dosing across various cancer types [322, 323]. Clinical trials primarily focus on patients with cancers retaining wild-type TP53, as the mechanism of action depends on p53 reactivation. MDM2 inhibitors have shown some clinical activity, especially in hematological malignancies such as AML and chronic lymphocytic leukemia (CLL) [321]. For instance, a phase I study of RG7112 in hematologic malignancies assessed dose and safety [324], and subsequent phase Ib trials explored RG7112 in combination with Ara-C for AML treatment [325], as well as monotherapy for relapsed/refractory solid tumors [326]. Another phase Ib study evaluated RG7112 with Dox in advanced soft tissue sarcoma [327]. APG-115, a potent small-molecule MDM2 inhibitor and immune modulator, demonstrated promising antitumor activity [328]. Subsequent phase I trials in patients with advanced solid tumors assessed its safety, pharmacokinetics, pharmacodynamics, and antitumor effects [328]. A Chinese study (CTR20170975) similarly focused on APG-115 in advanced soft tissue sarcomas [329]. Further research suggests that APG-115 enhances programmed death ligand 1 (PD-L1) immunotherapy efficacy in thyroid cancer [330]. Additionally, other small-molecule MDM2 inhibitors, including JNJ-26854165 (Serdemetan), ALRN-6924, RG7388 (Idasanutlin), MK-8242, SAR-405838, CGM097, DS3032b (Milademetan), Siremadlin (HDM-201), AMG232, RO6839921, and BI907828, are under investigation in clinical trials to assess their therapeutic potential (Table 5) [321, 331-341]. However, resistance to MDM2 inhibitors represents a significant obstacle. The mechanisms of resistance can be categorized broadly as p53-dependent or p53-independent [342]. P53-dependent resistance involves alterations in the p53 pathway, such as TP53 mutations or dysregulation of p53 target genes [343]. P53-independent resistance entails activation of alternative signaling pathways that circumvent p53 reactivation [344]. Furthermore, combination therapies pairing MDM2 inhibitors with chemotherapy, targeted therapies, or immunotherapy offer a promising avenue to enhance efficacy [345]. Understanding these mechanisms is critical for developing strategies to overcome resistance.
IAP inhibitors
Inhibitor of Apoptosis Proteins (IAPs) are key regulators of programmed cell death, and their inhibitors are emerging as potential cancer therapeutics. Small-molecule IAP inhibitors mimic the endogenous IAP antagonist Smac/DIABLO. They bind with high affinity to cellular IAP1 (cIAP1), cIAP2, and X-linked IAP (XIAP), subsequently inducing their proteasome-dependent degradation [346]. Specifically, the IAP antagonist GDC-0152 entered phase I clinical trials to evaluate safety, tolerability, and pharmacokinetics in patients with advanced solid tumors [347, 348]. Preclinical studies demonstrate that GDC-0152 binds the XIAP BIR3 domain and the BIR domains of cIAP1/cIAP2, promotes cIAP1 degradation, and reduces the viability of BC cells [346, 347]. GDC-0152 can also modulate ABCB1-ATPase activity and suppress BIRC5 expression, mechanisms associated with overcoming multidrug resistance [348]. LCL161, another Smac mimetic, has undergone evaluation as a single agent and in combination therapies [349, 350]. For instance, LCL161 has been investigated in patients with intermediate or high-risk myelofibrosis who have failed or are intolerant to JAK inhibitors [351]. A phase I study in Japanese patients with advanced solid tumors combined LCL161 with paclitaxel, while data suggest an increased risk of infection with the combination [349]. Currently, several IAP inhibitors are in clinical trials, including AT-406 (NCT04122625, NCT03871959, NCT02022098, NCT03270176, NCT01078649), TL-32711(NCT01940172), APG-1387 (NCT03386526), AEG40826 (NCT00708006), and BI-891065 (NCT04138823, NCT03166631), demonstrating promising antitumor activity (Table 5) [29, 352-354]. Despite their promise, challenges remain with IAP inhibitor development, including toxicity/tolerability issues, optimal patient selection strategies, primary and acquired resistance mechanisms, and potential off-target effects [355]. Further research is essential to fully realize the clinical potential of targeting IAPs in cancer therapy.
Drug development targeting DUBs
DUBs play crucial roles in cellular processes such as protein homeostasis, DNA repair, signal transduction, and epigenetic regulation. Dysregulation of DUB activity is implicated in diverse pathologies, including cancers, autoimmune disorders, chronic inflammation, and neurodegenerative diseases [356]. Moreover, DUBs possess well-defined catalytic sites, most of which contain a catalytic cysteine. Consequently, DUBs are emerging as promising targets for drug discovery [357]. DUB inhibitors can promote the degradation of oncogenic proteins, particularly those stabilized by DUBs and resistant to direct targeting [356]. Specific DUBs, such as USP7, have been identified as potential targets in malignancies, including hematological cancers [358]. Based on the types of target enzymes they act upon, the current small molecule inhibitors of DUBs are classified as: USP family inhibitors, UCH family inhibitors, JAMM family inhibitors, MJD family inhibitors, OUT family inhibitors, and SENP family inhibitors [35]. Among them, the USP family is the most extensively studied in preclinical research. For instance, HBX19818 and P22077 (USP inhibitors targeting USP7, USP14, and USP22) suppress cancer cell proliferation and enhance the efficacy of conventional therapies like Dox [359, 360]. Additionally, targeting DUBs can reverse chemoresistance in cancer cells, offering a strategy for more effective treatment [361, 362]. Despite growing interest in DUB biological function and potential as therapeutic targets, few selective small-molecule inhibitors and no approved drugs currently exist [363].
Several small molecules targeting oncogenic DUBs have been identified, with some demonstrating promising anticancer activity and advancing into clinical trials (Table 5) [364, 365]. VLX1570, a selective inhibitor of USP14 and UCHL5 derived from b-AP15, exhibited significant antitumor effects in murine models of Waldenstrom macroglobulinemia (WM) by modulating BCR signaling and CXCR4 expression [366]. However, its clinical development encountered challenges. In a phase I/II clinical trial (NCT02372240), dose-limiting toxicities were observed in patients with MM during dose escalation, leading to trial termination in 2017 [367]. Perifosine, an oral alkylphospholipid that targets UCHL3, shows significant activity against BC cells both in vitro and in vivo [368]. In a phase I clinical trial of recurrent/refractory pediatric solid tumors, the Perifosine combination of AKT and mTOR inhibitors was safe and feasible [369]. However, in a phase II clinical trial of patients with metastatic BC, only 19% of the patients experienced stable disease after 2 months of treatment [368]. KSQ-4279, an inhibitor of USP1, displays strong synergistic activity with PARP inhibitors in BRCA-mutant cancers [370]. In a phase 1 trial (NCT05240898), KSQ-4279 showed promising efficacy and safety both as monotherapy and in combination with Olaparib or Carboplatin. Additionally, other USP1 inhibitors such as TNG348, XL309, SIM0501, and HSK39775 are also progressing through clinical trials [35, 364]. Notably, TNG348 development was halted due to significant liver toxicity observed in patients treated beyond eight weeks, leading to termination of its phase 1/2 trial (NCT06065059). XL309 has demonstrated preclinical efficacy in BRCA1-mutated TNBC and is being evaluated in an ongoing phase 1 trial (NCT05932862) for safety and preliminary antitumor activity. Furthermore, HSK39775 and SIM0501 are under investigation in phase 1 trials (NCT06331559, NCT06314373) to assess safety and initial efficacy in advanced solid tumors. Collectively, these findings underscore the therapeutic potential of DUB-targeting agents while highlighting critical challenges in development, particularly the need for rigorous safety monitoring during early-phase clinical investigations.
Conclusions and perspectives
EMT is a crucial process in cancer metastasis, facilitating tumor progression, invasion, and drug resistance [371]. Cells undergoing EMT frequently exhibit diminished sensitivity to chemotherapeutic agents, which contributes to treatment failure and disease relapse [260]. Dysregulation of E3 ligases and DUBs contributes to the progression of EMT [372], which involves abnormal activation of EMT-TFs and the EMT-associated signaling pathways [50]. Furthermore, accumulating evidence demonstrates that ubiquitination-mediated regulation of EMT significantly influences metastasis and chemoresistance in tumors. In this review, we comprehensively reviewed the mechanisms by which E3 ligases and DUBs regulate EMT and further emphasize the significance of ubiquitination-regulated EMT in tumor metastasis and chemoresistance. Furthermore, some preclinical and clinical evidence indicate that drugs targeting E3 ligases or DUBs have reversed EMT-induced metastasis and resistance in cancer.
Although drug development targeting ubiquitination factors holds significant promise for cancer therapy, the development of E3 or DUB inhibitors has encountered substantial challenges. The primary reason is due to the high structural and functional diversity of these enzymes, which complicates achieving inhibitor specificity and avoiding off-target effects that could lead to toxicity. Moreover, ubiquitination dynamics are influenced by cancer-specific mutations, microenvironmental factors, and heterogeneity in the EMT process, further exacerbating the complexity of inhibitor development. To address these challenges, future research should pursue innovative screening and development strategies, including: (1) Utilizing high-throughput assays and advanced cellular models to enable sensitive quantification of DUB activity and inhibition; (2) developing dual-action inhibitors targeting ubiquitination and EMT effectors simultaneously to counteract treatment resistance, building on recent progress in characterizing selective compounds against E3 ligases or DUBs; (3) validating predictive biomarkers in clinical cohorts for patient stratification and therapeutic optimization; (4) Employing emerging technologies, such as proximity-based labeling approaches for mapping ubiquitin dynamics, to resolve unanswered questions regarding ubiquitin signaling specificity; (5) identifying specific ubiquitination-EMT networks across diverse cancer types to clarify the mechanisms of ubiquitination-dependent EMT regulation in chemoresistance, as well as examining the universality and variability of this mechanism across different tumor types.
In conclusion, ubiquitination regulates tumor EMT through various mechanisms, thereby influencing tumor metastasis and treatment resistance. Targeting specific E3 ligases or DUBs reverses the EMT process, leading to sensitizing tumor cells to chemotherapeutic agents and suppressing distant metastasis. These findings provide novel insights into the mechanisms underlying tumor chemoresistance. Consequently, the development of E3 and DUB inhibitors offers a promising strategy for mitigating chemoresistance and metastasis in clinical oncology.
Abbreviations
5-FU: 5-Fluorouracil; ACF7: actin crosslinking factor 7; AML: acute myeloid leukemia; ATXN1: ataxin-1; AXL: AXL receptor tyrosine kinase; AR: androgen receptor; AXIN: axis inhibitory protein; BC: breast cancer; BCR: B cell receptor; BIRC5: baculoviral IAP repeat containing 5; BRCA1: breast cancer susceptibility gene 1; CAFs: cancer-associated fibroblasts; ccRCC: clear cell renal cell carcinoma; CDK4/6: cyclin-dependent kinase 4/6; CHIP: C-terminus of HSC70-interacting protein; CIC: cancer-initiating cell; CLL: chronic lymphocytic leukemia; CPT1C: carnitine palmitoyltransferase 1C; CRC: colorectal cancer; CRLs: cullin-RING ligases; CSC: cancer stem cell; CXCR4: C-X-C motif chemokine receptor 4; CRL4-CRBN: cullin4-RING E3 ubiquitin ligase-cereblon complex; DUB: deubiquitinating enzyme; Dox: doxorubicin; EBV: epstein-barr virus; EMT: epithelial-mesenchymal transition; ER: estrogen receptor; ESCC: esophageal squamous cell carcinoma; FAT10: HLA-F adjacent transcript 10; FBX: F-box protein; FBXW: F-box and WD repeat domain containing; FBXL: F-box and leucine-rich repeat protein; FZD: frizzled; GBM: glioblastoma; GC: gastric cancer; Gli: glioma-associated oncogene homolog; GSK-3β: glycogen synthase kinase-3β; GSPT1: G1-to-S phase transition 1; HCC: hepatocellular carcinoma; HECT: homologous to E6AP carboxyl terminus; HECTD1: HECT domain E3 ubiquitin protein ligase 1; HERC3: HECT and RLD domain containing E3 ubiquitin protein ligase 3; Hh: hedgehog; HIF: hypoxia-inducible Factor; HRD+: homologous recombination deficiency positive; IAP: inhibitor of apoptosis protein; iCCA: intrahepatic cholangiocarcinoma; KIRC: kidney renal clear cell carcinoma; LC: lung cancer; LOX: lysyl oxidase; LUAD: lung adenocarcinoma; MARCH: membrane-associated RING-CH protein; MDM2: murine double minute 2; MGD: molecular glue degrader; MIB1: MIB E3 ubiquitin protein ligase 1; MM: multiple myeloma; MMPs: matrix metalloproteinases; mCRPC: metastatic castration-resistant prostate cancer; NAE: NEDD8-activating enzyme; NEDD4L: neural precursor cell expressed developmentally downregulated 4-Like; NEDD8: neural precursor cell expressed developmentally downregulated 8; NICD: notch intracellular domain; NSCLC: non-small cell lung cancer; OC: ovarian cancer; OGA: O-GlcNAcase; OS: osteosarcoma; OSCC: oral squamous cell carcinoma; OTU: ovarian tumor-related protease; OTUD6B: ovarian tumor domain-containing 6B; PARK2: parkin RBR E3 ubiquitin protein ligase; PC: pancreatic cancer; PCa: prostate cancer; PD-L1: programmed cell death ligand 1; PELI1: pellino1; PROTAC: proteolysis-targeting chimera; PSMD14: proteasome non-ATPase regulatory subunit 14; PTEN: phosphatase and tensin homolog; PTM: post-translational modification; RBR: RING-Between-RING; RBPJ: recombination signal binding protein for immunoglobulin Kappa J region; RCC: renal cell carcinoma; RING: really interesting new gene; RNF: ring finger protein; SCLC: small cell lung cancer; SMAC: second mitochondrial-Derived activator of caspases; SIAH1: seven in absentia homolog 1; SIP: siah-interacting protein; SMAD: small mother against decapentaplegic; Smo: smoothened; Snail: snail family transcriptional repressor; SPOP: speckle-type POZ protein; STAT3: signal transducer and activator of transcription 3; STAMBPL1: STAM binding protein like 1; STUB1: STIP1 homology and U-box containing protein 1; SUZ12: suppressor of zeste 12; TAK1: transforming growth factor β-activated kinase 1; TβRI: TGF-β type I receptor; TβRII: TGF-β type II receptor; TGF-β: transforming growth factor β; TMZ: temozolomide; TNBC: triple-negative breast cancer; TRIM: tripartite motif; TRAF6: TNF receptor associated factor 6; Twist: twist family BHLH transcription factor; Ub: Ubiquitin; UBR5: ubiquitin ligase E3 component N-recognition protein 5; UCHL5: ubiquitin carboxyl-terminal hydrolase isozyme L5; UPS: ubiquitin-proteasome system; USP: ubiquitin-specific protease; VHL: von hippel-lindau; WM: waldenström macroglobulinemia; WWP2: WW domain containing E3 ubiquitin protein ligase 2; XIAP: X-linked inhibitor of apoptosis protein; ZEB1/2: zinc finger E-box binding homeobox 1/2; β-TrCP: β-transducin repeat-containing protein; UCHs: ubiquitin carboxy-terminal hydrolase; MINDYs: motif interacting with ubiquitin-containing novel DUB; JAMMs: JAMM/MPN domain-associated metallopeptidases; MJDs: machado-joseph domain proteases; ZUFSP: Zinc finger and UFSP domain protein; SPSB3: SPRY Domain-Containing SOCS Box Protein 3; MARCH2: membrane associated ring-CH-type finger 2.
Acknowledgements
We thank Hubei University of Technology for financial support for this review.
Funding
This research was funded by the National Key R&D Program of China (2023YFC2507900 to JT), Hubei Natural Science Foundation of China (2024AFB218 to SX), National Natural Science Foundation of China (82273970 to JT, 32270768 to CZ, 32571378 to CZ, and 82370715 to XC), Science and Technology Talent Program of Hubei (2024DJA037 to JT), and Innovation Group Project of Hubei Province (No. 2023AFA026 to JT).
Author contributions
Conceptualization, SX, LT, XG, and WQ; writing—original draft preparation, SX, YY, ML, RZ, HL, DG, QZ, and XC; writing—review and editing, SX, XG, XX, and DS; investigation, ML, XG, HL, XX, YY, and DS; visualization, WQ, ML, RZ, XG, and DG; project administration QZ, XC, CZ, and JT; supervision, CZ and JT. All authors have read and agreed to the published version of the manuscript.
Competing interests
The authors have declared that no competing interest exists.
References
1. Swatek KN, Komander D. Ubiquitin modifications. Cell research. 2016;26:399-422
2. Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159-80
3. Agrata R, Komander D. Ubiquitin-A structural perspective. Molecular cell. 2025;85:323-46
4. Meyer HJ, Rape M. Enhanced protein degradation by branched ubiquitin chains. Cell. 2014;157:910-21
5. Akutsu M, Dikic I, Bremm A. Ubiquitin chain diversity at a glance. Journal of cell science. 2016;129:875-80
6. Yang Q, Zhao J, Chen D, Wang Y. E3 ubiquitin ligases: styles, structures and functions. Molecular biomedicine. 2021;2:23
7. Ren J, Yu P, Liu S, Li R, Niu X, Chen Y. et al. Deubiquitylating Enzymes in Cancer and Immunity. Adv Sci (Weinh). 2023;10:e2303807
8. Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156-72
9. Li X, Chen L, Peng X, Zhan X. Progress of tumor-associated macrophages in the epithelial-mesenchymal transition of tumor. Front Oncol. 2022;12:911410
10. Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850
11. Bill R, Christofori G. The relevance of EMT in breast cancer metastasis: Correlation or causality? FEBS Lett. 2015;589:1577-87
12. Zhang K, Zhang M, Yao Q, Han X, Zhao Y, Zheng L. et al. The hepatocyte-specifically expressed lnc-HSER alleviates hepatic fibrosis by inhibiting hepatocyte apoptosis and epithelial-mesenchymal transition. Theranostics. 2019;9:7566-82
13. Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol Cancer. 2016;15:18
14. Liao TT, Yang MH. Revisiting epithelial-mesenchymal transition in cancer metastasis: the connection between epithelial plasticity and stemness. Mol Oncol. 2017;11:792-804
15. Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST. et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472-6
16. Rashid M, Hari K, Thampi J, Santhosh NK, Jolly MK. Network topology metrics explaining enrichment of hybrid epithelial/mesenchymal phenotypes in metastasis. PLoS Comput Biol. 2022;18:e1010687
17. Xu Z, Zhang Y, Dai H, Han B. Epithelial-Mesenchymal Transition-Mediated Tumor Therapeutic Resistance. Molecules. 2022 27
18. Yang H, Xu F, Chen Y, Tian Z. Structural N-glycoproteomics characterization of cell-surface N-glycosylation of MCF-7/ADR cancer stem cells. J Chromatogr B Analyt Technol Biomed Life Sci. 2023;1219:123647
19. Voutsadakis IA. The ubiquitin-proteasome system and signal transduction pathways regulating Epithelial Mesenchymal transition of cancer. J Biomed Sci. 2012;19:67
20. Jin Y, Zhang Y, Li B, Zhang J, Dong Z, Hu X. et al. TRIM21 mediates ubiquitination of Snail and modulates epithelial to mesenchymal transition in breast cancer cells. Int J Biol Macromol. 2019;124:846-53
21. Hong KS, Ryu KJ, Kim H, Kim M, Park SH, Kim T. et al. MSK1 promotes colorectal cancer metastasis by increasing Snail protein stability through USP5-mediated Snail deubiquitination. Exp Mol Med. 2025;57:820-35
22. Ito K, Harada I, Martinez C, Sato K, Lee E, Port E. et al. MARCH2, a Novel Oncogene-regulated SNAIL E3 Ligase, Suppresses Triple-negative Breast Cancer Metastases. Cancer Res Commun. 2024;4:946-57
23. Fu Z, Yu W, Wang H, Chen X. Overexpression of RNF187 induces cell EMT and apoptosis resistance in NSCLC. J Cell Physiol. 2019;234:14161-9
24. Rodriguez-Alonso A, Casas-Pais A, Roca-Lema D, Grana B, Romay G, Figueroa A. Regulation of Epithelial-Mesenchymal Plasticity by the E3 Ubiquitin-Ligases in Cancer. Cancers (Basel). 2020;12:3093
25. Celia-Terrassa T, Kang Y. How important is EMT for cancer metastasis? PLoS Biol. 2024;22:e3002487
26. Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nature reviews Molecular cell biology. 2009;10:755-64
27. Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319-31
28. Liu W, Tang X, Qi X, Fu X, Ghimire S, Ma R. et al. The Ubiquitin Conjugating Enzyme: An Important Ubiquitin Transfer Platform in Ubiquitin-Proteasome System. Int J Mol Sci. 2020;21:2894
29. Liu F, Chen J, Li K, Li H, Zhu Y, Zhai Y. et al. Ubiquitination and deubiquitination in cancer: from mechanisms to novel therapeutic approaches. Mol Cancer. 2024;23:148
30. Buetow L, Huang DT. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol. 2016;17:626-42
31. Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550-63
32. Fang Y, Fu D, Shen XZ. The potential role of ubiquitin c-terminal hydrolases in oncogenesis. Biochim Biophys Acta. 2010;1806:1-6
33. Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363-97
34. Lim KH, Ramakrishna S, Baek KH. Molecular mechanisms and functions of cytokine-inducible deubiquitinating enzymes. Cytokine & growth factor reviews. 2013;24:427-31
35. Liu P, Chen Z, Guo Y, He Q, Pan C. Recent advances in small molecule inhibitors of deubiquitinating enzymes. Eur J Med Chem. 2025;287:117324
36. Dewson G, Eichhorn PJA, Komander D. Deubiquitinases in cancer. Nature reviews Cancer. 2023;23:842-62
37. Elu N, Osinalde N, Ramirez J, Presa N, Rodriguez JA, Prieto G. et al. Identification of substrates for human deubiquitinating enzymes (DUBs): An up-to-date review and a case study for neurodevelopmental disorders. Semin Cell Dev Biol. 2022;132:120-31
38. Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140:384-96
39. Shen J, Hong L, Chen L. Ubiquitin-specific protease 14 regulates ovarian cancer cisplatin-resistance by stabilizing BCL6 oncoprotein. Biochem Biophys Res Commun. 2020;524:683-8
40. Nieszporek A, Skrzypek K, Adamek G, Majka M. Molecular mechanisms of epithelial to mesenchymal transition in tumor metastasis. Acta Biochim Pol. 2019;66:509-20
41. Tomecka P, Kunachowicz D, Gorczynska J, Gebuza M, Kuznicki J, Skinderowicz K. et al. Factors Determining Epithelial-Mesenchymal Transition in Cancer Progression. Int J Mol Sci. 2024;25:8972
42. Sravani A, Thomas J. Targeting epithelial-mesenchymal transition signaling pathways with Dietary Phytocompounds and repurposed drug combinations for overcoming drug resistance in various cancers. Heliyon. 2025;11:e41964
43. Pan G, Liu Y, Shang L, Zhou F, Yang S. EMT-associated microRNAs and their roles in cancer stemness and drug resistance. Cancer Commun (Lond). 2021;41:199-217
44. Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nature reviews Cancer. 2007;7:415-28
45. Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155-66
46. Wang Y, Shi J, Chai K, Ying X, Zhou BP. The Role of Snail in EMT and Tumorigenesis. Current cancer drug targets. 2013;13:963-72
47. Singh M, Yelle N, Venugopal C, Singh SK. EMT: Mechanisms and therapeutic implications. Pharmacol Ther. 2018;182:80-94
48. Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E. et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267-78
49. Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H. et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nature cell biology. 2017;19:518-29
50. Basu B, Ghosh MK. Ubiquitination and deubiquitination in the regulation of epithelial-mesenchymal transition in cancer: Shifting gears at the molecular level. Biochim Biophys Acta Mol Cell Res. 2022;1869:119261
51. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178-96
52. Hao Y, Baker D, Ten Dijke P. TGF-beta-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int J Mol Sci. 2019;20:2767
53. Derynck R, Muthusamy BP, Saeteurn KY. Signaling pathway cooperation in TGF-β-induced epithelial-mesenchymal transition. Current Opinion in Cell Biology. 2014;31:56-66
54. Katsuno Y, Lamouille S, Derynck R. TGF-beta signaling and epithelial-mesenchymal transition in cancer progression. Curr Opin Oncol. 2013;25:76-84
55. Sheikh KA, Amjad M, Irfan MT, Anjum S, Majeed T, Riaz MU. et al. Exploring TGF-β Signaling in Cancer Progression: Prospects and Therapeutic Strategies. OncoTargets and therapy. 2025;18:233-62
56. Moon JH, Lee SH, Lim YC. Wnt/beta-catenin/Slug pathway contributes to tumor invasion and lymph node metastasis in head and neck squamous cell carcinoma. Clin Exp Metastasis. 2021;38:163-74
57. Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6392-7
58. Matsuno Y, Coelho AL, Jarai G, Westwick J, Hogaboam CM. Notch signaling mediates TGF-beta1-induced epithelial-mesenchymal transition through the induction of Snai1. Int J Biochem Cell Biol. 2012;44:776-89
59. Teglund S, Toftgard R. Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim Biophys Acta. 2010;1805:181-208
60. Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454-72
61. Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. 2017;11:28-39
62. Castaneda M, den Hollander P, Kuburich NA, Rosen JM, Mani SA. Mechanisms of cancer metastasis. Semin Cancer Biol. 2022;87:17-31
63. Shi M, Zhang R, Lyu H, Xiao S, Guo D, Zhang Q. et al. Long non-coding RNAs: Emerging regulators of invasion and metastasis in pancreatic cancer. J Adv Res. 2025 S2090-1232: 00073-6
64. Morin C, Baudin-Baillieu A, Van Long FN, Isaac C, Bidou L, Arbes H. et al. Intricate ribosome composition and translational reprogramming in epithelial-mesenchymal transition. Proc Natl Acad Sci U S A. 2024;121:e2408114121
65. Feng P, Zhu L, Jie J, Yang P, Sheng N, Chen X. et al. Cannabidiol inhibits invasion and metastasis in colorectal cancer cells by reversing epithelial-mesenchymal transition through the Wnt/beta-catenin signaling pathway. J Cancer Res Clin Oncol. 2023;149:3587-98
66. Hussein ZH, Hassawi BA, Ibraheem Q. Aberrant beta-Catenin Expression and Its Association with Epithelial-Mesenchymal Transition and Clinical Outcomes of Colorectal Cancer. Cureus. 2024;16:e53104
67. Akhmetkaliyev A, Alibrahim N, Shafiee D, Tulchinsky E. EMT/MET plasticity in cancer and Go-or-Grow decisions in quiescence: the two sides of the same coin? Mol Cancer. 2023;22:90
68. Yang HL, Chang YH, Pandey S, Bhat AA, Vadivalagan C, Lin KY. et al. Antrodia camphorata and coenzyme Q(0), a novel quinone derivative of Antrodia camphorata, impede HIF-1alpha and epithelial-mesenchymal transition/metastasis in human glioblastoma cells. Environ Toxicol. 2023;38:1548-64
69. Zhang G, Xiao Y, Tan J, Liu H, Fan W, Li J. Elevated SLC1A5 associated with poor prognosis and therapeutic resistance to transarterial chemoembolization in hepatocellular carcinoma. J Transl Med. 2024;22:543
70. Li S, Qi S, Li Y, Zhang C, Sun L, Liu C. et al. MARVELD3 inhibits the epithelial-mesenchymal transition and cell migration by suppressing the Wnt/beta-catenin signaling pathway in non-small cell lung cancer cells. Thorac Cancer. 2023;14:1045-58
71. He H, Li Y, Wang Y, Li M. Geniposide Inhibits Non-Small Cell Lung Cancer by Regulating Proliferation, Apoptosis, Invasion, Migration, Epithelial-Mesenchymal Transition, and Cancer Stem-Like Cell Property Via Wnt/beta-Catenin Pathway. Biochem Genet. 2025;5:11030
72. Haslehurst AM, Koti M, Dharsee M, Nuin P, Evans K, Geraci J. et al. EMT transcription factors snail and slug directly contribute to cisplatin resistance in ovarian cancer. BMC Cancer. 2012;12:91
73. Molina-Ortiz P, Villarejo A, MacPherson M, Santos V, Montes A, Souchelnytskyi S. et al. Characterization of the SNAG and SLUG domains of Snail2 in the repression of E-cadherin and EMT induction: modulation by serine 4 phosphorylation. PLoS One. 2012;7:e36132
74. Liu Y, Zhou H, Zhu R, Ding F, Li Y, Cao X. et al. SPSB3 targets SNAIL for degradation in GSK-3beta phosphorylation-dependent manner and regulates metastasis. Oncogene. 2018;37:768-76
75. Park SM, Park SH, Ryu KJ, Kim IK, Han H, Kim HJ. et al. Downregulation of CHIP promotes ovarian cancer metastasis by inducing Snail-mediated epithelial-mesenchymal transition. Mol Oncol. 2019;13:1280-95
76. Kao SH, Wang WL, Chen CY, Chang YL, Wu YY, Wang YT. et al. GSK3beta controls epithelial-mesenchymal transition and tumor metastasis by CHIP-mediated degradation of Slug. Oncogene. 2014;33:3172-82
77. Lv X, Liu S, Liu C, Li Y, Zhang T, Qi J. et al. TRIB3 promotes pulmonary fibrosis through inhibiting SLUG degradation by physically interacting with MDM2. Acta Pharm Sin B. 2023;13:1631-47
78. Lim SO, Kim H, Jung G. p53 inhibits tumor cell invasion via the degradation of snail protein in hepatocellular carcinoma. FEBS Lett. 2010;584:2231-6
79. Xie W, Jiang Q, Wu X, Wang L, Gao B, Sun Z. et al. IKBKE phosphorylates and stabilizes Snail to promote breast cancer invasion and metastasis. Cell Death Differ. 2022;29:1528-40
80. Wu ZQ, Li XY, Hu CY, Ford M, Kleer CG, Weiss SJ. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic Breast Cancer 1, Early Onset (BRCA1) repression. Proc Natl Acad Sci U S A. 2012;109:16654-9
81. Zhang Y, Zhang X, Ye M, Jing P, Xiong J, Han Z. et al. FBW7 loss promotes epithelial-to-mesenchymal transition in non-small cell lung cancer through the stabilization of Snail protein. Cancer Lett. 2018;419:75-83
82. Vinas-Castells R, Frias A, Robles-Lanuza E, Zhang K, Longmore GD, Garcia de Herreros A. et al. Nuclear ubiquitination by FBXL5 modulates Snail1 DNA binding and stability. Nucleic Acids Res. 2014;42:1079-94
83. Wu W, Ding H, Cao J, Zhang W. FBXL5 inhibits metastasis of gastric cancer through suppressing Snail1. Cell Physiol Biochem. 2015;35:1764-72
84. Song L, Guo J, Chang R, Peng X, Li J, Xu X. et al. LKB1 obliterates Snail stability and inhibits pancreatic cancer metastasis in response to metformin treatment. Cancer Sci. 2018;109:1382-92
85. Lander R, Nordin K, LaBonne C. The F-box protein Ppa is a common regulator of core EMT factors Twist, Snail, Slug, and Sip1. J Cell Biol. 2011;194:17-25
86. Kim SH, Ryu KJ, Hong KS, Kim H, Han H, Kim M. et al. ERK3 Increases Snail Protein Stability by Inhibiting FBXO11-Mediated Snail Ubiquitination. Cancers (Basel). 2023;16:105
87. Qiao X, Lin J, Shen J, Chen Y, Zheng L, Ren H. et al. FBXO28 suppresses liver cancer invasion and metastasis by promoting PKA-dependent SNAI2 degradation. Oncogene. 2023;42:2878-91
88. Zou S, Ma C, Yang F, Xu X, Jia J, Liu Z. FBXO31 Suppresses Gastric Cancer EMT by Targeting Snail1 for Proteasomal Degradation. Mol Cancer Res. 2018;16:286-95
89. Xu M, Zhu C, Zhao X, Chen C, Zhang H, Yuan H. et al. Atypical ubiquitin E3 ligase complex Skp1-Pam-Fbxo45 controls the core epithelial-to-mesenchymal transition-inducing transcription factors. Oncotarget. 2015;6:979-94
90. Yang J, Liao Y, Wang B, Cui L, Yu X, Wu F. et al. EDARADD promotes colon cancer progression by suppressing E3 ligase Trim21-mediated ubiquitination and degradation of Snail. Cancer Lett. 2023;577:216427
91. Ma X, Ma X, Qiu Y, Zhu L, Lin Y, You Y. et al. TRIM50 suppressed hepatocarcinoma progression through directly targeting SNAIL for ubiquitous degradation. Cell Death Dis. 2018;9:608
92. Wang X, De Geyter C, Jia Z, Peng Y, Zhang H. HECTD1 regulates the expression of SNAIL: Implications for epithelial-mesenchymal transition. Int J Oncol. 2020;56:1186-98
93. Jeon YK, Kim CK, Hwang KR, Park HY, Koh J, Chung DH. et al. Pellino-1 promotes lung carcinogenesis via the stabilization of Slug and Snail through K63-mediated polyubiquitination. Cell Death Differ. 2017;24:469-80
94. Liu SS, Qi J, Teng ZD, Tian FT, Lv XX, Li K. et al. Resistomycin attenuates triple-negative breast cancer progression by inhibiting E3 ligase Pellino-1 and inducing SNAIL/SLUG degradation. Signal Transduct Target Ther. 2020;5:133
95. Kuang J, Min L, Liu C, Chen S, Gao C, Ma J. et al. RNF8 Promotes Epithelial-Mesenchymal Transition in Lung Cancer Cells via Stabilization of Slug. Mol Cancer Res. 2020;18:1638-49
96. Lee JH, Jung SM, Yang KM, Bae E, Ahn SG, Park JS. et al. A20 promotes metastasis of aggressive basal-like breast cancers through multi-monoubiquitylation of Snail1. Nat Cell Biol. 2017;19:1260-73
97. Sonego M, Pellarin I, Costa A, Vinciguerra GLR, Coan M, Kraut A. et al. USP1 links platinum resistance to cancer cell dissemination by regulating Snail stability. Sci Adv. 2019;5:eaav3235
98. Zhang Y, Zuo D, Qiu J, Li K, Niu Y, Yuan Y. et al. NXN suppresses metastasis of hepatocellular carcinoma by promoting degradation of Snail through binding to DUB3. Cell Death Dis. 2022;13:676
99. Wei C, Zhao X, Zhang H, Wang L. USP2 promotes cell proliferation and metastasis in choroidal melanoma via stabilizing Snail. J Cancer Res Clin Oncol. 2023;149:9263-76
100. Fan L, Chen Z, Wu X, Cai X, Feng S, Lu J. et al. Ubiquitin-Specific Protease 3 Promotes Glioblastoma Cell Invasion and Epithelial-Mesenchymal Transition via Stabilizing Snail. Mol Cancer Res. 2019;17:1975-84
101. Li R, Shang R, Li S, Ren Y, Shen L, Yang L. et al. LOXL3-promoted hepatocellular carcinoma progression via promotion of Snail1/USP4-mediated epithelial-mesenchymal transition. Environ Toxicol. 2022;37:2540-51
102. Guan T, Yang X, Liang H, Chen J, Chen Y, Zhu Y. et al. Deubiquitinating enzyme USP9X regulates metastasis and chemoresistance in triple-negative breast cancer by stabilizing Snail1. J Cell Physiol. 2022;237:2992-3000
103. Luo Y, Zhu Q, Xiang S, Wang Q, Li J, Chen X. et al. Downregulated circPOKE promotes breast cancer metastasis through activation of the USP10-Snail axis. Oncogene. 2023;42:3236-51
104. Wang W, Wang J, Yan H, Zhang K, Liu Y. Upregulation of USP11 promotes epithelial-to-mesenchymal transition by deubiquitinating Snail in ovarian cancer. Oncol Rep. 2019;41:1739-48
105. Zhang T, Zheng J, Qiao L, Zhao W. Deubiquitinase USP13 promotes the epithelial-mesenchymal transition and metastasis in gastric cancer by maintaining Snail protein. Pathol Res Pract. 2022;229:153705
106. Feng L, Zhang J, Sun M, Qiu F, Chen W, Qiu W. Tumor Suppressor LINC02487 Inhibits Oral Squamous Cell Carcinoma Cell Migration and Invasion Through the USP17-SNAI1 Axis. Front Oncol. 2020;10:559808
107. Huang F, Zheng C, Huang L, Lin C, Wang J. USP18 directly regulates Snail1 protein through ubiquitination pathway in colorectal cancer. Cancer Cell Int. 2020;20:346
108. Li L, Zhou H, Zhu R, Liu Z. USP26 promotes esophageal squamous cell carcinoma metastasis through stabilizing Snail. Cancer Lett. 2019;448:52-60
109. Lambies G, Miceli M, Martinez-Guillamon C, Olivera-Salguero R, Pena R, Frias CP. et al. TGFbeta-Activated USP27X Deubiquitinase Regulates Cell Migration and Chemoresistance via Stabilization of Snail1. Cancer Res. 2019;79:33-46
110. Liu Z, Yu Y, Zhou S, Zhang X, Zhou Z. Inhibition of Pard3 promotes breast cancer metastasis via the USP28 mediated deubiquitination of Snail1. Heliyon. 2023;9:e22599
111. Qian W, Li Q, Wu X, Li W, Li Q, Zhang J. et al. Deubiquitinase USP29 promotes gastric cancer cell migration by cooperating with phosphatase SCP1 to stabilize Snail protein. Oncogene. 2020;39:6802-15
112. Sun K, Liao S, Yao X, Yao F. USP30 promotes the progression of breast cancer by stabilising Snail. Cancer Gene Ther. 2024;31:472-83
113. Ma C, Tian Z, Wang D, Gao W, Qian L, Zang Y. et al. Ubiquitin-specific Protease 35 Promotes Gastric Cancer Metastasis by Increasing the Stability of Snail1. Int J Biol Sci. 2024;20:953-67
114. Cai J, Li M, Wang X, Li L, Li Q, Hou Z. et al. USP37 Promotes Lung Cancer Cell Migration by Stabilizing Snail Protein via Deubiquitination. Front Genet. 2019;10:1324
115. Yoon JY, Seo SU, Woo SM, Kwon TK. USP41 Enhances Epithelial-Mesenchymal Transition of Breast Cancer Cells through Snail Stabilization. Int J Mol Sci. 2023 24
116. Choi BJ, Park SA, Lee SY, Cha YN, Surh YJ. Hypoxia induces epithelial-mesenchymal transition in colorectal cancer cells through ubiquitin-specific protease 47-mediated stabilization of Snail: A potential role of Sox9. Sci Rep. 2017;7:15918
117. Zhou H, Liu Y, Zhu R, Ding F, Cao X, Lin D. et al. OTUB1 promotes esophageal squamous cell carcinoma metastasis through modulating Snail stability. Oncogene. 2018;37:3356-68
118. Gao Y, Tang J, Ma X, Zhang C, Huang L, Che J. et al. OTUD4 regulates metastasis and chemoresistance in melanoma by stabilizing Snail1. J Cell Physiol. 2023;238:2546-55
119. Guo X, Zhu R, Luo A, Zhou H, Ding F, Yang H. et al. EIF3H promotes aggressiveness of esophageal squamous cell carcinoma by modulating Snail stability. J Exp Clin Cancer Res. 2020;39:175
120. Ma X, Qi W, Yang F, Pan H. Deubiquitinase JOSD1 promotes tumor progression via stabilizing Snail in lung adenocarcinoma. Am J Cancer Res. 2022;12:2323-36
121. Zhu R, Liu Y, Zhou H, Li L, Li Y, Ding F. et al. Deubiquitinating enzyme PSMD14 promotes tumor metastasis through stabilizing SNAIL in human esophageal squamous cell carcinoma. Cancer Lett. 2018;418:125-34
122. Meng J, Ai X, Lei Y, Zhong W, Qian B, Qiao K. et al. USP5 promotes epithelial-mesenchymal transition by stabilizing SLUG in hepatocellular carcinoma. Theranostics. 2019;9:573-87
123. Ouchida AT, Kacal M, Zheng A, Ambroise G, Zhang B, Norberg E. et al. USP10 regulates the stability of the EMT-transcription factor Slug/SNAI2. Biochem Biophys Res Commun. 2018;502:429-34
124. Li W, Shen M, Jiang YZ, Zhang R, Zheng H, Wei Y. et al. Deubiquitinase USP20 promotes breast cancer metastasis by stabilizing SNAI2. Genes Dev. 2020;34:1310-5
125. Lin Y, Wang Y, Shi Q, Yu Q, Liu C, Feng J. et al. Stabilization of the transcription factors slug and twist by the deubiquitinase dub3 is a key requirement for tumor metastasis. Oncotarget. 2017;8:75127-40
126. Wei Z, Babkirk K, Chen S, Pei M. Epithelial-to-mesenchymal transition transcription factors: New strategies for mesenchymal tissue regeneration. Cytokine Growth Factor Rev. 2025;83:99-124
127. Chu PY, Hu FW, Yu CC, Tsai LL, Yu CH, Wu BC. et al. Epithelial-mesenchymal transition transcription factor ZEB1/ZEB2 co-expression predicts poor prognosis and maintains tumor-initiating properties in head and neck cancer. Oral Oncol. 2013;49:34-41
128. Zhao X, Han Z, Liu R, Li Z, Mei L, Jin Y. FBXO11 Mediates Ubiquitination of ZEB1 and Modulates Epithelial-to-Mesenchymal Transition in Lung Cancer Cells. Cancers (Basel). 2024;16:3269
129. Li X, Yuan J, Song C, Lei Y, Xu J, Zhang G. et al. Deubiquitinase USP39 and E3 ligase TRIM26 balance the level of ZEB1 ubiquitination and thereby determine the progression of hepatocellular carcinoma. Cell Death Differ. 2021;28:2315-32
130. Chen A, Wong CS, Liu MC, House CM, Sceneay J, Bowtell DD. et al. The ubiquitin ligase Siah is a novel regulator of Zeb1 in breast cancer. Oncotarget. 2015;6:862-73
131. Yang H, Lu X, Liu Z, Chen L, Xu Y, Wang Y. et al. FBXW7 suppresses epithelial-mesenchymal transition, stemness and metastatic potential of cholangiocarcinoma cells. Oncotarget. 2015;6:6310-25
132. Li N, Babaei-Jadidi R, Lorenzi F, Spencer-Dene B, Clarke P, Domingo E. et al. An FBXW7-ZEB2 axis links EMT and tumour microenvironment to promote colorectal cancer stem cells and chemoresistance. Oncogenesis. 2019;8:13
133. Wang J, Hu M, Min J, Li X. A positive feedback loop of SRSF9/USP22/ZEB1 promotes the progression of ovarian cancer. Cancer Biol Ther. 2024;25:2427415
134. Li J, Xiao X, Wang H, Wang W, Ou Y, Wang Z. et al. CDK4/6-USP51 axis regulates lung adenocarcinoma metastasis through ZEB1. Cancer Gene Ther. 2022;29:1181-92
135. Liu YJ, Zeng SH, Zhang W, Li JP, Yin Y, Zhuang YW. et al. USP51/ZEB1/ACTA2 axis promotes mesenchymal phenotype in gastric cancer and is associated with low cohesion characteristics. Pharmacol Res. 2023;188:106644
136. Song C, Peng J, Wei Y, Shao J, Chen X, Zhang X. et al. USP18 promotes tumor metastasis in esophageal squamous cell carcinomas via deubiquitinating ZEB1. Exp Cell Res. 2021;409:112884
137. Lin JJ, Lu YC. Ubiquitin-specific protease 21 promotes tumorigenicity and stemness of colorectal cancer by deubiquitinating and stabilizing ZEB1. World J Gastrointest Oncol. 2024;16:1006-18
138. Ye DX, Wang SS, Huang Y, Wang XJ, Chi P. USP43 directly regulates ZEB1 protein, mediating proliferation and metastasis of colorectal cancer. J Cancer. 2021;12:404-16
139. Huang Q, Zheng S, Gu H, Yang Z, Lu Y, Yu X. et al. The deubiquitinase BRCC3 increases the stability of ZEB1 and promotes the proliferation and metastasis of triple-negative breast cancer cells. Acta Biochim Biophys Sin (Shanghai). 2024;56:564-75
140. Sun L, Yu J, Guinney J, Qin B, Sinicrope FA. USP10 Regulates ZEB1 Ubiquitination and Protein Stability to Inhibit ZEB1-Mediated Colorectal Cancer Metastasis. Mol Cancer Res. 2023;21:578-90
141. Qin Q, Xu Y, He T, Qin C, Xu J. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res. 2012;22:90-106
142. Wei C, Liu Y, Liu X, Cheng J, Fu J, Xiao X. et al. The speckle-type POZ protein (SPOP) inhibits breast cancer malignancy by destabilizing TWIST1. Cell Death Discov. 2022;8:389
143. Guo W, You X, Xu D, Zhang Y, Wang Z, Man K. et al. PAQR3 enhances Twist1 degradation to suppress epithelial-mesenchymal transition and metastasis of gastric cancer cells. Carcinogenesis. 2016;37:397-407
144. Zhong J, Ogura K, Wang Z, Inuzuka H. Degradation of the transcription factor Twist, an oncoprotein that promotes cancer metastasis. Discov Med. 2013;15:7-15
145. Yang WH, Su YH, Hsu WH, Wang CC, Arbiser JL, Yang MH. Imipramine blue halts head and neck cancer invasion through promoting F-box and leucine-rich repeat protein 14-mediated Twist1 degradation. Oncogene. 2016;35:2287-98
146. Lee HJ, Li CF, Ruan D, Powers S, Thompson PA, Frohman MA. et al. The DNA Damage Transducer RNF8 Facilitates Cancer Chemoresistance and Progression through Twist Activation. Mol Cell. 2016;63:1021-33
147. Shao J, Feng Q, Jiang W, Yang Y, Liu Z, Li L. et al. E3 ubiquitin ligase RBX1 drives the metastasis of triple negative breast cancer through a FBXO45-TWIST1-dependent degradation mechanism. Aging (Albany NY). 2022;14:5493-510
148. Xu J, Guo R, Wen N, Li L, Yi Y, Chen J. et al. FBXO3 stabilizes USP4 and Twist1 to promote PI3K-mediated breast cancer metastasis. PLoS Biol. 2023;21:e3002446
149. Cai H, Ke ZB, Chen JY, Li XD, Zhu JM, Xue YT. et al. Ubiquitin-specific protease 5 promotes bladder cancer progression through stabilizing Twist1. Oncogene. 2024;43:703-13
150. Zhao B, Huo W, Yu X, Shi X, Lv L, Yang Y. et al. USP13 promotes breast cancer metastasis through FBXL14-induced Twist1 ubiquitination. Cell Oncol (Dordr). 2023;46:717-33
151. Cai X, Feng S, Zhang J, Qiu W, Qian M, Wang Y. USP18 deubiquitinates and stabilizes Twist1 to promote epithelial-mesenchymal transition in glioblastoma cells. Am J Cancer Res. 2020;10:1156-69
152. Guan T, Li M, Song Y, Chen J, Tang J, Zhang C. et al. Phosphorylation of USP29 by CDK1 Governs TWIST1 Stability and Oncogenic Functions. Adv Sci (Weinh). 2023;10:e2205873
153. Zhu Y, Qu C, Hong X, Jia Y, Lin M, Luo Y. et al. Trabid inhibits hepatocellular carcinoma growth and metastasis by cleaving RNF8-induced K63 ubiquitination of Twist1. Cell Death Differ. 2019;26:306-20
154. Zhang Y, Yang Y, Qi X, Cui P, Kang Y, Liu H. et al. SLC14A1 and TGF-beta signaling: a feedback loop driving EMT and colorectal cancer metachronous liver metastasis. J Exp Clin Cancer Res. 2024;43:208
155. Zou J, Zhou X, Ma Y, Yu R. Losartan ameliorates renal interstitial fibrosis through metabolic pathway and Smurfs-TGF-beta/Smad. Biomed Pharmacother. 2022;149:112931
156. Chandhoke AS, Karve K, Dadakhujaev S, Netherton S, Deng L, Bonni S. The ubiquitin ligase Smurf2 suppresses TGFbeta-induced epithelial-mesenchymal transition in a sumoylation-regulated manner. Cell Death Differ. 2016;23:876-88
157. Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem. 2000;275:36818-22
158. Wu N, Jiang M, Liu H, Chu Y, Wang D, Cao J. et al. LINC00941 promotes CRC metastasis through preventing SMAD4 protein degradation and activating the TGF-beta/SMAD2/3 signaling pathway. Cell Death Differ. 2021;28:219-32
159. Chang TM, Fang WY, Hsu HP, Chu PY, Jiang SS, Huang KW. et al. PCK2 promotes invasion and epithelial-to-mesenchymal transition in triple-negative breast cancer by promoting TGF-beta/SMAD3 signaling through inhibiting TRIM67-mediated SMAD3 ubiquitination. Cancer Biol Ther. 2025;26:2478670
160. Wu W, Wang Z, Zhang H, Zhang X, Tian H. circGRHPR inhibits aberrant epithelial-mesenchymal transformation progression of lung epithelial cells associated with idiopathic pulmonary fibrosis. Cell Biol Toxicol. 2024;40:7
161. Li H, Li J, Chen L, Qi S, Yu S, Weng Z. et al. HERC3-Mediated SMAD7 Ubiquitination Degradation Promotes Autophagy-Induced EMT and Chemoresistance in Glioblastoma. Clin Cancer Res. 2019;25:3602-16
162. Zhang Z, He G, Lv Y, Liu Y, Niu Z, Feng Q. et al. HERC3 regulates epithelial-mesenchymal transition by directly ubiquitination degradation EIF5A2 and inhibits metastasis of colorectal cancer. Cell Death Dis. 2022;13:74
163. Chen Y, Wang DD, Wu YP, Su D, Zhou TY, Gai RH. et al. MDM2 promotes epithelial-mesenchymal transition and metastasis of ovarian cancer SKOV3 cells. Br J Cancer. 2017;117:1192-201
164. Zhang Y, Li QS, Liu HL, Tang HT, Yang HL, Wu DQ. et al. MKRN1 promotes colorectal cancer metastasis by activating the TGF-beta signalling pathway through SNIP1 protein degradation. J Exp Clin Cancer Res. 2023;42:219
165. Gai Z, Zhou G, Gui T, Itoh S, Oikawa K, Uetani K. et al. Trps1 haploinsufficiency promotes renal fibrosis by increasing Arkadia expression. J Am Soc Nephrol. 2010;21:1468-76
166. Chen H, Yang T, Lei Z, Wang L, Yang H, Tong X. et al. RNF111/Arkadia is regulated by DNA methylation and affects TGF-beta/Smad signaling associated invasion in NSCLC cells. Lung Cancer. 2015;90:32-40
167. Kim JH, Ham S, Lee Y, Suh GY, Lee YS. TTC3 contributes to TGF-beta(1)-induced epithelial-mesenchymal transition and myofibroblast differentiation, potentially through SMURF2 ubiquitylation and degradation. Cell Death Dis. 2019;10:92
168. Han D, Wang L, Chen B, Zhao W, Liang Y, Li Y. et al. USP1-WDR48 deubiquitinase complex enhances TGF-beta induced epithelial-mesenchymal transition of TNBC cells via stabilizing TAK1. Cell Cycle. 2021;20:320-31
169. Pu JY, Zhang Y, Wang LX, Wang J, Zhou JH. Inhibition of Ubiquitin-specific Protease 4 Attenuates Epithelial-Mesenchymal Transition of Renal Tubular Epithelial Cells via Transforming Growth Factor Beta Receptor Type I. Curr Med Sci. 2022;42:1000-6
170. Qiu C, Liu Y, Mei Y, Zou M, Zhao Z, Ye M. et al. Ubiquitin-speci fi c protease 4 promotes metastasis of hepatocellular carcinoma by increasing TGF-beta signaling-induced epithelial-mesenchymal transition. Aging (Albany NY). 2018;10:2783-99
171. Song C, Liu W, Li J. USP17 is upregulated in osteosarcoma and promotes cell proliferation, metastasis, and epithelial-mesenchymal transition through stabilizing SMAD4. Tumour Biol. 2017;39:1010428317717138
172. Yuan T, Chen Z, Yan F, Qian M, Luo H, Ye S. et al. Deubiquitinating enzyme USP10 promotes hepatocellular carcinoma metastasis through deubiquitinating and stabilizing Smad4 protein. Mol Oncol. 2020;14:197-210
173. Zhang J, van Dinther M, Thorikay M, Gourabi BM, Kruithof BPT, Ten Dijke P. Opposing USP19 splice variants in TGF-beta signaling and TGF-beta-induced epithelial-mesenchymal transition of breast cancer cells. Cell Mol Life Sci. 2023;80:43
174. Zhao Y, Wang X, Wang Q, Deng Y, Li K, Zhang M. et al. USP2a Supports Metastasis by Tuning TGF-beta Signaling. Cell Rep. 2018;22:2442-54
175. Wu X, Liu M, Zhu H, Wang J, Dai W, Li J. et al. Ubiquitin-specific protease 3 promotes cell migration and invasion by interacting with and deubiquitinating SUZ12 in gastric cancer. J Exp Clin Cancer Res. 2019;38:277
176. Xie F, Zhou X, Li H, Su P, Liu S, Li R. et al. USP8 promotes cancer progression and extracellular vesicle-mediated CD8+ T cell exhaustion by deubiquitinating the TGF-beta receptor TbetaRII. EMBO J. 2022;41:e108791
177. Garcia DA, Baek C, Estrada MV, Tysl T, Bennett EJ, Yang J. et al. USP11 Enhances TGFbeta-Induced Epithelial-Mesenchymal Plasticity and Human Breast Cancer Metastasis. Mol Cancer Res. 2018;16:1172-84
178. Kit Leng Lui S, Iyengar PV, Jaynes P, Isa Z, Pang B, Tan TZ. et al. USP26 regulates TGF-beta signaling by deubiquitinating and stabilizing SMAD7. EMBO Rep. 2017;18:797-808
179. Xue W, Yang L, Chen C, Ashrafizadeh M, Tian Y, Sun R. Wnt/beta-catenin-driven EMT regulation in human cancers. Cell Mol Life Sci. 2024;81:79
180. Rocha MR, Castillo-Medina YK, de Lima Coelho BM, Rios LLL, Morgado-Diaz JA. Wnt/beta-catenin pathway as a link between therapy resistance-driven epithelial-mesenchymal transition and stemness in colorectal cancer. Cell Biol Int. 2025;49:154-60
181. Silva-Garcia O, Valdez-Alarcon JJ, Baizabal-Aguirre VM. Wnt/beta-Catenin Signaling as a Molecular Target by Pathogenic Bacteria. Front Immunol. 2019;10:2135
182. Giebel N, de Jaime-Soguero A, Garcia Del Arco A, Landry JJM, Tietje M, Villacorta L. et al. USP42 protects ZNRF3/RNF43 from R-spondin-dependent clearance and inhibits Wnt signalling. EMBO Rep. 2021;22:e51415
183. Zhu L, Shi H, Li P, Zhang P. RNF43 Suppressed Triple-Negative Breast Cancer Progression by Inhibiting Wnt/beta-Catenin Pathway. Ann Clin Lab Sci. 2023;53:21-9
184. Peng T, Wonganan O, Zhang Z, Yu J, Xi R, Cao Y. et al. A 2-Benzylmalonate Derivative as STAT3 Inhibitor Suppresses Tumor Growth in Hepatocellular Carcinoma by Upregulating beta-TrCP E3 Ubiquitin Ligase. Int J Mol Sci. 2021;22:3354
185. Xuan Z, Chen C, Sun H, Yang K, Li J, Fu M. et al. NDR1/FBXO11 promotes phosphorylation-mediated ubiquitination of beta-catenin to suppress metastasis in prostate cancer. Int J Biol Sci. 2024;20:4957-77
186. Wang J, He A, Huang Z, Lu H, Xiong J, Wu R. et al. NCAPG promotes epithelial-mesenchymal transition through SIP-dependent ubiquitination regulation of beta-catenin in hepatocellular carcinoma. Cell Signal. 2025;134:111953
187. Chen SY, Liu PQ, Qin DX, Lv H, Zhou HQ, Xu Y. E3 ubiquitin ligase NEDD4L inhibits epithelial-mesenchymal transition by suppressing the beta-catenin/HIF-1alpha positive feedback loop in chronic rhinosinusitis with nasal polyps. Acta Pharmacol Sin. 2024;45:831-43
188. Liu Y, Jiang Q, Liu X, Lin X, Tang Z, Liu C. et al. Cinobufotalin powerfully reversed EBV-miR-BART22-induced cisplatin resistance via stimulating MAP2K4 to antagonize non-muscle myosin heavy chain IIA/glycogen synthase 3beta/beta-catenin signaling pathway. EBioMedicine. 2019;48:386-404
189. Wu H, Lu XX, Wang JR, Yang TY, Li XM, He XS. et al. TRAF6 inhibits colorectal cancer metastasis through regulating selective autophagic CTNNB1/beta-catenin degradation and is targeted for GSK3B/GSK3beta-mediated phosphorylation and degradation. Autophagy. 2019;15:1506-22
190. Liao L, Duan L, Guo Y, Zhou B, Xu Q, Zhang C. et al. TRIM46 upregulates Wnt/beta-catenin signaling by inhibiting Axin1 to mediate hypoxia-induced epithelial-mesenchymal transition in HK2 cells. Mol Cell Biochem. 2022;477:2829-39
191. Zhang L, Qin B, Zou B, Wang S, Quan X, Wang J. et al. Knockdown of TRIM15 inhibits the proliferation, migration and invasion of esophageal squamous cell carcinoma cells through inactivation of the Wnt/beta-catenin signaling pathway. J Bioenerg Biomembr. 2021;53:213-22
192. Deng B, Zhang S, Zhang Y, Miao Y, Meng X, Guo K. Knockdown of Tripartite Motif Containing 28 suppresses the migration, invasion and epithelial-mesenchymal transition in ovarian carcinoma cells through down-regulation of Wnt/beta-catenin signaling pathway. Neoplasma. 2017;64:893-900
193. Kuang J, Li L, Guo L, Su Y, Wang Y, Xu Y. et al. RNF8 promotes epithelial-mesenchymal transition of breast cancer cells. J Exp Clin Cancer Res. 2016;35:88
194. Oh E, Kim JY, Sung D, Cho Y, Lee N, An H. et al. Inhibition of ubiquitin-specific protease 34 (USP34) induces epithelial-mesenchymal transition and promotes stemness in mammary epithelial cells. Cell Signal. 2017;36:230-9
195. Liu J, Wang Y, Zhang S, Sun L, Shi Y. ADAM9 deubiquitination induced by USP22 suppresses proliferation, migration, invasion, and epithelial-mesenchymal transition of trophoblast cells in preeclampsia. Placenta. 2024;146:50-7
196. Xue S, Wu W, Wang Z, Lu G, Sun J, Jin X. et al. USP5 Promotes Metastasis in Non-Small Cell Lung Cancer by Inducing Epithelial-Mesenchymal Transition via Wnt/beta-Catenin Pathway. Front Pharmacol. 2020;11:668
197. Basu B, Karmakar S, Basu M, Ghosh MK. USP7 imparts partial EMT state in colorectal cancer by stabilizing the RNA helicase DDX3X and augmenting Wnt/beta-catenin signaling. Biochim Biophys Acta Mol Cell Res. 2023;1870:119446
198. Zeng Q, Li Z, Zhao X, Guo L, Yu C, Qin J. et al. Ubiquitin-specific protease 7 promotes osteosarcoma cell metastasis by inducing epithelial-mesenchymal transition. Oncol Rep. 2019;41:543-51
199. Zhou M, Gao Y, Wu S, Chen J, Ding J, Wang Y. et al. Ubiquitin-specific protease 7 prevents recurrent spontaneous abortion by targeting enhancer of zeste homolog 2 to regulate trophoblast proliferation, apoptosis, migration, and invasion through the Wnt/beta-catenin pathwaydagger. Biol Reprod. 2023;109:204-14
200. Wang A, Wang Y, Ma Q, Chen X. The carcinogenesis of esophageal squamous cell cancer is positively regulated by USP13 through WISP1 deubiquitination. Biofactors. 2025;51:e2139
201. Zhong M, Zhou L, Fang Z, Yao YY, Zou JP, Xiong JP. et al. Ubiquitin-specific protease 15 contributes to gastric cancer progression by regulating the Wnt/beta-catenin signaling pathway. World J Gastroenterol. 2021;27:4221-35
202. Liu Y, Ma J, Lu S, He P, Dong W. USP25 promotes hepatocellular carcinoma progression by interacting with TRIM21 via the Wnt/beta-catenin signaling pathway. Chin Med J (Engl). 2023;136:2229-42
203. Zhu S, Hou S, Lu Y, Sheng W, Cui Z, Dong T. et al. USP36-Mediated Deubiquitination of DOCK4 Contributes to the Diabetic Renal Tubular Epithelial Cell Injury via Wnt/beta-Catenin Signaling Pathway. Front Cell Dev Biol. 2021;9:638477
204. Wang J, Dong Y, Wei Z, Zhang Y, Wu N, Zhang C. et al. Deubiquitinase OTUB2 promotes intrahepatic cholangiocarcinoma progression by stabilizing the CTNNB1-ZEB1 axis. Exp Cell Res. 2023;425:113537
205. Shi Q, Xue C, Zeng Y, Yuan X, Chu Q, Jiang S. et al. Notch signaling pathway in cancer: from mechanistic insights to targeted therapies. Signal Transduct Target Ther. 2024;9:128
206. Cheng Y, Sun Q, Chen Y, Wang J, Chen Y, Yang Y. et al. DTX3 suppresses bladder cancer cell invasion and metastasis by inhibiting the Notch signaling pathway. Int Immunopharmacol. 2025;153:114529
207. Zhang Z, Lu YX, Liu F, Sang L, Shi C, Xie S. et al. lncRNA BREA2 promotes metastasis by disrupting the WWP2-mediated ubiquitination of Notch1. Proc Natl Acad Sci U S A. 2023;120:e2206694120
208. Ehrmann AS, Quijada-Alamo M, Close V, Guo M, Carracoi V, Perez-Carretero C. et al. NOTCH1 signaling is dysregulated by loss of the deubiquitinase USP28 with del(11q), uncovering USP28 inhibition as novel therapeutic target in CLL. Leukemia. 2025;39:1892-904
209. Ma J, Fang B, Zeng F, Ma C, Pang H, Cheng L. et al. Down-regulation of miR-223 reverses epithelial-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Oncotarget. 2015;6:1740-9
210. Jiang J, Ren H, Xu Y, Wudu M, Wang Q, Liu Z. et al. TRIM67 Promotes the Proliferation, Migration, and Invasion of Non-Small-Cell Lung Cancer by Positively Regulating the Notch Pathway. J Cancer. 2020;11:1240-9
211. Wang H, Huang Q, Xia J, Cheng S, Pei D, Zhang X. et al. The E3 Ligase MIB1 Promotes Proteasomal Degradation of NRF2 and Sensitizes Lung Cancer Cells to Ferroptosis. Mol Cancer Res. 2022;20:253-64
212. Kang AR, An HT, Ko J, Kang S. Ataxin-1 regulates epithelial-mesenchymal transition of cervical cancer cells. Oncotarget. 2017;8:18248-59
213. Chen S, He Z, Cai K, Zhang Y, Zhu H, Pang C. et al. TRIM59/RBPJ positive feedback circuit confers gemcitabine resistance in pancreatic cancer by activating the Notch signaling pathway. Cell Death Dis. 2024;15:932
214. Seymour L, Nuru N, Johnson KR, Gutierrez JMV, Njoku VT, Darie CC. et al. Roles of Post-Translational Modifications of Transcription Factors Involved in Breast Cancer Hypoxia. Molecules. 2025;30:645
215. Hapke RY, Haake SM. Hypoxia-induced epithelial to mesenchymal transition in cancer. Cancer Lett. 2020;487:10-20
216. Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ. et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295-305
217. Tang C, Liu T, Wang K, Wang X, Xu S, He D. et al. Transcriptional regulation of FoxM1 by HIF-1alpha mediates hypoxia-induced EMT in prostate cancer. Oncol Rep. 2019;42:1307-18
218. Paz-Gomez D, Castillejos-Lopez M, Romero Y, Flores-Soto E, Romero-Martinez BS, Vazquez-Perez JA. et al. The Hypoxia-Retinoid Axis in Idiopathic Pulmonary Fibrosis: Multifaceted Etiology and Therapeutic Potential. Int J Mol Sci. 2025 26
219. Brown BA, Myers PJ, Adair SJ, Pitarresi JR, Sah-Teli SK, Campbell LA. et al. A Histone Methylation-MAPK Signaling Axis Drives Durable Epithelial-Mesenchymal Transition in Hypoxic Pancreatic Cancer. Cancer Res. 2024;84:1764-80
220. Liang J, Yao N, Deng B, Li J, Jiang Y, Liu T. et al. GINS1 promotes ZEB1-mediated epithelial-mesenchymal transition and tumor metastasis via beta-catenin signaling in hepatocellular carcinoma. J Cell Physiol. 2024;239:e31237
221. Liu J, Huang B, Xiu Z, Zhou Z, Liu J, Li X. et al. PI3K/Akt/HIF-1alpha signaling pathway mediates HPV-16 oncoprotein-induced expression of EMT-related transcription factors in non-small cell lung cancer cells. J Cancer. 2018;9:3456-66
222. Ma X, Tan Z, Zhang Q, Ma K, Xiao J, Wang X. et al. VHL Ser65 mutations enhance HIF2alpha signaling and promote epithelial-mesenchymal transition of renal cancer cells. Cell Biosci. 2022;12:52
223. Pauzaite T, Nathan JA. A closer look at the role of deubiquitinating enzymes in the Hypoxia Inducible Factor pathway. Biochem Soc Trans. 2024;52:2253-65
224. Schokrpur S, Hu J, Moughon DL, Liu P, Lin LC, Hermann K. et al. CRISPR-Mediated VHL Knockout Generates an Improved Model for Metastatic Renal Cell Carcinoma. Sci Rep. 2016;6:29032
225. Liu X, Zhang X, Peng Z, Li C, Wang Z, Wang C. et al. Deubiquitylase OTUD6B Governs pVHL Stability in an Enzyme-Independent Manner and Suppresses Hepatocellular Carcinoma Metastasis. Adv Sci (Weinh). 2020;7:1902040
226. Guo K, Wei Y, Wang Z, Zhang X, Zhang X, Liu X. et al. Deubiquitylase OTUD6B stabilizes the mutated pVHL and suppresses cell migration in clear cell renal cell carcinoma. Cell Death Dis. 2022;13:97
227. Youssef E, Zhao S, Purcell C, Olson GL, El-Deiry WS. Targeting the SMURF2-HIF1alpha axis: a new frontier in cancer therapy. Front Oncol. 2024;14:1484515
228. Wang Y, Liu X, Wang M, Wang Y, Wang S, Jin L. et al. UBE3B promotes breast cancer progression by antagonizing HIF-2alpha degradation. Oncogene. 2023;42:3394-406
229. Czerwinski A, Sidlowski P, Mooers E, Liu Y, Teng RJ, Pritchard K Jr. et al. Stub1 Acetylation by CBP/p300 Attenuates Chronic Hypoxic-driven Pulmonary Hypertension by Suppressing HIF-2alpha. Am J Respir Cell Mol Biol. 2025;02:0353
230. Gao A, Zhang M, Zhu SQ, Zou S, Chen H, Li X. et al. DNA polymerase iota promotes EMT and metastasis of esophageal squamous cell carcinoma by interacting with USP7 to stabilize HIF-1alpha. Cell Death Dis. 2024;15:171
231. Zhang Z, Yu X, Wen L, Wang J, Li Z, Zhang Y. et al. USP9X integrates TGF-beta and hypoxia signalings to promote ovarian cancer chemoresistance via HIF-2alpha-maintained stemness. Cell Death Dis. 2025;16:312
232. Zhang Y, Beachy PA. Cellular and molecular mechanisms of Hedgehog signalling. Nat Rev Mol Cell Biol. 2023;24:668-87
233. Singh R, Ray A. Therapeutic potential of hedgehog signaling in advanced cancer types. Int Rev Cell Mol Biol. 2024;386:49-80
234. Tang C, Mei L, Pan L, Xiong W, Zhu H, Ruan H. et al. Hedgehog signaling through GLI1 and GLI2 is required for epithelial-mesenchymal transition in human trophoblasts. Biochim Biophys Acta. 2015;1850:1438-48
235. Zhang Q, Jiang J. Regulation of Hedgehog Signal Transduction by Ubiquitination and Deubiquitination. Int J Mol Sci. 2021;22:13338
236. Uspienski T, Niewiadomski P. The Proteasome and Cul3-Dependent Protein Ubiquitination Is Required for Gli Protein-Mediated Activation of Gene Expression in the Hedgehog Pathway. Cells. 2024;13:1496
237. Bordin F, Terriaca G, Apostolico A, Di Fiore A, Mir FT, Bellardinelli S. et al. SMURF1 and SMURF2 directly target GLI1 for ubiquitination and proteasome-dependent degradation. Cell Death Discov. 2024;10:498
238. Qin T, Li B, Feng X, Fan S, Liu L, Liu D. et al. Abnormally elevated USP37 expression in breast cancer stem cells regulates stemness, epithelial-mesenchymal transition and cisplatin sensitivity. J Exp Clin Cancer Res. 2018;37:287
239. Wu Q, Liu R, Yang Y, Peng J, Huang J, Li Z. et al. USP5 promotes tumorigenesis by activating Hedgehog/Gli1 signaling pathway in osteosarcoma. Am J Cancer Res. 2024;14:1204-16
240. Zhou Z, Yao X, Li S, Xiong Y, Dong X, Zhao Y. et al. Deubiquitination of Ci/Gli by Usp7/HAUSP Regulates Hedgehog Signaling. Dev Cell. 2015;34:58-72
241. Zhou Z, Yao X, Pang S, Chen P, Jiang W, Shan Z. et al. The deubiquitinase UCHL5/UCH37 positively regulates Hedgehog signaling by deubiquitinating Smoothened. J Mol Cell Biol. 2018;10:243-57
242. Debaugnies M, Rodriguez-Acebes S, Blondeau J, Parent MA, Zocco M, Song Y. et al. RHOJ controls EMT-associated resistance to chemotherapy. Nature. 2023;616:168-75
243. de Santana DA, Braga PR, Camillo-Coutinho CM, Freitas VS, Cury PR, Ribeiro DA. et al. E-CADERIN, N-CADERIN, SLUG, SNAIL, and TWIST contribute to epithelial-mesenchymal transition in salivary gland tumors. J Oral Pathol Med. 2024;53:193-200
244. Yu Q, Zhou BP, Wu Y. The regulation of snail: on the ubiquitin edge. Cancer Cell Microenviron. 2017;4:e1567
245. Wan QK, Li TT, Liu BB, He B. USP5 promotes tumor progression by stabilizing SLUG in bladder cancer. Oncol Lett. 2024;28:572
246. Silvestrini VC, Thome CH, Albuquerque D, de Souza Palma C, Ferreira GA, Lanfredi GP. et al. Proteomics analysis reveals the role of ubiquitin specific protease (USP47) in Epithelial to Mesenchymal Transition (EMT) induced by TGFbeta2 in breast cells. J Proteomics. 2020;219:103734
247. Ma X, Wan R, Wen Y, Liu T, Song Y, Zhu Y. Deubiquitinating enzyme OTUD4 regulates metastasis in triple-negative breast cancer by stabilizing Snail1. Exp Cell Res. 2024;434:113864
248. Wu Y, Wang Y, Lin Y, Liu Y, Wang Y, Jia J. et al. Dub3 inhibition suppresses breast cancer invasion and metastasis by promoting Snail1 degradation. Nat Commun. 2017;8:14228
249. Wu L, Zhao N, Zhou Z, Chen J, Han S, Zhang X. et al. PLAGL2 promotes the proliferation and migration of gastric cancer cells via USP37-mediated deubiquitination of Snail1. Theranostics. 2021;11:700-14
250. Parfenyev SE, Daks AA, Shuvalov OY, Fedorova OA, Pestov NB, Korneenko TV. et al. Dualistic role of ZEB1 and ZEB2 in tumor progression. Biol Direct. 2025;20:32
251. Sirera J, Sarlak S, Teisseire M, Carminati A, Nicolini VJ, Savy C. et al. Disrupting USP39 deubiquitinase function impairs the survival and migration of multiple myeloma cells through ZEB1 degradation. J Exp Clin Cancer Res. 2024;43:335
252. Zhang Z, Li J, Ou Y, Yang G, Deng K, Wang Q. et al. CDK4/6 inhibition blocks cancer metastasis through a USP51-ZEB1-dependent deubiquitination mechanism. Signal Transduct Target Ther. 2020;5:25
253. Wang Y, Li C, Jiang T, Yin Y, Wang Y, Zhao H. et al. A comprehensive exploration of twist1 to identify a biomarker for tumor immunity and prognosis in pan-cancer. Medicine (Baltimore). 2024;103:e37790
254. Li F, Hu Q, He T, Xu J, Yi Y, Xie S. et al. The Deubiquitinase USP4 Stabilizes Twist1 Protein to Promote Lung Cancer Cell Stemness. Cancers (Basel). 2020;12:1582
255. Chen J, Wu Z, Deng W, Tang M, Wu L, Lin N. et al. USP51 promotes non-small cell lung carcinoma cell stemness by deubiquitinating TWIST1. J Transl Med. 2023;21:453
256. Wang X, Eichhorn PJA, Thiery JP. TGF-beta, EMT, and resistance to anti-cancer treatment. Semin Cancer Biol. 2023;97:1-11
257. Tolue Ghasaban F, Moghbeli M. Long non-coding RNAs as the pivotal regulators of epithelial mesenchymal transition through WNT/beta-catenin signaling pathway in tumor cells. Pathol Res Pract. 2024;263:155683
258. Sun L, Xing J, Zhou X, Song X, Gao S. Wnt/beta-catenin signalling, epithelial-mesenchymal transition and crosslink signalling in colorectal cancer cells. Biomed Pharmacother. 2024;175:116685
259. Gu Y, Yang R, Zhang Y, Guo M, Takehiro K, Zhan M. et al. Molecular mechanisms and therapeutic strategies in overcoming chemotherapy resistance in cancer. Mol Biomed. 2025;6:2
260. De Las Rivas J, Brozovic A, Izraely S, Casas-Pais A, Witz IP, Figueroa A. Cancer drug resistance induced by EMT: novel therapeutic strategies. Arch Toxicol. 2021;95:2279-97
261. Umar H, Wahab HA, Attiq A, Amjad MW, Bukhari SNA, Ahmad W. Platinum-based targeted chemotherapies and reversal of cisplatin resistance in non-small cell lung cancer (NSCLC). Mutat Res. 2024;828:111856
262. Kurokawa M, Ise N, Omi K, Goishi K, Higashiyama S. Cisplatin influences acquisition of resistance to molecular-targeted agents through epithelial-mesenchymal transition-like changes. Cancer Sci. 2013;104:904-11
263. Chen R, Wang J, Huang S, Hu X, He X, Zhang T. et al. Carnitine palmitoyltransferase 1C promotes EMT-associated cisplatin resistance in non-small cell lung cancer cells. Cancer Biol Med. 2025;22:48-66
264. Feng S, Yang G, Yang H, Liang Z, Zhang R, Fan Y. et al. NEDD4 is involved in acquisition of epithelial-mesenchymal transition in cisplatin-resistant nasopharyngeal carcinoma cells. Cell Cycle. 2017;16:869-78
265. Xiao G, Li Y, Wang M, Li X, Qin S, Sun X. et al. FBXW7 suppresses epithelial-mesenchymal transition and chemo-resistance of non-small-cell lung cancer cells by targeting snai1 for ubiquitin-dependent degradation. Cell Prolif. 2018;51:e12473
266. Su Y, Ma Y, Wang Y, Xu P, Guo M, Cao H. et al. Downregulated CCND3 Is a Key Event Driving Lung Adenocarcinoma Metastasis during Acquired Cisplatin Resistance. International journal of biological sciences. 2025;21:708-24
267. Liu Z, Wu Y, Tao Z, Ma L. E3 ubiquitin ligase Hakai regulates cell growth and invasion, and increases the chemosensitivity to cisplatin in non-small-cell lung cancer cells. International journal of molecular medicine. 2018;42:1145-51
268. Jing C, Li X, Zhou M, Zhang S, Lai Q, Liu D. et al. The PSMD14 inhibitor Thiolutin as a novel therapeutic approach for esophageal squamous cell carcinoma through facilitating SNAIL degradation. Theranostics. 2021;11:5847-62
269. Zhou F, Du C, Xu D, Lu J, Zhou L, Wu C. et al. Knockdown of ubiquitin-specific protease 51 attenuates cisplatin resistance in lung cancer through ubiquitination of zinc-finger E-box binding homeobox 1. Molecular medicine reports. 2020;22:1382-90
270. Dou N, Hu Q, Li L, Wu Q, Li Y, Gao Y. USP32 promotes tumorigenesis and chemoresistance in gastric carcinoma via upregulation of SMAD2. International Journal of Biological Sciences. 2020;16:1648-57
271. Liu JH, Yang HL, Deng ST, Hu Z, Chen WF, Yan WW. et al. The small molecule chemical compound cinobufotalin attenuates resistance to DDP by inducing ENKUR expression to suppress MYH9-mediated c-Myc deubiquitination in lung adenocarcinoma. Acta Pharmacol Sin. 2022;43:2687-95
272. Li J, Gao R, Zhang J. USP22 Contributes to Chemoresistance, Stemness, and EMT Phenotype of Triple-Negative Breast Cancer Cells by egulating the Warburg Effect via c-Myc Deubiquitination. Clinical breast cancer. 2023;23:162-75
273. Yun X, Zhang K, Wang J, Pangeni RP, Yang L, Bonner M. et al. Targeting USP22 Suppresses Tumorigenicity and Enhances Cisplatin Sensitivity Through ALDH1A3 Downregulation in Cancer-Initiating Cells from Lung Adenocarcinoma. Molecular cancer research: MCR. 2018;16:1161-71
274. Capdevila J, Elez E, Peralta S, Macarulla T, Ramos FJ, Tabernero J. Oxaliplatin-based chemotherapy in the management of colorectal cancer. Expert Rev Anticancer Ther. 2008;8:1223-36
275. Zhu W, Wu C, Liu Z, Zhao S, Huang J. OTU deubiquitinase, ubiquitin aldehyde binding 2 (OTUB2) modulates the stemness feature, chemoresistance, and epithelial-mesenchymal transition of colon cancer via regulating GINS complex subunit 1 (GINS1) expression. Cell Commun Signal. 2024;22:420
276. Bisht A, Avinash D, Sahu KK, Patel P, Das Gupta G, Kurmi BD. A comprehensive review on doxorubicin: mechanisms, toxicity, clinical trials, combination therapies and nanoformulations in breast cancer. Drug Deliv Transl Res. 2025;15:102-33
277. Mirzaei S, Abadi AJ, Gholami MH, Hashemi F, Zabolian A, Hushmandi K. et al. The involvement of epithelial-to-mesenchymal transition in doxorubicin resistance: Possible molecular targets. Eur J Pharmacol. 2021;908:174344
278. Long L, Xiang H, Liu J, Zhang Z, Sun L. ZEB1 mediates doxorubicin (Dox) resistance and mesenchymal characteristics of hepatocarcinoma cells. Exp Mol Pathol. 2019;106:116-22
279. Han X, Liu F, Zhang C, Ren Z, Li L, Wang G. Corrigendum to: SIAH1/ZEB1/IL-6 axis is involved in doxorubicin (Dox) resistance of osteosarcoma cells. Biol Chem. 2019;400:1241
280. Yu J, Zhang W, Gao F, Liu YX, Chen ZY, Cheng LY. et al. FBW7 increases chemosensitivity in hepatocellular carcinoma cells through suppression of epithelial-mesenchymal transition. Hepatobiliary Pancreat Dis Int. 2014;13:184-91
281. Ding J, Zhao Z, Song J, Luo B, Huang L. MiR-223 promotes the doxorubicin resistance of colorectal cancer cells via regulating epithelial-mesenchymal transition by targeting FBXW7. Acta Biochim Biophys Sin (Shanghai). 2018;50:597-604
282. Xu X, Liu J, Shen C, Ding L, Zhong F, Ouyang Y. et al. The role of ubiquitin-specific protease 14 (USP14) in cell adhesion-mediated drug resistance (CAM-DR) of multiple myeloma cells. European journal of haematology. 2017;98:4-12
283. Wu Y, Zhang Y, Wang D, Zhang Y, Zhang J, Zhang Y. et al. USP29 enhances chemotherapy-induced stemness in non-small cell lung cancer via stabilizing Snail1 in response to oxidative stress. Cell death & disease. 2020;11:796
284. Tu X, Li C, Sun W, Tian X, Li Q, Wang S. et al. Suppression of Cancer Cell Stemness and Drug Resistance via MYC Destabilization by Deubiquitinase USP45 Inhibition with a Natural Small Molecule. Cancers (Basel). 2023;15:930
285. Jia Y, Xie J. Promising molecular mechanisms responsible for gemcitabine resistance in cancer. Genes Dis. 2015;2:299-306
286. Du Y, Yang Z, Shi H, Chen Z, Chen R, Zhou F. et al. E3 ubiquitin ligase UBR5 promotes gemcitabine resistance in pancreatic cancer by inducing O-GlcNAcylation-mediated EMT via destabilization of OGA. Cell Death Dis. 2024;15:340
287. Zhang J, Chen C, Geng Q, Li H, Wu M, Chan B. et al. ZNF263 cooperates with ZNF31 to promote the drug resistance and EMT of pancreatic cancer through transactivating RNF126. J Cell Physiol. 2024;239:e31259
288. Zhu J, Zhao J, Luo C, Zhu Z, Peng X, Zhu X. et al. FAT10 promotes chemotherapeutic resistance in pancreatic cancer by inducing epithelial-mesenchymal transition via stabilization of FOXM1 expression. Cell Death Dis. 2022;13:497
289. Zhang WL, Zhang JH, Wu XZ, Yan T, Lv W. miR-15b promotes epithelial-mesenchymal transition by inhibiting SMURF2 in pancreatic cancer. International journal of oncology. 2015;47:1043-53
290. Ashrafizadeh M, Zarrabi A, Hushmandi K, Kalantari M, Mohammadinejad R, Javaheri T. et al. Association of the Epithelial-Mesenchymal Transition (EMT) with Cisplatin Resistance. Int J Mol Sci. 2020;21:4002
291. Qin Y, Han S, Yu Y, Qi D, Ran M, Yang M. et al. Lenvatinib in hepatocellular carcinoma: Resistance mechanisms and strategies for improved efficacy. Liver Int. 2024;44:1808-31
292. Wu H, Gao W, Chen P, Wei Y, Zhao H, Wang F. Research progress of drug resistance mechanism of temozolomide in the treatment of glioblastoma. Heliyon. 2024;10:e39984
293. Lee SY. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016;3:198-210
294. Kuang J, Duan T, Gao C, Liu C, Chen S, Zhu LY. et al. RNF8 depletion attenuates hepatocellular carcinoma progression by inhibiting epithelial-mesenchymal transition and enhancing drug sensitivity. Acta biochimica et biophysica Sinica. 2023;55:661-71
295. Huang S, Qin X, Fu S, Hu J, Jiang Z, Hu M. et al. STAMBPL1/TRIM21 Balances AXL Stability Impacting Mesenchymal Phenotype and Immune Response in KIRC. Adv Sci (Weinh). 2024: e2405083.
296. Han S, Wang R, Zhang Y, Li X, Gan Y, Gao F. et al. The role of ubiquitination and deubiquitination in tumor invasion and metastasis. Int J Biol Sci. 2022;18:2292-303
297. Sampson C, Wang Q, Otkur W, Zhao H, Lu Y, Liu X. et al. The roles of E3 ubiquitin ligases in cancer progression and targeted therapy. Clin Transl Med. 2023;13:e1204
298. Martinez-Iglesias O, Casas-Pais A, Castosa R, Diaz-Diaz A, Roca-Lema D, Concha A. et al. Hakin-1, a New Specific Small-Molecule Inhibitor for the E3 Ubiquitin-Ligase Hakai, Inhibits Carcinoma Growth and Progression. Cancers (Basel). 2020;12:1340
299. Zhu Y, Kang N, Zhang L, Tao J, Xue W, Li H. et al. Targeting and degradation of OTUB1 by Erianin for antimetastasis in esophageal squamous cell carcinoma. Phytomedicine. 2024;135:155969
300. Li J, Kou Y, Zhang X, Xiao X, Ou Y, Cao L. et al. Biochanin A inhibits lung adenocarcinoma progression by targeting ZEB1. Discover oncology. 2022;13:138
301. Hou R, Li Y, Luo X, Zhang W, Yang H, Zhang Y. et al. ENKUR expression induced by chemically synthesized cinobufotalin suppresses malignant activities of hepatocellular carcinoma by modulating beta-catenin/c-Jun/MYH9/USP7/c-Myc axis. Int J Biol Sci. 2022;18:2553-67
302. Xu G, Zhou Q, Qi J, Li Z, Yin L, Li Z. et al. Resveratrol-derived inhibitors of the E3 ubiquitin ligase PELI1 inhibit the metastasis of triple-negative breast cancer. Eur J Med Chem. 2024;265:116060
303. Li F, Zhou Y, Lin X, Zhang Y, Hu Q, Zhao E. et al. A novel USP4 inhibitor that suppresses colorectal cancer stemness by promoting beta-catenin and Twist1 degradation. J Transl Med. 2025;23:114
304. Liu Z, Zhao T, Li Z, Sun K, Fu Y, Cheng T. et al. Discovery of [1,2,3]triazolo[4,5-d]pyrimidine derivatives as highly potent, selective, and cellularly active USP28 inhibitors. Acta Pharm Sin B. 2020;10:1476-91
305. Madhukar G, Haque MA, Khan S, Kim JJ, Danishuddin. E3 ubiquitin ligases and their therapeutic potential in disease Management. Biochem Pharmacol. 2025;236:116875
306. Jiang H, Xiong H, Gu SX, Wang M. E3 ligase ligand optimization of Clinical PROTACs. Front Chem. 2023;11:1098331
307. Wang B, Cao S, Zheng N. Emerging strategies for prospective discovery of molecular glue degraders. Curr Opin Struct Biol. 2024;86:102811
308. Ramachandran S, Ciulli A. Building ubiquitination machineries: E3 ligase multi-subunit assembly and substrate targeting by PROTACs and molecular glues. Curr Opin Struct Biol. 2021;67:110-9
309. Kaur SD, Bedi N, Kumar D, Kapoor DN. PROTACs: Promising approach for anticancer therapy. Cancer Lett. 2023;556:216065
310. Gao X, III HAB, Vuky J, Dreicer R, Sartor AO, Sternberg CN. et al. Phase 1/2 study of ARV-110, an androgen receptor (AR) PROTAC degrader, in metastatic castration-resistant prostate cancer (mCRPC). Journal of Clinical Oncology. 2022;40:17
311. Shore ND, Shen J, Devitt ME, Lu H, Alicea J, Parameswaran J. et al. Phase 1b study of bavdegalutamide, an androgen receptor PROTAC degrader, combined with abiraterone in patients with metastatic prostate cancer. Journal of Clinical Oncology. 2022;40:TPS5106
312. Petrylak DP, Gao X, Vogelzang NJ, Garfield MH, Taylor I, Moore MD. et al. First-in-human phase I study of ARV-110, an androgen receptor (AR) PROTAC degrader in patients (pts) with metastatic castrate-resistant prostate cancer (mCRPC) following enzalutamide (ENZ) and/or abiraterone (ABI). Journal of Clinical Oncology. 2020;38:3500
313. Hamilton EP, Schott AF, Nanda R, Lu H, Keung CF, Gedrich R. et al. ARV-471, an estrogen receptor (ER) PROTAC degrader, combined with palbociclib in advanced ER+/human epidermal growth factor receptor 2-negative (HER2-) breast cancer: Phase 1b cohort (part C) of a phase 1/2 study. Journal of Clinical Oncology. 2022;40:TPS1120
314. Campone M, Ma CX, Laurentiis MD, Iwata H, Hurvitz SA, Wander SA. et al. VERITAC-2: A global, randomized phase 3 study of ARV-471, a proteolysis targeting chimera (PROTAC) estrogen receptor (ER) degrader, vs fulvestrant in ER+/human epidermal growth factor receptor 2 (HER2)- advanced breast cancer. Journal of Clinical Oncology. 2023;41:TPS1122
315. He X, Weng Z, Zou Y. Progress in the controllability technology of PROTAC. Eur J Med Chem. 2024;265:116096
316. Li Y, Wu Y, Gao S, Sun T, Jiang C. PROTAC delivery in tumor immunotherapy: Where are we and where are we going? J Control Release. 2025;378:116-44
317. Hu Y, Yan Y, Wang J, Hou J, Lin Q. Molecular glue degrader for tumor treatment. Front Oncol. 2024;14:1512666
318. Surka C, Jin L, Mbong N, Lu CC, Jang IS, Rychak E. et al. CC-90009, a novel cereblon E3 ligase modulator, targets acute myeloid leukemia blasts and leukemia stem cells. Blood. 2021;137:661-77
319. Feng Y, Zhang Z, Tang W, Dai Y. Gel/hydrogel-based in situ biomaterial platforms for cancer postoperative treatment and recovery. Exploration (Beijing). 2023;3:20220173
320. Chinnam M, Xu C, Lama R, Zhang X, Cedeno CD, Wang Y. et al. MDM2 E3 ligase activity is essential for p53 regulation and cell cycle integrity. PLoS Genet. 2022;18:e1010171
321. Konopleva M, Martinelli G, Daver N, Papayannidis C, Wei A, Higgins B. et al. MDM2 inhibition: an important step forward in cancer therapy. Leukemia. 2020;34:2858-74
322. Zhu H, Gao H, Ji Y, Zhou Q, Du Z, Tian L. et al. Targeting p53-MDM2 interaction by small-molecule inhibitors: learning from MDM2 inhibitors in clinical trials. J Hematol Oncol. 2022;15:91
323. Wang S, Chen FE. Small-molecule MDM2 inhibitors in clinical trials for cancer therapy. Eur J Med Chem. 2022;236:114334
324. Andreeff M, Kelly KR, Yee K, Assouline S, Strair R, Popplewell L. et al. Results of the Phase I Trial of RG7112, a Small-Molecule MDM2 Antagonist in Leukemia. Clin Cancer Res. 2016;22:868-76
325. Yee K, Martinelli G, Assouline S, Kasner M, Vey N, Kelly KR. et al. Phase 1b Study Of The MDM2 Antagonist RG7112 In Combination With 2 Doses/Schedules Of Cytarabine. Blood. 2013;122:498
326. Kurzrock R, Blay J-Y, Nguyen BB, Wagner AJ, Maki RG, Schwartz GK. et al. A phase I study of MDM2 antagonist RG7112 in patients (pts) with relapsed/refractory solid tumors. Journal of Clinical Oncology. 2012;30:e13600
327. Chawla SP, Blay J-Y, Italiano A, Gutierrez M, Cesne AL, Gomez-Roca CA. et al. Phase Ib study of RG7112 with doxorubicin (D) in advanced soft tissue sarcoma (ASTS). Journal of Clinical Oncology. 2013;31:10514
328. Zhang X, Wen X, Peng R, Pan Q, Weng D, Ma Y. et al. A first-in-human phase I study of a novel MDM2/p53 inhibitor alrizomadlin in advanced solid tumors. ESMO Open. 2024;9:103636
329. Zhang X, Wen X, Yang C, Zeng S, Men L, Wang H. et al. A phase I study of a novel MDM2-P53 antagonist APG-115 in Chinese patients with advanced soft tissue sarcomas. Journal of Clinical Oncology. 2019;37:3124
330. Liang L, Chen Z, Lei D, Mo C, Lan D, Ke J. et al. APG-115 Induces SLC7A11-Mediated Ferroptosis and Upregulates PD-L1 Expression in Thyroid Cancer. ACS Omega. 2025;10:31099-107
331. Fang Y, Liao G, Yu B. Small-molecule MDM2/X inhibitors and PROTAC degraders for cancer therapy: advances and perspectives. Acta Pharm Sin B. 2020;10:1253-78
332. Chargari C, Leteur C, Angevin E, Bashir T, Schoentjes B, Arts J. et al. Preclinical assessment of JNJ-26854165 (Serdemetan), a novel tryptamine compound with radiosensitizing activity in vitro and in tumor xenografts. Cancer Lett. 2011;312:209-18
333. Saleh MN, Patel MR, Bauer TM, Goel S, Falchook GS, Shapiro GI. et al. Phase 1 Trial of ALRN-6924, a Dual Inhibitor of MDMX and MDM2, in Patients with Solid Tumors and Lymphomas Bearing Wild-type TP53. Clin Cancer Res. 2021;27:5236-47
334. Daver NG, Dail M, Garcia JS, Jonas BA, Yee KWL, Kelly KR. et al. Venetoclax and idasanutlin in relapsed/refractory AML: a nonrandomized, open-label phase 1b trial. Blood. 2023;141:1265-76
335. Ravandi F, Gojo I, Patnaik MM, Minden MD, Kantarjian H, Johnson-Levonas AO. et al. A phase I trial of the human double minute 2 inhibitor (MK-8242) in patients with refractory/recurrent acute myelogenous leukemia (AML). Leuk Res. 2016;48:92-100
336. de Weger VA, de Jonge M, Langenberg MHG, Schellens JHM, Lolkema M, Varga A. et al. A phase I study of the HDM2 antagonist SAR405838 combined with the MEK inhibitor pimasertib in patients with advanced solid tumours. Br J Cancer. 2019;120:286-93
337. Uy GL, Assouline S, Young AM, Blotner S, Higgins B, Chen LC. et al. Phase 1 study of the MDM2 antagonist RO6839921 in patients with acute myeloid leukemia. Invest New Drugs. 2020;38:1430-41
338. Moschos SJ, Sandhu S, Lewis KD, Sullivan RJ, Puzanov I, Johnson DB. et al. Targeting wild-type TP53 using AMG 232 in combination with MAPK inhibition in Metastatic Melanoma; a phase 1 study. Invest New Drugs. 2022;40:1051-65
339. Stein EM, DeAngelo DJ, Chromik J, Chatterjee M, Bauer S, Lin CC. et al. Results from a First-in-Human Phase I Study of Siremadlin (HDM201) in Patients with Advanced Wild-Type TP53 Solid Tumors and Acute Leukemia. Clin Cancer Res. 2022;28:870-81
340. Sekiguchi N, Kasahara S, Miyamoto T, Kiguchi T, Ohno H, Takagi T. et al. Phase I dose-escalation study of milademetan in patients with relapsed or refractory acute myeloid leukemia. Int J Hematol. 2023;117:68-77
341. Bauer S, Demetri GD, Halilovic E, Dummer R, Meille C, Tan DSW. et al. Pharmacokinetic-pharmacodynamic guided optimisation of dose and schedule of CGM097, an HDM2 inhibitor, in preclinical and clinical studies. Br J Cancer. 2021;125:687-98
342. Haronikova L, Bonczek O, Zatloukalova P, Kokas-Zavadil F, Kucerikova M, Coates PJ. et al. Resistance mechanisms to inhibitors of p53-MDM2 interactions in cancer therapy: can we overcome them? Cell Mol Biol Lett. 2021;26:53
343. Cinatl J, Speidel D, Hardcastle I, Michaelis M. Resistance acquisition to MDM2 inhibitors. Biochem Soc Trans. 2014;42:752-7
344. Thomasova D, Mulay SR, Bruns H, Anders HJ. p53-independent roles of MDM2 in NF-kappaB signaling: implications for cancer therapy, wound healing, and autoimmune diseases. Neoplasia. 2012;14:1097-101
345. Alaseem AM. Advancements in MDM2 inhibition: Clinical and pre-clinical investigations of combination therapeutic regimens. Saudi Pharm J. 2023;31:101790
346. Cossu F, Milani M, Mastrangelo E, Lecis D. Targeting the BIR Domains of Inhibitor of Apoptosis (IAP) Proteins in Cancer Treatment. Comput Struct Biotechnol J. 2019;17:142-50
347. Flygare JA, Beresini M, Budha N, Chan H, Chan IT, Cheeti S. et al. Discovery of a potent small-molecule antagonist of inhibitor of apoptosis (IAP) proteins and clinical candidate for the treatment of cancer (GDC-0152). J Med Chem. 2012;55:4101-13
348. Lin IL, Lin YT, Chang YC, Kondapuram SK, Lin KH, Chen PC. et al. The SMAC mimetic GDC-0152 is a direct ABCB1-ATPase activity modulator and BIRC5 expression suppressor in cancer cells. Toxicol Appl Pharmacol. 2024;485:116888
349. Morita S, Minami H, Mitsuma A, Toyoda M, Kiyota N, Ando Y. A phase I study of LCL161, a novel oral pan-inhibitor of apoptosis protein (IAP) antagonist, in Japanese patients with advanced solid tumors. Asia Pac J Clin Oncol. 2022;18:e427-e34
350. Johnson ML, Patel MR, Aljumaily R, Jones SF, Burris Iii HA, Spigel DR. A Phase Ib Dose-Escalation Study of LCL161 Plus Oral Topotecan for Patients With Relapsed/Refractory Small Cell Lung Cancer and Select Gynecologic Malignancies. Oncologist. 2023;28:640
351. Pemmaraju N, Carter BZ, Bose P, Jain N, Kadia TM, Garcia-Manero G. et al. Final results of a phase 2 clinical trial of LCL161, an oral SMAC mimetic for patients with myelofibrosis. Blood Adv. 2021;5:3163-73
352. Li Q, Zhang W. Progress in Anticancer Drug Development Targeting Ubiquitination-Related Factors. Int J Mol Sci. 2022;23:15104
353. Tao Y, Sun XS, Pointreau Y, Le Tourneau C, Sire C, Kaminsky MC. et al. Extended follow-up of a phase 2 trial of xevinapant plus chemoradiotherapy in high-risk locally advanced squamous cell carcinoma of the head and neck: a randomised clinical trial. Eur J Cancer. 2023;183:24-37
354. Noonan AM, Bunch KP, Chen JQ, Herrmann MA, Lee JM, Kohn EC. et al. Pharmacodynamic markers and clinical results from the phase 2 study of the SMAC mimetic birinapant in women with relapsed platinum-resistant or -refractory epithelial ovarian cancer. Cancer. 2016;122:588-97
355. Ye Q, Zhuang XZ, Li J, Zhou X. Targeting the inhibitors of apoptosis proteins (IAPs) to combat drug resistance in cancers. Front Pharmacol. 2025;16:1562167
356. Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018;17:57-78
357. Xian Y, Ye J, Tang Y, Zhang N, Peng C, Huang W. et al. Deubiquitinases as novel therapeutic targets for diseases. MedComm (2020). 2024;5:e70036
358. Lei H, Wang J, Hu J, Zhu Q, Wu Y. Deubiquitinases in hematological malignancies. Biomark Res. 2021;9:66
359. Fan YH, Cheng J, Vasudevan SA, Dou J, Zhang H, Patel RH. et al. USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis. 2013;4:e867
360. Zhang W, Zhang J, Xu C, Zhang S, Bian S, Jiang F. et al. Ubiquitin-specific protease 7 is a drug-able target that promotes hepatocellular carcinoma and chemoresistance. Cancer Cell Int. 2020;20:28
361. Chen H, Zhu X, Sun R, Ma P, Zhang E, Wang Z. et al. Ubiquitin-specific protease 7 is a druggable target that is essential for pancreatic cancer growth and chemoresistance. Invest New Drugs. 2020;38:1707-16
362. Lei H, Xu HZ, Shan HZ, Liu M, Lu Y, Fang ZX. et al. Targeting USP47 overcomes tyrosine kinase inhibitor resistance and eradicates leukemia stem/progenitor cells in chronic myelogenous leukemia. Nat Commun. 2021;12:51
363. Varca AC, Casalena D, Chan WC, Hu B, Magin RS, Roberts RM. et al. Identification and validation of selective deubiquitinase inhibitors. Cell Chem Biol. 2021;28:1758-71
364. Gao H, Xi Z, Dai J, Xue J, Guan X, Zhao L. et al. Drug resistance mechanisms and treatment strategies mediated by Ubiquitin-Specific Proteases (USPs) in cancers: new directions and therapeutic options. Mol Cancer. 2024;23:88
365. Bakkar M, Khalil S, Bhayekar K, Kushwaha ND, Samarbakhsh A, Dorandish S. et al. Ubiquitin-Specific Protease Inhibitors for Cancer Therapy: Recent Advances and Future Prospects. Biomolecules. 2025;15:240
366. Paulus A, Akhtar S, Samuel K, Yousaf H, Cogen D, Jiang J. et al. VLX1570, a First in Class Dub Inhibitor, Modulates BCR Signaling and CXCR4 Expression and Demonstrates Significant In Vivo Antitumor Activity in a Murine Model of Human Waldenstrom Macroglobulinemia. Blood. 2015;126:703
367. Rowinsky EK, Paner A, Berdeja JG, Paba-Prada C, Venugopal P, Porkka K. et al. Phase 1 study of the protein deubiquitinase inhibitor VLX1570 in patients with relapsed and/or refractory multiple myeloma. Invest New Drugs. 2020;38:1448-53
368. Leighl NB, Dent S, Clemons M, Vandenberg TA, Tozer R, Warr DG. et al. A Phase 2 study of perifosine in advanced or metastatic breast cancer. Breast Cancer Res Treat. 2008;108:87-92
369. Becher OJ, Gilheeney SW, Khakoo Y, Lyden DC, Haque S, De Braganca KC. et al. A phase I study of perifosine with temsirolimus for recurrent pediatric solid tumors. Pediatr Blood Cancer. 2017;64:26409
370. Cadzow L, Brenneman J, Tobin E, Sullivan P, Nayak S, Ali JA. et al. The USP1 Inhibitor KSQ-4279 Overcomes PARP Inhibitor Resistance in Homologous Recombination-Deficient Tumors. Cancer Res. 2024;84:3419-34
371. Nie F, Sun X, Sun J, Zhang J, Wang Y. Epithelial-mesenchymal transition in colorectal cancer metastasis and progression: molecular mechanisms and therapeutic strategies. Cell Death Discov. 2025;11:336
372. Huang Y, Hong W, Wei X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J Hematol Oncol. 2022;15:129
Author contact
![]() Corresponding authors: Cefan Zhou (cefanedu.cn) and Jingfeng Tang (tangjingfengedu.cn). National “111” Center for Cellular Regulation and Molecular Pharmaceutics, Key Laboratory of Fermentation Engineering (Ministry of Education), Hubei University of Technology, Wuhan 430074, P. R. China; Tel.: +86 027-5975-0472; Fax: +86 027-5975-0472.
Corresponding authors: Cefan Zhou (cefanedu.cn) and Jingfeng Tang (tangjingfengedu.cn). National “111” Center for Cellular Regulation and Molecular Pharmaceutics, Key Laboratory of Fermentation Engineering (Ministry of Education), Hubei University of Technology, Wuhan 430074, P. R. China; Tel.: +86 027-5975-0472; Fax: +86 027-5975-0472.

 Global reach, higher impact
Global reach, higher impact