10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2025; 21(14):6156-6178. doi:10.7150/ijbs.117838 This issue Cite
Research Paper
Dysregulation of SRSF3/circSAMD4/CIRBP Axis Promotes Iodinated Contrast-induced Acute Kidney Injury
Department of Nephrology, The Second Xiangya Hospital of Central South University; Hunan Key Laboratory of Kidney Disease and Blood Purification, Changsha 410011, China.
Received 2025-5-19; Accepted 2025-9-18; Published 2025-9-29
Abstract
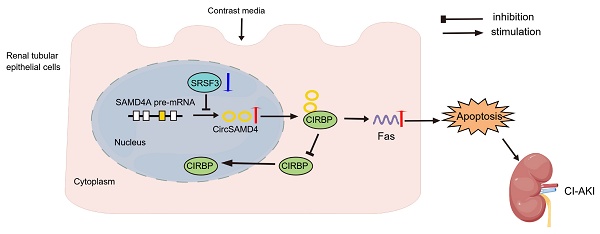
Iodinated contrast agents are a common cause of contrast-induced acute kidney injury (CI-AKI), yet the underlying mechanisms remain unclear. We found that circSAMD4 is markedly upregulated in renal tubular epithelial cells (RTECs) from iohexol-induced CI-AKI mice and patients diagnosed with acute tubular injury (ATI). Silencing circSAMD4 alleviated kidney injury and tubular cell death in CI-AKI mice, whereas its overexpression promoted apoptosis in iohexol-treated RTECs. Mechanistically, circSAMD4 binds to cold-inducible RNA-binding protein (CIRBP) and inhibits its nuclear import. Renal tubule-specific Cirbp deletion mitigated CI-AKI, while CIRBP overexpression abolished the protective effects of circSAMD4 knockdown against iohexol-induced apoptosis. CircSAMD4 upregulation in iohexol-treated RTECs was driven by serine/arginine-rich splicing factor 3 (SRSF3) downregulation. Similar molecular alterations in clinical samples correlated with kidney function decline. These findings identify the SRSF3/circSAMD4/CIRBP axis as a novel pathogenic mechanism in CI-AKI and highlight circSAMD4 as a promising therapeutic target.
Keywords: Iodinated contrast media, acute kidney injury, circSAMD4, CIRBP, SRSF3
Introduction
Iodinated contrast-induced acute kidney injury (CI-AKI) is defined as a rapid decline in renal function following the intravascular administration of iodinated contrast media, after excluding other potential causes [1]. This kind of disease represents a significant healthcare burden, accounting for approximately 11% of hospitalized AKI cases [2]. Large-scale randomized controlled trials have demonstrated that traditional therapies, including sodium bicarbonate and oral N-acetylcysteine have not shown significant benefits in patients with CI-AKI [3]. Previous studies have suggested that iodinated contrast media contribute to renal injury by altering renal hemodynamics and directly inducing damage or apoptosis in renal tubular epithelial cells (RTECs) [4]. However, the precise pathogenic mechanisms underlying CI-AKI remain poorly understood. Thus, elucidating these molecular mechanisms is greatly beneficial for identifying novel therapeutic targets.
Circular RNAs (circRNAs), a class of noncoding RNAs, are characterized by a single-stranded, covalently closed-loop structure with high stability and conservation in mammalian cells [5, 6]. This kind of RNA has diverse biological functions, including acting as microRNA and protein sponges [7-9], serving as scaffolds for enzyme-substrate interactions [10], and functioning as templates for protein translation [11-13]. Emerging evidence indicates that circRNAs play critical regulatory roles in kidney diseases [14-16]. In AKI models induced by sepsis or ischemia-reperfusion injury (IRI), aberrant circRNA expression has been implicated in pathological cascades such as dysregulated intracellular signaling, oxidative stress, apoptosis, excessive inflammation, and tissue damage [16]. Although circRNA dysregulation has also been reported in drug-induced AKI, its pathogenic mechanisms remain largely unexplored [16]. In our previous study using an iohexol-induced CI-AKI rat model, we identified 38 differentially expressed circRNAs compared to saline-treated controls, highlighting altered circRNA-associated competing endogenous RNA (ceRNA) networks [17]. Subsequently, another study reported that circRNA-associated ceRNA networks were also involved in the protective effects of icariin in CI-AKI [18]. However, the critical circRNAs that participate in CI-AKI pathogenesis remain elusive.
Cold-inducible RNA-binding protein (CIRBP) is a glycine-rich RNA-binding protein initially identified for its role in cold stress [19]. CIRBP regulates various cellular processes, including proliferation and apoptosis [20]. Under stress conditions, such as mild cold shock, ultraviolet irradiation, or hypoxia, CIRBP relocates from the nucleus to the cytoplasm, stabilizing specific mRNAs and promoting their translation [21]. Emerging evidence implicates CIRBP in renal pathophysiology and AKI. In IRI-induced AKI, CIRBP exacerbates tissue damage by modulating inflammatory and oxidative responses [22], whereas a CIRBP-derived peptide can attenuate inflammation in septic AKI [23]. Conversely, CIRBP may protect the kidney after hypothermic cardiovascular surgery by suppressing PHD3/HIF-1α signaling [24]. However, its role in CI-AKI remains unexplored.
Serine/arginine-rich splicing factor 3 (SRSF3) is an evolutionarily conserved RNA-binding protein that primarily regulates alternative splicing [25]. In Drosophila, SR proteins have been shown to inhibit circular RNA production, such as Laccase2 circRNA [26]. Recent studies highlight SRSF3 as a key regulator of both N6-methyladenosine (m6A) modifications in long non-coding RNAs and circRNAs [27, 28]. Functionally, SRSF3 has been implicated in liver diseases, ischemic stroke, and acute myocardial infarction [29-31]. However, its role in AKI has not yet been explored.
In this study, we demonstrated that upregulation of circSAMD4 exacerbated iodinated contrast-induced cell apoptosis through inhibiting nuclear import of CIRBP, while downregulation of SRSF3 mediated the increase of circSAMD4 expression in RTECs. Notably, these molecular alterations were also observed in RTECs from patients with biopsy-confirmed acute tubular injury (ATI), where their dysregulated expression correlated with kidney dysfunction. Collectively, these findings establish the dysregulation of the SRSF3/circSAMD4/CIRBP axis as a novel pathogenic mechanism in CI-AKI and highlight circSAMD4 as a potential therapeutic target.
Methods
Establishment of CI-AKI mice model and and cisplatin-induced AKI mice model
The Animal Ethical and Welfare Committee of Central South University approved the protocols used in all animal investigation (approval number: CSU-2023-0021). C57BL/6 mice (6-8 weeks, male) were bred in the Experimental Animal Center of Central South University and were fed under specific pathogen-free conditions. The in vivo induction method for CI-AKI, adapted from previous studies [32-35], is illustrated in Figure 1A. Briefly, following administration of pentobarbital (60 mg/kg), mice received right nephrectomy. Three weeks post-surgery, the following interventions were performed sequentially: 24 h water deprivation, intravenous administration of furosemide (10 mL/kg; Harvest Pharmaceutical, Shanghai, China), followed by iohexol (10 mL/kg; Omnipaque, 350 mg iodine/mL; GE Healthcare, Shanghai, China) or normal saline via tail vein injection after a 20-min interval. Mice were euthanized 24 h after iohexol or saline administration for the collection and storage of serum and kidney tissues.
For cisplatin-induced AKI mice model, C57BL/6 mice (8 weeks, male) were injected by i.p. injection with a single dose of cisplatin at 30 mg/kg [36]. Control mice were injected with normal saline. Serum and kidney tissues were collected at 48 hours after cisplatin injection.
Analysis of renal function and histopathology
Renal function was evaluated by detecting serum creatinine (SCr) levels using an automatic biochemical analyzer at the Department of Clinical Laboratory Medicine. Histological examination was performed to assess tubular injury. Kidney tissues were processed by fixation in 4% paraformaldehyde, followed by paraffin embedding and sectioning at a thickness of 4 μm for hematoxylin and eosin (H&E) staining. Tubular injury was evaluated by examining at least ten randomly selected fields from the renal cortex and outer medulla. The extent of tubular injury was graded on a scale of 0 to 4, according to the percentage of damaged tubules: 0 (no damage), 1 (<25%), 2 (25-50%), 3 (50-75%), and 4 (>75%) [37].
Microarray analysis
Total RNAs were extracted from kidney cortex of mice with or without iohexol treatment. The samples were prepared and microarray hybridization was processed in line with Arraystar's protocols (KangChen Biotech, Shanghai, China). Briefly, total RNAs were subjected to RNase R exonuclease, leading to the elimination of linear RNAs and enrichment of circRNAs. Subsequently, the enriched circRNAs were amplified and transcribed into fluorescent cRNA using a random priming method (Arraystar Super RNA Labeling Kit; Arraystar). The labeled cRNAs were hybridized onto the Arraystar Mouse circRNA Array v2 (8x15K, Arraystar). The array images were scanned using an Agilent Scanner G2505C and analyzed with Agilent Feature Extraction software (version 11.0.1.1). R software limma package was employed for subsequent data processing. The identification of dysregulated expressed circRNAs was conducted using dual criteria: |log2 fold change| > 1 and P < 0.05.
RNA sequencing
Total RNAs were extracted from iohexol-induced proximal RTEC line (HK-2 cells) after treatments and underwent quality assessment. For sequencing library preparation, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations in an Illumina proprietary buffer. First-strand cDNA was synthesized using random hexamers and SuperScript II, followed by second-strand synthesis with DNA Polymerase I and RNase H. Remaining overhangs were converted to blunt ends using exonuclease/polymerase activities. After end repair and A-tailing, Illumina PE adapter oligonucleotides were ligated to the cDNA fragments. The library was purified using the AMPure XP system (Beckman Coulter) and enriched by PCR. The final library was quantified using the Agilent high sensitivity DNA assay on a Bioanalyzer 2100 (Agilent). Sequencing was performed on the NovaSeq 6000 platform (Illumina) by Personalbio (Shanghai, China).
Gene Ontology (GO) analysis
The GO project provides a controlled vocabulary to describe gene and gene product attributes in any organism (http://www.geneontology.org) [38]. GO analysis was performed using the R package TopGO (version 2.32.0; http://www.bioconductor.org/packages/release/bioc/html/topGO.html). GO annotations were obtained from the GO.db package (version 3.5.0; https://bioconductor.org/packages/3.6/data/annotation/html/GO.db.html), using the release dated 2017-11-01. GO enrichment analysis was performed to explore the functions of genes producing these dysregulated expressed circRNAs. The criterion for determining significant function was set at P < 0.05.
Identification strategy for conserved mouse-human circRNAs
To identify evolutionarily conserved circRNAs between mice and humans, we employed a multi-step screening approach. First, we identified differentially expressed circRNAs in the CI-AKI mouse model using dual criteria (fold change > 2 and P < 0.05) and restricted our analysis to exon-derived circRNAs ranging from 200 to 2000 nucleotides in length. We then mapped these circRNAs to their corresponding gene symbols using circBase (http://www.circbase.org/). Next, we identified human homologous genes and their associated hsa_circ_RNAs [39], selecting human circRNAs of comparable length to their mouse counterparts. Sequence conservation was assessed using BLAST alignment (https://blast.ncbi.nlm.nih.gov/Blast.cgi), with stringent criteria of > 85% sequence identity and > 95% query coverage. Finally, we validated the conservation status of the identified circRNA pairs using circBank (http://www.circbank.cn/).
Virus infection
The in vivo function of circSamd4 was investigated using an adeno-associated virus 9 (AAV9)-mediated knockdown approach. AAV9 vectors containing either circSamd4 short hairpin RNA (shRNA) or scrambled control shRNA (AAV9-NC) (Hanbio Biotechnology Co., Ltd., Shanghai, China) was diluted to 1 × 10¹² viral genomes/mL in PBS. The circSamd4 shRNA sequence was 5'-GCACGAGAAUCAUUAACCAAU-3'. Multiple sites in the left renal cortex were injected with either AAV9-circSamd4 shRNA or AAV9-NC using an insulin syringe. The mice were subjected to CI-AKI induction after one week post injection.
Generation of tubule-specific Cirbp knockout mice
Cirbp homozygous floxed (Cirbpfl/fl) mice and Ggt1-Cre mice (both C57BL/6 background) were provided by Cyagen Biosciences Inc. (Suzhou, China). Cirbpfl/fl mice were crossed with Ggt1-Cre mice to produce tubule-specific Cirbp knockout mice (Ggt1-Cre+/Cirbpfl/fl). Their Ggt1-Cre-/Cirbpfl/fl littermates served as controls. Genotyping was performed by PCR analysis of tail-derived genomic DNA (gDNA) using a Mouse Direct PCR Kit (Bimake, Houston, TX). All PCR primer sequences are provided in Table S1.
Plasmid construction
The circSAMD4 and CIRBP overexpression plasmids were generated using pCD25-ciR vector (Geneseed, Guangzhou, China) and CV702 vector (Genechem, Shanghai, China), respectively. Additionally, the truncated CIRBP (Δ1-90) and truncated CIRBP (Δ91-172) overexpression plasmids were also generated using CV702 vector (Genechem, Shanghai, China).
Cell culture and treatments
The immortalized proximal RTEC line (HK-2 cells) initially from ATCC and 293T cells were maintained in DMEM/F-12 and DMEM respectively, both containing 10% FBS and 1% antibiotics (Gibco, Carlsbad, CA). Based on our previous study [40], HK-2 cells were exposed to 200 mg iodine/mL iohexol for 6 h.
For loss-of-function studies, HK-2 cells were transfected with circSAMD4 small interfering RNA (siRNA) (Hanbio Biotechnology, Shanghai, China), SRSF3 siRNA, or CIRBP siRNA (Tsingke Biotechnology, Beijing, China), or negative control using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) as instructed by the manufacturer. The sequences of all siRNAs used are detailed in Table S2.
For gain-of-function experiments, all plasmid transfections, including corresponding empty vectors, were conducted with Lipofectamine 3000 (Invitrogen, Carlsbad, CA) as recommended by the manufacturer.
Isolation and culture of human primary RTECs
Human primary RTECs were harvested from the normal renal cortical tissue located at a minimum distance of 4 cm from tumor margins. The cortical tissue was mechanically dissected and enzymatically digested using Hanks' balanced salt solution containing collagenase II (1 mg/mL; Sigma-Aldrich, St. Louis, MO). Subsequently, three kinds of sieves with mesh diameters of 100 μm, 70 μm, and 40 μm were used to filter the mixture in turn. The final 40 μm filtrate was collected and centrifuged. The cell pellet was seeded into culture dishes. RTECs were confirmed by immunofluorescence staining for Cytokeratin 18 (A01357-1, BOSTER, Wuhan, China). For some experiments, human primary RTECs were stimulated by 200 mg iodine/mL iohexol for 6 h.
RNA Extraction and qRT-PCR
Total RNAs from kidney tissues and cells was extracted with TRIzol reagents (TaKaRa, Dalian, China). Next, RNA was reversely transcribed to complementary DNA (cDNA) utilizing PrimeScript™ RT reagent Kit (TaKaRa, Dalian, China). qRT-PCR was performed using the TB Green Premix Ex Taq™ II (TaKaRa, Dalian, China) on a LightCycler 96 Real-Time PCR System. All primer sequences are provided in Table S1. Gene expression was determined using primer amplification curves, melting curves, and CT values.
RNA Fluorescence in Situ Hybridization (FISH) analysis
The Ribo™ FISH kit (Ribobio, Guangzhou, China) was used to perform RNA-FISH experiments on the basis of the instructions in test kit. The specific probes spanning the splice junction site of circSAMD4 or circSamd4 were labelled with Cy3 and synthesized by RiboBio, which also provided the Cy3-labeled U6 and 18S probes. For in vitro experiments, HK-2 cells or human primary RTECs grown on slides were fixed, permeabilized, and pre-hybridized. The cells were then incubated with hybridization buffer containing specific probes overnight at 37 °C under dark conditions, followed by washing with hybridization washing solutions and PBS. Nuclei were counterstained with DAPI. For in vivo FISH, paraffin-embedded mouse or human kidney sections were deparaffinized, hydrated, and permeabilized with proteinase K (GenePharma, Shanghai, China) prior to hybridization. Staining intensity was quantified using a blinded approach. From each human specimen, six random microscopic fields at 20× magnification were selected for quantification of circSAMD4-positive regions using ImageJ software.
RNase R treatment
RNA samples extracted from HK-2 cells or human primary RTECs were incubated with 3 U/μg RNase R (Geneseed, Guangzhou, China) at 37 °C for 30 min [41]. The expression levels of circSAMD4, linear SAMD4A, and GAPDH were then analyzed by qRT-PCR following RNase R digestion.
TUNEL staining
Cell death in kidney after CI-AKI was evaluated utilizing by TUNEL assay (Roche Diagnostics, Basel, Switzerland). In brief, after deparaffinization and permeabilization with 0.3% Triton X-100, kidney sections were stained with the TUNEL reaction assay at 37 °C for 1 h under dark conditions. The sections were counterstained with DAPI and imaged under fluorescence microscopy. Ten random fields per section were analyzed to quantify TUNEL-positive cells.
Immunoblot analysis
Renal tissues or cells were lysed by RIPA buffer mixed with 1% protease inhibitor (cwbio, Beijing, China). Equal amounts of protein were separated by sodium dodecyl sulfate polyacrylamide gel and transferred to polyvinylidene fluoride membrane. After blocking with 5% bovine serum albumin (BSA) for 1 h, membranes were then incubated with primary antibodies overnight at 4 °C, followed by corresponding secondary antibodies for 1 h at room temperature. The specific primary antibodies described below were used: cleaved caspase-3 (9664, Cell Signaling Technology, Danvers, MA), SRSF3 (ab198291, Abcam, Cambridge, UK), CIRBP (ab191885 or ab246510, Abcam, Cambridge, UK), Fas (4233, Cell Signaling Technology, Danvers, MA) and GAPDH (10494-1-AP, Proteintech, Wuhan, China). The second antibodies were purchased from Servicebio (GB23303, Wuhan, China).
Flow cytometry analysis
Annexin V - Alexa Fluor 647 / propidium iodide (PI) Apoptosis detection Kit was purchased from 4 A Biotech (Beijing, China) to evaluate cell apoptosis. Concisely, cells were collected using 0.25% EDTA-free trypsin and washed with cold PBS. Following a 10-min incubation with Annexin V (5 μL) at room temperature, PI solution (10 μL, 20 μg/mL) was added. Flow cytometric analysis was conducted within 1 h, with subsequent data processing using FlowJo software. Apoptotic cells were calculated as the sum of Annexin V single-positive (Q3) and Annexin V/PI double-positive (Q2) populations.
RNA immunoprecipitation (RIP) assay
The RIP assay was conducted with a commercial kit (bes5101, BersinBio, Guangzhou, China). Concisely, the cells were collected, lysed, and then reacted with an anti-IgG antibody, or anti-SRSF3 antibody (RN080PW, MBL, Tokyo, Japan), or anti-FLAG antibody (F1804, Sigma-Aldrich, USA) at 4 °C for 16 h. The immune complexes were then captured with Protein A/G magnetic beads at 4 °C for 1 h and washed with polysome washing buffer. RNA was eluted by incubating the samples with polysome elution buffer at 55 °C for 1 h. The eluted RNA was purified using TRIzol Reagent and analyzed by qRT-PCR as described above.
Immunofluorescence staining
Cells were washed with PBS, then fixed in 4% paraformaldehyde for 5 min at room temperature, followed by permeabilization with pre-cooled methanol for 10 min at -20 °C. After a 1-h blocking step at room temperature, cells were incubated overnight at 4 °C with primary antibodies against CIRBP (10209-2-AP, Proteintech, Wuhan, China) or Cytokeratin 18 (A01357-1, Boster, Wuhan, China). The cells were then incubated with secondary antibodies - either Alexa-Fluor 488 (ab150077, Abcam, Cambridge, UK) or CoraLite594 (SA00013-4, Proteintech, Wuhan, China) - for 1 h at 37 °C. Nuclei were counterstained with DAPI (SouthernBiotech, Birmingham, AL). For quantification, nuclear and cytoplasmic fluorescence intensities were measured in individual cell using ImageJ software, and their ratio was calculated.
For in vivo analyses, kidney sections (4 µm) were deparaffinized, rehydrated through graded alcohols, and subjected to citrate-based antigen retrieval. After permeabilization and blocking with 5% BSA, sections were incubated with primary antibodies against CIRBP (10209-2-AP, Proteintech, Wuhan, China) or SRSF3 (RN080PW, MBL, Tokyo, Japan) overnight at 4 °C, followed by secondary antibody and DAPI staining. Ten random fields per section were imaged, and ImageJ software was employed to calculate relative fluorescence intensity.
RNA FISH- immunofluorescence examination
In vitro: cells were hybridized with Cy3-labeled probes specific for circSAMD4, washed with hybridization buffer and PBS, and then blocked for 1 hour at room temperature. Cells were incubated overnight at 4 °C with a primary antibody against CIRBP (10209-2-AP, Proteintech, Chicago, USA), followed by incubation with an Alexa Fluor 488-conjugated secondary antibody for 1 hour at 37 °C. Nuclei were counterstained with DAPI, and images were captured using fluorescence microscopy.
In vivo: kidney sections were blocked for 1 hour at room temperature and then incubated with fluorescein-conjugated markers for 2 hours at room temperature, including anti-lotus tetragonolobus lectin (LTL) (FL-1321), anti-peanut agglutinin (PNA) (FL-1071), and anti-dolichos biflorus agglutinin (DBA) (FL-1031) (all from VECTOR Laboratories, CA, USA). Nuclei were counterstained with DAPI, and images were acquired using fluorescence microscopy.
Nuclear and cytoplasmic protein extraction
Nuclear and cytoplasmic proteins were isolated from HK-2 cells using the Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, Shanghai, China). Briefly, adherent cells were harvested through PBS washing, scraping, and centrifugation. Cytoplasmic protein extraction involved sequential treatment with phenylmethanesulfonyl fluoride -containing reagents A and B: samples were subjected to vigorous vortex after adding reagent A and then incubated on ice for 10-15 min. Subsequently, after adding reagent B, the sample was subjected to two vigorous vortex steps for 5 s each, separated by a 1-min incubation on ice. The sample was then centrifuged to collect the supernatant containing cytoplasmic proteins. Nuclear proteins were isolated by adding nuclear protein extraction reagent, subjecting the sample to repeated vortex and ice-bath cycles for 30 min. The nuclear protein fraction was obtained by collecting the supernatant following centrifugation.
Immunohistochemistry analysis
Kidney biopsy sections (4 μm) were subjected to deparaffinization and gradient ethanol rehydration, followed by citrate-mediated antigen retrieval and hydrogen peroxide treatment. After being blocked with 5% BSA, the sections were then incubated overnight at 4 °C with primary antibodies against either CIRBP (10209-2-AP, Proteintech, Wuhan, China) or SRSF3 (RN080PW, MBL, Tokyo, Japan), followed by secondary antibody incubation. Immunoreactivity was visualized using diaminobenzidine (Beijing Zhongshan Jinqiao Biotechnology, Beijing, China), and sections were counterstained with hematoxylin (Servicebio, Wuhan, China). Ten random fields per section were captured for quantitative analysis, and ImageJ software was employed to measure the percentage of positive staining area.
Patient's information
Renal biopsy specimens with biopsy-confirmed ATI were collected from the Second Xiangya Hospital, Central South University. Patient demographics and clinical data are shown in Table S3 and Table S4. Normal kidney tissues adjacent to renal cell carcinoma served as controls. The Ethics Committee of the Second Xiangya Hospital, Central South University approved this study (approval number: LYF20230056), which complied with the Declaration of Helsinki principles. The study also obtained appropriate informed consent from all participants.
Statistical analysis
Quantitative data are presented as mean ± standard deviation (SD). Statistical analyses were performed using GraphPad Prism 8 software (San Diego, CA). Data normality was assessed using the Shapiro-Wilk test. For normally distributed data, differences between two groups were analyzed using unpaired two-tailed Student's t-test. Multiple group comparisons were conducted using one-way ANOVA with Tukey's multiple comparisons test or two-way ANOVA with Tukey's multiple comparisons test. Correlations between variables were evaluated using Pearson's correlation coefficient, and their relationships were further assessed by linear regression analysis. A value of P < 0.05 was considered statistically significant. All experiments were performed independently at least three times.
Results
CircSamd4 and its human homolog circSAMD4 were up-regulated in RTECs in CI-AKI
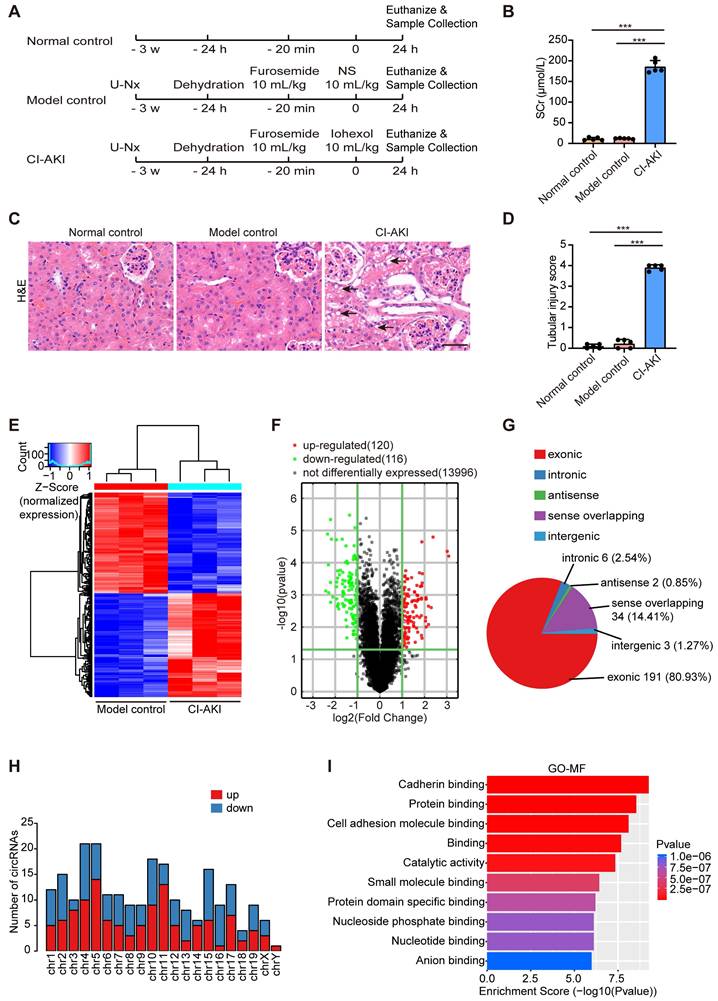
First, we established a CI-AKI murine model as evidenced by elevated SCr levels and distinctive tubular morphological injuries (Figure 1A-D). Then, Arraystar Mouse CircRNA Microarray analysis was performed to reveal distinct circRNA expression patterns in renal tissues between the CI-AKI group and the model control group. As shown in Figure 1E and F, the screening identified 236 significantly altered circRNA transcripts, comprising 120 upregulated and 116 downregulated transcripts in CI-AKI tissues compared to model controls. Analysis of genomic origins showed that exonic sequences accounted for 80.93% of the altered circRNAs, while sense overlapping regions contributed 14.41%. The remaining circRNAs aligned with intronic regions, intergenic regions, and antisense regions (Figure 1G). These differentially expressed circRNAs were primarily located on chromosomes 4 and 5 (Figure 1H). Next, using GO analysis, we annotated these circRNAs by examining the biological functions of their host genes. Molecular function analysis of source genes for the enhanced circRNAs revealed significant enrichment in protein binding and protein domain specific binding, suggesting that these circRNAs may influence CI-AKI pathogenesis through interactions with specific proteins (Figure 1I).
Subsequently, we selected circRNAs (fold change > 2; nucleotide length > 200 and < 2000; exon-derived) from the microarray data. The top six circRNAs, ranked by sequence conservation identity, were shown in Table S5. Of these candidates, five circRNAs exhibited elevated expression levels, while one showed reduced abundance (Figure 2A). The characteristics of these conserved circRNAs between human and mouse were as follows: identical parental gene symbols; similar sequence lengths; sequence identity of at least 85% upon BLAST comparison; and verification of conservation in the circBank database. Among these circRNAs, we further focused on circSamd4 (circID: mmu_circRNA_005305), which is derived from the mouse gene Samd4, and its human homolog circSAMD4 (circID: hsa_circ_0004846), considering their highest identity (93%) and the highest expression abundance in renal cortical tissues (Figure S1). Consistent with the microarray data, iohexol injection significantly upregulated circSamd4 expression in mouse kidney tissues (Figure 2B). Furthermore, FISH assay visually demonstrated the upregulation of circSamd4 levels mainly in the cytoplasm of RTECs of CI-AKI mice (Figure 2C). To further examine the localization of circSamd4, we performed co-staining with segment-specific tubular markers: LTL for proximal tubules, PNA for distal tubules, and DBA for collecting ducts. As shown in Figure S2, circSamd4 was extensively co-localized in renal tubules, particularly in proximal tubules, with minimal expression in collecting ducts.
Differential circRNA expression profiles in a CI-AKI mouse model. Male C57BL/6J mice were subjected to the following procedure: unilateral nephrectomy (U-Nx) with a 3-week recovery period, followed by sequential interventions of 24-hour water deprivation, tail vein injection of furosemide (10 mL/kg), and after 20 minutes, tail vein administration of either iohexol (10 mL/kg; CI-AKI group) or normal saline (NS; model control group). Serum and kidney tissues were harvested 24 hours after the final injection. (A) Schematic illustration of experimental groups: CI-AKI, model control, and normal control. (B) Serum creatinine (SCr) levels (n = 5). (C and D) Representative images of hematoxylin and eosin (H&E) staining and pathological score in kidney tissues. The black arrow indicated vacuolar degeneration of tubular epithelial cells (n = 5). Scale bar: 50 μm. (E and F) Hierarchical clustering and volcano plot analysis of differentially expressed circRNAs in kidney samples from model control and CI-AKI groups using Arraystar Mouse CircRNA Microarray (cutoff criteria: |log2 fold change| > 1, P < 0.05). (G) Distribution of genomic origins of the 236 differentially expressed circRNAs. (H) Chromosomal distribution of differentially expressed circRNAs. (I) Gene Ontology (GO) analysis showing enriched molecular functions (MF) for the host genes of upregulated circRNAs. All quantitative data are presented as mean ± SD (B and D). Statistical analysis was performed using one-way ANOVA with Tukey's multiple comparisons test (B and D). ***P < 0.001.
CircSamd4 and its human homolog circSAMD4 were up-regulated in RTECs in CI-AKI. (A) Heatmap showing expression patterns of six conserved circRNAs (five upregulated, one downregulated). (B) CircSamd4 (mmu_circ_005305) expression in kidney tissues from normal control, model control, and CI-AKI groups, measured by qRT-PCR (n = 6). (C) RNA fluorescence in situ hybridization (FISH) analysis showing circSamd4 expression and subcellular localization in kidney tissues after iohexol treatment. The white arrows indicated circSamd4-positive signals. Scale bar: 20 μm. (D) Schematic representation of circSAMD4 genomic location and splicing pattern. CircSAMD4 was formed by back-splicing of exon 3 of the human SAMD4A gene. (E) Cell morphology and representative immunofluorescence images of Cytokeratin 18 in HK-2 cells and human primary RTECs. Scale bar: 50 μm. (F and G) Agarose gel electrophoresis showing amplification of circSAMD4 from complementary DNA (cDNA) but not from genomic DNA (gDNA) using divergent primers, while convergent primers amplified linear SAMD4A from both templates in HK-2 cells and human primary RTECs. (H and I) Relative abundance of circSAMD4, linear SAMD4A, and GAPDH in HK-2 cells and human primary RTECs with or without RNase R treatment, assessed by qRT-PCR (n = 4). (J and K) CircSAMD4 (hsa_circ_0004846) expression in HK-2 cells and human primary RTECs after 6-hour iohexol (200 mg iodine/mL) exposure (n = 4). (L and M) RNA FISH analysis of circSAMD4 subcellular localization in HK-2 cells and human primary RTECs. Nuclei were counterstained with DAPI; U6 and 18S served as nuclear and cytoplasmic markers, respectively. Scale bar: 10 μm. All quantitative data are presented as mean ± SD (B, and H-K). Statistical analysis was performed using one-way ANOVA with Tukey's multiple comparisons test (B) or unpaired two-tailed Student's t-test (H-K). ***P < 0.001.
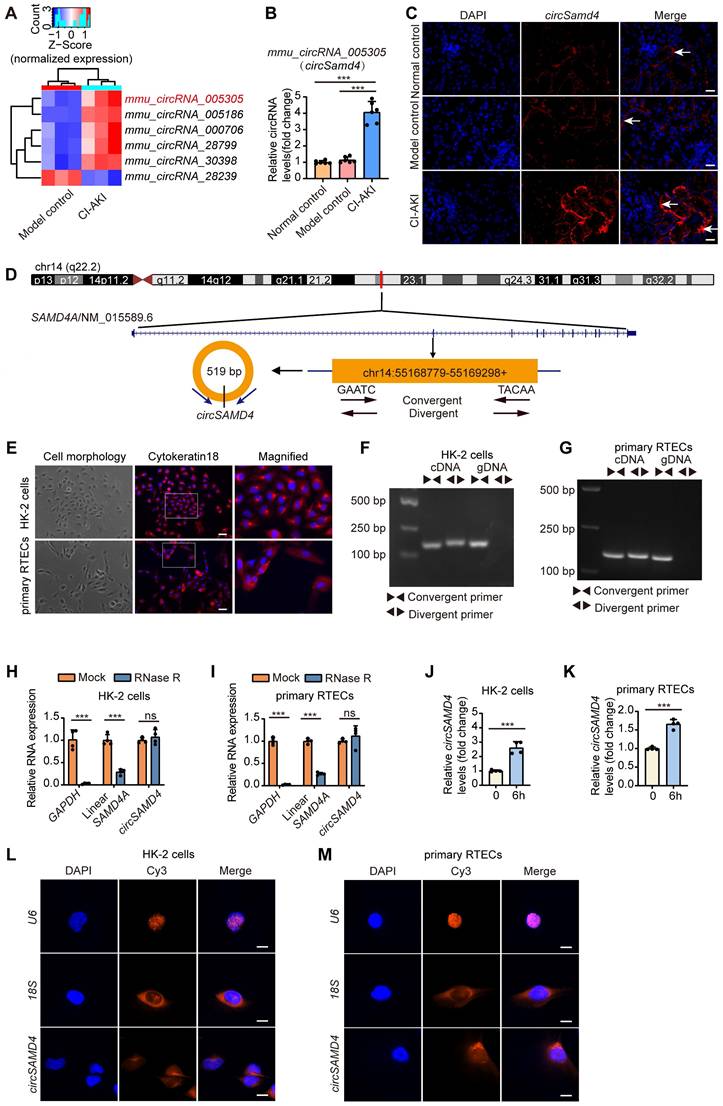
Then, we further explored the characteristics of circSAMD4. As shown in Figure 2D, circSAMD4 was derived from exon 3 of the human SAMD4A gene (NM_015589.6 transcript) and was located in the 55168779-55169298 region of chromosome 14. Both HK-2 cells and human primary RTECs were identified by Cytokeratin 18 expression using immunofluorescence staining (Figure 2E). Nucleic acid electrophoresis demonstrated that circSAMD4 was exclusively detected using divergent primers on cDNA templates, while remaining undetectable in gDNA samples. However, SAMD4A mRNA was successfully amplified by convergent primers regardless of template source (cDNA or gDNA) in both cell types (Figure 2F and G). Additionally, the circular configuration of circSAMD4 was further validated by its stability during RNase R treatment, unlike the significant degradation observed for both GAPDH and linear SAMD4A transcripts exposed to the same enzyme (Figure 2H and I). Exposure of HK-2 cells to iohexol for 6 h significantly increased circSAMD4 expression, a trend that was also observed in iohexol-treated human primary RTECs (Figure 2J and K). FISH assays further revealed that circSAMD4 predominantly localized to the cytoplasm of HK-2 cells and human primary RTECs (Figure 2L and M). Collectively, these findings demonstrated that both circSamd4 and its human homolog circSAMD4 were up-regulated in RTECs in CI-AKI.
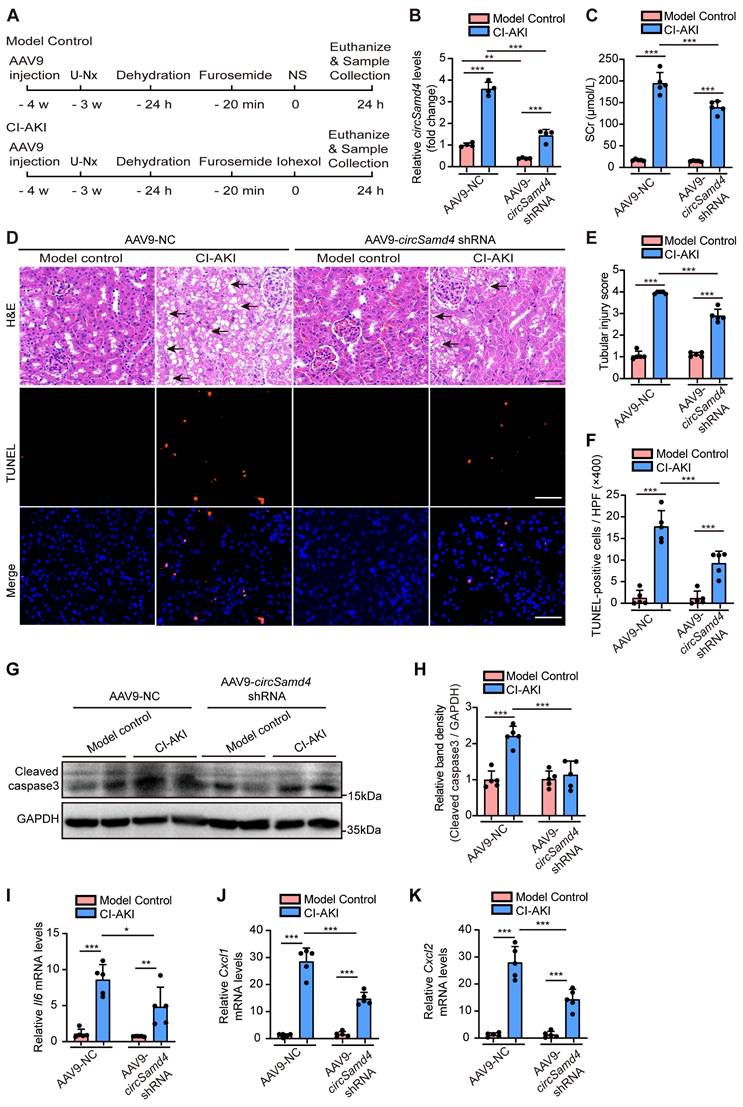
Inhibition of circSamd4 in mice ameliorated CI-AKI
To explore the role of circSamd4 in CI-AKI pathogenesis, we administered AAV9 carrying circSamd4 shRNA into the left renal cortex of mice via in situ renal injection, followed by the construction of a CI-AKI model (Figure 3A). Figure 3B indicates the significant reduction of circSamd4 in mice following local injection of AAV9-circSamd4 shRNA into the renal cortex. AAV9-circSamd4 shRNA did not induce renal functional or structural damage in model control mice, but it remarkably reduced SCr levels and improved renal tubular morphology in CI-AKI mice (Figure 3C-E). TUNEL assays further showed a marked decrease in cell death in renal tissues of CI-AKI mice treated with AAV9-circSamd4 shRNA compared to those treated with AAV9-NC (Figure 3D and F). Consistently, the inhibition of circSamd4 expression significantly reversed the accumulation of cleaved caspase-3 in kidney cortical tissues of CI-AKI mice (Figure 3G and H). Additionally, mRNA levels of the inflammatory cytokine Il6 and chemokines Cxcl1 and Cxcl2 were significantly elevated in the renal cortical tissues of CI-AKI mice, whereas circSamd4 inhibition markedly reduced their expression (Figure 3I-K). Taken together, these data revealed the pathological role of circSamd4 in CI-AKI.
CircSAMD4 aggravated iohexol-induced apoptosis in RTECs in vitro
As RTECs apoptosis exerted a vital effect on CI-AKI [35, 42], we then investigated the effect of circSAMD4 on the apoptosis of HK-2 cells induced by iohexol. Transfection with siRNA targeting the splice junction site of circSAMD4 effectively inhibited circSAMD4 expression without affecting SAMD4A mRNA levels (Figure 4A). Exposure to iohexol markedly elevated cleaved caspase-3 levels in HK-2 cells; however, this effect was significantly attenuated following circSAMD4 knockdown (Figure 4B and C). Consistently, flow cytometry analysis indicated silencing circSAMD4 notably reduced the apoptosis rate of iohexol-induced HK-2 cells (Figure 4D and E).
In turn, we investigated the effects of circSAMD4 overexpression on apoptosis of HK-2 cells stimulated by iohexol. CircSAMD4 overexpression notably elevated circSAMD4 levels without altering SAMD4A mRNA levels in HK-2 cells (Figure 4F). As we expected, immunoblot analysis demonstrated that overexpressing circSAMD4 obviously enhanced cleaved caspase-3 levels in iohexol-treated HK-2 cells (Figure 4G and H). Similarly, flow cytometry analysis reiterated that circSAMD4 overexpression markedly increased apoptosis rate in iohexol-stimulated HK-2 cells (Figure 4I and J). In summary, the above results consistently indicated that circSAMD4 exacerbated apoptosis in iohexol-stimulated HK-2 cells.
CircSamd4 knockdown protected against CI-AKI in mice. Male C57BL/6J mice received multi-site injections of AAV9-circSamd4 shRNA or AAV9-NC into the left renal cortex. Subsequently, the mice were subjected to CI-AKI or model control establishment. Briefly, one week after the injection, the mice underwent unilateral nephrectomy (U-Nx) and were allowed a 3-week recovery period. The mice then received sequential interventions: 24-hour water deprivation, furosemide administration via tain vein injection (10 mL/kg), and iohexol (10 mL/kg) or normal saline (NS) injection via tail vein after 20 minutes. Serum and kidney tissues were harvested 24 hours after iohexol or NS administration. (A) Schematic illustration of the experimental design. (B) Relative circSamd4 levels of kidney tissues detected by qRT-PCR (n = 4). (C) Serum creatinine (SCr) levels (n = 5). (D) Representative images of hematoxylin and eosin (H&E) and TUNEL staining in kidney tissues. The black arrow indicated vacuolar degeneration of tubular epithelial cells. Scale bar: 50 μm. (E) Tubular injury score in kidney tissues (n = 5). (F) Quantification of TUNEL-positive cells in kidney tissues (n = 5). (G and H) Immunoblot and densitometric analyses of cleaved caspase-3 expression in kidney cortex (n = 5). (I-K) qRT-PCR analysis of Il6, Cxcl1, and Cxcl2 mRNA levels (n = 5). All quantitative data are presented as mean ± SD, and analyzed by two-way ANOVA with Tukey's multiple comparisons test (B, C, E, F, H-K). *P < 0.05, **P < 0.01, ***P < 0.001.
CircSAMD4 aggravated apoptosis in HK-2 cells treated by iohexol. (A) Relative expression of circSAMD4 and linear SAMD4A in HK-2 cells transfected with circSAMD4-targeting siRNAs (n = 4). (B and C) Immunoblot and densitometric analyses of cleaved caspase-3 in HK-2 cells. Cells were transfected with circSAMD4 siRNA or control siRNA for 48 hours, followed by 6-hour treatment with iohexol (200 mg iodine/mL) (n = 4). (D and E) Flow cytometric analysis of apoptosis in HK-2 cells. Cells were treated as in (B), then stained with Annexin V-Alexa Fluor 647 and PI (n = 4). (F) Relative expression of circSAMD4 and linear SAMD4A in HK-2 cells transfected with circSAMD4 overexpression plasmid (n = 4). (G and H) Immunoblot and densitometric analyses of cleaved caspase-3 in HK-2 cells. Cells were transfected with circSAMD4 overexpression plasmid or empty vector for 48 hours, followed by 6-hour treatment with iohexol (200 mg iodine/mL) (n = 4). (I and J) Flow cytometric analysis of apoptosis in HK-2 cells. Cells were treated as in (G), then stained with Annexin V-Alexa Fluor 647 and PI (n = 4). All quantitative data are presented as mean ± SD (A, C, E, F, H, and J). Statistical analysis was performed using unpaired two-tailed Student's t-test (A and F) or two-way ANOVA with Tukey's multiple comparisons test (C, E, H and J). *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant.
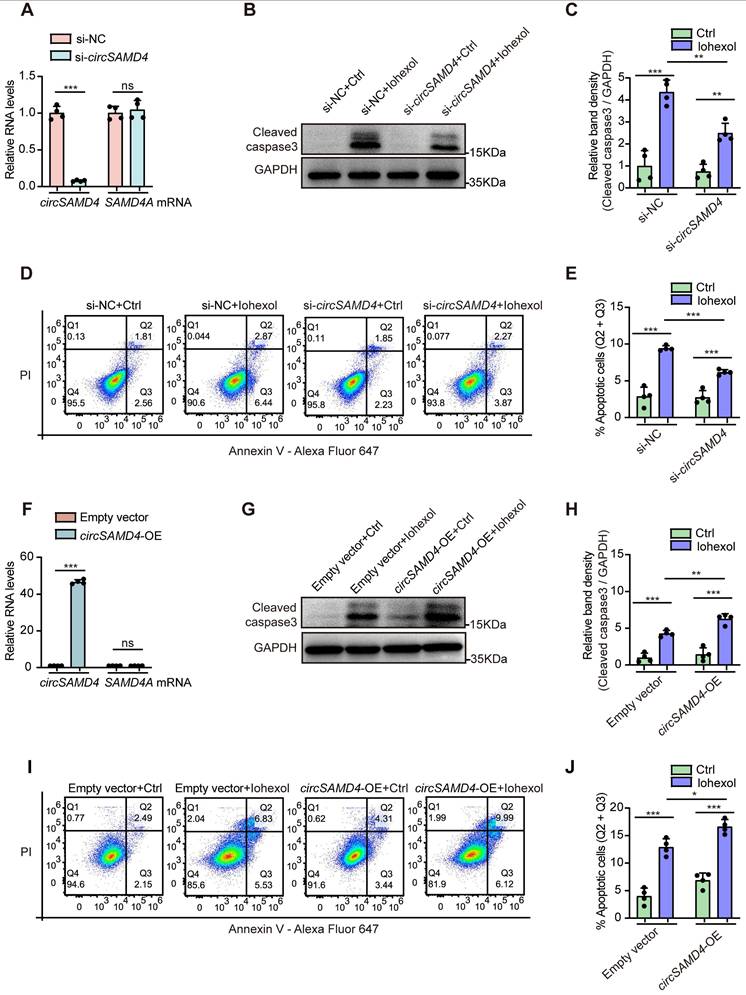
Knockdown of circSAMD4 prevented cytoplasmic accumulation of CIRBP in RTECs during exposure to iohexol
To further elucidate the mechanisms by which circSAMD4 contributed to CI-AKI, we sought to identify its downstream targets. A prior study identified proteins interacting with circSAMD4 using mass spectrometry following affinity pull-down assays in both human and mouse myoblasts [41]. Among them, CIRBP showed strong apoptosis regulation function in sepsis-associated or IRI-induced AKI [20]. Thus, we suspected that CIRBP maybe serve as a potential downstream effector of circSAMD4 in RTECs during CI-AKI pathogenesis. By performing RIP assay, we found that the relative enrichment of circSAMD4 in the anti-FLAG group was significantly higher than that in the IgG group (Figure 5A). This result confirmed the interaction between circSAMD4 and CIRBP. In addition, circSAMD4 and CIRBP showed significant co-localization in iohexol-treated HK-2 cells (Figure S3). To investigate the underlying mechanism, we generated CIRBP variants lacking residues 1-90 or 91-172 (Figure 5B). RIP assays revealed a marked reduction in circSAMD4 binding to the Δ91-172 variant compared with full-length CIRBP (Figure 5C), indicating that residues 91-172 are critical for the interaction. These results consistently confirm the specific interaction between circSAMD4 and CIRBP.
Meanwhile, we found that iohexol injection significantly upregulated Cirbp transcript and its protein levels in the renal cortical regions of mice. However, circSamd4 knockdown had no significant effects on Cirbp mRNA or protein levels in renal cortical regions from CI-AKI mice (Figure 5D-F). Similarly, inhibiting circSAMD4 expression did not significantly alter CIRBP mRNA or its protein levels in iohexol-stimulated HK-2 cells (Figure 5G-I). In turn, we investigated the effect of low CIRBP levels on circSAMD4 expression by transfection with CIRBP siRNA3 (Figure S4). Similarly, knockdown of CIRBP did not significantly affect circSAMD4 expression in iohexol-induced HK-2 cells (Figure 5J). These results demonstrated that circSAMD4 could not directly regulate the expression of CIRBP.
Many evidences have indicated that circRNAs can perform its function by altering the distribution of target proteins [43, 44]. We then investigated the effect of circSAMD4 on the subcellular location of CIRBP in RTECs in CI-AKI. Immunofluorescence staining showed that iohexol injection dramatically increased the cytoplasm distribution of CIRBP in RTECs of kidney tissues, but this effect was significantly suppressed by knockdown of circSamd4 expression (Figure 5K). Consistently, immunofluorescence detection further confirmed that circSAMD4 knockdown inhibited cytoplasmic accumulation of CIRBP in iohexol-induced HK-2 cells (Figure 5L and Figure S5). In vitro subcellular fractionation analysis revealed a decrease in the cytoplasm and the increase in the nucleus of CIRBP protein in iohexol-treated HK-2 cells with circSAMD4 knockdown (Figure 5M and N). Our findings demonstrated that circSAMD4 bound to CIRBP, and that circSAMD4 silencing prevented CIRBP cytoplasmic accumulation in RTECs in CI-AKI.
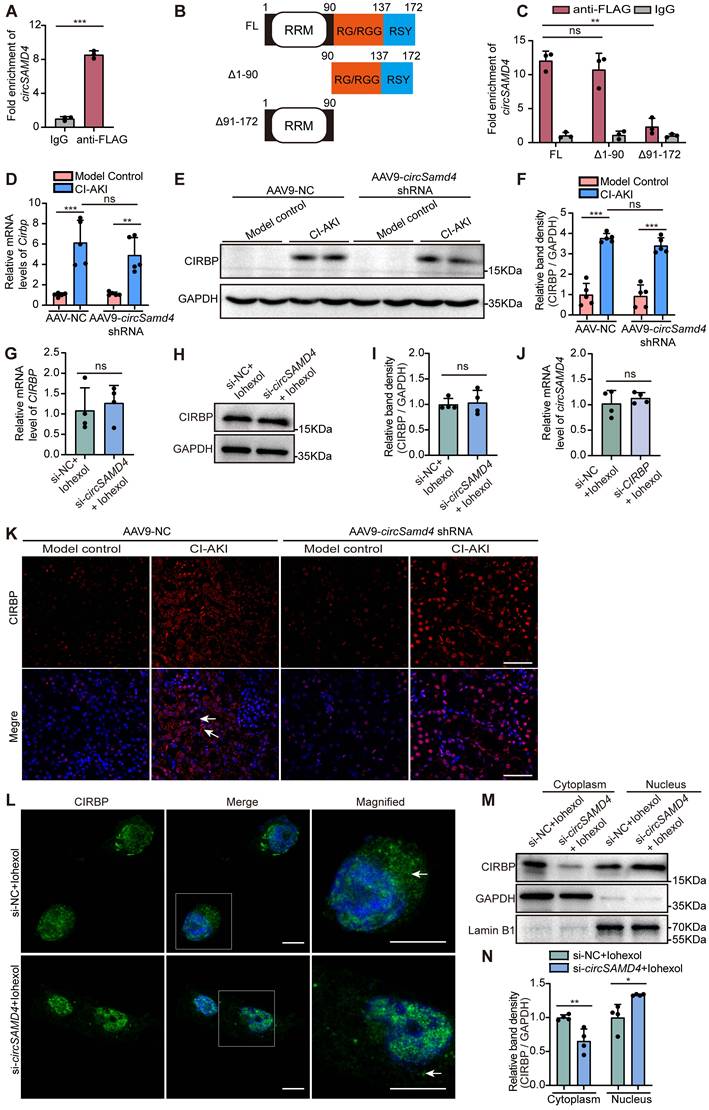
Renal tubule-specific Cirbp deletion in mice ameliorated CI-AKI
In the following experiment, we established renal tubule-specific Cirbp knockout mice (Ggt1-Cre+/Cirbp fl/fl, Figure 6A) to deeply explore the role of CIRBP in RTECs in CI-AKI. Tail genotyping, qRT-PCR assay and immunoblot analysis confirmed the successful generation of the knockout mice (Figure 6B-E). Scr levels and H&E staining clearly indicated that renal tubule-specific Cirbp deletion significantly improved renal function and tubular morphology in CI-AKI mice (Figure 6F-H). Consistently, TUNEL assays revealed a marked decrease in cell death in renal tissues of Ggt1-Cre+/Cirbp fl/fl mice with CI-AKI compared to Ggt1-Cre-/Cirbp fl/fl mice with CI-AKI (Figure 6G and I). Similarly, renal tubule-specific Cirbp deletion remarkably reduced accumulation of cleaved caspase-3 in renal cortical regions of CI-AKI mice (Figure 6J and K). In addition, renal tubule-specific Cirbp deletion significantly reduced the production of inflammatory cytokines Il6 and chemokines Cxcl1 and Cxcl2 (Figure 6L-N). These results revealed CIRBP as a pathogenic mediator in RTECs during CI-AKI progression.
CircSAMD4 knockdown promoted the nuclear import of CIRBP in RTECs in CI-AKI. (A) 293T cells were co-transfected with circSAMD4 and CIRBP-FLAG plasmids for 48 hours. The RNA immunoprecipitation (RIP) assay was performed using anti-FLAG antibody followed by qRT-PCR to detect the enrichment of circSAMD4. Anti-IgG antibody was served as a control (n = 3). (B) Scheme of CIRBP illustrating its domain organization and deletion of individual domains. (C) The interaction between circSAMD4 and CIRBP was validated by RIP assay in 293T cells transfected with full length (FL) or truncated CIRBP plasmids (n = 3). (D) Relative Cirbp mRNA levels in kidney tissues from mice treated with AAV9-circSamd4 shRNA and CI-AKI (n = 5). (E and F) Immunoblot and densitometric analyses of CIRBP protein in renal cortex homogenates from mice treated with AAV9-circSamd4 shRNA and CI-AKI (n = 5). (G) Relative CIRBP mRNA levels analyzed by qRT-PCR in HK-2 cells. Cells were transfected with circSAMD4 siRNA or control siRNA for 48 hours, followed by 6-hour treatment with iohexol (200 mg iodine/mL) (n = 4). (H and I) Immunoblot and densitometric analyses of whole-cell CIRBP expression in HK-2 cells treated as described in (G) (n = 4). (J) qRT-PCR analysis of circSAMD4 expression in HK-2 cells transfected with CIRBP siRNA3 or control siRNA for 48 hours, followed by iohexol treatment (200 mg iodine/mL, 6 hours) (n = 4). (K) Representative immunofluorescence images of CIRBP in kidney sections from mice treated with AAV9-circSamd4 shRNA and CI-AKI. The arrows point to the cells with cytoplasmic CIRBP. (L) Representative immunofluorescence images showing CIRBP (green) localization and nuclear DAPI staining (blue) in HK-2 cells treated as described in (G). The arrows indicate cytoplasmic CIRBP in iohexol-treated cells. Scale bar: 10 μm. (M and N) Immunoblot and densitometric analyses of nuclear and cytoplasmic CIRBP expression in HK-2 cells treated as described in (G), using GAPDH and Lamin B1 as cytoplasmic and nuclear controls, respectively (n = 4). All quantitative data are presented as mean ± SD (A, C, D, F, G, I, J, and N). Statistical analysis was performed using unpaired two-tailed Student's t-test (A, G, I, J, and N), one-way ANOVA with Tukey's multiple comparisons test (C) or two-way ANOVA with Tukey's multiple comparisons test (D and F). *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant.
Knockdown of CIRBP ameliorated iohexol-induced apoptosis in RTECs in vitro
We next evaluated the effects of CIRBP knockdown on apoptosis in HK-2 cells exposed to iohexol. Inhibition of CIRBP protein level notably reduced accumulation of cleaved caspase-3 (Figure 7A and B), and decreased the apoptosis rate in iohexol-stimulated HK-2 cells (Figure 7C and D). Collectively, our findings indicated CIRBP promoted apoptosis in HK-2 cells exposed to iohexol.
CIRBP overexpression abrogated the beneficial effects by circSAMD4 suppression in iohexol-treated RTECs in vitro
Then, we assessed whether the pro-apoptotic effect of circSAMD4 in iohexol-stimulated HK-2 cells was mediated through a CIRBP-dependent mechanism. Immunoblot analysis confirmed that the CIRBP overexpression notably elevated CIRBP protein levels in HK-2 cells (Figure 7E and F). As shown in Figure 7G and H, knockdown of circSAMD4 could markedly decrease iohexol-caused accumulation of the cleaved caspase-3, and this effect was reversed by CIRBP overexpression. Consistently, flow cytometry analysis further verified these findings (Figure 7I and J). In summary, these results revealed that circSAMD4 promoted apoptosis in iohexol-stimulated HK-2 cells through CIRBP-dependent mechanisms.
CIRBP suppression attenuates apoptosis by downregulating Fas in iohexol-induced HK-2 cells
To explore the mechanism underlying the pro-apoptotic function of CIRBP, we performed RNA sequencing in iohexol-treated HK-2 cells transfected with CIRBP siRNA or control siRNA. Among the downregulated genes following CIRBP knockdown, FAS mRNA was significantly reduced (Figure S6A). Fas, a transmembrane protein of the TNF family, mediates inflammation and apoptosis upon binding to Fas ligand [45]. CIRBP knockdown decreased both FAS mRNA and protein levels in iohexol-treated HK-2 cells (Figure S6B-D), and renal tubule-specific Cirbp deletion similarly reduced Fas mRNA in CI-AKI mouse kidneys (Figure S6E). RIP assay confirmed the binding between CIRBP and FAS mRNA (Figure S6F). These findings indicate that CIRBP suppression attenuates apoptosis through downregulating Fas in iohexol-induced HK-2 cells.
SRSF3 negatively regulated circSAMD4 production
Previous studies revealed that the formation of circRNAs relied on the spliceosome machinery, including cis- and trans-regulatory elements. Reverse intronic complementary sequences facilitated spatial proximity of the 5′- and 3′-termini of an exon or of consecutive exons to induce “head-to-tail” splicing [46]. However, no intronic complementary sequences existed in the flanking sequences of the back-splicing junction site of circSAMD4, extending 5000 bp in both upstream and downstream directions. Thus, we suspect that RNA binding proteins (RBPs) maybe closely relate to the regulation of circSAMD4 expression. As three RBPs—SRSF3, heterogeneous nuclear ribonucleoprotein M (HNRNPM), and RNA-binding motif protein 3 (RBM3) showed the function for mediating RNA splicing [47-51], we accordingly investigated their roles in regulating circSAMD4 formation by using siRNA knockdown assay. SRSF3 knockdown significantly increased circSAMD4 expression but did not obviously alter linear SAMD4A expression in HK-2 cells (Figure 8A-C). In contrast, silencing either HNRNPM or RBM3 did not significantly affect the expression of circSAMD4 and its linear transcript SAMD4A (Figure S7A and B). These results demonstrated that SRSF3 is a key regulator of circSAMD4 formation. Moreover, bioinformatic analysis using the CATRAPID tool indicated a SRSF3 binding site in mature circSAMD4 (Table S6). Subsequently, RIP experiments were performed to assess the interaction between SRSF3 and circSAMD4. The relative enrichment of circSAMD4 in the anti-SRSF3 group was obviously higher compared to the IgG group (Figure 8D).
Renal tubule-specific Cirbp deletion in mice ameliorated CI-AKI. (A) The experimental scheme of generating renal tubule cell-specific Cirbp (Ggt1-Cre+/Cirbp fl/fl) knockout mice by Cre-loxP recombinant system. (B) Genotype confirmation by PCR analysis of mouse tail DNA. (C) qRT-PCR analysis of Cirbp mRNA levels in renal cortex from Ggt1-Cre+/Cirbp fl/fl and Ggt1-Cre-/Cirbp fl/fl mice (n = 6). (D and E) Immunoblot and densitometric analyses of CIRBP expression in kidney cortex from Ggt1-Cre+/Cirbp fl/fl and Ggt1-Cre-/Cirbp fl/fl mice (n = 6). (F) Serum creatinine (SCr) levels from Ggt1-Cre+/Cirbp fl/fl and Ggt1-Cre-/Cirbp fl/fl mice with CI-AKI (n = 6). (G) Representative images of hematoxylin and eosin (H&E) staining and TUNEL staining in kidney sections from Ggt1-Cre+/Cirbp fl/fl and Ggt1-Cre-/Cirbp fl/fl mice with CI-AKI. The black arrow indicated vacuolar degeneration of tubular epithelial cells. Scale bar: 50 μm. (H) Tubular injury score in kidney tissues (n = 6). (I) Quantification of TUNEL-positive cells in kidney sections from Ggt1-Cre+/Cirbp fl/fl and Ggt1-Cre-/Cirbp fl/fl mice with CI-AKI (n = 6). (J and K) Immunoblot and densitometric analyses of cleaved caspase-3 expression in kidney cortex from Ggt1-Cre+/Cirbp fl/fl and Ggt1-Cre-/Cirbp fl/fl mice with CI-AKI (n = 6). (L-N) qRT-PCR analysis of Il6, Cxcl1, and Cxcl2 mRNA levels (n = 6). All quantitative data are presented as mean ± SD (C, E, F, H, I and K-N). Statistical analysis was performed using unpaired two-tailed Student's t-test (C and E) or two-way ANOVA with Tukey's multiple comparisons test (F, H, I and K-N). *P < 0.05, ***P < 0.001.
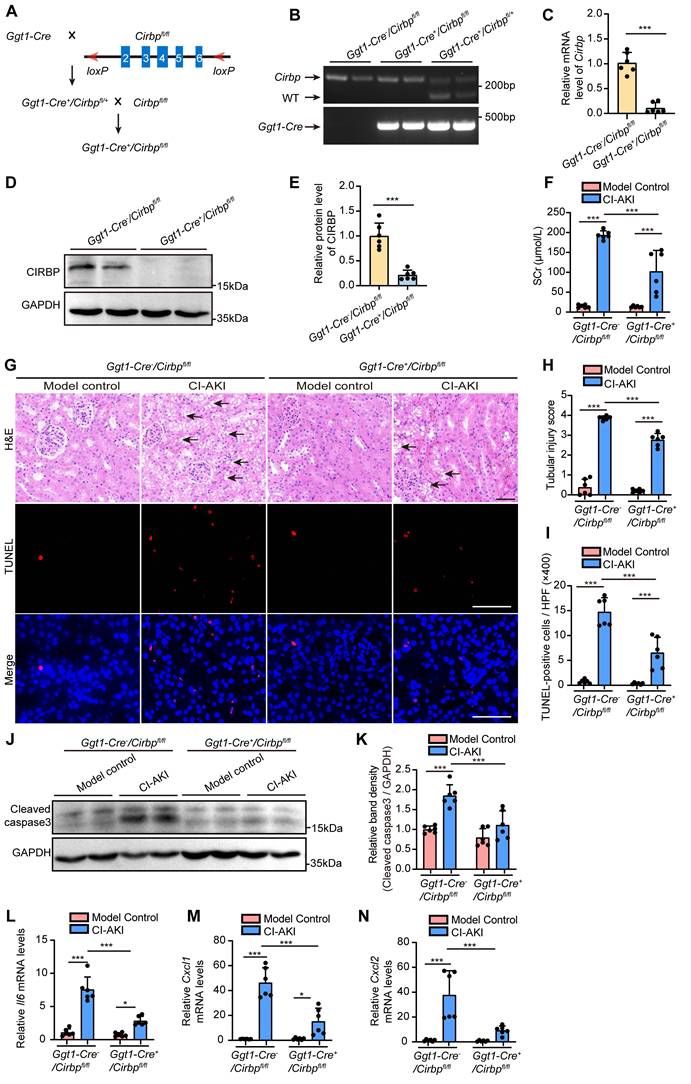
CIRBP mediated the pro-apoptotic effect of circSAMD4 in iohexol-treated HK-2 cells. (A and B) Immunoblot and densitometric analyses of cleaved caspase-3 expression in HK-2 cells. Cells were transfected with CIRBP siRNA3 or control siRNA for 48 hours, followed by 6-hour treatment with iohexol (200 mg iodine/mL) (n = 4). (C and D) Flow cytometric analysis of apoptosis in HK-2 cells. Cells were treated as in (A), then stained with Annexin V-Alexa Fluor 647 and PI (n = 4). (E and F) Immunoblot and densitometric analyses of CIRBP expression in HK-2 cells transfected with CIRBP-FLAG plasmid or empty vector (n = 4). (G and H) Immunoblot and densitometric analyses of cleaved caspase-3 expression in HK-2 cells. Cells were co-transfected with circSAMD4 siRNA (or control siRNA) and CIRBP-FLAG plasmid (or empty vector plasmid) for 48 hours, followed by 6-hour treatment with iohexol (200 mg iodine/mL) (n = 4). (I and J) Flow cytometric analysis of apoptosis in HK-2 cells. Cells were treated as in (G), then stained with Annexin V-Alexa Fluor 647 and PI (n = 4). All quantitative data are presented as mean ± SD (B, D, F, H and J). Statistical analysis was performed using two-way ANOVA with Tukey's multiple comparisons test (B and D), or unpaired two-tailed Student's t-test (F), or one-way ANOVA with Tukey's multiple comparisons test (H and J). *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant.
Inhibition of SRSF3 mediated the upregulation of circSAMD4. (A and B) Immunoblot and densitometric analyses of SRSF3 expression in HK-2 cells transfected with three different SRSF3-targeting siRNAs (n = 4). (C) Relative expression of circSAMD4, linear SAMD4A and SRSF3 in HK-2 cells transfected with three different SRSF3-targeting siRNA (n = 4). (D) 293T cells were transfected with circSAMD4 plasmid for 48 hours. The RIP assay was performed using anti-SRSF3 antibody followed by qRT-PCR to detect the enrichment of circSAMD4. Anti-IgG antibody was served as a control (n = 3). (E and F) Immunoblot and densitometric analyses of SRSF3 expression in kidney cortex from CI-AKI mice (n = 6). (G and H) Representative immunofluorescence images and quantification of SRSF3 staining in kidney sections from CI-AKI mice (n = 6). (I and J) Immunoblot and densitometric analyses of SRSF3 expression in HK-2 cells stimulated by iohexol for 6 hours (n = 4). (K and L) Immunoblot and densitometric analyses of cleaved caspase-3 expression in HK-2 cells. Cells were transfected with SRSF3 siRNA3 or control siRNA for 48 hours, followed by 6-hour treatment with iohexol (200 mg iodine/mL) (n = 4). (M and N) Flow cytometric analysis of apoptosis in HK-2 cells. Cells were treated as in (K), then stained with Annexin V-Alexa Fluor 647 and PI (n = 4). All quantitative data are presented as mean ± SD (B-D, F, H, J, L and N). Statistical analysis was performed using one-way ANOVA with Tukey's multiple comparisons test (B, C, F and H), or unpaired two-tailed Student's t-test (D and J), or two-way ANOVA with Tukey's multiple comparisons test (L and N). *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant.
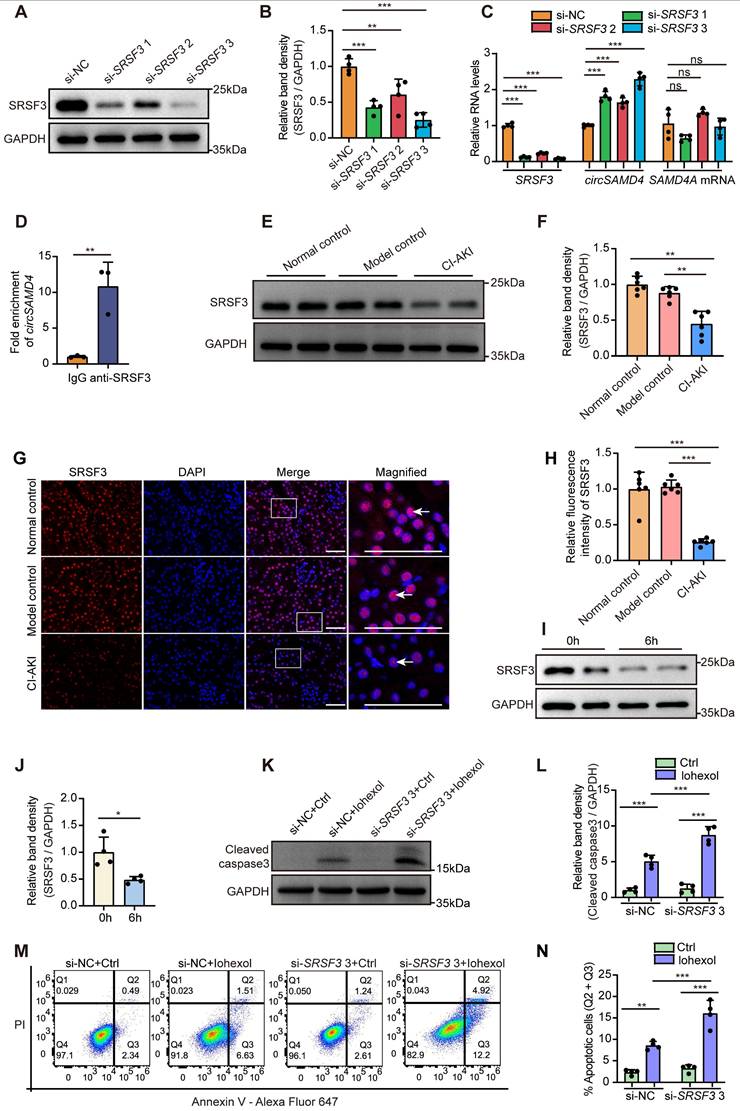
Next, we investigated SRSF3's expression and role in RTECs in CI-AKI. Immunoblot analysis revealed a reduction of SRSF3 expression in kidney tissues of CI-AKI mice (Figure 8E and F). Immunofluorescence detection further confirmed that iohexol remarkably decreased SRSF3 protein levels primarily in the nuclei of RTECs (Figure 8G and H). Consistently, in vitro exposure to iohexol markedly reduced SRSF3 protein levels in HK-2 cells (Figure 8I and J). Given that siRNA3 exhibited the most potent inhibitory effect on SRSF3 protein levels, we utilized this siRNA for subsequent functional studies. SRSF3 knockdown significantly enhanced both cleaved caspase-3 levels (Figure 8K and L) and the apoptosis rate in iohexol-stimulated HK-2 cells (Figure 8M and N). Collectively, above data suggested that SRSF3 bound to circSAMD4; SRSF3 knockdown up-regulated circSAMD4 expression, thereby exacerbating apoptosis in iohexol-stimulated HK-2 cells.
CircSAMD4 knockdown abrogated the pro-apoptotic effects by SRSF3 suppression in iohexol-treated RTECs in vitro
Then, we assessed whether the pro-apoptotic effect of SRSF3 suppression in iohexol-stimulated HK-2 cells was mediated through a circSAMD4-dependent mechanism. As shown in Figure S8A and B, SRSF3 suppression could markedly increase iohexol-caused accumulation of the cleaved caspase-3, and this effect was reversed by circSAMD4 knockdown. Consistently, flow cytometry analysis further verified these findings (Figure S8C and D). In summary, these results revealed that circSAMD4 knockdown reverse the pro-apoptotic effects of SRSF3 depletion in iohexol-treated RTECs.
Aberrant expression of SRSF3, circSamd4, and CIRBP in RTECs in cisplatin-induced AKI
To evaluate whether the dysregulation of SRSF3, circSamd4, and CIRBP extends beyond CI-AKI, we established a cisplatin-induced AKI mouse model (Figure S9A-C). RNA FISH and RT-PCR analyses confirmed renal accumulation of circSamd4 (Figure S9D-E). Immunofluorescence revealed upregulation and cytoplasmic accumulation of CIRBP in RTECs, whereas SRSF3 expression was reduced (Figure S9F-G), a finding further validated by immunoblotting (Figure S9H-I). These results suggest that aberrant expression of SRSF3, circSamd4, and CIRBP may represent a conserved feature across different types of AKI, although additional studies are required to fully confirm this.
Altered expression of SRSF3, circSAMD4 and CIRBP in RTECs from human patients with biopsy-diagnosed ATI
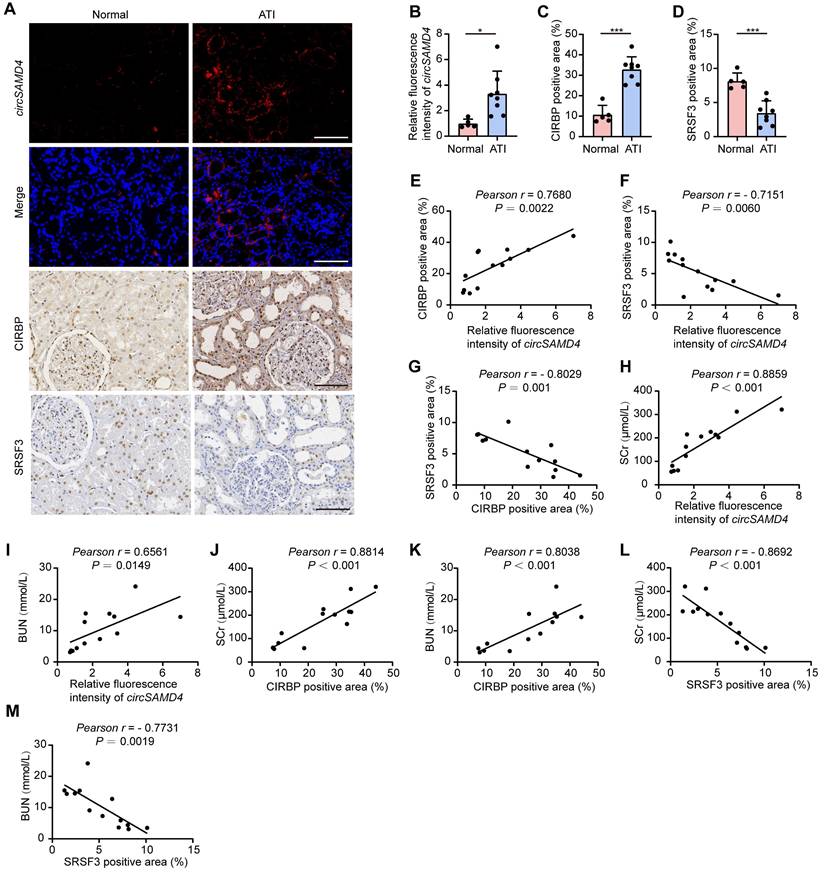
To evaluate the clinical significance of SRSF3, circSAMD4 and CIRBP in AKI, we examined their expression in kidney tissues from patients with biopsy-diagnosed ATI. RNA FISH analysis revealed a significant elevation in circSAMD4 expression in RTECs from patients with biopsy-confirmed ATI (Figure 9A and B). We performed co-staining of circSAMD4 with segment-specific tubular markers, including LTL for proximal tubules, PNA for distal tubules, and DBA for collecting ducts. As shown in Figure S10, circSAMD4 was extensively co-localized in renal tubules, particularly in proximal tubules, with minimal expression in collecting ducts. Immunohistochemical staining showed upregulation and cytoplasmic accumulation of CIRBP in RTECs from patients with biopsy-confirmed ATI, whereas SRSF3 expression was reduced (Figure 9A, C and D). Additionally, circSAMD4 expression positively correlated with CIRBP protein levels, whereas both of them negatively correlated with SRSF3 expression (Figure 9E-G). Moreover, both of the levels of circSAMD4 and CIRBP showed positive correlation with peak SCr and blood urea nitrogen levels in patients (Figure 9H-K). Conversely, SRSF3 expression negatively correlated with kidney function in patients (Figure 9L and M).
Discussion
Iodinated contrast media are a leading cause of AKI in hospitalized patients, yet the underlying mechanisms remain poorly understood. This study identifies the upregulation of circSAMD4 as a pivotal driver of CI-AKI. Mechanistically, we demonstrate that circSAMD4 inhibits nuclear import of CIRBP, while downregulation of SRSF3 promotes the expression of circSAMD4. Notably, depletion of Cirbp in RTECs significantly alleviates CI-AKI. Importantly, similar alterations in these molecules are observed in RTECs derived from patients with ATI, where their dysregulated expression correlates with the extent of kidney injury. Collectively, our findings establish the SRSF3/circSAMD4/CIRBP axis as a novel pathogenic pathway in CI-AKI and position circSAMD4 as a promising therapeutic target for CI-AKI.
Altered expression of SRSF3, circSAMD4 and CIRBP in RTECs correlated with kidney injury in patients pathologically diagnosed with ATI. (A-D) Representative RNA fluorescence in situ hybridization (FISH) staining images and quantification of circSAMD4 expression in renal biopsy specimens from patients pathologically diagnosed with ATI (n = 8) and para-carcinoma kidney sections (n = 5). Representative immunohistochemistry images and quantitative analysis of CIRBP and SRSF3 expression from patients pathologically diagnosed with ATI (n = 8) and para-carcinoma kidney sections (n = 5). Scale bar: 100 μm. (E) Correlation between circSAMD4 expression and CIRBP expression in all participants (n = 13). (F) Correlation between circSAMD4 expression and SRSF3 expression in all participants (n = 13). (G) Correlation between CIRBP expression and SRSF3 expression in all participants (n = 13). (H and I) Correlation between circSAMD4 expression and peak serum creatinine (SCr) and blood urea nitrogen (BUN) levels in all participants (n = 13). (J and K) Correlation between CIRBP expression and peak SCr and BUN levels in all participants (n = 13). (L and M) Correlation between SRSF3 expression and peak SCr and BUN levels in all participants (n = 13). All quantitative data are presented as mean ± SD (B-M). Statistical analysis was performed using unpaired two-tailed Student's t-test (B-D), or Pearson's correlation coefficient r with two-tailed P-value (E-M). *P < 0.05, ***P < 0.001.
Recent studies have demonstrated that circRNAs play important roles in the development of AKI [11, 14, 52]; however, the critical circRNAs involved in the development of CI-AKI remain unidentified. Through circRNA microarray analysis, we identified 236 differentially expressed circRNAs in kidney tissues of CI-AKI mice compared to control mice (Figure 1). As the biological properties of circRNAs become increasingly understood, their evolutionary conservation across species has garnered significant attention [6]. Given that sequence conservation often implies functional relevance [53], we prioritized the six circRNAs with the highest sequence identity between mouse and human. Among these, circSamd4 and its human homolog, circSAMD4, exhibited the highest sequence identity (93%). Our findings provided several evidence supporting that upregulation of circSAMD4 contributed critically to cell death in CI-AKI: (1) circSAMD4 was upregulated in RTECs of both murine models and cellular models of CI-AKI (Figure 2). (2) functionally, circSamd4 knockdown in the kidney cortex mitigated renal dysfunction, tubular injury, and cell death in CI-AKI mice (Figure 3). Similar findings were observed in iohexol-treated HK-2 cells (Figure 4). (3) notably, increased circSAMD4 expression correlated positively with kidney dysfunction in biopsy-confirmed ATI patients (Figure 9). Collectively, these findings support circSAMD4 (or circSamd4) as a key contributor to CI-AKI pathogenesis, and highlight its potential as a novel clinical biomarker for CI-AKI diagnosis.
Current evidence suggests that circRNAs regulate cellular processes by interacting with proteins and modulating their nucleocytoplasmic distribution [44]. In this study, we identified that circSAMD4 interacted with CIRBP, and found that inhibiting circSAMD4 promoted the nuclear translocation of CIRBP (Figure 5). CIRBP contains two non-classical nuclear localization signals (RG/RGG- and RSY-rich regions, residues 91-172) that are essential for nuclear import [54]. The interaction with circSAMD4 was significantly reduced in the Δ91-172 variant (Figure 5), suggesting that circSAMD4 binds to this region to inhibit CIRBP nuclear import.
Under several cellular stress, CIRBP can relocalize to the cytoplasm, where it modulates various cellular processes, including apoptosis and proliferation [20]. It is widely expressed across a variety of tissues and cell types, including those in the kidney [55]. Notably, previous studies have reported conflicting roles of CIRBP in different AKI models [20]. Cen et al. reported Cirbp knockout protected against IRI by reducing apoptosis and inflammation in mice [22], whereas another study demonstrated that CIRBP exerted renoprotective effects in AKI induced by deep hypothermic circulatory arrest [24]. However, these previous studies used global Cirbp knockout models in vivo, which affected various cell types within the kidney. By using a renal proximal tubule-specific Cirbp depletion murine model, we demonstrated that Cirbp deletion in RTECs ameliorated CI-AKI in mice (Figure 6), and this finding was further supported by in vitro experiments (Figure 7). RNA sequencing revealed that CIRBP knockdown significantly reduced FAS mRNA, a key mediator of apoptosis and inflammation in RTECs during AKI [56-59]. Consistently, CIRBP suppression decreased FAS mRNA and protein in iohexol-treated HK-2 cells and in vivo. RIP assays confirmed CIRBP binds to FAS mRNA (Figure S6). Collectively, our results suggest that CIRBP promotes apoptosis in CI-AKI by upregulating Fas.
SRSF3, a member of the serine/arginine-rich protein family, is involved in multiple RNA processing events, including alternative splicing [47, 60], RNA export and polyadenylation [61,62], protein translation [63,64], and pri-miRNA processing [65]. In this study, we present evidence supporting the crucial role of SRSF3 in regulating circSAMD4 expression in CI-AKI (Figure 8). Our findings revealed that SRSF3 knockdown significantly increased circSAMD4 expression without notably affecting SAMD4A mRNA levels in HK-2 cells. Furthermore, bioinformatics analysis identified a putative SRSF3 binding site within mature circSAMD4, and RIP assays confirmed the interaction between SRSF3 and circSAMD4 (Figure 8). These findings align with prior research in which mass spectrometry analysis identified SRSF3 as a circSAMD4-associated protein following the affinity pull-down of circSAMD4 in myoblasts [41]. Similarly, it has been reported in Drosophila that SR proteins inhibit the production of Laccase2 circular RNA [26]. These proteins may modulate spliceosome assembly, splicing kinetics, or circular RNA stability [26]. Notably, SRSF3 knockdown did not alter the half-life of circSAMD4 (Figure S11), indicating that it does not affect circSAMD4 stability. We therefore propose that SRSF3 may regulate SAMD4A pre-mRNA splicing via binding to flanking introns or exons. Further studies are needed to elucidate the precise mechanism.
This study also showed that SRSF3 knockdown exacerbated iohexol-induced cell apoptosis (Figure 8), and decreased SRSF3 levels correlated with more severe kidney dysfunction in biopsy-confirmed ATI patients (Figure 9), indicating a protective role for SRSF3 in RTECs during CI-AKI. Notably, emerging evidence also supports protective effects of SRSF3 in acute injuries of other organs, including ischemic cerebral infarction and myocardial infarction [30, 31]. For instance, cardiomyocyte-specific Srsf3 deletion in mice resulted in severe systolic dysfunction and mortality within eight days [31]. Collectively, the findings from others and us support an important role of SRSF3 in acute tissue injuries.
This study has direct clinical relevance by identifying the SRSF3/circSAMD4/CIRBP axis as a key driver of CI-AKI, linking molecular mechanisms in mouse models to patients with ATI. Dysregulation of circSAMD4 correlates with kidney injury, and modulating circSAMD4 or CIRBP levels can mitigate tubular cell death, highlighting potential therapeutic strategies. These findings provide a promising foundation for developing interventions to prevent or reduce CI-AKI in clinical practice, where effective treatments are currently limited.
Conclusion
This study identifies dysregulation of the SRSF3/circSAMD4/CIRBP axis as a novel pathogenic mechanism in CI-AKI. Specifically, circSAMD4 upregulation exacerbates iodinated contrast-induced injury by inhibiting nuclear import of CIRBP and triggering cell death, while SRSF3 downregulation contributes to increased circSAMD4 expression in RTECs. These findings advance our understanding of CI-AKI pathogenesis and highlight circSAMD4 as a promising therapeutic target and diagnostic biomarker with significant translational potential for improving CI-AKI management. Despite these compelling findings, further studies are needed to determine whether dysregulation of the SRSF3/circSAMD4/CIRBP axis represents a common pathogenic mechanism underlying AKI. Additionally, we confirm the dysregulation of this axis in RTECs derived from biopsy-confirmed ATI patients, with altered expression levels correlating with the severity of kidney injury. Future research should explore whether these molecules—individually or in combination—can serve as reliable biomarkers for the early diagnosis or prognosis of AKI.
Abbreviations
AAV9: adeno-associated virus 9; AKI: acute kidney injury; ATI: acute tubular injury; BSA: bovine serum albumin; CI-AKI: contrast-induced acute kidney injury; cDNA: complementary DNA; CIRBP: cold-inducible RNA-binding protein; circRNAs: circular RNAs; DBA: dolichos biflorus agglutinin; FISH: fluorescence in situ hybridization; gDNA: genomic DNA; GO: gene ontology; H&E: hematoxylin and eosin; HNRNPM: heterogeneous nuclear ribonucleoprotein M; IRI: ischemia-reperfusion injury; PI: propidium iodide; PNA: peanut agglutinin; RBM3: RNA binding motif protein 3; RBP: RNA binding protein; RIP: RNA immunoprecipitation; RTECs: renal tubular epithelial cells; SAMD4A: sterile alpha motif domain-containing protein 4A; SCr: serum creatinine; shRNA: short hairpin RNA; siRNA: small interfering RNA; SRSF3: serine-rich splicing factor 3; LTL: lotus tetragonolobus lectin.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
The authors acknowledge the grants from the National Natural Science Foundation of China (No.82270732). We appreciate the assistance of Figdraw in drawing the graphical abstract.
Data availability
The datasets analyzed in the current study are available from the corresponding author upon reasonable request.
Author contributions
Xi Wu and Shaobin Duan conceived and designed this study and wrote the manuscript. Xi Wu, Ting Wu and Xiufeng Wang conducted the experiments. Xi Wu, Meiyu Zeng, Ting Wu, and Zhiwen Liu performed the data collection and analysis. Chengyuan Tang, Juan Cai, Anqun Chen, Guochun Chen and Yu Liu advised on the experimental design and manuscript writing.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fliser D, Laville M, Covic A, Fouque D, Vanholder R, Juillard L. et al. A European Renal Best Practice (ERBP) position statement on the Kidney Disease Improving Global Outcomes (KDIGO) clinical practice guidelines on acute kidney injury: part 1: definitions, conservative management and contrast-induced nephropathy. Nephrol Dial Transplant. 2012;27:4263-72
2. Nash K, Hafeez A, Hou S. Hospital-acquired renal insufficiency. Am J Kidney Dis. 2002;39:930-6
3. Weisbord SD, Gallagher M, Jneid H, Garcia S, Cass A, Thwin SS. et al. Outcomes after Angiography with Sodium Bicarbonate and Acetylcysteine. N Engl J Med. 2018;378:603-14
4. Fähling M, Seeliger E, Patzak A, Persson PB. Understanding and preventing contrast-induced acute kidney injury. Nat Rev Nephrol. 2017;13:169-80
5. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381-8
6. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-57
7. Cheng X, Ai K, Yi L, Liu W, Li Y, Wang Y. et al. The mmu_circRNA_37492/hsa_circ_0012138 function as potential ceRNA to attenuate obstructive renal fibrosis. Cell Death Dis. 2022;13:207
8. Liu X, Jiang L, Zeng H, Gao L, Guo S, Chen C. et al. Circ-0000953 deficiency exacerbates podocyte injury and autophagy disorder by targeting Mir665-3p-Atg4b in diabetic nephropathy. Autophagy. 2024;20:1072-97
9. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S. et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361-9
10. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846-58
11. Ouyang X, He Z, Fang H, Zhang H, Yin Q, Hu L. et al. A protein encoded by circular ZNF609 RNA induces acute kidney injury by activating the AKT/mTOR-autophagy pathway. Mol Ther. 2023;31:1722-38
12. Liu H, Fang D, Zhang C, Zhao Z, Liu Y, Zhao S. et al. Circular MTHFD2L RNA-encoded CM-248aa inhibits gastric cancer progression by targeting the SET-PP2A interaction. Mol Ther. 2023;31:1739-55
13. Wang P, Huang Z, Peng Y, Li H, Lin T, Zhao Y. et al. Circular RNA circBNC2 inhibits epithelial cell G2-M arrest to prevent fibrotic maladaptive repair. Nat Commun. 2022;13:6502
14. van Zonneveld AJ, Kölling M, Bijkerk R, Lorenzen JM. Circular RNAs in kidney disease and cancer. Nat Rev Nephrol. 2021;17:814-26
15. van Zonneveld AJ, Zhao Q, Rotmans JI, Bijkerk R. Circulating non-coding RNAs in chronic kidney disease and its complications. Nat Rev Nephrol. 2023;19:573-86
16. So BYF, Yap DYH, Chan TM. Circular RNAs in Acute Kidney Injury: Roles in Pathophysiology and Implications for Clinical Management. Int J Mol Sci. 2022;23(15):8509
17. Cheng W, Li XW, Xiao YQ, Duan SB. Non-coding RNA-Associated ceRNA Networks in a New Contrast-Induced Acute Kidney Injury Rat Model. Mol Ther Nucleic Acids. 2019;17:102-12
18. Lin Y, Zhu G, Li X, Yu H, Luo Y, Lin J. et al. Icariin and Competing Endogenous RNA Network: A Potential Protective Strategy Against Contrast-Induced Acute Kidney Injury. Drug Des Devel Ther. 2022;16:2343-63
19. Nishiyama H, Itoh K, Kaneko Y, Kishishita M, Yoshida O, Fujita J. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J Cell Biol. 1997;137:899-908
20. Seufert L, Benzing T, Ignarski M, Müller RU. RNA-binding proteins and their role in kidney disease. Nat Rev Nephrol. 2022;18:153-70
21. De Leeuw F, Zhang T, Wauquier C, Huez G, Kruys V, Gueydan C. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp Cell Res. 2007;313:4130-44
22. Cen C, Yang WL, Yen HT, Nicastro JM, Coppa GF, Wang P. Deficiency of cold-inducible ribonucleic acid-binding protein reduces renal injury after ischemia-reperfusion. Surgery. 2016;160:473-83
23. Zhang F, Brenner M, Yang WL, Wang P. A cold-inducible RNA-binding protein (CIRP)-derived peptide attenuates inflammation and organ injury in septic mice. Sci Rep. 2018;8:3052
24. Zhang P, Bai L, Tong Y, Guo S, Lu W, Yuan Y. et al. CIRP attenuates acute kidney injury after hypothermic cardiovascular surgery by inhibiting PHD3/HIF-1α-mediated ROS-TGF-β1/p38 MAPK activation and mitochondrial apoptotic pathways. Mol Med. 2023;29:61
25. Ajiro M, Jia R, Yang Y, Zhu J, Zheng ZM. A genome landscape of SRSF3-regulated splicing events and gene expression in human osteosarcoma U2OS cells. Nucleic Acids Res. 2016;44:1854-70
26. Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S. et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168-82
27. Wang ZW, Pan JJ, Hu JF, Zhang JQ, Huang L, Huang Y. et al. SRSF3-mediated regulation of N6-methyladenosine modification-related lncRNA ANRIL splicing promotes resistance of pancreatic cancer to gemcitabine. Cell Rep. 2022;39:110813
28. Qin C, Xu F, Yue B, Zhong J, Chai Z, Wang H. SRSF3 and hnRNP A1-mediated m6A-modified circCDK14 regulates intramuscular fat deposition by acting as miR-4492-z sponge. Cell Mol Biol Lett. 2025;30:26
29. Li Y, Wang T, Liao Q, Luo X, Wang X, Zeng S. et al. Loss of Splicing Factor SRSF3 Impairs Lipophagy Through Ubiquitination and Degradation of Syntaxin17 in Hepatocytes. J Lipid Res. 2023;64:100342
30. Cui L, Zhao S, Liu H. SRSF3 Alleviates Ischemic Cerebral Infarction Damage by Activating the PI3K/AKT Pathway. Dev Neurosci. 2024;46:308-18
31. Ortiz-Sánchez P, Villalba-Orero M, López-Olañeta MM, Larrasa-Alonso J, Sánchez-Cabo F, Martí-Gómez C. et al. Loss of SRSF3 in Cardiomyocytes Leads to Decapping of Contraction-Related mRNAs and Severe Systolic Dysfunction. Circ Res. 2019;125:170-83
32. Lin Q, Li S, Jiang N, Shao X, Zhang M, Jin H. et al. PINK1-parkin pathway of mitophagy protects against contrast-induced acute kidney injury via decreasing mitochondrial ROS and NLRP3 inflammasome activation. Redox Biol. 2019;26:101254
33. Zhang Z, Shao X, Jiang N, Mou S, Gu L, Li S. et al. Caspase-11-mediated tubular epithelial pyroptosis underlies contrast-induced acute kidney injury. Cell Death Dis. 2018;9:983
34. Lin Q, Li S, Jiang N, Jin H, Shao X, Zhu X. et al. Inhibiting NLRP3 inflammasome attenuates apoptosis in contrast-induced acute kidney injury through the upregulation of HIF1A and BNIP3-mediated mitophagy. Autophagy. 2021;17:2975-90
35. Shen J, Wang L, Jiang N, Mou S, Zhang M, Gu L. et al. NLRP3 inflammasome mediates contrast media-induced acute kidney injury by regulating cell apoptosis. Sci Rep. 2016;6:34682
36. Li J, Sun X, Yang N, Ni J, Xie H, Guo H. et al. Phosphoglycerate mutase 5 initiates inflammation in acute kidney injury by triggering mitochondrial DNA release by dephosphorylating the pro-apoptotic protein Bax. Kidney Int. 2023;103:115-33
37. Tang C, Han H, Yan M, Zhu S, Liu J, Liu Z. et al. PINK1-PRKN/PARK2 pathway of mitophagy is activated to protect against renal ischemia-reperfusion injury. Autophagy. 2018;14:880-97
38. Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600-7
39. Cao Y, Mi X, Zhang D, Wang Z, Zuo Y, Tang W. Transcriptome sequencing of circular RNA reveals a novel circular RNA-has_circ_0114427 in the regulation of inflammation in acute kidney injury. Clin Sci (Lond). 2020;134:139-54
40. Liu Q, Duan SB, Wang L, Luo XQ, Wang HS, Deng YH. et al. Apelin-13 alleviates contrast-induced acute kidney injury by inhibiting endoplasmic reticulum stress. Ren Fail. 2023;45:2179852
41. Pandey PR, Yang JH, Tsitsipatis D, Panda AC, Noh JH, Kim KM. et al. circSamd4 represses myogenic transcriptional activity of PUR proteins. Nucleic Acids Res. 2020;48:3789-805
42. Romano G, Briguori C, Quintavalle C, Zanca C, Rivera NV, Colombo A. et al. Contrast agents and renal cell apoptosis. Eur Heart J. 2008;29:2569-76
43. Li B, Xi W, Bai Y, Liu X, Zhang Y, Li L. et al. FTO-dependent m(6)A modification of Plpp3 in circSCMH1-regulated vascular repair and functional recovery following stroke. Nat Commun. 2023;14:489
44. Chen C, Yuan W, Zhou Q, Shao B, Guo Y, Wang W. et al. N6-methyladenosine-induced circ1662 promotes metastasis of colorectal cancer by accelerating YAP1 nuclear localization. Theranostics. 2021;11:4298-315
45. Wajant H. The Fas signaling pathway: more than a paradigm. Science. 2002;296:1635-6
46. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134-47
47. Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 2010;24:1073-4
48. Jeong S. SR Proteins: Binders, Regulators, and Connectors of RNA. Mol Cells. 2017;40:1-9
49. Ho JS, Di Tullio F, Schwarz M, Low D, Incarnato D, Gay F. et al. HNRNPM controls circRNA biogenesis and splicing fidelity to sustain cancer cell fitness. Elife. 2021;10:e59654
50. Dong W, Dai ZH, Liu FC, Guo XG, Ge CM, Ding J. et al. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine. 2019;45:155-67
51. Erkelenz S, Grzonka M, Papadakis A, Schaal H, Hoeijmakers JHJ, Gyenis Á. Rbm3 deficiency leads to transcriptome-wide splicing alterations. RNA Biol. 2024;21:1-13
52. Peng X, Li H, Zhang W, Zhang D. Discovery and verification of mmu_Circ_26986/hsa_Circ_0072463 as a potential biomarker and intervention target for sepsis-associated acute kidney injury. Cell Mol Life Sci. 2024;81:154
53. Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S. et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58:870-85
54. Bourgeois B, Hutten S, Gottschalk B, Hofweber M, Richter G, Sternat J. et al. Nonclassical nuclear localization signals mediate nuclear import of CIRBP. Proc Natl Acad Sci U S A. 2020;117:8503-14
55. Corre M, Lebreton A. Regulation of cold-inducible RNA-binding protein (CIRBP) in response to cellular stresses. Biochimie. 2024;217:3-9
56. Schelling JR, Nkemere N, Kopp JB, Cleveland RP. Fas-dependent fratricidal apoptosis is a mechanism of tubular epithelial cell deletion in chronic renal failure. Lab Invest. 1998;78:813-24
57. Raff MC. Social controls on cell survival and cell death. Nature. 1992;356:397-400
58. Hughes J, Johnson RJ. Role of Fas (CD95) in tubulointerstitial disease induced by unilateral ureteric obstruction. Am J Physiol. 1999;277:F26-32
59. Ortiz-Arduan A, Danoff TM, Kalluri R, González-Cuadrado S, Karp SL, Elkon K. et al. Regulation of Fas and Fas ligand expression in cultured murine renal cells and in the kidney during endotoxemia. Am J Physiol. 1996;271:F1193-201
60. Majerciak V, Lu M, Li X, Zheng ZM. Attenuation of the suppressive activity of cellular splicing factor SRSF3 by Kaposi sarcoma-associated herpesvirus ORF57 protein is required for RNA splicing. RNA. 2014;20:1747-58
61. Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell. 2001;7:899-905
62. Maciolek NL, McNally MT. Serine/arginine-rich proteins contribute to negative regulator of splicing element-stimulated polyadenylation in rous sarcoma virus. J Virol. 2007;81:11208-17
63. Kim J, Park RY, Chen JK, Kim J, Jeong S, Ohn T. Splicing factor SRSF3 represses the translation of programmed cell death 4 mRNA by associating with the 5'-UTR region. Cell Death Differ. 2014;21:481-90
64. Rahimian R, Guruswamy R, Boutej H, Cordeau P Jr, Weng YC, Kriz J. Targeting SRSF3 restores immune mRNA translation in microglia/macrophages following cerebral ischemia. Mol Ther. 2024;32:783-99
65. Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844-58
Author contact
![]() Corresponding author: Professor Shaobin Duan, Department of Nephrology, the Second Xiangya Hospital of Central South University, 139 Renmin Road, Changsha 410011, China. Email: duansb528edu.cn.
Corresponding author: Professor Shaobin Duan, Department of Nephrology, the Second Xiangya Hospital of Central South University, 139 Renmin Road, Changsha 410011, China. Email: duansb528edu.cn.

 Global reach, higher impact
Global reach, higher impact