Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2025; 21(15):7104-7117. doi:10.7150/ijbs.125134 This issue Cite
Review
Proteasome: Role in T Cell Function Regulation
1. Cancer Center, Faculty of Health Sciences, University of Macau, Macau SAR, China.
2. Center for Precision Medicine Research and Training, Faculty of Health Sciences, University of Macau, Macau SAR, China.
3. MoE Frontiers Science Center for Precision Oncology, University of Macau, Macau SAR, China.
Received 2025-9-12; Accepted 2025-10-27; Published 2025-11-5
Abstract
The proteasome plays a pivotal role in proteostasis and is deeply involved in various cellular processes. Currently, three proteasome inhibitors have been used for clinical therapies of liquid cancers with favorable efficacy, however they fail to achieve ideal efficiency in clinical trials for solid cancers without a clear clue. Recent studies have unveiled that beyond its canonical role in ubiquitin-mediated protein degradation, the proteasome also elicits a multifaceted influence on T cell fate, steering it through antigen processing, metabolic reprogramming, and the prevention of exhaustion. The proteasome inhibitors may affect tumor progression through their critical role in modulating T cell-mediated antitumor immunity, an understanding of which may solve the mystery underlying the poor efficacy of the proteasome inhibitors for solid cancers and unlock novel strategies for precision immunotherapy. This review will summarize the current knowledge of how proteasome activity weaves its threads through thymic selection, T cell aging, activation, differentiation, and immune evasion. Moreover, we will explore how cutting-edge technologies-CRISPR editing, single-cell proteomics, and AI-driven drug design can expand the application of the proteasome inhibitors in the treatment of cancer and autoimmune diseases.
Keywords: proteasome, proteasome inhibitors, autoimmune diseases, anti-tumor immunity
1. Introduction
In 2004, the Nobel Prize in Chemistry was awarded to Prof. Aaron Ciechanover, Prof. Avram Hershko, and Prof. Irwin Rose for their groundbreaking discovery of ubiquitin-mediated protein degradation, a process primarily mediated by the proteasome. Recent advancements have unveiled that the proteasome's degradation capabilities extend beyond ubiquitin-tagged proteins, encompassing even those devoid of ubiquitin labels [1]. This degradation of non-ubiquitinated proteins is orchestrated by Midnolin, which engages in direct interactions with proteasome subunits [1, 2]. The proteasome, with its unique and indispensable functions, plays a pivotal role in proteostasis and is deeply involved in key cellular processes, including cell cycle regulation, DNA damage repair, cell death regulation, drug resistance, inflammatory responses, immune responses, and more [3-9].
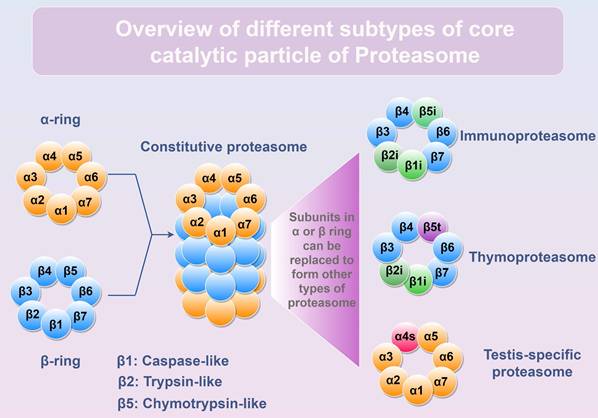
Proteasome is a huge protein complex that manifests in diverse subtypes (26S, 30S, or Hybrid proteasome) through the interplay of core catalytic particle (CP, including Constitutive proteasome, Immunoproteasome, Thymoproteasome, and Testis-specific proteasome) with various regulatory particles (RP, including PA700, PA28αβ, PA28γ, PA200, and PI31) [10]. By binding with different RPs, the assembled proteasome exhibits distinct substrate specificity and degradation rates [11]. Within the constitutive 20S CP, there exist 4 heptameric rings arranged in the order of α1-7/β1-7/β1-7/α1-7, with the β ring responsible for degrading cargo delivered to proteasome. Among the seven β-subunits (β1-β7), three-β1/PSMB6, β2/PSMB7, and β5/PSMB5-possess proteolytic capacity (Figure 1). However, in immune cells, these three catalytic subunits are replaced by (β1i/LMP2, β2i/MECL1, and β5i/LMP7) to form immunoproteasome [12]. Notably, upon exposure to interferon (IFN)-γ, tumor necrosis factor (TNF)-α or other stimuli, the expression of β1i, β2i, and β5i is induced and incorporated into newly synthesized 20S CP in normal cells, leading to the assembly of the immunoproteasome and the execution of its biological functions (Table 1). The immunoproteasome is more efficient at generating antigen peptides for loading onto major histocompatibility complex class I (MHC-I) than the constitutive proteasome [13], thereby possessing greater potency in activating T cell-mediated immunity.
Stimuli that can induce the expression of immunoproteasome subunit
| Stimuli | Impact | Ref. |
|---|---|---|
| IFN-γ | Induces the transcription and translation of immunoproteasome subunits, PA28αβ, and PA28γ | [14] |
| TNF-α | Induces the transcription and translation of immunoproteasome subunits. Often synergy with IFN-γ. | [15] |
| Retinoic acid | Induces the transcription and translation of immunoproteasome subunits. | [16] |
| Nitric oxide | Induces the expression of immunoproteasome subunits (β1i and β5i). | [17] |
| Toll-like receptor agonists | Induces the expression of immunoproteasome subunits (β5i). | [18] |
| Type I interferons | Induces the transcription and translation of immunoproteasome subunits | [19] |
| Mammalian target of rapamycin (mTOR) signaling | Promotes immunoproteasome formation via PRAS40 phosphorylation | [20] |
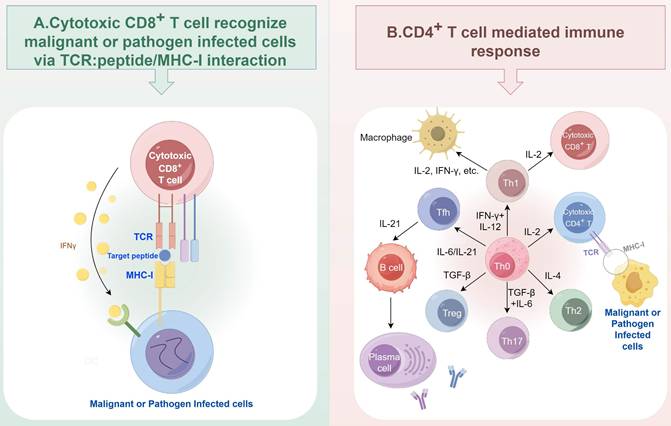
T cell mediated immune response is a cornerstone of adaptive immunity. Activated CD8+ T cells, known as cytotoxic lymphocytes (CTLs), rely on their T cell receptor (TCR) to recognize and eliminate the malignant cells or pathogen- infected cells via recognizing the peptide presented by MHC-I molecules (Figure 2A). CD4+ T cells are another major subtype of T cells, which are composed of several major subtypes, including Th1, Th2, Th17, regulatory T (Treg), T follicular helper (Tfh), cytotoxic CD4+ T cells, and so on. The different subtypes of CD4+ T cells can be differentiated from Th0 cells upon the exposure of different cytokines [21]. They mainly secrete cytokines to assist the activation of other immune cells, including macrophages and B cells (Figure 2B). Each subset of CD4+ T cells can have both anti-tumor and pro-tumor functions [22]. For example, the IFN-γ released by Th1 cells can recruit and enhance the proliferation and cytotoxicity of CD8+ T cells [23], while the sustained exposure to this cytokine can lead to tumor growth and metastasis [24]. In anti-cancer immunity, defects in antigen processing and presentation represent a key mechanism by which tumor cells escape surveillance by both cytotoxic CD8+ T cells and CD4+ T helper cells [25-27]. During the TCR signal-mediated T cell activation process, proteasome also plays critical roles. Proteasome can be a negative regulator for TCR signal by degrading T cell specific tyrosine kinase p56lck [28], and T cell development essential adaptor protein SLP-76 [29] to suppress T cell activation. The proteasome also acts as a positive regulator. Since TCR activation increases the demand for protein turnover, the proteasome is essential for maintaining T cell function [30].
Different subtypes of core catalytic particle of Proteasome. The α1-7 or β1-7 subunits form α-ring or β-ring, respectively. The core catalytic particle is assembled by two α rings and two β rings. Subunits inside α or β ring can be replaced to generate different types of proteasomes. β1, β2, and β5 subunits are the proteolytic subunits inside the constitutive proteasome. In immune cells, β1i, β2i, and β5i can replace β1, β2, and β5 to form immunoproteasome. In thymus, β1i, β2i, and β5t replace β1, β2, and β5 to form thymoproteasome. In testis, α4s subunit can replace α4 to form testis-specific proteasome. Figure was generated at FigDraw.
T cell mediated immune response. (A) Cytotoxic CD8+ T cells eliminate malignant or pathogen infected cells via TCR: peptide/MHC-I interaction. (B) Major subtypes of CD4+ T cells and their interaction with macrophages and B cells. Figure was generated at FigDraw.
Beyond its well-recognized role in antigen generation to facilitate T cell-mediated immunity, the proteasome also critically regulates T cell survival and differentiation, as highlighted by recent research. Especially, the approval of proteasome inhibitors in clinical practice for cancer treatment marked a milestone, spurring exploration of their immunomodulatory functions. In this review, we summarize current understanding of proteasome-mediated regulation in T cell immunity and evaluate the therapeutic potential of proteasome inhibitors for immune system modulation.
2. Functions of proteasome to regulate T cell fitness, development and differentiation
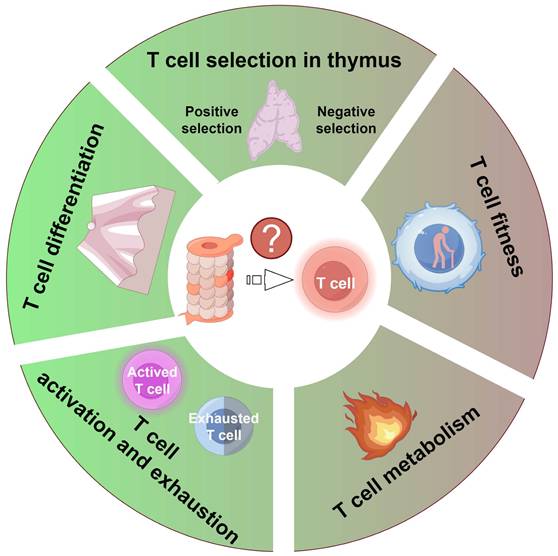
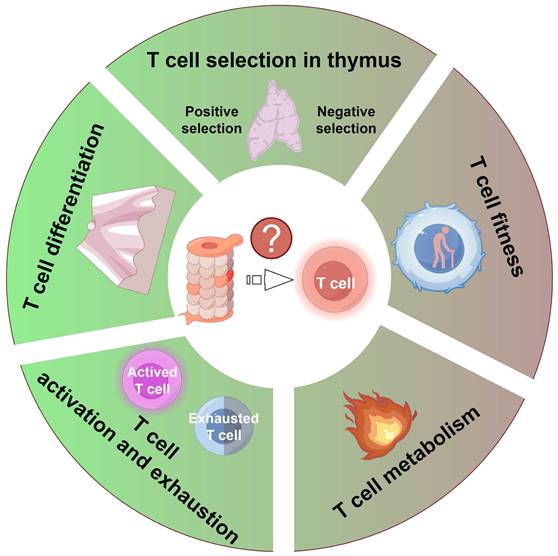
Given the proteasome's indispensable role in maintaining cellular homeostasis and regulating diverse signaling pathways, its activity critically governs T cell function at multiple levels. This section discusses how proteasome activity modulates T cell-mediated immune responses (Figure 3).
2.1 Proteasome and thymic selection process
In the intricate microcosm of the thymus, T cells undergo both positive and negative selection, a journey that begins in the cortex and traverses into the medulla [31]. Within cortical thymic epithelial cells (cTEC), the β5t/PSMB11 subunit together with the β1i/LMP2, β2i/MECL1 forms thymoproteasome [32]. Notably, the incorporation of β5t into the thymoproteasome results in a decrease in chymotrypsin-like activity, as compared to proteasomes incorporating β5 or β5i [33]. This specialized thymoproteasome generates unique cleavage motifs, which are loaded onto low-affinity TCR ligands, thereby facilitating the specific positive selection of functionally competent CD8+ T cells [33, 34]. However, aberrant expression of thymoproteasome outside of thymus cause the dysfunction of CD8+ T cells homeostasis, accumulation of exhausted or memory CD8+ T cells, and autoreactive T cell response [35]. In addition, the polymorphisms of PSMB11, especially G49S mutation, have been implicated in influencing T cell selection [36]. After navigating the positive selection gauntlet in the cortex, T cells proceed to undergo negative selection in the medulla, a process mediated by medullary thymic epithelial cells (mTECs). These cells express β5i/LMP7, contributing to the formation of the immunoproteasome. Utilizing the lymphocytic choriomeningitis virus (LCMV) infection model, it was found that mice lack of LMP7 cannot generate glycoprotein (GP)118-125-specific CTL response [37]. Thus, proteasome displays a critical role in T cell development in thymus and the generation of cytotoxic T cell repertoire.
Proteasome activity exerts a pivotal influence on T cell function by modulating several crucial aspects: 1) the positive and negative selection of T cells within the thymus; 2) the fitness of T cells due to proteostasis dysfunction; 3) T cell metabolism; 4) the activation and exhaustion processes of T cells; and 5) the differentiation process of T cells. Figure was generated at FigDraw.
2.2 Proteasome and T cell fitness
Dysfunction of proteostasis, a hallmark of aging, has been implicated in the diminished functionality of T cells [38, 39]. One pivotal factor contributing to this loss of proteostasis is the gradual decline in proteasome activity with age. In human T cells, the aging dependent proteasome activity loss results in decline of IL-2 receptor expression in response to TNF-α stimulation [40]. Moreover, CD4+ T cells treated with proteasome inhibitors exhibited a senescence-associated phenotype through accumulating p21 and p27 to induce cell cycle arrest and stabilizing p53 to induce cell apoptosis [30, 41]. Of note, the specific knockout of Rpn13, a proteasome subunit with ubiquitin binding function, in T cells results in an increased frequency of PD-1+/CD44High/CD4+ T cells upon TCR stimulation, indicating the onset of T cell senescence [30]. Collectively, these studies suggested a potent anti-aging role of proteasome in T cells, which is crucial for the combating of age-related decline of T cell function.
In addition, proteasome also acts as a critical but double-edged sword in T cell leukemogenesis, whose effect is determined by which key regulatory proteins it targets for destruction [42, 43]. Wang et al. reveal that Hsp90 chaperone can prevent the degradation of oncogenic driver intracellular Notch1 (ICD1) by proteasome and therefore promote cancer progression. Thus, inhibiting Hsp90 re-arms the proteasome to destroy the ICD1 [42]. Conversely, Jiang et al. show that the Aurora B kinase (AURKB) mediated c-Myc phosphorylation at serine 67 competes with its phosphorylation at threonine 58, which serves as a signal for c-Myc degradation. Ultimately, the AURKB-c-Myc axis promotes T cell leukemogenesis [43].
2.3 Proteasome and T cell metabolism
A study conducted by Widjaja et al. revealed that proteasome activity is directly linked to CD8+ T lymphocyte metabolism [44]. Inhibition of proteasome activity in CD8+ T cells results in the stabilization of c-Myc, a transcription factor critical for the controlling of glycolysis [44]. This groundbreaking research underscores the proteasome's pivotal role in directly modulating T cell metabolism by regulating the stability of crucial proteins integral to energy production and metabolic pathways.
2.4 Proteasome and T cell activation or exhaustion
2.4.1 Proteasome regulating CD4+ T cell function
The suppression of proteasome in CD4+ T cells results in the loss of activation-associated and functional receptor expression on the cell surface, including CD25, CD28, CD27, CD120b, CD95 and CD134. Additionally, it impairs the production of cytokines such as IFN-γ, TNF-α, IL-4 and IL-5 upon dendritic cell activation. This phenomenon is attributed to the inhibition of nuclear translocation and abundance of nuclear factor of activated T cells (NFAT) c2, ultimately disrupting T cell function [41]. As proteasome directly control de novo protein synthesis via decreasing intracellular ATP level [45], it is plausible that the inhibition of proteasome activity hinders the translation of these cytokines. Moreover, studies have observed impaired proteasome activity, cell cycle arrest, and cell apoptosis in T cells following the introduction of a β5i specific mutation (G197V) along with Psmb5 specific knockout [46]. Interestingly, another study utilizing an in vitro CD4+ T cell activation model indicated that inhibition of proteasome activity can reduce the percentage of activation induced cell death through reducing the activity of nuclear factor-κB (NF-κB) [47]. Collectively, these studies illuminate the proteasome's multifaceted role in preserving the functional integrity of T cells, underscoring its intricate involvement in regulating various aspects of T cell biology.
2.4.2 Proteasome regulating CD8+ T cell function
For CD8+ T cells, it was found that Bortezomib treatment of tumor-bearing mice resulted in elevated levels of Notch signal components in lymphoid tissues. This, in turn, upregulated the secretion of IFN-γ and the expression of effector molecules such as perforin, granzyme B, and the T-box transcription factor eomesodermin [48]. Importantly, Bortezomib treatment could rescue the dysfunction state of CD8+ T cells in tumor microenvironment by enhancing the crosstalk between Notch intracellular domain and NF-κB [48]. Moreover, the proteasome plays a crucial role in the degradation of the RNA-binding protein Regnase-1 in T cells. Regnase-1 facilitates the transcription of c-Rel, Ox40, and IL-2 transcripts, thereby acting as a restraint on T cell activation [49].
In addition, it was recently reported that the hyperaction of translation in CD8+ T cells leads to increased proteotoxic stress and drives T cell exhaustion [50-52]. As a result, pharmacological activation of proteasome was reported able to prevent T cells exhaustion, a status often observed when undergoing chronic infections or combating with cancer, and increase their capacity to fight against cancer [53]. The mechanisms behind that are exhausted T cells experience high level of oxidative stress to accumulate oxidized proteins, which will inevitability impair their fitness and function [53]. Therefore, the activation of proteasome relieves proteotoxic stress inside T cells when undergoing TCR stimulation and prevents the entry into exhaustion status. Utilizing a high-throughput screening strategy, a novel molecule named compound A was found able to bolster immunoproteasome activity and antigen presentation to increase the anti-tumor efficacy of both allogenic and autologous T cells [54]. Collectively, these studies revealed that the proteasome could either activate T cells or induce T cells exhaustion based on different contextual conditions.
2.5 Proteasome and T cell differentiation
The proteasome was also found to play a critical role in regulating the differentiation and fate decisions of T cells. The E3 ligase Cul5 was revealed as a critical factor to determine the decision of CD4+ T cell differentiated into Treg or Th cells by regulating the IL-4 receptor signaling [55]. CD8+ T cells undergo asymmetric segregation during their initial division upon encountering microbial infections [56], resulting in daughter cells exhibiting varied proteasome activity [44]. Analysis of CD8+ T cells derived from Listeria monocytogenes infected host showed the daughter cells with low proteasome activity (IL-2RaHigh/CD62LLow) tend to differentiate into terminal effector fate, whereas the daughter cells (IL-2RaLow/CD62LHigh) with high proteasome activity will tend toward a long-lived memory state. Proteasome activation diminishes Myc expression, steering T cell differentiation towards the memory phenotype and disrupting the balance between effector and memory T cell populations [44]. Furthermore, the age-related decline in proteasome activity contributes to the differentiation of T cells into effectors rather than memory cells [39].
In the auto-immune or inflammatory disease scenario, the proteasome's activity is also involved. For example, in the experimental Sjögren's syndrome model, LMP7 is observed to be highly expressed within Th17 cells [57]. Consequently, inhibiting proteasome activity via Bortezomib suppresses the differentiation of Th17 cells, thereby alleviating the syndrome's manifestations [57]. Furthermore, the repression of LMP7 has also been found to mitigate the inflammatory phenotype in the brain associated with LCMV-induced meningitis [58]. In the case of Sézary syndrome, it was found that dysfunction of proteasome in T cell led to the accumulation of cytotoxic T-lymphocyte antigen-4 (CTLA-4) and GATA binding protein 3 (GATA3). This accumulation, in turn, facilitates Th2 differentiation and impairs T cell proliferation and responsiveness [59]. Furthermore, treating the CD4+ T cell mediated experimental autoimmune neuritis (EAN) model with PR-957 to inhibit the immunoproteasome reduced the syndrome of this disease via suppressing the differentiation of Th17 cells [60]. Similarly, in a colitis model induced by DSS, it was discovered that proteasome inhibition abolished the differentiation capacity of Th17 cells by impeding the activation of naïve T cells. Notably, this inhibition did not impact the survival of already differentiated Th17 and Treg cells [61]. These findings underscore the intricate involvement of proteasome activity in the pathogenesis of autoimmune and inflammatory diseases, highlighting the role of proteasome in maintaining homeostasis.
3. Proteasome modulation to shape T cell response
Given the importance of proteasome in T cells as described above, the prospect of harnessing proteasome inhibitors or activators for therapeutic interventions in cancer or T cell-related autoimmune diseases becomes an alluring endeavor. To this end, we have revisited the current repertoire proteasome inhibitors and activators and summarized them in Table 2.
List of known proteasome inhibitors and activators
| Category | Name | Type | Status | Ref. |
|---|---|---|---|---|
| Inhibitor | Bortezomib | Dipeptide boronic acid/Reversible inhibitor | Approved to treat multiple myeloma and mantle cell lymphoma | [62] |
| Carfilzomib | Epoxyketone-peptide/Irreversible inhibitor | Approved to treat multiple myeloma and mantle cell lymphoma | [63] | |
| Ixazomib | Dipeptide boronic acid/Reversible inhibitor | Approved to treat multiple myeloma and mantle cell lymphoma | [64] | |
| MG132 | Peptide derivates/Reversible inhibitor | Preclinical | [65] | |
| MG262 | Peptide derivates/Reversible inhibitor | Preclinical | [66] | |
| Marizomib | β-lactone-γ-lactam /Irreversible inhibitor | Phase III clinical trial to treat glioblastoma | [67, 68] | |
| Oprozomib | Tripeptide epoxyketone/Irreversible inhibitor | Phase Ib/II clinical trial to treat multiple myeloma | [69, 70] | |
| Delanzomib | Boronic acid peptide/Reversible inhibitor | Phase I/II clinical trial to treat multiple myeloma | [71, 72] | |
| Epoxomicin | Epoxyketone/Irreversible inhibitor | Preclinical | [73] | |
| Lactacystin | Cyclic α,α-disubstituted amino acid including a γ-lactam-β-lactone/Irreversible inhibitor | Preclinical | [74] | |
| Disulfiram/Cu | Likely targets protein cysteine not at the binding site | Approved to treat chronic alcoholism | [75] | |
| Epigallocatechin-3-gallate | Polyphenol/Irreversible inhibitor | Preclinical | [76] | |
| Beta-hydroxy beta-methylbutyrate | Leucine metabolite with unidentified mechanism to inhibit proteasome activity | Preclinical | [77] | |
| PR-957 | Epoxyketone-peptide/ Irreversible immunoproteasome inhibitor | Preclinical | [60] | |
| PI31 | Endogenous 20S proteasome binding protein | - | [78] | |
| Activator | PA28α/β | Endogenous RP | - | [10] |
| PA28γ | Endogenous RP | - | [10] | |
| PA200 | Endogenous RP | - | [10] | |
| cAMP | Activate PKA to phosphorylate Rpn6/PSMD11 | - | [79] | |
| cGMP | Activate PKG to increase phosphorylation on proteasome subunits | - | [80] | |
| ZFAND5/ZNF216 | Interact with 19S proteasome to induce conformation change | - | [81, 82] | |
| Ursolic acid | Triterpenoid | Preclinical | [83] | |
| Betulinic acid | Triterpenoid | Preclinical | [84] | |
| Oleuropein | Triterpenoid | Preclinical | [85] | |
| MK-886 | Indole-based compound | Preclinical | [86] | |
| TCH-155 | Imidazolines | Preclinical | [87] | |
| Low concentration (0.04-0.08%) SDS | Synthetic organosulfate salt/Likely via inducing gate opening of the proteasome | Preclinical | [88] | |
| Chlorpromazine | Phenothiazines | Approved to treat mental health condition | [89] | |
| Pyrazolones | Five-membered lactam ring | Preclinical | [90] | |
| Fluspirilene | Allosteric modulate proteasome active site | Approved to treat schizophrenia | [91] |
3.1 Direct modulation of T cell function by proteasome inhibitors
Given the widespread clinical and preclinical use of proteasome inhibitors, this review focuses on their role in modulating T cell responses. As monotherapy, these inhibitors have been investigated for cancer and autoimmune disease treatment due to their capacity to either augment or suppress T cell activity. In tumor bearing mouse models, Bortezomib treatment did not affect the total number of lymphocytes, and dendritic cells [92]. Also, this treatment did not reduce the levels of costimulatory molecules (CD80, CD86, and CD40) in the dendritic cells and its ability to present MHC associated antigen to T cells. Instead, Bortezomib treatment increased the phosphorylation of NF-κB p65 in CD8+ T cells, which induce the expression of CD3ζ and IL-2 receptor-α and maintain the section of IFN-γ, suggesting the activation of CD8+ T cell function in vivo. In the context of adoptive T cell therapy, Bortezomib was combined with adoptive T-cell immunotherapy to treat the Rag2-/- tumor bearing mice and observed reduced lung metastasis and increased survival rate [92]. Notably, two independent studies revealed the co-treatment with proteasome inhibitors (Bortezomib or Carfilzomib) boost the efficiency of anti-B cell maturation antigen (anti-BCMA) Chimeric antigen receptor T (CAR-T) cell in multiple myeloma animal model and human patients via blocking the proteasome mediated degradation of BCMA to stabilize its in cell membrane [93, 94]. In melanoma, Bortezomib treatment sensitizes cancer cells to the attack of antigen specific cytotoxic T lymphocytes (CTL). Mechanistically, it was found that Bortezomib treatment induced the accumulation of NOXA to stimulate the release of second mitochondria-derived activator of caspase (SMAC) from mitochondrial upon the attack from effector proteins caspase 8 and granzyme B release from CTL [95]. In malignant mesothelioma, Bortezomib treatment induced the generation and recruitment of functional CD4+ and CD8+ T cells [96]. These studies highlight the potential of proteasome inhibitors to enhance antitumor T-cell function in cancer immunotherapy.
3.2 Indirect modulation of T cell function by proteasome inhibitors
In addition to their direct modulation of T cell function, recent research has illuminated the indirect influence that proteasome activity regulators exert on T cell performance by reshaping the tumor microenvironment. The suboptimal outcomes observed with proteasome inhibitors in the treatment of solid tumors [97] have prompted a shift towards exploring the combination of proteasome inhibitors with other drugs, which have achieved variably enhanced therapeutic effects (Table 3 and Figure 4), and stimulated the activation of the immune system to fight against solid tumors [98-102].
Effective proteasome inhibitors centered drug combinations to treat solid tumors
| Proteasome inhibitor | Additional drug | Tumor type | Treatment outcome | Ref. |
|---|---|---|---|---|
| Bortezomib | TM | Breast cancer | Inhibit breast cancer (4T1, EMT6, HP10069, and Py8119) growth in immunocompetent mice through activating CD8+ T cell | [99] |
| Plerixafor | Breast cancer | |||
| Cisplatin | Breast cancer | Inhibit MDA-MB-231 tumor growth in nude mice relied on their direct killing effects on tumor cells (Phase I clinical trial) | [8] | |
| Nedaplatin | Breast cancer | Inhibit MCF7 tumor growth in zebrafish xenografts | [103] | |
| PD0166285 | Liver Cancer | Induction of pyroptosis to inhibit the growth of liver cancer in immunocompetent mice | [104] | |
| Vorinostat | Cervical cancer | Induce the generation of anti-specific CD8+ T to inhibit tumor growth in immunocompetent mice | [101] | |
| Cisplatin | Lung cancer | Synergistically inhibit lung cancer cell growth in vitro | [105] | |
| Gefitinib | Lung cancer | |||
| Gemcitabine | Lung cancer | |||
| Vinorelbine | Lung cancer | |||
| Paclitaxel, carboplatin, and radiation therapy | Lung cancer | Lung cancer patients treated with those combinations showed increased overall survival (Phase I/II clinical trial) | [106] | |
| Carfilzomib | Lopinavir | Renal cell carcinoma | Synergistically inhibit renal cell carcinoma cell growth in vitro though induction unfolded protein response | [107] |
| Nelfinavir | Renal cell carcinoma | |||
| HRS-4642 | KRAS G12D-mutant cancer | Synergistically inhibit the growth of pancreatic cancer, colorectal cancer, and non-small lung cancer through Notch4 inhibition and interferon alpha response activation (Phase I clinical trial) | [100] | |
| Irinotecan | Small cell lung cancer | Objective clinical response was 19% (Phase Ib clinical trial) | [108] | |
| ONX 0912 | Head and neck cancer | Induce cell death via suppression of Mcl-1 or autophagy | [109] | |
| CUDC-101 | Thyroid cancer | Induce cell death via induction of p21 expression | [110] | |
| Ixazomib | Dinaciclib | Hepatocellular carcinoma | Synergistically inhibit the growth of human patient derived organoids and xenografts models | [111] |
Representative proteasome inhibitor centered combination therapy for activating T cell mediated immune response. The combination of TM with bortezomib or plerixafor with bortezomib induced the generation of cytotoxic, antigen-specific CD8+ T cells in breast cancer. Figure was generated at FigDraw.
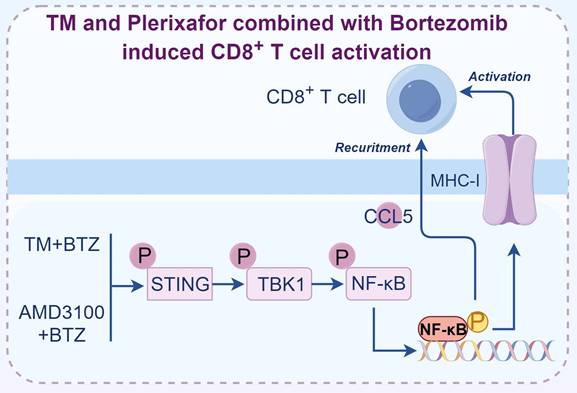
Realizing the combination of proteasome inhibitors with different drugs illicits variable effect, we suspected that the drug pairs used by different studies might not be optimal. To select the drug combinations with higher therapeutic efficacy, we employed a drug combination screening approach to select FDA-approved drugs that can augment the efficacy of Bortezomib [99]. To this end, we selected both BRCA1 wild-type and mutant breast cancer cells to do screening with the aim to identify effective drug pairs. After screening the 115 FDA-approved drugs, we found only two drugs, TM and Plerixafor, were able to significantly enhance the efficacy of Bortezomib in inhibiting the growth of breast cancer cell lines, tumor slices, and organoids. Interestingly, when we applied these drug combinations for breast cancer treatment in vivo, the tumor inhibition effects were only observed in immune intact mice but not the immunodeficient mice, highlighting the importance of T cells in mediating the killing effects of drug combinations [99]. The killing effects of drug combinations were compromised when utilizing anti-CD8α antibody to deplete CD8+ T cells in immune intact mice, suggesting drug combinations treatment induces the generation and expansion of cytotoxic CD8+ T cells, which are responsible for the inhibition of tumor growth. To explore the detailed mechanisms that mediate the activation of CD8+ T cells, we analyzed the drug combinations treated tumor tissues and found tumor microenvironment was reshaped through CCL5 mediated CD8+ T cell recruitment. In the meantime, we showed that the cancer cells displayed high levels of MHC-I upon drug combination treatment mediated by the activation of STING/TBK1/NF-κB pathway [99]. Taken together, we provided a strategy that inhibiting proteasome activity in cancer cells promotes the production of MHC-I and CCL5, which facilitates the activation of CD8+ T cells.
4. Challenges in targeting proteasome for therapeutic purposes
The clinical use of proteasome inhibitors for cancer treatment provided a proof-of-concept foundation for targeting proteasome to treat human diseases. Future efforts are focused on optimizing these strategies for broader clinical use, with a nuanced understanding of their complex immunomodulatory effects. This involves a delicate balancing act: enhancing antitumor immunity while mitigating the disruption of immune homeostasis.
4.1 Debates on proteasome inhibition in disease treatment
Debates persist regarding the therapeutic efficacy and immunomodulatory effects of proteasome inhibitors across diseases. In autoimmune contexts, their application remains contentious. For rheumatoid arthritis (RA), proteasome inhibitors suppress proinflammatory cytokine release by inhibiting NF-κB activation, yet their long-term disease control efficacy is incompletely established [112, 113]. Another debate is the “bounce-back effect” of applying proteasome inhibitors for disease treatment [114]. For example, the specific immunoproteasome inhibitors were initially believed to deplete of pathogenic immune cells but not non-hematopoietic cells, however, it was recently found those drugs did not lead to immune cell death as anticipated [115]. Subsequent studies revealed that compensatory upregulation of constitutive proteasomes confers cellular protection against immunoproteasome inhibitor-induced apoptosis [115].
In the context of cancer therapy, proteasome inhibitors paradoxically may suppress antigen presentation and NF-κB signaling-potentially dampening host immunity and limiting synergy with immunotherapies [116]. Nevertheless, recent evidence demonstrates that tumor cell-targeted proteasome inhibition triggers antitumor CD8+ T cell responses [99-101, 104]. This suggests cell type-specific outcomes: proteasome inhibition in malignant versus immune cells exerts divergent effects on CD8+ T cell functionality.
4.2 Challenges in targeting proteasome for therapeutic purpose
Although proteasome inhibitors have been approved for the treatment of multiple myeloma and mantle T cell lymphoma in clinical practice, their utilization in cancer therapy continues to confront several hurdles. Firstly, the currently used proteasome inhibitors, such as Bortezomib and Carfilzomib, can target both the constitutive and immunoproteasome. The lack of specificity of proteasome inhibitors results in adverse effects, such as peripheral neuropathy due to their impact on normal cells, thereby limiting their therapeutic efficacy [117]. Furthermore, when these inhibitors are applied to treat cancer cells, they may inhibit the function of cytotoxic immune cells such as nature killer cells, which also rely on protein turnover to produce cytokines [41]. Secondly, both intrinsic and acquired resistance to proteasome inhibitors treatment have been observed. In multiple myeloma, some patients are initially resistant or become resistant to treatment of proteasome inhibitors [118]. Various resistance mechanisms were involved, including Wnt signal activation, ABCB1 overexpression, and the “bounce-back effect” caused by proteasome inhibitor treatment [114, 119, 120]. Thirdly, the three approved proteasome inhibitors in clinical only demonstrate treatment efficacy in liquid tumors but not in solid tumors. This is partly due to insufficient delivery of proteasome inhibitors to solid tumor sites, resulting from their poor penetration and limited tumor blood supply, which compromises efficacy [97]. Therefore, identifying novel proteasome inhibitors and exploring proteasome inhibitor-centered drug combination therapies effective against solid tumors is essential. For example, the combination of TM+Bortezomib or Plerixafor+Bortezomib (Figure 4), which significantly inhibit tumor growth in immunocompetent mouse models [99], warrant clinical evaluation to assess their efficacy in human patients. Last but not least, the ethical considerations of proteasome inhibitors for disease treatment, especially the off-label usage of proteasome inhibitors and how to select the appropriate patients for treatment, still require further discussion.
4.3 New strategies to deal with the current challenges faced by proteasome inhibitors centered therapy
To address these aforementioned challenges, extensive research has been conducted to explore and develop novel strategies. Regarding the first challenge, increased efforts have been directed towards the discovery of subunit- or subtype-specific proteasome inhibitors, aiming to mitigate side effects while maximizing therapeutic efficacy [121-123]. Given that current proteasome inhibitors primarily target the β5/PSMB5 subunit, researchers have developed a β2/PSMB7-specific inhibitor, which has demonstrated the ability to sensitize cancer cells to β5/PSMB5 inhibition [121, 122]. To avoid the cross effects of proteasome modulators on different types of cells, it is necessary to develop cell-type specific delivery systems to targeted delivery of reagents to induce death in tumor cells and inhibitory immune cells, while preserving and enhancing the function of tumor killing immune cells. Given that proteasome plays distinct functions in the activation to exhaustion process of T cells [47, 50, 51], the function of T cell should be assessed at first before the selection of proteasome inhibitors or activators for disease treatment to achieve optimal treatment outcomes. To counteract ABCB1 overexpression mediated drug resistance, proteasome inhibitors were combined with ABCB1 inhibitors (Verapamil, Nelfinavir, and Lopinavir) to block the efflux of proteasome inhibitors to the extracellular space, which will definitely increase the concentration of proteasome inhibitors inside the cells to eliminate the cancer cells [119, 120]. Addressing the “bounce-back effect” where cancer cells develop resistance to proteasome inhibitors, researchers have identified promising combinations. Specifically, the integration of Cisplatin [124], N-glycanase 1 (NGLY1) inhibitors [125], and bromodomain extra-terminal (BET) inhibitors [126] has shown the capacity to suppress the transcriptional activity of Nuclear Factor, Erythroid 2 Like 1 (NFE2L1, Nrf1). This suppression completely reverses the resistance effect and elicits robust anti-tumor responses from proteasome inhibitors. In response to the insufficient accumulation of proteasome inhibitors in solid tumors, the development of innovative drug delivery strategies or novel inhibitors with superior tumor targeting capabilities is imperative [127, 128]. To deal with patient selection challenge, PTEN mutation has emerged as a biomarker predictive of cholangiocarcinoma sensitivity to proteasome inhibitors. Consequently, clinical trials using proteasome inhibitors for cholangiocarcinoma treatment have yielded encouraging results [129, 130]. In our recent study, we also proposed that cancer patients with low STAT3/PRAKK1 or STAT3/PRAKK2 ratio tend to have better response to Bortezomib treatment compared with those with high ratio. This suggests that measuring and calculating the gene ratio of these two genes may serve as novel predictive biomarkers for Bortezomib treatment outcomes in cancer patients [99]. Despite the significant challenges, identifying specific biomarkers for predicting proteasome inhibitor treatment outcomes remains a crucial endeavor. At last, the tumor immunomicroenvironment reshaping capability of proteasome modulators lays a foundation to combine proteasome-centered therapies with immune checkpoint inhibitors, CAR-T cell therapy, and HDAC inhibitors. In conclusion, the therapeutic landscape of proteasome modulation is rapidly advancing from simple inhibition to sophisticated immunomodulation. By leveraging the above-described approaches, we can unlock the full potential of proteasome modulators to treat cancer and autoimmune diseases with greater efficacy and precision.
5. Conclusion
In this review, we have systematically delineated the current understanding of proteasome's activity in orchestrating T cell function. We have delved into the role of proteasome inhibitors in modulating the anti-tumor effects of T cells, explored combination strategies to augment the therapeutic efficacy of proteasome inhibitors in cancer treatment, and examined their impact on T cell activation. Given the proteasome's indispensable and unique function within cells, as well as its pivotal role in the pathogenesis of numerous human diseases, modulating proteasome activity stands out as an alluring therapeutic strategy, promising to pave the way for personalized medicine. The proteasome plays a critical role in modulating T cell function, and its inhibition holds promise as a strategy to forestall T cell dysfunction, thereby overcoming a formidable hurdle in cancer immunotherapy. As research in this domain progresses, the immunoproteasome is poised to remain a key target for regulating T cell function and treating a spectrum of diseases, including cancer, autoimmune disorders, and neurological conditions [131]. The development of more specific immunoproteasome inhibitors and an in-depth understanding of their long-term immunological effects will be indispensable in advancing this field. As our insights into the proteasome's mediation of immune crosstalk deepen, targeting these intricate interactions emerges as a viable therapeutic approach for personalized immunotherapies. Personalized proteasome-targeted therapies, meticulously tailored to individual genetic profiles, immune statuses, and disease characteristics, hold the potential to usher in more efficacy and less toxic treatment modalities in the future. The continued exploration of proteasome research in immunology is expected to broaden our understanding of immune regulation and disease pathogenesis, heralding new horizons in therapeutic interventions.
5.1 Future perspectives on proteasome and T cell mediated immunity
5.1.1 Emerging technology to study the function of proteasome function
To gain a deeper understanding of the diverse roles of the proteasome in various cell populations, the adoption of innovative technologies is imperative. Firstly, CRISPR-Cas technology, with its conventional gene knockout/knock-in capabilities and cutting-edge base editing methods, allows for precise manipulation of genes associated with the proteasome system in T cells. This precision editing accelerates our comprehension of proteasome activity or specific gene mutations within the proteasome influence T cell function [132-135]. Secondly, the Mass spectrometry-based proteomics approach offers a means to elucidate the dynamic proteome of the proteasome complex under varying conditions. This could provide novel insights into the mechanisms regulating proteasome function [136]. Thirdly, the emerging of activity-based probes facilitates the identification of the spatial localization of the proteasome within cells and tissues [137]. Furthermore, high-throughput functional single-cell proteomics emerges as a potent tool to dissect cellular heterogeneity and study proteasome activity across all cells within the tumor microenvironment [138-140]. The combination of activity-based probes and single-cell proteomics enables researchers to dissect variations in proteasome activity among different immune cell subsets and elucidate its role in anti-tumor immunity. Building upon these technologies, increasing research focus has been directed towards understanding the complexity of the tumor microenvironment and how specific cell types influence the function of other cell types, particularly the interplay among various cell types. As proteasome modulators treatment could potentially affect the function of different cells and thus their immune outcomes, it is imperative to employ novel ex vivo co-culture systems such as 3D-tumor slice culture [141, 142], air-liquid culture [143], and patients derived organoid culture systems [144]. These systems allow for the study of the fate of each cell type when exposed to these drugs and how T cell function is affected in complex scenarios. Finally, the emergence of artificial intelligence and machine learning methods holds promise for designing more specific proteasome inhibitors. Future inhibitors could target only the constitutive proteasome while sparing the immunoproteasome, and these methods could predict the interplay between proteasome function and T cell response using current multi-omics data. These cutting-edge technologies and advancements will undoubtedly aid in deciphering the intricate interactions between proteasome function and T cell response.
5.1.2 Future directions in proteasome and T cell research
Future research in proteasome and T cell research is likely to focus on several key areas. A recent study utilizes mRNA vaccines to express fusion peptide of antigen and a proteasome-targeting peptide that can be processed by proteasome to present antigen on the cell surface. By utilizing this mRNA vaccine, they observed very strong CD8+ T cell mediated immune response [145]. In the context of CAR-T cell therapies for cancer, there is a need to further explore the optimal treatment strategy including treatment dosage, frequency, and treatment route of proteasome inhibitors when combined with the BCMA targeted CAR-T cells [93, 94, 146]. This also involves extending these findings and treatment strategy to other CAR-T cells in addition to the BCMA targeted CAR-T cells. For the complex function of proteasome in T cell development, differentiation, survival, activation, and exhaustion, more efforts are needed to clearly define the impact of proteasome activity on different types and their developmental stages of T cells and also the optimal strategy for intervention. Furthermore, a deep understanding of the complex dynamics of proteasome in tumor microenvironment and the role of the proteasome in immune activation and evasion will be crucial for the development of these new therapeutic approaches.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (82030094 and 82573786); the Chair Professor Grant (CPG2025-00035-FHS); the Multi-Year Research Grant (MYRG) 2023-00029-FHS and 2024-00073-FHS of the University of Macau; and the Science and Technology Development Fund, Macau SAR (0009/2022/AKP, 0054/2023/RIA1, and 0193/2024/AGJ, 0129/2024/RIA2).
Declaration of generative AI and AI-assisted technologies
During the preparation of this work, the author(s) used DeepSeek in order to polish the language. After using this tool or service, the author(s) reviewed and edited the content as needed and take(s) full responsibility for the content of the publication.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Gu X, Nardone C, Kamitaki N, Mao A, Elledge SJ, Greenberg ME. The midnolin-proteasome pathway catches proteins for ubiquitination-independent degradation. Science. 2023;381:eadh5021
2. Nardone C, Gao J, Seo H-S, Mintseris J, Ort L, Yip MCJ. et al. Structural basis for the midnolin-proteasome pathway and its role in suppressing myeloma. Molecular Cell. 2025;85:2597-609.e11
3. King RW, Deshaies RJ, Peters J-M, Kirschner MW. How Proteolysis Drives the Cell Cycle. Science. 1996;274:1652-9
4. Arkinson C, Dong KC, Gee CL, Martin A. Mechanisms and regulation of substrate degradation by the 26S proteasome. Nature Reviews Molecular Cell Biology. 2025;26:104-22
5. Rousseau A, Bertolotti A. Regulation of proteasome assembly and activity in health and disease. Nature Reviews Molecular Cell Biology. 2018;19:697-712
6. Bernassola F, Ciechanover A, Melino G. The ubiquitin proteasome system and its involvement in cell death pathways. Cell Death & Differentiation. 2010;17:1-3
7. Kammerl IE, Meiners S. Proteasome function shapes innate and adaptive immune responses. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2016;311:L328-L36
8. Shao F, Lyu X, Miao K, Xie L, Wang H, Xiao H. et al. Enhanced Protein Damage Clearance Induces Broad Drug Resistance in Multitype of Cancers Revealed by an Evolution Drug-Resistant Model and Genome-Wide siRNA Screening. Advanced Science. 2020;7:2001914
9. Adams J. The proteasome: structure, function, and role in the cell. Cancer Treatment Reviews. 2003;29:3-9
10. Budenholzer L, Cheng CL, Li Y, Hochstrasser M. Proteasome Structure and Assembly. Journal of Molecular Biology. 2017;429:3500-24
11. Stadtmueller BM, Hill CP. Proteasome Activators. Molecular Cell. 2011;41:8-19
12. Sijts EJAM, Kloetzel PM. The role of the proteasome in the generation of MHC class I ligands and immune responses. Cellular and Molecular Life Sciences. 2011;68:1491-502
13. Murata S, Takahama Y, Kasahara M, Tanaka K. The immunoproteasome and thymoproteasome: functions, evolution and human disease. Nature Immunology. 2018;19:923-31
14. Aki M, Shimbara N, Takashina M, Akiyama K, Kagawa S, Tamura T. et al. Interferon-γ Induces Different Subunit Organizations and Functional Diversity of Proteasomes1. The Journal of Biochemistry. 1994;115:257-69
15. Hallermalm K, Seki K, Wei C, Castelli C, Rivoltini L, Kiessling R. et al. Tumor necrosis factor-α induces coordinated changes in major histocompatibility class I presentation pathway, resulting in increased stability of class I complexes at the cell surface. Blood. 2001;98:1108-15
16. Yang XW, Wang P, Liu JQ, Zhang H, Xi WD, Jia XH. et al. Coordinated regulation of the immunoproteasome subunits by PML/RARα and PU.1 in acute promyelocytic leukemia. Oncogene. 2014;33:2700-8
17. Kotamraju S, Matalon S, Matsunaga T, Shang T, Hickman-Davis JM, Kalyanaraman B. Upregulation of immunoproteasomes by nitric oxide: Potential antioxidative mechanism in endothelial cells. Free Radical Biology and Medicine. 2006;40:1034-44
18. Dimasuay KG, Schaunaman N, Berg B, Cervantes D, Kruger E, Heppner FL. et al. Airway epithelial immunoproteasome subunit LMP7 protects against rhinovirus infection. Scientific Reports. 2022;12:14507
19. Shin E-C, Seifert U, Kato T, Rice CM, Feinstone SM, Kloetzel P-M. et al. Virus-induced type I IFN stimulates generation of immunoproteasomes at the site of infection. The Journal of Clinical Investigation. 2006;116:3006-14
20. Yun Young S, Kim Kwan H, Tschida B, Sachs Z, Noble-Orcutt Klara E, Moriarity Branden S. et al. mTORC1 Coordinates Protein Synthesis and Immunoproteasome Formation via PRAS40 to Prevent Accumulation of Protein Stress. Molecular Cell. 2016;61:625-39
21. Tay RE, Richardson EK, Toh HC. Revisiting the role of CD4+ T cells in cancer immunotherapy—new insights into old paradigms. Cancer Gene Therapy. 2021;28:5-17
22. Montauti E, Oh DY, Fong L. CD4+ T cells in antitumor immunity. Trends in Cancer. 2024;10:969-85
23. Saad MK, Rupen D, Andrew C, Alexandra L, Gavin PD, Allegra P. et al. Impact of CD4 T cells on intratumoral CD8 T-cell exhaustion and responsiveness to PD-1 blockade therapy in mouse brain tumors. Journal for ImmunoTherapy of Cancer. 2022;10:e005293
24. Ni C, Ma P, Qu L, Wu F, Hao J, Wang R. et al. Accelerated tumour metastasis due to interferon-γ receptor-mediated dissociation of perivascular cells from blood vessels. The Journal of Pathology. 2017;242:334-46
25. Jhunjhunwala S, Hammer C, Delamarre L. Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nature Reviews Cancer. 2021;21:298-312
26. Oh DY, Fong L. Cytotoxic CD4+ T cells in cancer: Expanding the immune effector toolbox. Immunity. 2021;54:2701-11
27. Wu X, Li T, Jiang R, Yang X, Guo H, Yang R. Targeting MHC-I molecules for cancer: function, mechanism, and therapeutic prospects. Molecular Cancer. 2023;22:194
28. Choi YB, Son M, Park M, Shin J, Yun Y. SOCS-6 Negatively Regulates T Cell Activation through Targeting p56lck to Proteasomal Degradation*. Journal of Biological Chemistry. 2010;285:7271-80
29. Wang X, Li J-P, Chiu L-L, Lan J-L, Chen D-Y, Boomer J. et al. Attenuation of T Cell Receptor Signaling by Serine Phosphorylation-mediated Lysine 30 Ubiquitination of SLP-76 Protein *. Journal of Biological Chemistry. 2012;287:34091-100
30. Arata Y, Watanabe A, Motosugi R, Murakami R, Goto T, Hori S. et al. Defective induction of the proteasome associated with T-cell receptor signaling underlies T-cell senescence. Genes to Cells. 2019;24:801-13
31. Takaba H, Takayanagi H. The Mechanisms of T Cell Selection in the Thymus. Trends in Immunology. 2017;38:805-16
32. Murata S, Sasaki K, Kishimoto T, Niwa S-i, Hayashi H, Takahama Y. et al. Regulation of CD8+ T Cell Development by Thymus-Specific Proteasomes. Science. 2007;316:1349-53
33. Sasaki K, Takada K, Ohte Y, Kondo H, Sorimachi H, Tanaka K. et al. Thymoproteasomes produce unique peptide motifs for positive selection of CD8+ T cells. Nature Communications. 2015;6:7484
34. Xing Y, Jameson SC, Hogquist KA. Thymoproteasome subunit-β5T generates peptide-MHC complexes specialized for positive selection. Proceedings of the National Academy of Sciences. 2013;110:6979-84
35. Tomaru U, Konno S, Miyajima S, Kimoto R, Onodera M, Kiuchi S. et al. Restricted Expression of the Thymoproteasome Is Required for Thymic Selection and Peripheral Homeostasis of CD8<sup>+</sup> T Cells. Cell Reports. 2019;26:639-51.e2
36. Nitta T, Kochi Y, Muro R, Tomofuji Y, Okamura T, Murata S. et al. Human thymoproteasome variations influence CD8 T cell selection. Science Immunology. 2017;2:eaan5165
37. Basler M, Mundt S, Groettrup M. The immunoproteasome subunit LMP7 is required in the murine thymus for filling up a hole in the T cell repertoire. European Journal of Immunology. 2018;48:419-29
38. Mittelbrunn M, Kroemer G. Hallmarks of T cell aging. Nature Immunology. 2021;22:687-98
39. Gressler AE, Leng H, Zinecker H, Simon AK. Proteostasis in T cell aging. Seminars in Immunology. 2023;70:101838
40. Ponnappan U, Zhong M, Trebilcock GU. Decreased Proteasome-Mediated Degradation in T Cells from the Elderly: A Role in Immune Senescence. Cellular Immunology. 1999;192:167-74
41. Berges C, Haberstock H, Fuchs D, Miltz M, Sadeghi M, Opelz G. et al. Proteasome inhibition suppresses essential immune functions of human CD4+ T cells. Immunology. 2008;124:234-46
42. Wang Z, Hu Y, Xiao D, Wang J, Liu C, Xu Y. et al. Stabilization of Notch1 by the Hsp90 Chaperone is Crucial for T-Cell Leukemogenesis. Clinical Cancer Research. 2017;23:3834-46
43. Jiang J, Wang J, Yue M, Cai X, Wang T, Wu C. et al. Direct Phosphorylation and Stabilization of MYC by Aurora B Kinase Promote T-cell Leukemogenesis. Cancer Cell. 2020;37:200-15.e5
44. Widjaja CE, Olvera JG, Metz PJ, Phan AT, Savas JN, de Bruin G. et al. Proteasome activity regulates CD8+ T lymphocyte metabolism and fate specification. The Journal of Clinical Investigation. 2017;127:3609-23
45. Osana S, Kitajima Y, Naoki S, Takada H, Murayama K, Kano Y. et al. Little involvement of recycled-amino acids from proteasomal proteolysis in de novo protein synthesis. Biochemical and Biophysical Research Communications. 2022;634:40-7
46. Shinebaatar E, Morimoto J, Koga R, Nguyen TN, Sasaki Y, Yonemura S. et al. Proteasome dysfunction in T cells causes immunodeficiency via cell cycle disruption and apoptosis. International Immunology. 2025: dxaf021.
47. Cui H, Matsui K, Omura S, Schauer SL, Matulka RA, Sonenshein GE. et al. Proteasome regulation of activation-induced T cell death. Proceedings of the National Academy of Sciences. 1997;94:7515-20
48. Thounaojam MC, Dudimah DF, Pellom Jr ST, Uzhachenko RV, Carbone DP, Dikov MM. et al. Bortezomib enhances expression of effector molecules in anti-tumor CD8 + T lymphocytes by promoting Notch-nuclear factor-κB crosstalk. Oncotarget; Vol 6, No 32. 2015
49. Mao R, Yang R, Chen X, Harhaj EW, Wang X, Fan Y. Regnase-1, a rapid response ribonuclease regulating inflammation and stress responses. Cellular & Molecular Immunology. 2017;14:412-22
50. Wang Y, Ma A, Song N-J, Shannon AE, Amankwah YS, Chen X. et al. Proteotoxic stress response drives T cell exhaustion and immune evasion. Nature. 2025
51. Liu Y, Ni H, Li J, Yang J, Sekielyk I, Snow BE. et al. LARP4-mediated hypertranslation drives T cell dysfunction in tumors. Nature Immunology. 2025;26:1488-500
52. Marchingo JM, Cantrell DA. Protein synthesis, degradation, and energy metabolism in T cell immunity. Cellular & Molecular Immunology. 2022;19:303-15
53. de Blas A, Andreatta M, Antoñana-Vildosola A, Velasco P, Egia-Mendikute L, Jimenez-Lasheras B. et al. Abstract 1354: Proteasome activation prevents T cell exhaustion and improves antitumor activity. Cancer Research. 2023;83:1354 -
54. Rana PS, Ignatz-Hoover JJ, Guo C, Mosley AL, Malek E, Federov Y. et al. Immunoproteasome Activation Expands the MHC Class I Immunopeptidome, Unmasks Neoantigens, and Enhances T-cell Anti-Myeloma Activity. Molecular Cancer Therapeutics. 2024;23:1743-60
55. Kumar B, Field NS, Kim DD, Dar AA, Chen Y, Suresh A. et al. The ubiquitin ligase Cul5 regulates CD4+ T cell fate choice and allergic inflammation. Nature Communications. 2022;13:2786
56. Chang John T, Ciocca Maria L, Kinjyo I, Palanivel Vikram R, McClurkin Courtney E, DeJong Caitlin S. et al. Asymmetric Proteasome Segregation as a Mechanism for Unequal Partitioning of the Transcription Factor T-bet during T Lymphocyte Division. Immunity. 2011;34:492-504
57. Xiao F, Lin X, Tian J, Wang X, Chen Q, Rui K. et al. Proteasome inhibition suppresses Th17 cell generation and ameliorates autoimmune development in experimental Sjögren's syndrome. Cellular & Molecular Immunology. 2017;14:924-34
58. Mundt S, Engelhardt B, Kirk CJ, Groettrup M, Basler M. Inhibition and deficiency of the immunoproteasome subunit LMP7 attenuates LCMV-induced meningitis. European Journal of Immunology. 2016;46:104-13
59. Gibson HM, Mishra A, Chan DV, Hake TS, Porcu P, Wong HK. Impaired Proteasome Function Activates GATA3 in T Cells and Upregulates CTLA-4: Relevance for Sézary Syndrome. Journal of Investigative Dermatology. 2013;133:249-57
60. Liu H, Wan C, Ding Y, Han R, He Y, Xiao J. et al. PR-957, a selective inhibitor of immunoproteasome subunit low-MW polypeptide 7, attenuates experimental autoimmune neuritis by suppressing Th17-cell differentiation and regulating cytokine production. The FASEB Journal. 2017;31:1756-66
61. Oliveri F, Mink D, Muchamuel T, Basler M. Immunoproteasome Inhibition Impairs Differentiation but Not Survival of T Helper 17 Cells. Cells. 2025
62. Kane RC, Bross PF, Farrell AT, Pazdur R. Velcade®: U.S. FDA Approval for the Treatment of Multiple Myeloma Progressing on Prior Therapy. The Oncologist. 2003;8:508-13
63. Parlati F, Lee SJ, Aujay M, Suzuki E, Levitsky K, Lorens JB. et al. Carfilzomib can induce tumor cell death through selective inhibition of the chymotrypsin-like activity of the proteasome. Blood. 2009;114:3439-47
64. Kupperman E, Lee EC, Cao Y, Bannerman B, Fitzgerald M, Berger A. et al. Evaluation of the Proteasome Inhibitor MLN9708 in Preclinical Models of Human Cancer. Cancer Research. 2010;70:1970-80
65. Jensen TJ, Loo MA, Pind S, Williams DB, Goldberg AL, Riordan JR. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129-35
66. Mezquita J, Mezquita B, Pau M, Mezquita C. Down-regulation of Flt-1 gene expression by the proteasome inhibitor MG262. Journal of Cellular Biochemistry. 2003;89:1138-47
67. Singh AV, Palladino MA, Lloyd GK, Potts BC, Chauhan D, Anderson KC. Pharmacodynamic and efficacy studies of the novel proteasome inhibitor NPI-0052 (marizomib) in a human plasmacytoma xenograft murine model. British Journal of Haematology. 2010;149:550-9
68. Roth P, Gorlia T, Reijneveld JC, de Vos F, Idbaih A, Frenel J-S. et al. Marizomib for patients with newly diagnosed glioblastoma: A randomized phase 3 trial. Neuro-Oncology. 2024;26:1670-82
69. Demo SD, Kirk CJ, Aujay MA, Buchholz TJ, Dajee M, Ho MN. et al. Antitumor activity of PR-171, a novel irreversible inhibitor of the proteasome. Cancer Res. 2007;67:6383
70. Ghobrial IM, Vij R, Siegel D, Badros A, Kaufman J, Raje N. et al. A Phase Ib/II Study of Oprozomib in Patients with Advanced Multiple Myeloma and Waldenström Macroglobulinemia. Clinical Cancer Research. 2019;25:4907-16
71. Piva R, Ruggeri B, Williams M, Costa G, Tamagno I, Ferrero D. et al. CEP-18770: A novel, orally active proteasome inhibitor with a tumor-selective pharmacologic profile competitive with bortezomib. Blood. 2008;111:2765-75
72. Vogl DT, Martin TG, Vij R, Hari P, Mikhael JR, Siegel D. et al. Phase I/II study of the novel proteasome inhibitor delanzomib (CEP-18770) for relapsed and refractory multiple myeloma. Leukemia & Lymphoma. 2017;58:1872-9
73. Meng L, Mohan R, Kwok BHB, Elofsson M, Sin N, Crews CM. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proceedings of the National Academy of Sciences. 1999;96:10403-8
74. Ostrowska H, Wojcik C, Omura S, Worowski K. Lactacystin, a Specific Inhibitor of the Proteasome, Inhibits Human Platelet Lysosomal Cathepsin A-like Enzyme. Biochemical and Biophysical Research Communications. 1997;234:729-32
75. Hasinoff BB, Patel D. Disulfiram is a slow-binding partial noncompetitive inhibitor of 20S proteasome activity. Archives of Biochemistry and Biophysics. 2017;633:23-8
76. Singh BN, Shankar S, Srivastava RK. Green tea catechin, epigallocatechin-3-gallate (EGCG): Mechanisms, perspectives and clinical applications. Biochemical Pharmacology. 2011;82:1807-21
77. Holecek M, Muthny T, Kovarik M, Sispera L. Effect of beta-hydroxy-beta-methylbutyrate (HMB) on protein metabolism in whole body and in selected tissues. Food and Chemical Toxicology. 2009;47:255-9
78. Rawson S, Walsh RM, Velez B, Schnell HM, Jiao F, Blickling M. et al. Yeast PI31 inhibits the proteasome by a direct multisite mechanism. Nature Structural & Molecular Biology. 2022;29:791-800
79. Lokireddy S, Kukushkin NV, Goldberg AL. cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proceedings of the National Academy of Sciences. 2015;112:E7176-E85
80. VerPlank JJS, Tyrkalska SD, Fleming A, Rubinsztein DC, Goldberg AL. cGMP via PKG activates 26S proteasomes and enhances degradation of proteins, including ones that cause neurodegenerative diseases. Proceedings of the National Academy of Sciences. 2020;117:14220-30
81. Lee D, Takayama S, Goldberg AL. ZFAND5/ZNF216 is an activator of the 26S proteasome that stimulates overall protein degradation. Proceedings of the National Academy of Sciences. 2018;115:E9550-E9
82. Lee D, Zhu Y, Colson L, Wang X, Chen S, Tkacik E. et al. Molecular mechanism for activation of the 26S proteasome by ZFAND5. Molecular Cell. 2023;83:2959-75.e7
83. Coleman RA, Trader DJ. Development and Application of a Sensitive Peptide Reporter to Discover 20S Proteasome Stimulators. ACS Combinatorial Science. 2018;20:269-76
84. Huang L, Ho P, Chen C-H. Activation and inhibition of the proteasome by betulinic acid and its derivatives. FEBS Letters. 2007;581:4955-9
85. Katsiki M, Chondrogianni N, Chinou I, Rivett AJ, Gonos ES. The Olive Constituent Oleuropein Exhibits Proteasome Stimulatory Properties In Vitro and Confers Life Span Extension of Human Embryonic Fibroblasts. Rejuvenation Research. 2007;10:157-72
86. Trader DJ, Simanski S, Dickson P, Kodadek T. Establishment of a suite of assays that support the discovery of proteasome stimulators. Biochimica et Biophysica Acta (BBA) - General Subjects. 2017;1861:892-9
87. Njomen E, Tepe JJ. Regulation of Autophagic Flux by the 20S Proteasome. Cell Chemical Biology. 2019;26:1283-94.e5
88. Tanaka K, Yoshimura T, Ichihara A. Role of Substrate in Reversible Activation of Proteasomes (Multi-Protease Complexes) by Sodium Dodecyl Sulfate1. The Journal of Biochemistry. 1989;106:495-500
89. Jones CL, Njomen E, Sjögren B, Dexheimer TS, Tepe JJ. Small Molecule Enhancement of 20S Proteasome Activity Targets Intrinsically Disordered Proteins. ACS Chemical Biology. 2017;12:2240-7
90. Trippier PC, Zhao KT, Fox SG, Schiefer IT, Benmohamed R, Moran J. et al. Proteasome Activation is a Mechanism for Pyrazolone Small Molecules Displaying Therapeutic Potential in Amyotrophic Lateral Sclerosis. ACS Chemical Neuroscience. 2014;5:823-9
91. Fiolek TJ, Keel KL, Tepe JJ. Fluspirilene Analogs Activate the 20S Proteasome and Overcome Proteasome Impairment by Intrinsically Disordered Protein Oligomers. ACS Chemical Neuroscience. 2021;12:1438-48
92. Shanker A, Pellom ST Jr, Dudimah DF, Thounaojam MC, de Kluyver RL, Brooks AD. et al. Bortezomib Improves Adoptive T-cell Therapy by Sensitizing Cancer Cells to FasL Cytotoxicity. Cancer Research. 2015;75:5260-72
93. Rieger L, Irlinger K, Füchsl F, Tietje M, Purcarea A, Barbian NM. et al. Boosting CAR T-Cell Efficacy by Blocking Proteasomal Degradation of Membrane Antigens. Blood. 2025: blood.2024027616.
94. Li J, Guo R, Li D, Yang J, Zhang Y, Gao H. et al. Bortezomib enhances the efficacy of BCMA CAR-T therapy through up-regulating BCMA expression in myeloma cells. International Immunopharmacology. 2025;148:114113
95. Seeger JM, Schmidt P, Brinkmann K, Hombach AA, Coutelle O, Zigrino P. et al. The Proteasome Inhibitor Bortezomib Sensitizes Melanoma Cells toward Adoptive CTL Attack. Cancer Research. 2010;70:1825-34
96. Benvenuto M, Angiolini V, Focaccetti C, Nardozi D, Palumbo C, Carrano R. et al. Antitumoral effects of Bortezomib in malignant mesothelioma: evidence of mild endoplasmic reticulum stress in vitro and activation of T cell response in vivo. Biology Direct. 2023;18:17
97. Huang Z, Wu Y, Zhou X, Xu J, Zhu W, Shu Y. et al. Efficacy of therapy with bortezomib in solid tumors: a review based on 32 clinical trials. Future Oncology. 2014;10:1795-807
98. Zhou J-B, Tang D, He L, Lin S, Lei JH, Sun H. et al. Machine learning model for anti-cancer drug combinations: Analysis, prediction, and validation. Pharmacological Research. 2023;194:106830
99. Tang D, Lin S, Zhou J, Lei JH, Shao F, Sun H. et al. Augment proteasome inhibitor efficacy activates CD8+ T cell-mediated antitumor immunity in breast cancer. Cell Reports Medicine. 2025;6:102211
100. Zhou C, Li C, Luo L, Li X, Jia K, He N. et al. Anti-tumor efficacy of HRS-4642 and its potential combination with proteasome inhibition in KRAS G12D-mutant cancer. Cancer Cell. 2024;42:1286-300.e8
101. Huang Z, Peng S, Knoff J, Lee SY, Yang B, Wu T-C. et al. Combination of proteasome and HDAC inhibitor enhances HPV16 E7-specific CD8+ T cell immune response and antitumor effects in a preclinical cervical cancer model. Journal of Biomedical Science. 2015;22:7
102. Dona AA, Tandoh T, Nigam L, Singer M, Caserta E, Murtadha M. et al. Proteasome inhibition enhances oncolytic reovirus therapy in multiple myeloma independently of its direct cytotoxic effects. Journal of Hematology & Oncology. 2025;18:1
103. Larsson P, Pettersson D, Olsson M, Sarathchandra S, Abramsson A, Zetterberg H. et al. Repurposing proteasome inhibitors for improved treatment of triple-negative breast cancer. Cell Death Discovery. 2024;10:57
104. Chen Z-L, Xie C, Zeng W, Huang R-Q, Yang J-E, Liu J-Y. et al. Synergistic induction of mitotic pyroptosis and tumor remission by inhibiting proteasome and WEE family kinases. Signal Transduction and Targeted Therapy. 2024;9:181
105. Sooman L, Gullbo J, Bergqvist M, Bergström S, Lennartsson J, Ekman S. Synergistic effects of combining proteasome inhibitors with chemotherapeutic drugs in lung cancer cells. BMC Research Notes. 2017;10:544
106. Zhao Y, Foster NR, Meyers JP, Thomas SP, Northfelt DW, Rowland KM. et al. A Phase I/II Study of Bortezomib in Combination with Paclitaxel, Carboplatin, and Concurrent Thoracic Radiation Therapy for Non-Small-Cell Lung Cancer: North Central Cancer Treatment Group (NCCTG)-N0321. Journal of Thoracic Oncology. 2015;10:172-80
107. Abt D, Besse A, Sedlarikova L, Kraus M, Bader J, Silzle T. et al. Improving the efficacy of proteasome inhibitors in the treatment of renal cell carcinoma by combination with the human immunodeficiency virus (HIV)-protease inhibitors lopinavir or nelfinavir. BJU International. 2018;121:600-9
108. Arnold SM, Chansky K, Leggas M, Thompson MA, Villano JL, Hamm J. et al. Phase 1b trial of proteasome inhibitor carfilzomib with irinotecan in lung cancer and other irinotecan-sensitive malignancies that have progressed on prior therapy (Onyx IST reference number: CAR-IST-553). Investigational New Drugs. 2017;35:608-15
109. Zang Y, Thomas SM, Chan ET, Kirk CJ, Freilino ML, DeLancey HM. et al. Carfilzomib and ONX 0912 Inhibit Cell Survival and Tumor Growth of Head and Neck Cancer and Their Activities Are Enhanced by Suppression of Mcl-1 or Autophagy. Clinical Cancer Research. 2012;18:5639-49
110. Zhang L, Boufraqech M, Lake R, Kebebew E. Carfilzomib potentiates CUDC-101-induced apoptosis in anaplastic thyroid cancer. Oncotarget; Vol 7, No 13. 2016
111. Lim JJ, Hooi L, Dan YY, Bonney GK, Zhou L, Chow PKH. et al. Rational drug combination design in patient-derived avatars reveals effective inhibition of hepatocellular carcinoma with proteasome and CDK inhibitors. Journal of Experimental & Clinical Cancer Research. 2022;41:249
112. Simmonds RE, Foxwell BM. Signalling, inflammation and arthritis: NF-κB and its relevance to arthritis and inflammation. Rheumatology. 2008;47:584-90
113. Tarjányi O, Olasz K, Rátky F, Sétáló G, Boldizsár F. Proteasome Inhibitors: Potential in Rheumatoid Arthritis Therapy? International Journal of Molecular Sciences. 2025
114. Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ. Transcription Factor Nrf1 Mediates the Proteasome Recovery Pathway after Proteasome Inhibition in Mammalian Cells. Molecular Cell. 2010;38:17-28
115. Ladi E, Everett C, Stivala CE, Daniels BE, Durk MR, Harris SF. et al. Design and Evaluation of Highly Selective Human Immunoproteasome Inhibitors Reveal a Compensatory Process That Preserves Immune Cell Viability. Journal of Medicinal Chemistry. 2019;62:7032-41
116. Huynh M, Pak C, Markovina S, Callander NS, Chng KS, Wuerzberger-Davis SM. et al. Hyaluronan and proteoglycan link protein 1 (HAPLN1) activates bortezomib-resistant NF-κB activity and increases drug resistance in multiple myeloma. Journal of Biological Chemistry. 2018;293:2452-65
117. Ettari R, Zappalà M, Grasso S, Musolino C, Innao V, Allegra A. Immunoproteasome-selective and non-selective inhibitors: A promising approach for the treatment of multiple myeloma. Pharmacology & Therapeutics. 2018;182:176-92
118. Ge M, Qiao Z, Kong Y, Liang H, Sun Y, Lu H. et al. Modulating proteasome inhibitor tolerance in multiple myeloma: an alternative strategy to reverse inevitable resistance. British Journal of Cancer. 2021;124:770-6
119. Chong KY, Hsu C-J, Hung T-H, Hu H-S, Huang T-T, Wang T-H. et al. Wnt pathway activation and ABCB1 expression account for attenuation of Proteasome inhibitor-mediated apoptosis in multidrug-resistant cancer cells. Cancer Biology & Therapy. 2015;16:149-59
120. Besse A, Stolze SC, Rasche L, Weinhold N, Morgan GJ, Kraus M. et al. Carfilzomib resistance due to ABCB1/MDR1 overexpression is overcome by nelfinavir and lopinavir in multiple myeloma. Leukemia. 2018;32:391-401
121. Marianne K, Juergen B, Paul PG, Emily SW, Anne CM, Tobias S. et al. The novel β2-selective proteasome inhibitor LU-102 synergizes with bortezomib and carfilzomib to overcome proteasome inhibitor resistance of myeloma cells. Haematologica. 2015;100:1350-60
122. Weyburne ES, Wilkins OM, Sha Z, Williams DA, Pletnev AA, de Bruin G. et al. Inhibition of the Proteasome β2 Site Sensitizes Triple-Negative Breast Cancer Cells to β5 Inhibitors and Suppresses Nrf1 Activation. Cell Chemical Biology. 2017;24:218-30
123. Jenkins TW, Fitzgerald JE, Park J, Wilson AM, Berry KL, Wong KS. et al. Highly specific Immunoproteasome inhibitor M3258 induces proteotoxic stress and apoptosis in KMT2A::AFF1 driven acute lymphoblastic leukemia. Scientific Reports. 2025;15:17284
124. Xiang Y, Jia M, Gao Y, Yang F, Wang T, Dai R. et al. Cisplatin Disrupts Proteasome Bounce-Back Effect through Suppressing ZEB1/Nfe2l1 in Cholangiocarcinoma. FBL. 2024 29
125. Tomlin FM, Gerling-Driessen UIM, Liu Y-C, Flynn RA, Vangala JR, Lentz CS. et al. Inhibition of NGLY1 Inactivates the Transcription Factor Nrf1 and Potentiates Proteasome Inhibitor Cytotoxicity. ACS Central Science. 2017;3:1143-55
126. Vangala JR, Potluri A, Radhakrishnan SK. BET Inhibitors Synergize with Carfilzomib to Induce Cell Death in Cancer Cells via Impairing Nrf1 Transcriptional Activity and Exacerbating the Unfolded Protein Response. Biomolecules. 2020
127. Gao G, Xu Y, Gan J, Cao X, Dong X, Fang M. et al. Biomimetic platelet nanoparticles encapsulated with proteasome inhibitor bortezomib for multiple myeloma treatment. APL Materials. 2023;11:121113
128. Yin D, Wu X, Chen X, Chen J-L, Xia X, Wang J. et al. Enhanced anticancer effect of carfilzomib by codelivery of calcium peroxide nanoparticles targeting endoplasmic reticulum stress. Materials Today Bio. 2025;32:101649
129. Zeng T-m, Jiang T-y, Yang G, Cheng Z, Lou C, Wei W. et al. Bortezomib in previously treated phosphatase and tension homology-deficient patients with advanced intrahepatic cholangiocarcinoma: An open-label, prospective and single-centre phase II trial. Clinical and Translational Medicine. 2024;14:e1675
130. Jiang T-Y, Pan Y-F, Wan Z-H, Lin Y-K, Zhu B, Yuan Z-g. et al. PTEN status determines chemosensitivity to proteasome inhibition in cholangiocarcinoma. Science Translational Medicine. 2020;12:eaay0152
131. Zerfas BL, Maresh ME, Trader DJ. The Immunoproteasome: An Emerging Target in Cancer and Autoimmune and Neurological Disorders. Journal of Medicinal Chemistry. 2020;63:1841-58
132. Ito Y, Inoue S, Kagoya Y. Gene editing technology to improve antitumor T-cell functions in adoptive immunotherapy. Inflammation and Regeneration. 2024;44:13
133. Schumann K, Lin S, Boyer E, Simeonov DR, Subramaniam M, Gate RE. et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proceedings of the National Academy of Sciences. 2015;112:10437-42
134. Schmidt R, Ward CC, Dajani R, Armour-Garb Z, Ota M, Allain V. et al. Base-editing mutagenesis maps alleles to tune human T cell functions. Nature. 2024;625:805-12
135. Chiesa R, Georgiadis C, Syed F, Zhan H, Etuk A, Gkazi Soragia A. et al. Base-Edited CAR7 T Cells for Relapsed T-Cell Acute Lymphoblastic Leukemia. New England Journal of Medicine. 2023;389:899-910
136. Kaake RM, Kao A, Yu C, Huang L. Characterizing the Dynamics of Proteasome Complexes by Proteomics Approaches. Antioxidants & Redox Signaling. 2014;21:2444-56
137. Rut W, Poręba M, Kasperkiewicz P, Snipas SJ, Drąg M. Selective Substrates and Activity-Based Probes for Imaging of the Human Constitutive 20S Proteasome in Cells and Blood Samples. Journal of Medicinal Chemistry. 2018;61:5222-34
138. Karagach S, Smollich J, Atrakchi O, Mohan V, Geiger T. High throughput single-cell proteomics of <em>in vivo</em> cells. Molecular & Cellular Proteomics.
139. Petrosius V, Aragon-Fernandez P, Üresin N, Kovacs G, Phlairaharn T, Furtwängler B. et al. Exploration of cell state heterogeneity using single-cell proteomics through sensitivity-tailored data-independent acquisition. Nature Communications. 2023;14:5910
140. Brinkerhoff H, Kang ASW, Liu J, Aksimentiev A, Dekker C. Multiple rereads of single proteins at single-amino acid resolution using nanopores. Science. 2021;374:1509-13
141. Xing F, Liu Y-C, Huang S, Lyu X, Su SM, Chan UI. et al. Accelerating precision anti-cancer therapy by time-lapse and label-free 3D tumor slice culture platform. Theranostics. 2021;11:9415-30
142. Lei JH, Lee M-H, Miao K, Huang Z, Yao Z, Zhang A. et al. Activation of FGFR2 Signaling Suppresses BRCA1 and Drives Triple-Negative Mammary Tumorigenesis That is Sensitive to Immunotherapy. Advanced Science. 2021;8:2100974
143. Feng Y, Zhao M, Wang L, Li L, Lei JH, Zhou J. et al. The heterogeneity of signaling pathways and drug responses in intrahepatic cholangiocarcinoma with distinct genetic mutations. Cell Death & Disease. 2024;15:34
144. Chen P, Zhang X, Ding R, Yang L, Lyu X, Zeng J. et al. Patient-Derived Organoids Can Guide Personalized-Therapies for Patients with Advanced Breast Cancer. Advanced Science. 2021;8:2101176
145. Ling J, Chen H, Huang M, Wang J, Du X. An mRNA vaccine encoding proteasome-targeted antigen enhances CD8+ T cell immunity. Journal of Controlled Release. 2025;381:113578
146. Sheykhhasan M, Ahmadieh-Yazdi A, Vicidomini R, Poondla N, Tanzadehpanah H, Dirbaziyan A. et al. CAR T therapies in multiple myeloma: unleashing the future. Cancer Gene Therapy. 2024;31:667-86
Author contact
![]() Corresponding author: Chu-Xia Deng; E-mail: cxdengedu.mo; Tel.: (853) 8822-4997; Fax: (853) 8822 2314.
Corresponding author: Chu-Xia Deng; E-mail: cxdengedu.mo; Tel.: (853) 8822-4997; Fax: (853) 8822 2314.

 Global reach, higher impact
Global reach, higher impact