10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(3):367-382. doi:10.7150/ijbs.17035 This issue Cite
Research Paper
Comparative Mitogenomic Analyses of Praying Mantises (Dictyoptera, Mantodea): Origin and Evolution of Unusual Intergenic Gaps
1. School of Life Sciences, Datong University, Datong 037009, China;
2. College of Ecology and Evolution, School of Life Sciences, Sun Yat-sen University, Guangzhou 510275, China;
3. State Key Laboratory of Biocontrol, Sun Yat-sen University, Guangzhou 510275, China;
4. College of Life Sciences, Shaanxi Normal University, Xi'an 710062, China.
* These authors contributed equally to this work.
Abstract
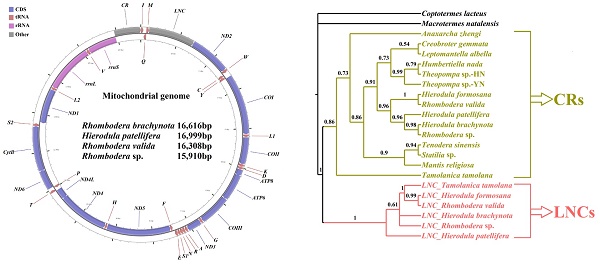
Praying mantises are a diverse group of predatory insects. Although some Mantodea mitogenomes have been reported, a comprehensive comparative and evolutionary genomic study is lacking for this group. In the present study, four new mitogenomes were sequenced, annotated, and compared to the previously published mitogenomes of other Mantodea species. Most Mantodea mitogenomes share a typical set of mitochondrial genes and a putative control region (CR). Additionally, and most intriguingly, another large non-coding region (LNC) was detected between trnM and ND2 in all six Paramantini mitogenomes examined. The main section in this common region of Paramantini may have initially originated from the corresponding control region for each species, whereas sequence differences between the LNCs and CRs and phylogenetic analyses indicate that LNC and CR are largely independently evolving. Namely, the LNC (the duplicated CR) may have subsequently degenerated during evolution. Furthermore, evidence suggests that special intergenic gaps have been introduced in some species through gene rearrangement and duplication. These gaps are actually the original abutting sequences of migrated or duplicated genes. Some gaps (G5 and G6) are homologous to the 5' and 3' surrounding regions of the duplicated gene in the original gene order, and another specific gap (G7) has tandem repeats. We analysed the phylogenetic relationships of fifteen Mantodea species using 37 concatenated mitochondrial genes and detected several synapomorphies unique to species in some clades.
Keywords: Mantodea, Mitochondrial genome, Control region (CR), Large non-coding region (LNC), Intergenic gap, Phylogeny.

 Global reach, higher impact
Global reach, higher impact