10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(7):835-851. doi:10.7150/ijbs.18868 This issue Cite
Research Paper
Down-regulation of aminopeptidase N and ABC transporter subfamily G transcripts in Cry1Ab and Cry1Ac resistant Asian corn borer, Ostrinia furnacalis (Lepidoptera: Crambidae)
1. State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China.
2. United States Department of Agriculture, Agricultural Research Service, Corn Insects & Crop Genetics Research Unit, Iowa State University, Ames, IA, 50011, USA
Tiantao Zhang and Brad S. Coates contributed equally to this work.
Received 2016-12-22; Accepted 2017-3-16; Published 2017-7-6
Abstract
The Asian corn borer (ACB), Ostrinia furnacalis (Lepidoptera: Crambidae), is a highly destructive pest of cultivated maize throughout East Asia. Bacillus thuringiensis (Bt) crystalline protein (Cry) toxins cause mortality by a mechanism involving pore formation or signal transduction following toxin binding to receptors along the midgut lumen of susceptible insects, but this mechanism and mutations therein that lead to resistance are not fully understood. In the current study, quantitative comparisons were made among midgut expressed transcripts from O. furnacalis susceptible (ACB-BtS) and laboratory selected strains resistant to Cry1Ab (ACB-AbR) and Cry1Ac toxins (ACB-AcR) when feeding on non-Bt diet. From a combined de novo transcriptome assembly of 83,370 transcripts, ORFs of ≥ 100 amino acids were predicted and annotated for 28,940 unique isoforms derived from 12,288 transcripts. Transcriptome-wide expression estimated from RNA-seq read depths predicted significant down-regulation of transcripts for previously known Bt resistance genes, aminopeptidase N1 (apn1) and apn3, as well as a putative ATP binding cassette transporter group G (abcg) gene in both ACB-AbR and -AcR (log2[fold-change] ≥ 1.36; P < 0.0001). The transcripts that were most highly differentially regulated in both ACB-AbR and -AcR compared to ACB-BtS (log2[fold-change] ≥ 2.0; P < 0.0001) included up- and down-regulation of serine proteases, storage proteins and cytochrome P450 monooxygenases, as well as up-regulation of genes with predicted transport function. This study predicted the significant down-regulation of transcripts for previously known Bt resistance genes, aminopeptidase N1 (apn1) and apn3, as well as abccg gene in both ACB-AbR and -AcR. These data are important for the understanding of systemic differences between Bt resistant and susceptible genotypes.
Keywords: Ostrinia furnacalis, Bacillus thuringiensis, resistance, midgut, transcriptome.
Introduction
The productivity of cultivated crop plants has increased in part due to technologies that effectively suppress the feeding damage caused by herbivorous insect pests. This has involved the widespread use of chemical pesticides and genetically modified crops that express transgenic insecticidal protein toxins derived from the soil bacterium, Bacillus thuringiensis (Bt; [1]). The widespread applications of chemical pesticides are implicated as likely causes for the increased incidence of functional resistance in insect populations [2], and has analogously occurred through selection pressures imposed by Bt crops [3]. The control of several insect species has been compromised due to the evolution of resistance to Bt toxins, and include the maize pests the African stem borer, Busseola fusca [4], the Fall armyworm, Spodoptera frugiperda [5-8], and the corn rootworm, Diabrotica virgifera virgifera [9, 10], as well as the cotton pests Helicoverpa armigera, H. zea, and Pectinophora gossypiella [11-15]. Other reports have documented a significant reduction in H. zea susceptibility towards the Cry1Ab toxin levels expressed by transgenic maize in the field [16]. Analogously, over the past decade the survivorship of field-collected western bean cutworm (WBC), Striacosta albicosta, on Cry1F toxin in bioassay laboratory has increased [17], where larvae show little growth inhibition even at high toxin exposures [18].
Investigations into the biochemical, molecular and genetic mechanism by which Bt resistance evolve has identified a great variety of different potential mechanisms [19-21], which are thought to inhibit the normal Bt mode of action [22, 23]. Membrane bound glycosylphosphatidylinositol (GPI) anchored aminopeptidase N (APN) was among the first proteins identified in the midgut of Lepidoptera capable of binding to Cry1A toxins in vitro [24, 25]. The involvement of apn genes in the Cry1A and Cry9C toxin modes of action was suggested following knockdown by RNA interference (RNAi) that resulted in reduced susceptibilities among lepidopteran larvae [26-28]. Implication of endogenous apn1 mutations in the evolution of Bt toxin resistance was shown to occur via the suppression apn1 transcription in Cry1Ca resistant S. exigua [29], Cry1Ac resistant T. ni [30], and Cry1Ab tolerant O. nubilalis [31]. Similarly, levels of apn3 transcripts were reduced in Cry1Ab resistant O. nubilalis [31] and Cry1Ac resistant O. furnacalis [32]. Initial indication that an ATP binding cassette (ABC) transporter may be involved in Bt toxin resistance traits came from QTL mapping results that linked insertion of a premature stop codon in abcc2 to Cry1Ac resistance in H. virescens [33]. Different sets of deletions and point mutations in abcc2 orthologs were subsequently associated with resistance of P. xylostella and T. ni to Cry1Ac [34], B. mori resistance to Cry1Ab [35], and O. nubilalis survival on transgenic Cry1Fa maize [36]. Furthermore, abcc2 and abcc3 transcripts were linked to Cry1Ac and Cry1Ca toxins resistance in S. exigua [37], as well as Cry1Ac resistance in the P. xylostella strain DBM1Ac-R [38]. The repertoire of ABC transporters likely involved in Bt toxin resistance has expanded to implicate the pigment transporting ABCG1, Pxwhite, that shows down-regulated transcription in Cry1Ac resistant P. xylostella [39]. Furthermore, a truncation of the abca2 gene of H. armigera was linked to Cry2Ab resistance [40], and spliceosome variants in H. armigera abcc2 are associated with Cry1Ac resistance [41].
A cell adhesion protein, cadherin (cad), was identified as an in vitro Cry1A toxin receptor in the midgut of M. sexta [42], and an ortholog from O. nubilalis analogously is capable of binding Cry1Ab, Cry1Ac and Cry1F toxins in vitro [43, 44] as well as Cry1A toxins in vivo [45]. A mutation caused by transposon insertion into the H. virescens cad provided the initial evidence for involvement in resistance mechanisms [46], and additional mutations were associated with Bt resistance traits among field-derived Pectinophora possypiella [12], H. armigera [47], and O. furnacalis [48]. Despite these prior findings, the cadherin locus is not linked to Cry1Ab or Cry1F resistance traits in O. nubilalis [31, 36], or Cry1Ac resistance in T. ni [49]. Analogously, isoforms of membrane-bound alkaline phosphatase (ALP) in the midgut of M. sexta and H. virescens bind the Cry1Ac toxin [50, 51], and the down-regulation of alp was subsequently associated with resistance to Cry1Ac in H. armigera and Cry1Fa in S. frugiperda [52] and Cry1Ac in P. xylostella [38]. Interestingly, Guo et al. demonstrated that alp down-regulation may be controlled by pathways involving a mitogen-activated protein kinase, MAPK4 [38]. Experiments on O. furnacalis show that although ALP is a putative receptor for Cry1Ac in the midgut of susceptible individuals, but neither toxin binding nor expression of alp transcripts were reduced among Cry1Ac resistant larvae [53].
The Asian corn borer, O. furnacalis (Guenée), is a highly destructive pest insect of cultivated maize and is endemic to parts of East Asia and Australia, including China [54, 55]. Although chemical insecticides and natural enemies are used for control [56, 57], these methods tend to show limited effectiveness due to difficulties in the timing of applications or environmental variation. Similar to other global regions, Bt transgenic crops hold great promise for the improvement of agricultural production in China, but concerns remain regarding potential evolution of resistance voiced nearly two decades ago [58] and now being observed amongst several insect pest populations [3, 59]. Laboratory strains of O. furnacalis have been selected for high levels of resistance to Cry1Ab [32, 60] and Cry1Ac [61], but a complete understanding of the genetic mechanisms of these resistance traits remain incomplete. Recent advances in next generation DNA sequencing technologies allow the analysis of variation in transcript abundance (gene expression) through the application of RNA sequencing (RNA-seq) [62], and have allowed the comparison gene regulation between Bt resistant and susceptible phenotypes [63, 64]. The following paper describes construction of a reference midgut transcriptome to which mapped RNA-seq read data were used to estimate gene expression differences among Bt resistant (Cry1Ab and Cry1Ac) and susceptible stains of O. furnacalis. These methods, in conjunction with secondary validation, demonstrate the capability of RNA-seq methods to identify candidate genes putatively involved in the evolution of Bt resistance traits. These data are important for the understanding of systemic differences between Bt resistant and susceptible genotypes, and how selection may result in fixed changes of expression level among numerous genes.
Materials and Methods
Insect resistance levels
Three O. furnacalis laboratory colonies were used in the current study; a Bt toxin susceptible colony (ACB-BtS), and two laboratory colonies selected for increased resistance to Cry1Ab (ACB-AbR) and a Cry1Ac toxins (ACB-AcR). Selection schemes were described previously for ACB-AbR [32, 60], and ACB-AcR [61]. In brief, O. furnacalis larvae were collected from maize fields throughout central China, and reared on artificial diet with a photoperiod of L:D=16:8 h and relative humidity at 70~80%. Larvae in ACB-AcR were initially exposed to 2.5 ng Cry1Ac toxin/g of artificial diet, and exposures steadily increased to levels that caused 40 (LC40) to 70% mortality (LC70) in subsequent generations. ACB-AcR was reared in subsequent generations a final concentration of 400 ng Cry1Ac/g artificial diet as described previously [65]. Selection was analogously replicated in order to generate the ACB-AbR colony [32, 60].
Resistance to Cry1Ab and Cry1Ac was respectively estimated among ACB-AbR and ACB-AcR larvae as the proportion of moribund larvae observed following a 7 d exposure to diet incorporated dose-response assays, and reported as a resistance ratio (RR) that is compared to the mortality among ACB-BtS larvae. Specifically, neonates from each strain were exposed to 9 concentrations of Cry1Ac (0, 1, 50, 100, 200, 400, 600, 800 and 1000 μg/g) and 9 concentrations of Cry1Ab (0, 0.1, 0.5, 1, 5, 10, 50, 100 and 200 μg/g) for 7 days to obtain 5% to 100% mortality. Bioassays at each toxin level were performed in triplicate (n = 48 neonates per replication), and tap water was used in the control treatment. The lethal concentration resulting in mortality among 50% of individuals (LC50) was calculated for each strain and each toxin using a Probit statistical model in Polo Plus (LeOra Software, USA). For each Bt toxin, a RR was calculated by dividing LC50 of the ACB-AbR or ACB-AcR strain by the LC50 estimated from the susceptible strain, ACB-BtS.
Midgut dissection and RNA extraction
Larvae from each of the ACB-AbR, ACB-AcR, and ACB-BtS colonies were fed on non-Bt toxin diet under standard rearing conditions, and 10 live 5th instars were randomly selected from each and placed on ice. The midgut tissues were dissected from each and the lumen was rapid cleaned with a solution of 0.7% NaCl (w/v) to remove debris, followed by submersion in liquid nitrogen. Dissected midguts were pooled by colony (n = 10 per pool), and total RNA was extracted separately from each the ACB-AbR, ACB-AcR and ACB-BtS pool using the Trizol reagent following manufacturer instructions (Invitrogen, CA). Resulting RNA samples were treated with DNase I (Promega, Madison, WI, USA) following manufacturer's instructions. Quantity and quality of RNA was assessed by denaturing gel electrophoresis and spectrophotometry on a Nanodrop 2000 (Thermo Scientific, Wilmington, DE, USA).
Midgut transcriptome sequencing, assembly and functional annotation
Three single-end cDNA libraries were constructed, one for each of the three midgut total RNA samples derived from ACB-BtS, ACB-AbR, and ACB-AcR. Purified total RNA from each sample was separately converted to complementary DNA (cDNA) using the ScriptSeq™ mRNA-seq Library Preparation Kit (Illumina, San Diego, CA) following the manufacturer's protocol. In brief, mRNA was isolated from 20 µg of purified total RNA using oligo (dT) magnetic beads, and the resulting mRNA fragmented by incubation with divalent cations at 94 °C for 5 min. First strand cDNA was generated by reverse transcription primed by random hexamers, followed by synthesis of the second-strand cDNA using RNase H and DNA polymerase I. After end-repair and ligation of Illumina sequencing and flow cell adaptors, the products were amplified by PCR and purified with QIAGEN MiniElute PCR Purification Kit (Qiagen, Venlo, Netherlands), and single-end 100 bp sequence read data was generated on an Illumina HiSeqTM 2000 platform at Novogen Co., Ltd. Beijing, China. Raw Illumina read data were submitted to the National Center for Biotechnology Information (NCBI) short read archive (SRA) database (Table 1; ACB-BtS: SRX751199, ACB-AbR: SRX751197, ACB-AcR: SRX750576) under BioProject PRJNA266299.
Lethal concentration (LC) and resistance ratio (RR) estimates for Cry1Ab and CryAc toxins among Ostrinia furnacalis larvae from ACB-AbR and ACB-AcR strains compared to those in the susceptible ACB-BtS strain.
| Bt toxin | ACB strain | LC50 (95% CI)* μg/g | RR |
|---|---|---|---|
| Cry1Ab | ACB-BtS | 0.36 (0.13-0.51) | / |
| ACB-AbR | 69.77 (49.10-98.66) | 193.80 | |
| Cry1Ac | ACB-BtS | 0.27 (0.19-0.34) | / |
| ACB-AcR | 967.76 (842.75-1092.81) | 3584.30 |
* 95% confidence interval
Read statistics and quality assessment was conducted using FastQC [66]. Contaminating Illumina adapters and nucleotides within a 20 bp window with a mean Phred 33 quality score less than 20 (q < 20) were trimmed from the raw data using Trimmomatic 0.32 [67]. The filtered high quality fastq read data from all libraries were separately used as input for the Trinity in silico read normalization script [68] using default parameters except a kmer size (--KMER_SIZE) was set to 25 and maximum coverage per kmer (--max_cov) limited to 30. Trinity r20140414 [69] was used to assemble contigs (Inchworm), cluster contigs (Chrysalis) and reconstruct transcript isoforms (Butterfly) according to Haas et al. [70]. The resulting contigs were submitted to the National Center for Biotechnology Information (NCBI) transcriptome shotgun assembly (TSA) database under RNA-seq Assembly project (r20131110). Open reading frames (ORFs) of ≥100 amino acids were predicted from Trinity fasta output using TransDecoder 1.0, and the resulting .cds file used as the reference for subsequent RNA-seq analyses. Putative functional annotations were obtained for all O. furnacalis derived proteins sequences obtained from the TransDecoder 1.0 .pep files using Blast2GO [71] as described by Garczynski et al. [72], except that BLASTp [73] was used to query the NCBI non-redundant (nr) protein database.
Estimates of differential-expression among midgut transcripts
Non-normalized trimmed single-end fastq read data derived from each Illumina RNA-seq library were each mapped separately to the ORF-containing transcripts identified by TransDecoder (.cds file) using Bowtie 2.2.1 [74] with default parameters and alignments output in sequence alignment map (SAM) file format. Each SAM file was converted to a binary sequence alignment map (BAM) format and counts of the number of reads aligned to each ORF-containing contig within indexed BAM files were output a tab-delimited text using SamTools [75]. The level of expression was estimated for each transcript among replicated ACB-BtS and ACB-AbR and ACB-AcR alignments using read counts as a proxy for transcript abundance [76], and implemented using the DESeq package [77] with the false discovery rate (FDR) set at 0.001 and quantile for removing insignificant estimates at 0.4. MA-plots comparing log2[fold-change] to mean of normalized read counts were generated, as well as a plot showing the distribution of p-values. Significance threshold were set at two levels 1) a moderate threshold of log2[fold-change] ≥ |1.0| and p-values ≤ 0.001, and 2) a high stringency threshold of log2[fold-change] ≥ |2.0| and p-values ≤ 0.001.
Nucleotide and derived protein sequences corresponding to each Trinity-defined transcript isoform (component; comp from Butterfly output) within putatively differentially expressed transcripts [log2[fold-change] ≥ |1.0| and p-values ≤ 0.001] were obtained from the reference transcriptome using the PERL script select Seqs.pl. Transcripts putatively annotated within our Blast2GO analyses as candidate Bt resistance genes were used as queries of all translated amino acid sequences using the tBLASTn algorithm (identity cutoffs ≥ 98% over ≥25 amino acids), and consensus transcripts were reconstructed used CAP3 [78]. Derived protein sequence of each CAP3 assembled contig was obtained using the Virtual Ribosome [79] and reading frame adjusted based on BLASTx search of the NCBI nr protein database. In all cases, the nearest ortholog from the model lepidopteran B. mori were downloaded in FASTA format. Additionally, homologous sequences from the O. nubilalis midgut transcriptome were identified by BLASTp query of a local database composed of translated fasta sequences obtained from Vellichirammal et al., [64]. Multiple sequence alignments performed among B. mori, O. nubilalis and O. furnacalis sequences with ClustalW v2.0 using default parameters [80]. Conserved protein primary and secondary structures were identified by search of the Conserved Domains Database (CDD) [81], and use of the Smart Modular Architecture Research Tool (SMART) [82], and Transmembrane protein topology Hidden Markov Model (TMHMM) prediction algorithm [83].
Three closely related aminopeptidase N paralogs are known within the genome of O. nubilalis (OnAPN3a, OnAPN3b, and OnAPN3c) that show high degrees of amino acid sequence similarity [31, 84]. Differentially expressed O. furnacalis transcripts annotated as encoding APN3-like proteins were aligned with Onapn3a, Onapn3b, and Onapn3c orthologs using the ClustalW algorithm (default parameters), and imported into the program Molecular Evolutionary and Genetic Analysis (MEGA) 6.0 [85]. Gluzincin aminopeptidase, GAMEN, and zinc binding motifs (HEX2HX18E) were identified from CDD and SMART output as described above. Putative GPI-anchors signals were predicted using GPI-SOM [86]. Phylogenetic inferences were made using the genetic distances with the Neighbor-joining (NJ) methods as well as by Maximum likelihood, and node support provided by 1,000 pseudoreplicates.
Quantitative RT-PCR (qRT-PCR) analysis was conducted to validate the abundance of O. furnacalis midgut transcripts that were annotated as candidate Bt resistance genes or that showed significant differential expression in RNA-seq experiments. The candidate Bt resistance genes alkaline phosphatase and cadherin which were not predicted to show significant up- or down-regulation from RNA-seq analyses were also tested by qRT-PCR. The O. furnacalis β-actin gene (accession number: GU301782) was chosen as a reference for normalizing qRT-PCR data. Oligonucleotide primers for qRT-PCR were designed with Primer Express 3.0 (ABI, USA) (Supplementary Data 1). Total RNA was extracted from midgut of ACB-AcR, ACB-AbR and ACB-BtS strains as described previously and 2.0 µg used in 1st strand cDNA synthesis. Each strain including 30 larvae, and each 10 larvae were treated as biological duplicates. Reactions were setup in triplicate using 20 µL reaction volumes containing 10 µL of SYBR Green Universal Master Mix II (Takara, Japan), 1.0 µL of PCR Forward Primer (10 μM), 1.0 µL of PCR Reverse Primer (10 μM), 0.5 µL of ROX reference dye II (50X), 1.0 µL of 1st strand cDNA as template, and 6.5 µL sterilized ultra-pure grade water. Amplification was performed using the ABI 7500 Fast Real-Time PCR System, at 95 °C (30 s), followed by 40 cycles at 95 °C (10 s) and 60 °C (20 s) (2 steps per cycle), and then melt-curve analysis to evaluate the specificity of the amplifications. Normalization of the experimental cycle threshold (CT) data relative to the β-actin reference gene was done as previously described [87, 88], and relative expression of normalized data was made to values from the ACB-BtS strain by the comparative 2-∆∆CT method [89]. The significance of any differences in estimated CT values between experimental unknowns was assessed by Nested ANOVA, and thresholds set at α = 0.05.
Structural variant predictions
The individual .BAM alignment generated for ACB-BtS, -AbR and AcR above were merged into as single file using the SamTools mpileup command. The resulting merged BAM file was interrogated for variant nucleotides with respect to the reference transcriptome using SamTools and BCFtools as described previously [90], and vcfutils.pl VarFilter options filtered the variant call format (.vcf) output file for read depths ≥ 20 and for variants  10 bp from any alignment gap. The vcf v4.1 file was further filtered for variant sites fixed differently between ACB-BtS and -AbR or -AcR, and mutations causing non-synonymous (amino acid) changes were predicted from annotated CDS data.
10 bp from any alignment gap. The vcf v4.1 file was further filtered for variant sites fixed differently between ACB-BtS and -AbR or -AcR, and mutations causing non-synonymous (amino acid) changes were predicted from annotated CDS data.
Results
Insect resistance levels
After 142 generations of selection for survival on Cry1Ab toxin diet, the ACB-AbR laboratory colony has an estimated RR of 193.8 compared to the susceptible ACB-BtS strain (Table 1), which is an increase compared to the RR of 106.8 previously reported after 51 generations of selection [60]. Analogously, the ACB-AcR strain shows an estimated 3,584.3-fold increase in Cry1Ac resistance compared to ACB-BtS after 144 generations of selection on Cry1Ac toxin, diet (Table 1).
Midgut transcriptome sequencing, assembly and functional annotation
A reference O. furnacalis midgut transcriptome was assembled from paired end mRNA-seq data derived from ACB-AbR, ACB-AcR and ACB-BtS strains reared on control artificial diet (without Bt toxin), and therefore was useful to investigate fixed (constitutive) differences in gene expression between strains. The combined set of libraries for each strain contained ≥ 41.2 million reads and ≥ 4,117 Mbp of sequence data (Table 2A). Following removal of adaptor, ambiguous and low-quality sequence data with a Phred 33 quality score < 20 (q < 20), ≥ 35.2 million reads and ≥ 3,400 Mbp of sequence data remained from each strain. Following read normalization, assembly of contigs, contig clustering, and isoform reconstruction using the Trinity pipeline, a combined midgut reference assembly of 83,370 O. furnacalis midgut-expressed transcripts was obtained. The assembled sequences were deposited in the NCBI Transcriptome Shotgun Assembly (TSA) database and linked to the BioProject PRJNA266299. These sequences do not represent unique genes, but instead contain a total count of transcript isoforms (alternative splice variants or closely-related paralogs) defined within 41,088 unique components (comp; defined by the program Chrysalis) within a set of transcripts by Butterfly (Table 2B; see Haas et al., [70] for additional details). From this reference assembly, a total of 12,288 unique coding sequences were identified from 28,940 components (isoforms groups; Table 2B).
A transcriptome was previously assembled from whole O. furnacalis larvae and used to predict putative DEGs between ACB-AbR and -BtS [32], but the combined reference assembly in this study represents the first midgut-specific genomic resource for this species, and provided the means by which to assess expression-level differences within midgut tissues. Nearly 60% of the 28,940 coding sequences predicted within the O. furnacalis midgut transcriptome did not receive any functional gene annotations. Although problematic for efforts to obtain putative homology-derived functions for our dataset, these data may represent potential genomic novelty what could be applied in future efforts to control crop damage by this insect. Regardless, annotation, GO enrichment and pathway analyses were possible for a number of transcripts (Table 3).
Summary for the Ostrinia furnacalis midgut transcriptome A) Illumina HiSeq2000 raw and processed read data, and B) the assembled reference sequences.
| Raw read data | Trimmed read data | |||||
|---|---|---|---|---|---|---|
| A) Illumina read data | ||||||
| Libraries | Count | Length (bp) | Count | Length (bp) | ||
| ACB-AbR.fastq | 52,243,872 | 100 | 44,502,745 | 96.4 | ||
| ACB-AcR.fastq | 47,658,725 | 100 | 40,650,855 | 96.4 | ||
| ACB-BtS.fastq | 41,167,042 | 100 | 35,229,832 | 96.5 | ||
| B) Assemblies | ||||||
| Midgut transcriptome | Count | Length (bp) | Components | Isoforms/comp | ||
| Reference, total1 | 83,370 | 617.9±554.2 | 41,088 | 2.00±3.45 | ||
| Reference, cds2,3 | 28,940 | 638.3±423.8 | 12,288 | 2.24±4.18 | ||
1. Assembly of pooled normalized Illumina read data from ACB-AbR, -AcR, and -BtS libraries. 2. Coding sequences in the reference assembly using Transdecoder (>100 amino acids). 3. Used as template for RNA-seq read mapping and estimation of differential expression.
Estimates of differential-expression among midgut transcripts
The goals of this study were to determine the transcripts and functional gene categories that are putatively involved in Cry1Ab and Cry1Ac resistance mechanisms. This was addressed by estimating transcript levels in the midgut of Cry1Ab and Cry1Ac resistant O. furnacalis based on counts of mapped RNA-seq read data, followed by application of statistical models to estimate quantities that differ significantly when compared to the midgut of susceptible larvae. Statistically significant fold-changes in transcript levels remain difficult to interpret within a biological context, but we follow the precedent of a large number of prior RNA-seq studies that assume that there is a biological consequence at the cellular or functional level. In addition, our data estimated differential expression among two colonies independently selected for Cry1Ab and Cry1Ac toxin resistance, wherein we interpreted constitutive up- or down-regulation in both colonies as strong evidence for involvement of those genes in the evolution of Cry1A toxin resistance.
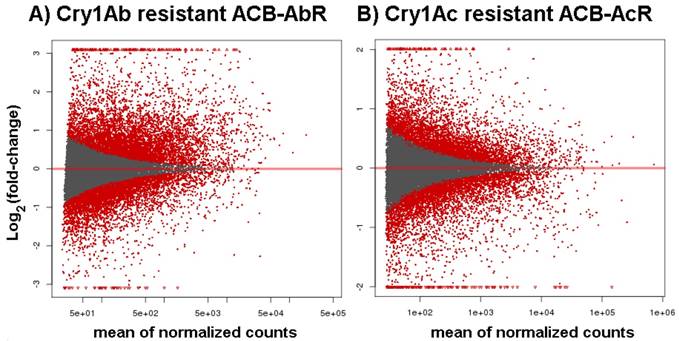
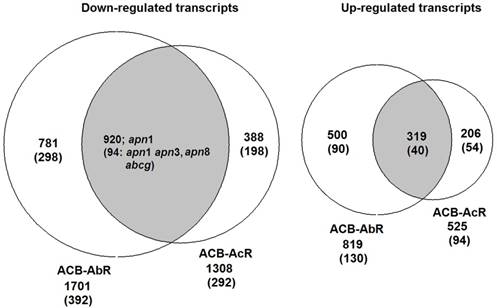
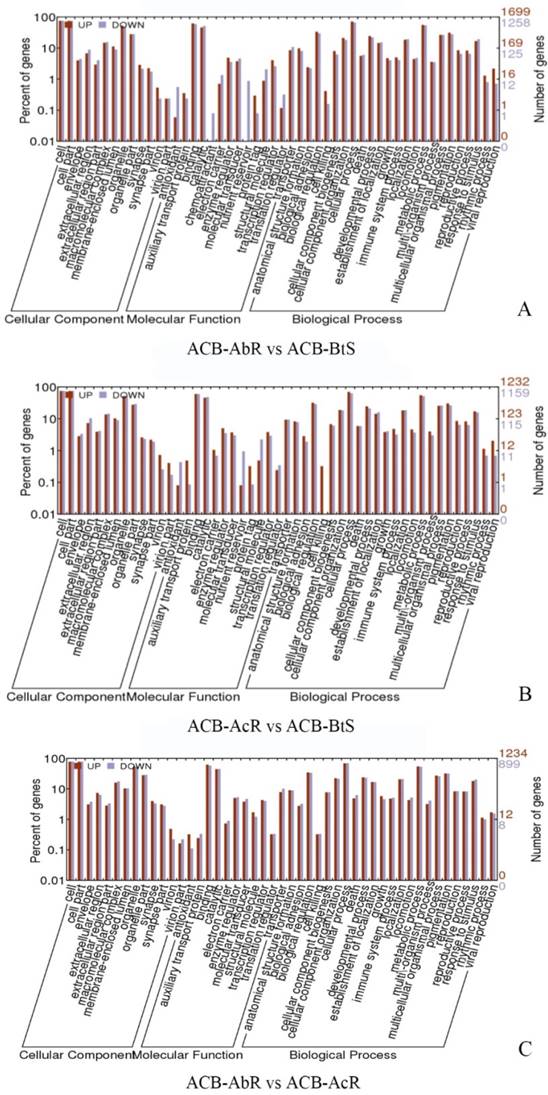
To assess the significance of any differences in estimated midgut transcript levels between resistant and susceptible larvae, the current study applied two criteria within our DESeq analysis; Threshold 1 of log2[fold-change] ≥ |1.0| and p-values ≤ 0.001 that identified ≤ 1,701 significantly up- or down-regulated transcripts between ACB-AbR or -AcR and ACB-BtS phenotypes (Fig. 1; Fig. 2), and ≤ 392 transcripts at a more stringent criteria of log2[fold-change] ≥ |2.0| and p-values ≤ 0.001 (Fig. 2; Supple Data 2). This comparison showed that a larger proportion of genes were significantly down-regulated compared to up-regulated, and down-regulated transcripts included those encoding aminopeptidase N paralogs and a member of the ABC transporter family (ABCG).
Additional genes highly down-regulated among Cry1Ab and Cry1Ac resistant larvae included those in putative functional categories involved in lipid metabolism and transport, binding, and stress response (Fig. 3). Some of the annotations that were made among the down-regulated transcripts for lipid storage and transport proteins (arylphorin, methionoe-rich protein), lipid metabolizing enzymes (e.g. lipases), and stress-response associated genes glutathione S-transferase (GST), cytochrome P450 monooxygenases, and heat shock proteins (HSP) 68- and 70-like (Supplementary Data 2). Several uridine diphosphates (UDP)-glucuronosyltransferase-like transcripts were also significantly repressed in the midgut of both Cry1Ab and Cry1Ac resistant O. furnacalis.
The statistics for the annotation of O. furnacalis midgut transcriptome across annotation resources. NA = no annotation(s) received.
| Database | ||||||||
|---|---|---|---|---|---|---|---|---|
| BlastX | BlastP | Swiss-Prot | eggNOG | GO | KEGG | Annotated | NA | |
| Unigene Number (%) | 28,641 (28.64) | 21,044 (21.04) | 32,683 (32.68) | 12,441 (12.44) | 2,364 (2.36) | 3,553 (35.54) | 39,869 (39.87) | 60,134 (60.13) |
MA plots of estimates change in transcript levels in the midgut of Ostrinia furnacalis from A) the Cry1Ab resistant ACB-AbR strain and B) the Cry1Ac resistant ACB-AcR strain. All data are log2 transformed ratios of transcripts between resistant and susceptible ACB-BtS strains scaled by the mean count of normalized reads assigned to the respective transcript.
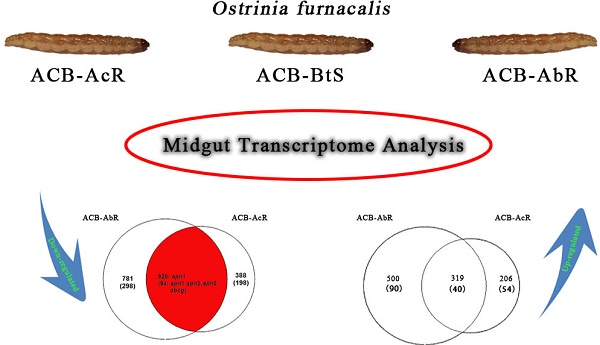
Venn diagrams showing number of transcripts the Cry1Ab resistant ACB-AbR colony and the Cry1Ac resistant ACB-AcR colony that are A) down-regulated and B) up-regulated in larval midgut tissues when compared to the susceptible ACB-BtS strain. Transcript numbers with statistically significant changes are shown the cutoffs of 1) log2(fold-change) > |1.0| and P < 0.001, and 2) log2(fold-change) > |2.0| and P < 0.001 are shown (the latter in parenthesis). The number of transcripts with significant changes compared to ACB-BtS shared between ACB-AbR and -AcR are represented in the grey overlapping regions. Regulation of previously known candidate Bt resistance genes are shown in the appropriate categories in each diagram.
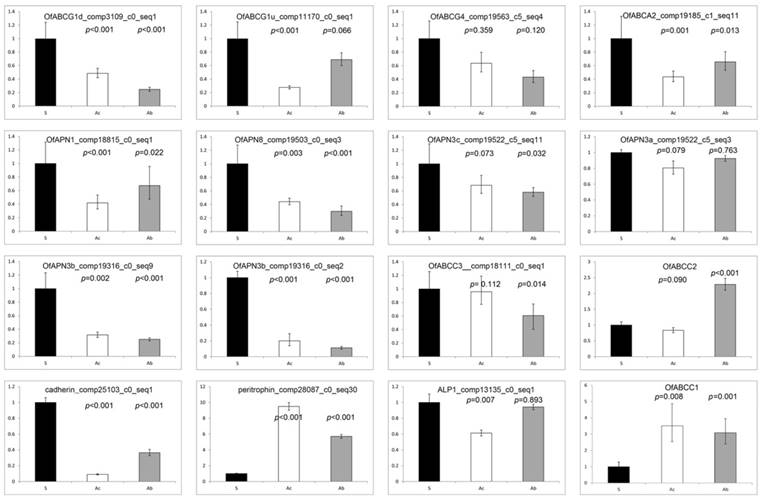
Also among the significantly down-regulated transcripts in the high stringency threshold group (log2[fold-change] ≤ -2.0 and p-values ≤ 0.001), several were annotated as previously known Bt resistance genes (see Introduction) including members of the aminopeptidase N gene family, apn3 paralogs and apn8, an ABC transporter in subgroup G, abcg, as well 10 serine protease genes. Sequence alignment of derived APN3 paralogs from species of Ostrinia indicated ≥ 96% intraspecific amino acid identity (Fig. 4A) and subsequent phylogenetic analyses resolved the association between paralogs with APN3a and 3c predicted to be the most recently diverged duplicates (Fig. 4B). Estimates of significantly reduced expression in RNA-seq data (Table 4) were corroborated by qRT-PCR results for Ofapn1 and Ofapn8, as well as for Ofapn3b, in the midgut of both ACB-AbR and -AcR (Fig. 5). Candidate Bt resistance genes were also annotated within the lower stringency threshold group of O. furnacalis transcripts (≥ 1.0 log2[fold-change] ≤ -1.0), and included those encoding ABC transporters from three different subgroups (A, C, and G), ABCA2, ABCC2, and ABCG4. The number of significantly up-regulated transcripts were smaller in number for both Cry1Ab and Cry1Ac resistant O. furnacalis compared to down-regulated genes (Fig. 2), and interestingly included two up-regulated ABC transporter subgroup G members (comp15850 and comp9932 (scarlet); Table 4).
Gene ontology (GO) classification of the different expression gene in ACB-AbR and ACB-BtS(A), ACB-AcR and ACB-BtS (B), ACB-AbR and ACB-AcR in O. furnacalis. The left Y-axis is the percentage of up and down expression among resistance and susceptible strains. The right Y-axis is the number of up and down expression among resistance and susceptible strains.
Assignment of aminopeptidase N3 (APN3) paralogs to three orthologous groups (APN3a, APN3b, and APN3c), each showing significant down-regulation among Cry1Ab and Cry1Ac resistant Ostrinia furnacalis (Table 4). A) Partial alignment of derived APN3 amino acid sequences from Ostrinia furnacalis and O. nubilalis. Variable residues are highlighted. Signal peptide encodes in a box, the gluzincin aminopeptidase motif underscored by "x", the histidine residues (H) of the Zn binding motif HEX2HX18E are underscored with "+", and missing data shown as "*". Phylogenetic reconstructions were conducted using the methods B) Neighbor-joining and C) Maximum likelihood. All node support provided from 1000 pseudoreplicates of the data.

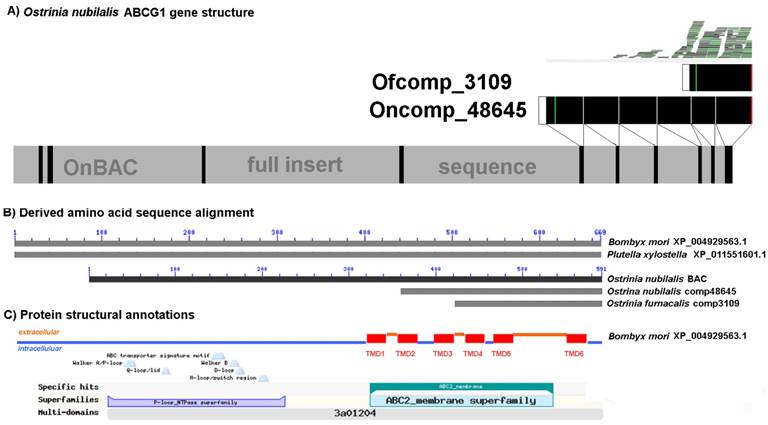
The putative ABCG1 encoded by O. furnacalis comp3109_c0_seq1:140-646(+) is the ABC transporter that was the highly down-regulated in both Cry1Ab and Cry1Ac resistant strains based on RNA-seq and qRT-PCR. The derived 168 amino acid sequence from the 646 bp O. furnacalis abcg1-like transcript (comp3109_c0_seq1:140-646(+)) shows ≥ 71% amino acid sequence identity with the putative ACBG1-like transporters from B. mori (XP_004929563.1; 667 aa; aligned positions 510 to 667) and P. xylostella (XP_011551601.1; 669 aa; aligned positions 502 to 669). These orthologs correspond to gene models KAIKOGA014788 on B. mori chromosome 12 and Px012058 on P. xylostella scaffold 47 (Fig. 6). Interestingly, the 386 aa sequence encoded by the B. mori gene model BGIBGA010555 spans the C-terminal half of XP_004929563.1 (positions 281 to 667) as well as Px012058 (positions 283 to 669). Similarly, the 2,289 bp the comp48645_c0_seq2 from an O. nubilalis midgut transcriptome [64] encodes a 345 aa long variant located at the C-terminus of the ortholog (Supplementary Data 3A). Annotation of the O. nubilalis OnBAC 35G22 full insert sequence (GenBank HTGS KX793137) using these data defined a putative 1,773 bp CDS fragment encoding a 590 aa partial OnABCG1 peptide, with the missing the N-terminus residing on another BAC (Fig. 6). Alignments to this genomic CDS showed that homology with truncated transcripts began at exon splice junctions, thus providing evidence that truncated copies are indeed splice variants (Fig. 6; Supplementary Data 4). Analogously, mapping of O. furnacalis RNA-seq reads to the 1,773 bp partial Ofabcg1 transcriptional unit on OnBAC 35G22 provided evidence that the Ofabcg1 comp3109_c0_seq1:140-646(+) transcript is the major isoform in the midgut. The above RNA-seq evidence and qRT-PCR data confirm that this Ofabcg1 isoform is down-regulated in Cry1Ab and Cry1Ac resistant larvae (Fig. 5). SMART and TMHMM secondary structural predictions indicated that the 168 aa OfABCG1 contains three C-terminal transmembrane domains (TMD4, TMD5, and TMD6) whereas the OnABCG1 homolog retains all six, but both have complete NBD deletions.
Estimates of transcripts levels (expression) among candidate Bacillus thuringiensis toxin resistance genes in the midgut of Ostrinia furnacalis based on RNA-seq read mapping to each Trinity component (comp). The transcripts significantly up- (+) or down-regulated (-) in two selected lines (ACB-AbR or -AcR) are based on log2[fold-change] estimates compared to the common susceptible stain (ACB-BtS). NS = not significant.
| Trinity component (transcript) | Annotation | ACB-AbR | ACB-AcR | gene |
|---|---|---|---|---|
| comp3109_c0_seq1:140-646(+) | ABC transporter G1 | -2.55(3.1e-21) | -3.54(1.8e-58) | OfABCG1d |
| comp9932_c0_seq1:509-967(+) | ABC transporter G1 | NS | +1.32(5.08e-05) | OfABCG1u |
| comp15850_c0_seq2:228-680(-) | ABC transporter G1S | +1.00(7.85e-05) | +1.08(5.18e-05) | Ofscarlet |
| comp17719_c0_seq4:1-1962(+) | ABC transporter G4 | -1.52(<0.0001) | -1.48(<0.0001) | OfABCG4 |
| comp19185_c1_seq11:1-807(-) | ABC transporter A2 | -1.30(4.12e-42) | NS | OfABCA2 |
| comp19455_c0_seq5:1-5130(+) | ABC transporter A2 | -1.26(<0.0001) | NS | OfABCA2 |
| comp16920_c0_seq5:3-1388(-) | ABC transporter A2 | -1.19(7.51e-89) | -1.01(1.81e-55) | OfABCA2 |
| comp19185_c1_seq2:783-1433(-) | ABC transporter A2 | -1.16(1.06e-33) | -1.04(2.97e-30) | OfABCA2 |
| comp18111_c0_seq1:196-2433(+) | ABC transporter C3 | -1.29(30.3e-130) | NS | OfABCC3 |
| comp16077_c0_seq4:2-3469(-) | ABC transporter C3 | -1.19(3.13e-162) | NS | OfABCC3 |
| comp19374_c4_seq1:579-2912(+) | Aminopeptidase N1 | -1.95(<0.0001) | -1.50(<0.0001) | OfAPN1 |
| comp18815_c0_seq1:194-3178(-) | Aminopeptidase N1 | -1.71(<0.0001) | -1.36(<0.0001) | OfAPN1 |
| comp19522_c5_seq3:173-997(-) | Aminopeptidase N3a | -2.95(<0.0001) | -2.29(,0.00001) | OfAPN3a |
| comp19316_c0_seq9:676-2964(+) | Aminopeptidase N3b | -2.78(<0.0001) | -2.06(<0.00001) | OfAPN3b |
| comp19316_c0_seq2:479-2614(+) | Aminopeptidase N3b | -2.63(<0.0001) | NS | OfAPN3b |
| comp19522_c5_seq11:1-474(-) | Aminopeptidase N3c | -3.67(<0.0001) | -2.62(<0.00001) | OfAPN3c |
| comp19316_c0_seq8:676-1932(+) | Aminopeptidase N3c | -2.73(<0.0001) | -2.11(<0.00001) | OfAPN3c |
| comp19503_c0_seq3:292-801(-) | Aminopeptidase N8 | -2.26(7.0E-87) | +2.07(4.3E-64) | OfAPN8 |
| comp19411_c8_seq1:3-434(-) | chymotrypsin | -4.27(<0.0001) | -4.41(<0.0001) | |
| comp12401_c0_seq1:1-579(-) | trypsin-like SP | -3.60(1.9e-299) | -5.11(<0.0001) | |
| comp14870_c3_seq2:396-728(+) | trypsin alkaline c | -3.50(1.59e-32) | -2.14( <0.0001) | |
| comp16028_c0_seq1:1-447(-) | trypsin-like SP | -3.11(<0.0001) | -3.70(<0.0001) | |
| comp19588_c4_seq4:1-753(+) | trypsin-like SP | -2.34(<0.0001) | -2.46(<0.0001) | |
| comp14501_c0_seq1:93-548 | trypsin alkaline c | -2.21(4.57e-14) | NS | |
| comp18273_c0_seq1:295-888(-) | trypsin-like SP | -2.05(<0.0001) | -2.42(<0.0001) | |
| comp19320_c6_seq4:2-748(+) | trypsin-like SP | -2.02(<0.0001) | -2.37(<0.0001) | |
| comp13807_c1_seq3:621-1112(+) | trypsin-like SP | -3.35(5.08e-32) | -5.07(<0.0001) | |
| comp17412_c0_seq2:220-786(+) | trypsin-like SP | -3.27(<0.0001) | -3.80(<0.0001) | |
| comp5178_c0_seq1:3-548(-) | trypsin-like SP | NS | -3.01(3.34e-23) | |
| comp18630_c10_seq1:120-701(-) | trypsin-like SP | NS | -2.76(<0.0001) | |
| comp15635_c1_seq3:174-1067(+) | chymotrypsin | -3.37(<0.0001) | -2.90(1.72e-23) | |
| comp19379_c9_seq4:444-881(+) | chymotrypsin | -2.45(<0.0001) | -2.19(<0.0001) | |
| comp9315_c0_seq2:3-521(+) | chymotrypsin | -2.24(6.84e-22) | NS | |
| comp11509_c0_seq1:286-633(-) | chymotrypsin | +4.29(2.61e-68) | +4.67(3.69e-68) | |
| comp11509_c0_seq2:504-851(-) | chymotrypsin | +3.84(3.87e-63) | +4.51(2.82e-67) | |
| comp5693_c0_seq2:2-433(+) | trypsin-beta-like | +3.77(9.3e_193) | +4.53(4.26e-210) | |
| comp16545_c0_seq2:933-1256(+) | chymotrypsin | +3.35(1.80e-39) | +4.35(1.73e-46) |
* Differentially expressed genes have a FDR value of ≤ 0.0001 and the absolute value of log2 (fold change) ≥ 1 or log2 (fold change) ≤ -1 between ACB-AbR or ACB-AcR and ACB-BtS.
OfABCG4-encoding transcripts are also predicted to be down-regulated in the midgut of Cry1Ab and Cry1Ac resistant larvae (Table 4; comp17719_co_seq4:1-1962(+)), which has the full OfABCG4 CDS (Supplementary Data 3B). In contrast, qRT-PCR results suggest that Ofabcg4 transcript levels are not significantly different in ACB-AbR or -AcR compared to ACB-BtS (Fig. 5). No Ofabcg4 transcription isoforms (variants) were identified. Four transcripts from a single OfABCA2-like CDS were differentially regulated among resistant strains, and most significantly reduced among Cry1Ab resistant larvae (Table 4). No O. furnacalis transcripts were found that encoded a full-length OfABCA2 peptide, but the entire sequence was reconstructed from fragments using CAP3 and shown to share > 97% identity with the translated sequence from O. nubilalis Oncomp46431_c0_seq2 (Supplementary Data 3C). Analogously, CAP3 reconstructed the complete CDS for the Ofabcc3 transcript that was significantly down-regulated in the ACB-AbR colony (Table 4; Supplementary Data 3D). ABC transporter functional domains were identified within all aligned peptides. Transcripts for abcc3 was the only ABC transporter family C member predicted to show significant differential regulation within RNA-seq analyses and only occurred via repression within the ACB-AbR colony (Table 4), which was supported by qRT-PCR data (Fig. 5). In contrast to our RNA-seq results, qRT-PCR indicated that Ofabcc1 and Ofabcc2 are up-regulated in both resistant strains, with the exception of Ofabcc2 in ACB-AbR.
Although the numbers of up-regulated transcripts are comparatively less in number, several genes in the same functional categories were shared with those that were down-regulated (Fig. 3). Specifically, these include a number of serine proteinases (Table 4) as well as and lipid storage and metabolizing proteins, and GST (Supplementary Data 2). Interestingly, transcripts encoding candidate Bt binding receptors a membrane-bound alkaline phosphatase (comp13277_c0_seq1:1-414(+)) was not significantly down-regulated in RNA-seq and corroborated by qRT-PCR (Fig. 5), in contrast cadherin was significantly down-regulated in qRT-PCR but not within RNA-seq results. Cry1Ab and Cry1Ac resistant larvae share the putative significant up-regulation of the ABCG-like scarlet gene, but was barely over the threshold (log2[fold-change] ≤ +1.08).
Structural variant predictions
A total of 12,487 variant sites with genotype quality score of 999 were predicted within transcripts across all RNA-seq alignments to the reference transcriptome (data not shown). Filtering of this .vcf output for homozygous variants resulted in 51 sites (50 SNPs and 1 indel), of which 30 were fixed between ACB-BtS and both -AbR and -AcR. Among the other sites, 20 were fixed between ACB-AbR compared to both -BtS and -AcR, and one SNP was fixed within ACB-AcR (Supplementary Data 5). Annotations defined 48 putative protein coding regions, in which SNPs predicted 5 non-synonymous changes. None of the fixed variant sites were predicted within transcripts from DEGs for any known candidate Bt resistance gene (Table 4).
Discussion
Insect resistance levels
Laboratory selected colonies remain effective models for the study of field-evolved resistance, in that mutations identified in selected colonies have also been detected among resistant phenotypes in the field [47, 91]. Furthermore, lab selected strains have been instrumental in the identification of genes involved in the Bt toxin mode of action as well as resistance mechanisms. Potential field relevance of this O. furnacalis trait conferring a 193.8-fold increase in resistance to Cry1Ab was demonstrated by neonate survival and development to pupation when feeding on silk tissues from the Cry1Ab-expressing transgenic event Mon810 [60], which compares to 100% mortality of an unselected field strain [92]. The ACB-AcR strain has a higher estimated Cry1Ac resistance ratio (3,584.3-fold increase compared to ACB-BtS) after 144 generations of selection (Table 1), but commercial maize hybrids expressing Cry1Ac toxin are not available in order to perform on-plant assays for survivorship. High fold-increases in resistance have analogously been estimated for these ACB-AbR and -AcR strains (Table 1) [32, 61], which agrees with prior evidence that chronic Bt toxin exposures can lead to incremental increases in the tolerance towards Bt toxins [93, 94]. Granted chronic low-level exposures such as those used for O. furnacalis and O. nubilalis may favor the selection of resistance traits that are polygenic and potentially influenced by epistasis (gene-gene interactions) [95]); a premise that was partially supported by the detection of more than one quantitative trait locus (QTL) influencing Cry1Ab tolerance in O. nubilalis [31] as well as maybe even for Cry1Ac resistance in H. virescens [46]. Moreover, these low-dose exposures may be pertinent for modeling Bt resistance traits that develop among lepidopteran species that feed on transgenic plant tissues with low Bt titres [96].
The Cry1Ab resistant ACB-AbR strain was reported to have a high degree of cross-resistance to Cry1Ac [60, 61], which is analogous to levels of cross-resistance to Cry1Ac among Cry1Ab selected O. nubilalis [97, 98] and Heliothis virescens [99]. These observations may be due to Cry1Ab and Cry1Ac sharing common midgut receptors [100, 101], that could be supported by results of competitive toxin binding assays [102] as well as immunoblotting data from O. nubilalis that demonstrate binding to similar midgut protein receptors [44, 103]. Surprisingly, ACB-AcR shows low cross-resistance towards Cry1Ab [104], but is analogous to the lack of cross-resistance to Cry1Ab among Cry1Ac selected P. xylostella [105]. This might suggest that although Cry1Ab and Cry1Ac may share some of the same midgut receptors, the molecular mechanism(s) involved in the development of these two O. furnacalis resistance traits may not be identical.
Midgut transcriptome sequencing, assembly and functional annotation
Sequencing of cDNAs and subsequent assembly of these data into transcripts is an effective means to discover novel genes [106], and has been greatly facilitated among non-model organisms by recent advances in high throughput RNA sequencing (RNA-seq) [107]. Due to complexities of assembling a heterogeneous mixture for differential splicing and allelic variants within RNA-seq data, the number of unique transcripts assembled in our O. furnacalis reference transcriptome (n = 83,370) is likely greater than the number of genes expressed in the midgut. Despite of this, analogous numbers of transcripts have been assembled and used successfully to estimate levels of differential expression among the non-model lepidopteran species Lymantria dispar [108], P. xylostella [63], and O. nubilalis [64]. Additional difficulties pertaining to gene annotation are often encountered with the study of non-model organisms. Specifically, our results showed that nearly 60% of the O. furnacalis midgut transcripts failed to receive any functional annotation, but are somewhat typical among non-model organisms [93]. The results may be influenced by the high proportion of transcripts belonging to the class of long non-coding RNAs (lncRNAs) that are transcribed as mRNA but do not encode open reading frames [109]. The assignment of lncRNA origins from numerous and widespread portions of mammalian genomes suggest that most regions show transcriptional activity [110], and are highly prevalent among RNA-seq data from insects [111]. In this era of high throughput sequencing computational annotations are relied upon to assign putative functional roles [112], and undoubtedly hinders the interpretation of the biological consequence of differentially transcription estimated from RNA-seq data [113]. Despite these shortfalls, the current study did not address any of the above stated gaps in knowledge, but instead used available genomic resources and empirical data to infer the possible functions of putative differentially expressed genes.
Estimates of differential-expression among midgut transcripts
RNA-seq experiments provide the opportunity to take a systems approach for evaluating global gene regulation, and can estimate significant genetic differences between phenotypes [114, 115]. Our statistical analysis of RNA-seq data and subsequent annotation of significantly up- and down-regulated genes demonstrate the over-representation of certain functional categories including binding, metabolism and transport in Bt resistant midgut tissues (Fig. 3). Closer interrogation of functional annotations show the presence of transcripts encoding peptides likely involved in lipid binding and transport, as well as lipid and protein metabolism. Since it remains difficult to investigate all of these differentially-regulated genes within the context of Bt resistance mechanisms, we used criteria that assumed genes and gene functional categories that were dysregulated between the two Cry1A toxin resistance traits to indicate the likely presence of a biochemical basis, or contrastingly, unique to once trait or the other. Moreover, the dearth of empirical evidence from non-model organisms in regards to defining gene function led both o the inability to derive putative functions for many transcripts as well as our confidence that all annotations are absolutely correct. Regardless, several transcripts estimated to be significant down-regulation in the midgut of both Cry1Ab and Cry1Ac resistant O. furnacalis, included members of large gene families previously identified as candidate Bt resistance genes; aminopeptidase N, ABC transporter subgroups, and serine proteases family members (Table 4). Upon the addition of qRT-PCR data, our results also suggested that the O. furnacalis cadherin (Ofcad) is also repressed in ACB-AbR and -AcR strains (Fig. 5) and agrees with previous qRT-PCR results from the ACB-AcR train [48]. Results from our study and prior research on ACB-AcR [48] are difficult to compare due to use of whole larval RNA as template in the latter, suggesting any conclusions might be confounded by tissue-specific differences. The lack of any reduction in the expression of O. furnacalis alkaline phosphatase is analogous to prior results using the ACB-AcR strain [53].
Down-regulation of aminopeptidase N transcripts in the midgut of Cry1Ab and Cry1Ac larvae: The involvement of APN receptors in the Bt mode of action and mechanisms of resistance is well documented (see Introduction). There are eight orthologous apn genes, apn1 thru apn8, which resulted from ancestral tandem duplication in the lepidopteran lineage [116-118], and confirmed to occur in O. nubilalis by genetic linkage and physical mapping [31]. Of these orthologs, apn1 and apn3 are identified as Cry1A toxin receptors in O. nubilalis [44] and O. furnacalis [43]. Experimental evidence also shows that the O. nubilalis apn1 (Onapn1) gene product expressed in the surface of cultured cells bind Cry1Ab toxins in vivo, and OnAPN3a and OnAPN8 peptides analogously bind Cry1Fa [84]. Results of the current study showed that Ofapn1, Ofapn8, as well as Ofapn3b are down-regulated in the midgut of larvae from both ACB-AbR and -AcR strains (Table 4). Association of reduced Ofapn1 transcript levels with Cry1Ab and Cry1Ac resistance may not be surprising given linkage to Cry1A resistance traits in O. nubilalis [31] and T. ni [30], or even our implication of Onapn3 due to prior evidence from O. nubilalis [31] and whole larval RNA-seq data from ACB-AcR [32]. Comparatively unique to all prior studies, our analysis allowed partitioning of RNA-seq data among the Ofapn3a, Ofapn3b, and Ofapn3c paralogs (Table 4; Fig. 4). Furthermore, our results from transcript-specific qRT-PCR suggest that Ofapn3b may be the paralog that is highly down-regulated among Cry1A toxin resistant Ostrinia. What remains interesting is that Onapn3 and Onapn8 bound Cry1Fa toxin but not Cry1Ab when expressed singularly in cell culture, whereas Onapn1 bound Cry1Ab and not Cry1Fa [84]. This presents a potential discordance between binding assay results and genetic linkage or association studies. Specifically, considering that Crava et al., [84] identified OnAPN3a and OnAPN8 protein products as likely receptors for Cry1Fa, but the involvement of APN8 and APN3a in Cry1A toxin resistance mechanisms remains uncertain. Since expression of APN8 was not quantified in the midgut of Cry1A resistant T. ni [30] or O. nubilalis [31], the novelty of this association in O. furnacalis is unknown.
qRT-PCR estimates of transcript abundances among candidate Bt resistance genes within O. furnacalis susceptible, ACB-AbR and ACB-AcR strains (sample size = 2 performed in triplicate for each treatment). Significance threshold set at p ≤ 0.05.
Annotation of Ostrinia furnacalis ATP-binding cassette subfamily G1 (ABCG1d) transcript (comp3109) significantly down-regulated in the midgut of Cry1Ab and Cry1Ac resistant larvae (Table 4). A) Gene structure of the ABCG1 homolog from an O. nubilalis BAC clone 35M22 (GenBank HTGS KX793137). B) Alignment of derived amino acid sequences from othologous in model species Bombyx mori (XP_004929563.1) and Plutella xylostella (XP_0111551601.1). The O. nubilalis transcript comp48645 was obtained from the transcriptome assembly by Vellichirammal et al. (2015). C) Annotation of lepidopteran orthologs with features typical of ATP binding cassette proteins, including an ATP binding domain and C-terminal transmembrane domains (TMD1 to TMD6). Full annotated alignment shown in Supplementary Data 4. Positions of ABC transporter signature motif [LSGGQ], phosphate binding Walker A [GX4GK(T/S)] and Walker B motifs [GX4Lh4D; h = hydrophobic residues], Q-loop, and H-motif are indicated.
Our evidence shows that several APN gene family members are down-regulated in Cry1Ab and Cry1Ac resistant larvae. This might suggest that > 1 APN receptor might affect toxin binding wherein sequential binding could expose novel receptor epitopes [22], or alternatively that binding is not always indicative of a role in the toxin modes of action [119]. Additionally, it was previously proposed that Onapn1 and Onapn3 may be part of a co-regulated gene network [31], wherein similar cis- and trans-acting factors may result in the coordinated transcription of gene family members. This premise may be partially supported by tissue specific expression of seven apns in B. mori [118], suggesting that tissue expression patterns may have been retained among gene duplicates. Thus is may be conceivable that multiple apn transcripts might be down-regulated as a result of a shared co-regulatory mechanism. Additional research is undoubtedly required in order to unravel the mechanisms that regulate apn expression in Lepidoptera, as well as how any perturbation in this transcriptional regulation can lead to the evolution of Bt resistance.
Down-regulation of transcripts for ABC transporters in the midgut of Cry1Ab and Cry1Ac resistant larvae: ATP-binding cassette (ABC) proteins are membrane-bound transporters implicated in the movement of solutes across lipid membranes and into the extracellular space [23, 120, 121]. Gene coding regions are comprised of full transporter (FT) versions that have paired nucleotide binding domain (NBD) and transmembrane domain (TMD) or a half transporter (HT) that have one each of the NDB and TMD, and hetero- or homodimerization of the latter is required for functionality [122]. Fifty one ABC transporter protein coding regions were identified in the B. mori genome and classified into within 8 subgroups by phylogenetic analysis (A-H) [123]. Bt resistance mechanisms have implicated a number of ABC transporters from subgroups A [124], C [34-36, 46] and G [39] (see Introduction for additional details), and interestingly members from these three subgroups were down-regulated in Cry1Ab and Cry1Ac resistant O. furnacalis (Table 4). Our qRT-PCR results confirm that the abundance of Ofabcg1 transcripts show the most significant proportional reduction in the midgut of Cry1Ab and Cry1Ac resistant larvae (Fig. 5). The ABCG subfamily is composed of HTs that have an inverted arrangement of NBD and TMDs, and the functions of a vast majority of insect orthologs still remains unknown [125].
Regardless, the Drosophila melanogaster ABCG1 white that functions in pigment transport into the eye [126], was linked to P. xylostella Cry1Ac resistance [39]. Also, regulation of 20-OH ecdysone response is regulated by the D. melanogaster ABCG1 E23 gene [127] and its ortholog from Tribolium castaneum [104]. As suggested by Guo et al., [39] ABCG1 HTs may form a heterodimers with other subfamily G members, such that the down-regulation of the Ofabcg1 may have a large number of functional consequences. Although extrapolated through vast evolutionary time periods, several human ABCG members are involved in lipid transport [128], and the prediction that putative lipid metabolic and transport genes are differentially regulated in Cry1A resistant O. furnacalis might possibly suggest an analogous role of OfABCG1 and G4 in species of insects. If found to be true the down-regulation of ABCG transporters could likely affect an array of cellular functions, such that the expression of genes that are functionally related may also be affected. Specifically, putative reduced lipid influx or efflux from cells may result in a decreased requirement for lipid metabolism and intracellular transport, and further suggest that interpretation of RNA-seq data could be confounded by effects that are downstream of causal genetic factor - in this case reduced ABCG1 and G4 transporter levels.
Even though ABCC2 can functional as a Cry1A toxin receptor [129], specific roles within Bt resistance mechanisms remain elusive. This could involve their direct binding with Bt toxins [23], where it is hypothesized that the extracellular regions of ABC transporters may be sufficient to initiate toxin oligimerization, membrane insertion and pre-pore formation [40], but could also require the involvement one or more additional receptors [33]. Alternatively, it was proposed that multiple potential Bt modes of action might exist all capable of forming pre-pores that can insert into the midgut epithelium, such that a number of receptors could be involved. This further suggests that mutations or changes in expression of one or more of these receptors (ALP, APNs, cadherin, and/or ABC transporters) may be capable of influencing Cry toxin resistance traits to different degrees. Indeed, additional research is required in order to decipher the individual roles as well as the interactions between Bt receptors within the framework of toxin modes of actions.
Structural variant predictions
Mutations that cause differences in the level of transcript of candidate Bt resistance genes have been associated with resistant strains of Lepidoptera (see Introduction; Table 4). Analogously, structural changes within several of these genes are predicted to cause frameshifts, translation of truncated proteins, or changes in functional amino acids that are proposed to impact resistance. There are 30 SNPs that are fixed differently within midgut expressed transcripts between resistant (ACB-AbR and -AcR) compared to ACB-BtS, and 21 others that are fixed in either ACB-AbR or -AcR. At the functional level, five SNPs lead to predicted changes in the protein sequence and do not exist within any previously identified candidate Bt resistance gene. These results might suggest that structural variation within proteins encoded by Cry1Ab or Cry1Ac resistant O. furnacalis, including candidate Bt resistance genes (ABC transporters, cadherin, alkaline phosphatase, amino peptidases, or proteases), may not contribute to the resistance traits within the colonies used in this study. Any potential linkage of the predicted fixed non-synonymous changes to Bt resistance remains unknown, but will likely be explored within future research.
Conclusions
This study quantitative compared the midgut transcripts among two resistance strain (ACB-AbR and ACB-AcR) and susceptible strain (ACB-BtS). The RNA-seq read depths predicted significant down-regulation of transcripts for previously known Bt resistance genes, aminopeptidase N1 (apn1) and apn3, as well as the abcg gene in both ACB-AbR and -AcR. In contrast, mutations impacting the structure of encoded candidate Bt resistance genes were not detected. These data are important for the understanding of systemic differences between Bt resistant and susceptible genotypes, and modes of adaption to Bt toxins.
Supplementary Material
Additional File 1Supplementary Data 1.
Supplementary Data 2.
Supplementary Data 3.
Supplementary Data 4.
Supplementary Data 5.
Acknowledgements
This work was supported by the “National Science and Technology Major Project” (2016zx08003-001). Data analysis and interpretation was supported by the United States Department of Agriculture, Agricultural Research Service (USDA-ARS; CRIS Project 5030-22000-018-00D) using USDA-ARS computational infrastructure (Ceres), and the Iowa Agriculture and Home Economics Experiment Station, Ames, IA (Project 3543).
Competing Interests
The authors have declared that no competing interest exists.
References
1. M. Legwaila G, K. Marokane T, Mojeremane W. Effects of Intercropping on the Performance of Maize and Cowpeas in Botswana. Int J Agric For. 2012;2:307-10
2. Mallet J. The evolution of insecticide resistance: Have the insects won? Trends Ecol Evol. 1989;4:336-40
3. Tabashnik BE, Brevault T, Carriere Y. Insect resistance to Bt crops: lessons from the first billion acres. Nat Biotechnol. 2013;31:510-21
4. van Rensburg JBJ. First report of field resistance by the stem borer, Busseola fusca (Fuller) to Bt-transgenic maize. S Afr J Plant Soil. 2007;24:147-51
5. Wang Z-G, Jin X, Bao X-G, Li X-F, Zhao J-H, Sun J-H. et al. Intercropping Enhances Productivity and Maintains the Most Soil Fertility Properties Relative to Sole Cropping. PLoS One. 2014;9:e113984
6. Farias JR, Andow DA, Horikoshi RJ, Sorgatto RJ, Fresia P, dos Santos AC. et al. Field-evolved resistance to Cry1F maize by Spodoptera frugiperda (Lepidoptera: Noctuidae) in Brazil. Crop Protect. 2014;64:150-8
7. Storer NP, Babcock JM, Schlenz M, Meade T, Thompson GD, Bing JW. et al. Discovery and characterization of field resistance to Bt maize: Spodoptera frugiperda (Lepidoptera: Noctuidae) in Puerto Rico. J Econ Entomol. 2010;103:1031-8
8. Monnerat R, Martins E, Macedo C, Queiroz P, Praça L, Soares CM. et al. Evidence of field-evolved resistance of Spodoptera frugiperda to Bt corn expressing Cry1F in Brazil that is still sensitive to modified Bt toxins. PLoS One. 2015;10:e0119544
9. Gassmann AJ, Petzold-Maxwell JL, Keweshan RS, Dunbar MW. Field-evolved resistance to bt maize by Western corn rootworm. PLoS One. 2011;6:e22629
10. Gassmann AJ, Petzold-Maxwell JL, Clifton EH, Dunbar MW, Hoffmann AM, Ingber DA. et al. Field-evolved resistance by western corn rootworm to multiple Bacillus thuringiensis toxins in transgenic maize. Proc Natl Acad Sci. 2014;111:5141-6
11. Gunning RV, Dang HT, Kemp FC, Nicholson IC, Moores GD. New resistance mechanism in Helicoverpa armigera threatens transgenic crops expressing Bacillus thuringiensis Cry1Ac toxin. Appl Environ Microb. 2005;71:2558-63
12. Dhurua S, Gujar GT. Field-evolved resistance to Bt toxin Cry1Ac in the pink bollworm, Pectinophora gossypiella (Saunders) (Lepidoptera: Gelechiidae), from India. Pest Manage Sci. 2011;67:898-903
13. Tabashnik BE, Carrière Y. Field-evolved resistance to Bt cotton: bollworm in the U.S. and pink bollworm in India. Southwest Entomol. 2010;35:417-24
14. Zhang X, Tiewsiri K, Kain W, Huang L, Wang P. Resistance of Trichoplusia ni to Bacillus thuringiensis toxin Cry1Ac is independent of alteration of the cadherin-like receptor for Cry toxins. PLoS One. 2012;7:e35991
15. Alvi AHK, Sayyed AH, Naeem M, Ali M. Field evolved resistance in Helicoverpa armigera (Lepidoptera: Noctuidae) to Bacillus thuringiensis toxin Cry1Ac in Pakistan. PLoS One. 2012;7:e47309
16. Xia HY, Zhao JH, Sun JH, Bao XG, Christie P, Zhang FS. et al. Dynamics of root length and distribution and shoot biomass of maize as affected by intercropping with different companion crops and phosphorus application rates. Field Crops Res. 2013;150:52-62
17. Ostrem J. Monitoring susceptibility of western bean cutworm (Lepidoptera: Noctuidae) field populations to Cry1F protein. J Econ Entomol. 2016
18. Dyer JM, Sappington TW, Coates BS. Evaluation of tolerance to Bacillus thuringiensis toxins among laboratory-reared western bean cutworm (Lepidoptera: Noctuidae). J Econ Entomol. 2013;106:2467-72
19. Heckel DG, Gahan LJ, Baxter SW, Zhao JZ, Shelton AM, Gould F. et al. The diversity of Bt resistance genes in species of Lepidoptera. J Invertebr Pathol. 2007;95:192-7
20. Xue Y, Xia H, Christie P, Zhang Z, Li L, Tang C. Crop acquisition of phosphorus, iron and zinc from soil in cereal/legume intercropping systems: a critical review. Ann Bot. 2016;117:363-77
21. Griffitts JS, Aroian RV. Many roads to resistance: how invertebrates adapt to Bt toxins. Bioessays. 2005;27:614-24
22. Bravo A, Gill SS, Soberón M. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon. 2007;49:423-35
23. Heckel DG. Learning the ABCs of Bt: ABC transporters and insect resistance to Bacillus thuringiensis provide clues to a crucial step in toxin mode of action. Pestic Biochem Phys. 2012;104:103-10
24. Garczynski SF, Adang MJ. Bacillus thuringiensis Cry1A(c) δ-endotoxin binding aminopeptidase in the Manduca sexta midgut has a glycosyl-phosphatidylinositol anchor. Insect Biochem Mol Biol. 1995;25:409-15
25. Knight PJ, Knowles BH, Ellar DJ. Molecular cloning of an insect aminopeptidase N that serves as a receptor for Bacillus thuringiensis CryIA(c) toxin. J Biol Chem. 1995;270:17765-70
26. Rajagopal R, Sivakumar S, Agrawal N, Malhotra P, Bhatnagar RK. Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as Bacillus thuringiensis toxin receptor. J Biol Chem. 2003;277:46849-51
27. Sivakumar S, Rajagopal R, Venkatesh GR, Srivastava A, Bhatnagar RK. Knockdown of aminopeptidase-N from Helicoverpa armigera larvae and in transfected Sf21 cells by RNA interference reveals its functional interaction with Bacillus thuringiensis insecticidal protein Cry1Ac. J Biol Chem. 2007;282:7312-9
28. Yang Y, Zhu YC, Ottea J, Husseneder C, Leonard BR, Abel C. et al. Molecular characterization and RNA interference of three midgut aminopeptidase N isozymes from Bacillus thuringiensis-susceptible and -resistant strains of sugarcane borer, Diatraea saccharalis. Insect Biochem Mol Biol. 2010;40:592-603
29. Herrero S, Gechev T, Bakker PL, Moar WJ, Maagd RAD. Bacillus thuringiensis Cry1Ca-resistant Spodoptera exigua lacks expression of one of four Aminopeptidase N genes. BMC Genomics. 2005;6:96
30. Tiewsiri K, Wang P. Differential alteration of two aminopeptidases N associated with resistance to Bacillus thuringiensis toxin Cry1Ac in cabbage looper. Proc Natl Acad Sci. 2011;108:14037-42
31. Coates BS, Johnson H, Kim K-S, Hellmich RL, Abel CA, Mason C. et al. Frequency of hybridization between Ostrinia nubilalis E- and Z-pheromone races in regions of sympatry within the United States. Ecol Evol. 2013;3:2459-70
32. Xu LN, Ling YH, Wang YQ, Wang ZY, Hu BJ, Zhou ZY. et al. Identification of differentially expressed microRNAs between Bacillus thuringiensis Cry1Ab-resistant and -susceptible strains of Ostrinia furnacalis. Sci Rep. 2015;5:15461
33. Gahan LJ, Pauchet Y, Vogel H, Heckel DG. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 2010;6:e1001248
34. Baxter S, Badenes-Pérez F, Morrison A, Vogel H, Crickmore N, Kain W. et al. Parallel evolution of Bacillus thuringiensis toxin resistance in lepidoptera. Genetics. 2011;189:675-9
35. Atsumi S, Miyamoto K, Yamamoto K, Narukawa J, Kawai S, Sezutsu H. et al. Single amino acid mutation in an ATP-binding cassette transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori. Proc Natl Acad Sci. 2012;109:1591-8
36. Coates BS, Siegfried BD. Linkage of an ABCC transporter to a single QTL that controls Ostrinia nubilalis larval resistance to the Bacillus thuringiensis Cry1Fa toxin. Insect Biochem Mol Biol. 2015;268:86-96
37. Park Y, González-Martínez RM, Navarro-Cerrillo G, Chakroun M, Kim Y, Ziarsolo P. et al. ABCC transporters mediate insect resistance to multiple Bt toxins revealed by bulk segregant analysis. BMC Biology. 2014;12:1-15
38. Guo Z, Kang S, Chen D, Wu Q, Wang S, Xie W. et al. MAPK signaling pathway alters expression of midgut ALP and ABCC genes and causes resistance to Bacillus thuringiensis Cry1Ac toxin in diamondback moth. Plos Genetics. 2015:11
39. Guo Z, Shi K, Xun Z, Xia J, Wu Q, Wang S. et al. Down-regulation of a novel ABC transporter gene (Pxwhite) is associated with Cry1Ac resistance in the diamondback moth, Plutella xylostella (L.). Insect Biochem Mol Biol. 2015;59:30-40
40. Tay WT, Mahon RJ, Heckel DG, Walsh TK, Downes S, James WJ. et al. Insect resistance to Bacillus thuringiensis toxin Cry2Ab is conferred by mutations in an abc transporter subfamily a protein. PloS Genetics. 2015:11
41. Xiao Y. Mis-splicing of the ABCC2 gene linked with Bt toxin resistance in Helicoverpa armigera. Sci Rep. 2014;4:6184
42. Vadlamudi RK, Weber E, Ji I, Ji TH Jr BL. Cloning and expression of a receptor for an insecticidal toxin of Bacillus thuringiensis. J Biol Chem. 1995;270:5490-4
43. Tan SY, Cayabyab BF, Alcantara EP, Huang F, He K, Nickerson KW. et al. Comparative binding of Cry1Ab and Cry1F Bacillus thuringiensis toxins to brush border membrane proteins from Ostrinia nubilalis, Ostrinia furnacalis and Diatraea saccharalis (Lepidoptera: Crambidae) midgut tissue. J Invertebr Pathol. 2013;114:234-40
44. Hua G, Masson L, Juratfuentes JL, Schwab G, Adang MJ. Binding analyses of Bacillus thuringiensis Cry delta-endotoxins using brush border membrane vesicles of Ostrinia nubilalis. Appl Environ Microb. 2001;67:872-9
45. Flannagan RD, Yu CG, Mathis JP, Meyer TE, Shi X, Siqueira HA. et al. Identification, cloning and expression of a Cry1Ab cadherin receptor from European corn borer, Ostrinia nubilalis (Hubner) (Lepidoptera: Crambidae). Insect Biochem Mol Biol. 2005;35:33-40
46. Gahan LJ, Gould F, Heckel DG. Identification of a gene associated with Bt resistance in Heliothis virescens. Science. 2001;293:857-60
47. Zhang H, Tian W, Zhao J, Jin L, Yang J, Liu C. et al. Diverse genetic basis of field-evolved resistance to Bt cotton in cotton bollworm from China. Proc Natl Acad Sci U S A. 2012;109:10275-80
48. Jin T, Chang X, Gatehouse A, Wang Z, Edwards M, He K. Down regulation and mutation of a cadherin gene associated with cry1ac resistance in the Asian corn borer, Ostrinia furnacalis (Guenée). Toxins. 2014;6:2676
49. Zhang X, Tiewsiri K, Kain W, Huang L, Wang P. Resistance of Trichoplusia ni to Bacillus thuringiensis toxin Cry1Ac is independent of alteration of the cadherin-like receptor for Cry toxins. PloS One. 2012;7:e35991
50. Mcnall RJ, Adang MJ. Identification of novel Bacillus thuringiensis Cry1Ac binding proteins in Manduca sexta midgut through proteomic analysis. Insect Biochem Mol Biol. 2003;33:999-1010
51. Jurat-Fuentes JL, Adang MJ. Characterization of a Cry1Ac-receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae. Eur J Biochem. 2004;271:3127-35
52. Jurat-Fuentes JL, Karumbaiah L, Jakka SRK, Ning C, Liu C, Wu K. et al. Reduced levels of membrane-bound alkaline phosphatase are common to lepidopteran strains resistant to Cry toxins from Bacillus thuringiensis. PLoS One. 2011:e17606
53. Jin T, Duan X, Bravo A, Soberón M, Wang Z, He K. Identification of an alkaline phosphatase as a putative Cry1Ac binding protein in Ostrinia furnacalis (Guenée). Pestic Biochem Phys. 2016;131:80-6
54. Huang Y, Takanashi T, Hoshizaki S, Tatsuki S, Honda H, Yoshiyasu Y. et al. Geographic variation in sex pheromone of Asian corn borer,Ostrinia furnacalis, in Japan. J Chem Ecol. 1998;24:2079-88
55. Nafus DM, Schreiner IH. Review of the biology and control of the Asian corn borer, Ostrinia furnacalis (Lep: Pyralidae). Int J Pest Manag. 1991;37:41-56
56. Feng C, Huang J, Song Q, Stanley D, Lü W, Zhang Y. et al. Parasitization by Macrocentrus cingulum (Hymenoptera: Braconidae) influences expression of prophenoloxidase in Asian corn borer Ostrinia furnacalis. Arch Insect Biochem Physiol. 2011;77:99-117
57. Wang ZY, He KL, Yan S. Large-scale augmentative biological control of Asian corn borer using Trichogramma in China: a success story. Second International Symposium on Biological Control of Arthropods. 2005:487-94
58. Gould F. Sustainability of transgenic insecticidal cultivars: integrating pest genetics and ecology. Annu Rev Entomol. 1998;43:701-26
59. Tabashnik BE, Carrière Y, Soberón M, Gao A, Bravo A. Successes and failures of transgenic Bt crops: global patterns of field-evolved resistance. 2015.
60. Xu L, Wang Z, Zhang J, He K, Ferry N, Gatehouse AMR. Cross-resistance of Cry1Ab-selected Asian corn borer to other Cry toxins. J Appl Entomol. 2010;134:429-38
61. Zhang T, He M, Gatehouse A, Wang Z, Edwards M, Li Q. et al. Inheritance patterns, dominance and cross-resistance of Cry1Ab- and Cry1Ac-selected Ostrinia furnacalis (Guenée). Toxins. 2014;6:2694-707
62. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57-63
63. Lei Y, Zhu X, Xie W, Wu Q, Wang S, Guo Z. et al. Midgut transcriptome response to a Cry toxin in the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Gene. 2014;533:180-7
64. Nanoth Vellichirammal N, Wang H, Eyun S-i, Moriyama E, Coates B, Miller N. et al. Transcriptional analysis of susceptible and resistant European corn borer strains and their response to Cry1F protoxin. BMC Genomics. 2015;16:558
65. Zhou DR, He KL, Wang ZY, Ye ZH, Wen LP, Gao YX. et al. Asian corn borer and its integrated management (In Chinese). Beijing, China: Golden Shield Press. 1995
66. Andrews S. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
67. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. BioInformatics. 2014;30:2114-20
68. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotech. 2011;29:644-52
69. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011:644-52
70. Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494-512
71. Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674-6
72. Garczynski SF, Coates BS, Unruh TR, Schaeffer S, Jiwan D, Koepke T. et al. Application of Cydia pomonella expressed sequence tags: Identification and expression of three general odorant binding proteins in codling moth. Insect Sci. 2013;20(5):559-574
73. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403-10
74. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Meth. 2012;9:357-9
75. Li H. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078-9
76. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106
77. Anders S, Huber W. Differential expression of RNA-Seq data at the gene level - the DESeq package. Embl. 2013
78. Huang X, Madan A. CAP3: A DNA Sequence Assembly Program. Genome Res. 1999;9:868-77
79. Wernersson R. Virtual Ribosome—a comprehensive DNA translation tool with support for integration of sequence feature annotation. Nucleic Acids Res. 2006;34:385-8
80. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H. et al. Clustal W and Clustal X version 2.0. BioInformatics. 2007;23:2947-8
81. Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M. et al. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 2005;33:D192-D6
82. Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:D257-D60
83. Krogh A, Larsson B, Heijne Gv, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes 1. J Mol Biol. 2001;305:567-80
84. Crava CM, Bel Y, Jakubowska AK, Ferré J, Escriche B. Midgut aminopeptidase N isoforms from Ostrinia nubilalis: Activity characterization and differential binding to Cry1Ab and Cry1Fa proteins from Bacillus thuringiensis. Insect Biochem Mol Biol. 2013;43:924-35
85. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596-9
86. Fankhauser N, Mäser P. Identification of GPI anchor attachment signals by a Kohonen self-organizing map. BioInformatics. 2005;21:1846-52
87. Bustin S, Benes V, Garson J, Hellemans J, Huggett J, Kubista M. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611-22
88. Nolan T, Hands R, Bustin S. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559-82
89. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[delta][delta]CT method. Methods. 2001;25:402-8
90. Coates BS, Siegfried BD. Linkage of an ABCC transporter to a single QTL that controls Ostrinia nubilalis larval resistance to the Bacillus thuringiensis Cry1Fa toxin. Insect Biochem Mol Biol. 2015;63:86-96
91. Fabrick JA, Ponnuraj J, Singh A, Tanwar RK, Unnithan GC, Yelich AJ. et al. Alternative splicing and highly variable cadherin transcripts associated with field-evolved resistance of pink bollworm to Bt cotton in India. PLoS One. 2014;9:e97900
92. He K, Wang Z, Zhou D, Wen L, Song Y, Yao Z. Evaluation of transgenic Bt corn for resistance to the Asian corn borer (Lepidoptera: Pyralidae). J Econ Entomol. 2003;96:935-40
93. Coates BS, Sumerford DV, Hellmich RL, Lewis LC. Mining an Ostrinia nubilalis midgut expressed sequence tag (EST) library for candidate genes and single nucleotide polymorphisms (SNPs). Insect Mol Biol. 2008;17:607-20
94. Pereira EJG, Lang BA, Storer NP, Siegfried BD. Selection for Cry1F resistance in the European corn borer and cross-resistance to other Cry toxins. Entomol Exp Appl. 2008;126:115-21
95. Groeters FR, Tabashnik BE. Roles of selection intensity, major genes, and minor genes in evolution of insecticide resistance. J Econ Entomol. 2000;93:1580-7
96. Mallet J, Porter P. Preventing insect adaptation to insect-resistant crops: are seed mixtures or refugia the best strategy? Proc R Soc Lond B Biol Sci. 1992;250:165-9
97. Siqueira H, Moellenbeck D, Spencer T, Siegfried B. Cross-resistance of Cry1Ab-selected Ostrinia nubilalis (Lepidoptera: Crambidae) to Bacillus thuringiensis delta-endotoxins. J Econ Entomol. 2014;97:1049-57
98. Crespo ALB, Rodrigo-Simón A, Siqueira HAA, Pereira EJG, Ferré J, Siegfried BD. Cross-resistance and mechanism of resistance to Cry1Ab toxin from Bacillus thuringiensis in a field-derived strain of European corn borer, Ostrinia nubilalis. J Invertebr Pathol. 2011;107:185-92
99. Gould F, Anderson A, Reynolds A, Bumgarner L, Moar W. Selection and genetic analysis of a Heliothis virescens (Lepidoptera: Noctuidae) strain with high levels of resistance to Bacillus thuringiensis toxins. J Econ Entomol. 1995;88:1545-59
100. Tabashnik BE, Liu Y-B, de Maagd RA, Dennehy TJ. Cross-resistance of pink bollworm (Pectinophora gossypiella) to Bacillus thuringiensis toxins. Appl Environ Microb. 2000;66:4582-4
101. Estela A, Escriche B, Ferré J. Interaction of Bacillus thuringiensis toxins with larval midgut binding sites of Helicoverpa armigera (Lepidoptera: Noctuidae). Appl Environ Microb. 2004;70:1378-84
102. Hernández-Rodríguez CS, Hernández-Martínez P, Van Rie J, Escriche B, Ferré J. Shared midgut binding sites for Cry1A.105, Cry1Aa, Cry1Ab, Cry1Ac and Cry1Fa proteins from Bacillus thuringiensis in two important corn pests, Ostrinia nubilalis and Spodoptera frugiperda. PLoS One. 2013;8:e68164
103. Li H, Gonzalez-Cabrera J, Oppert B, Ferre J, Higgins RA, Buschman LL. et al. Binding analyses of Cry1Ab and Cry1Ac with membrane vesicles from Bacillus thuringiensis-resistant and -susceptible Ostrinia nubilalis. Biochem Biophys Res Commun. 2004;323:52-7
104. Leonard P. Gianessi. The potential for worldwide crop production increase due to adoption of pesticides rice, wheat & maize. Crop Protection Research Institute: CropLife Foundation. 2013
105. Sayyed AH, Wright DJ. Cross-resistance and inheritance of resistance to Bacillus thuringiensis toxin Cry1Ac in diamondback moth (Plutella xylostella L) from lowland Malaysia. Pest Manag Sci. 2001;57:413-21
106. Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H. et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651-6
107. Vera JC, Wheat CW, Fescemyer HW, Frilander MJ, Crawford DL, Hanski I. et al. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol Ecol. 2008;17:1636-47
108. Sparks ME, Blackburn MB, Kuhar D, Gundersenrindal DE. Transcriptome of the Lymantria dispar (gypsy moth) larval midgut in response to infection by Bacillus thuringiensis. Plos One. 2013;8:43-51
109. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300-7
110. Carninci P, Kawai J. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559-63
111. Legeai F, Derrien T. Identification of long non-coding RNAs in insects genomes. Curr Opin Insect Sci. 2015;10:37-44
112. Skunca N, Altenhoff A, Dessimoz C. Quality of computationally inferred gene ontology annotations. PLoS Comput Biol. 2012;8:e1002533
113. Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods. 2011;8:469-77
114. Fang Z, Cui X. Design and validation issues in RNA-seq experiments. Brief Bioinform. 2011;12:280-7
115. Williams AG, Thomas S, Wyman SK, Holloway AK. RNA-seq data: challenges in and recommendations for experimental design and analysis. Current Protocols in Human Genetics: John Wiley & Sons, Inc. 2014
116. Field LM, James AA, Chang WXZ, Gahan LJ, Tabashnik BE, Heckel DG. A new aminopeptidase from diamondback moth provides evidence for a gene duplication event in Lepidoptera. Insect Mol Biol. 1999;8:171-7
117. Angelucci C, Barrett-Wilt GA, Hunt DF, Akhurst RJ, East PD, Gordon KHJ. et al. Diversity of aminopeptidases, derived from four lepidopteran gene duplications, and polycalins expressed in the midgut of Helicoverpa armigera: Identification of proteins binding the δ-endotoxin, Cry1Ac of Bacillus thuringiensis. Insect Biochem Mol Biol. 2008;38:685-96
118. Lin P, Cheng T, Jin S, Jiang L, Wang C, Xia Q. Structural, evolutionary and functional analysis of APN genes in the Lepidoptera Bombyx mori. Gene. 2014;535:303-11
119. Pigott CR, Ellar DJ. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol Mol Biol Rev. 2007;71:255-81
120. Dermauw W, Van Leeuwen T. The ABC gene family in arthropods: Comparative genomics and role in insecticide transport and resistance. Insect Biochem Mol Biol. 2014;45:89-110
121. Tabashnik BE. ABCs of Insect Resistance to Bt. Plos Genet. 2015:11
122. Pazoki A, Farokhi F, Pazoki Z. Corn seed varieties classification based on mixed morphological and color features using artificial neural networks. Res J Appl Sci Eng Technol. 2013;6:3506-13
123. Fenzi M, Jarvis DI, Arias Reyes LM, Latournerie Moreno L, Tuxill J. Longitudinal analysis of maize diversity in Yucatan, Mexico: influence of agro-ecological factors on landraces conservation and modern variety introduction. Plant Genet Resour. 2015;15:51-63
124. Tay WT, Mahon RJ, Heckel DG, Walsh TK, Downes S, James WJ. et al. Insect resistance to Bacillus thuringiensis toxin Cry2ab is conferred by mutations in an ABC transporter subfamily A protein. PLoS Genet. 2015:11
125. Dermauw W. ABC transporters in Arthropods: genomic comparison and role in insecticide transport and resistance. Insect Biochem Mol Biol. 2014:45
126. De Groote H. Maize yield losses from stem borers in Kenya. Int J Trop Insect Sci. 2002;22:89-96
127. Oliveira CM, Auad AM, Mendes SM, Frizzas MR. Crop losses and the economic impact of insect pests on Brazilian agriculture. Crop Protect. 2014;56:50-4
128. Bonnie L. Grant. Cross pollination of corn: preventing cross pollinating in corn. Gardening Know How Corn Cross Pollination Info. 2016
129. Degen T. High genetic variability of herbivore-induced volatile emission within a broad range of maize inbred lines. Plant Physiol. 2004;135:1928-38
Author contact
![]() Corresponding author: klhecn
Corresponding author: klhecn

 Global reach, higher impact
Global reach, higher impact