10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(8):946-956. doi:10.7150/ijbs.24121 This issue Cite
Research Paper
PTM-ssMP: A Web Server for Predicting Different Types of Post-translational Modification Sites Using Novel Site-specific Modification Profile
1. School of Information Science and Technology, University of Science and Technology of China, Hefei AH230027, China;
2. Centers for Biomedical Engineering, University of Science and Technology of China, Hefei AH230027, China.
* These authors contributed equally to this work
Abstract
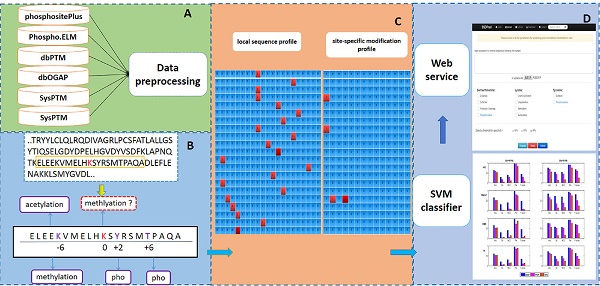
Protein post-translational modifications (PTMs) are chemical modifications of a protein after its translation. Owing to its play an important role in deep understanding of various biological processes and the development of effective drugs, PTM site prediction have become a hot topic in bioinformatics. Recently, many online tools are developed to prediction various types of PTM sites, most of which are based on local sequence and some biological information. However, few of existing tools consider the relations between different PTMs for their prediction task. Here, we develop a web server called PTM-ssMP to predict PTM site, which adopts site-specific modification profile (ssMP) to efficiently extract and encode the information of both proximal PTMs and local sequence simultaneously. In PTM-ssMP we provide efficient prediction of multiple types of PTM site including phosphorylation, lysine acetylation, ubiquitination, sumoylation, methylation, O-GalNAc, O-GlcNAc, sulfation and proteolytic cleavage. To assess the performance of PTM-ssMP, a large number of experimentally verified PTM sites are collected from several sources and used to train and test the prediction models. Our results suggest that ssMP consistently contributes to remarkable improvement of prediction performance. In addition, results of independent tests demonstrate that PTM-ssMP compares favorably with other existing tools for different PTM types. PTM-ssMP is implemented as an online web server with user-friendly interface, which is freely available at
Keywords: Post-translational modifications, web server, site-specific modification profile, prediction

 Global reach, higher impact
Global reach, higher impact