Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(3):712-727. doi:10.7150/ijbs.45885 This issue Cite
Review
Tight junctions and their regulation by non-coding RNAs
1. Department of Dermatology, Third Xiangya Hospital, Central South University, 138 Tongzipo Road, Changsha, Hunan 410013, P.R. China.
2. Institute of Chinese Materia Medica, Hunan Academy of Chinese Medicine, Yuehua Road, Changsha, Hunan 410013, P.R. China.
3. Department of Dermatology, Second Xiangya Hospital, Central South University, 139 Renminzhong Road, Changsha, Hunan 410013, P.R. China.
Received 2020-3-28; Accepted 2020-10-6; Published 2021-1-31
Abstract
Tight junction (TJ) is a “zippering up” junction structure located at the uppermost portion of adjacent epithelial/endothelial cells in organs and tissues. TJs maintain the relative stability of intracellular substances and functions by closing or opening intercellular pathways, coordinating the entry and exit of molecules of different sizes and charges, and regulating the permeability of paracellular barrier. TJs also prevent microbial invasion, maintain epithelial/endothelial cell polarity, and regulate cell proliferation. TJs are widely present in the skin and mucosal epithelial barriers, intestinal epithelial barrier, glomerular filtration barrier, bladder epithelial barrier, blood-brain barrier, brain-blood tumor barrier, and blood-testis barrier. TJ dysfunction in different organs can lead to a variety of diseases. In addition to signal pathways, transcription factors, DNA methylation, histone modification, TJ proteins can also be regulated by a variety of non-coding RNAs, such as micro-RNAs, long-noncoding RNAs, and circular RNAs, directly or indirectly. This review summarizes the structure of TJs and introduces the functions and regulatory mechanisms of TJs in different organs and tissues. The roles and mechanisms of non-coding RNAs in the regulation of TJs are also highlighted in this review.
Keywords: tight junction, micro-RNAs, long-noncoding RNAs, circular RNAs
Introduction
Mammals have four types of cell-cell junction that called tight junctions (TJs), adherens junctions (AJs), gap junctions (GJs), and desmosomes/hemidesmosomes [1]. According to functions, they can be divided into occluding junctions (TJs), anchoring junctions (AJs, desmosome/hemidesmosome), and communicating junctions (GJs) [2-4]. These junctions jointly regulate the paracellular or intra-/extracellular exchanges of material and information [5]. Among them, TJs are paracellular barriers that are essential for maintaining homeostasis in multicellular organisms and can quickly respond to stimuli, especially in the skin, visceral organs, and blood-brain barrier (BBB) [6]. As selective gates, TJs control the paracellular diffusion of ions and solutes, allow the passage of small soluble substances, restrict the exudation of macromolecular substances, such as proteins, and inhibit the entry of viruses and other microorganisms [5, 7, 8].
TJ structure
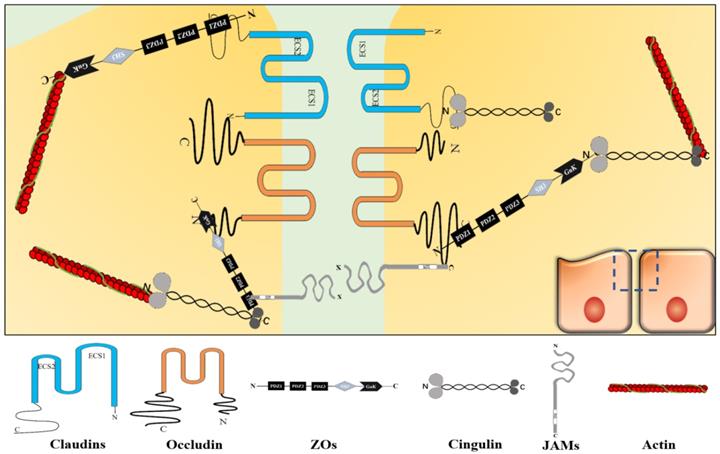
TJs are located in the topmost region of the lateral membrane [9]. A TJ is a reticular structure formed by the binding of transmembrane proteins (claudins, occludin, junctional adhesion molecule [JAM], etc.), cytoplasmic proteins (zonula occludens [ZOs], cingulin, etc.) and cytoskeletal proteins (actin, myosin, etc.). These transmembrane proteins polymerize on the cell membrane to form a “strand” structure, which calls TJ strand. Each TJ strand binds transversely to another one, which connects adjacent cells and forms a “zippering up” structure in the lateral to apical direction [6]. Besides, cytoplasmic proteins act as scaffold proteins that connect TJ strands and cytoskeletal proteins to maintain TJ functions [9] (Figure 1). Cytoplasmic protein components, such as afadin and atypical protein kinase C (aPKC), also reportedly assist in the TJ structure maintenance [10, 11].
TJ structure. Claudins, occludin, JAM, and etc. constitute the core components of TJ strand. Among them, claudins aggregate to form the backbone of the TJ strand. ZO protein family, cingulin, and etc. act as scaffold proteins to connect TJ strand and actin in TJ. TJ shows a reticular structure formed by the orderly binding of different functional proteins.
Claudins
Claudins constitute a protein family with at least 27 subtypes [12], their molecular weight is around 20-25 kDa [13]. Claudins are polymerized on the cell membrane into the backbone structure of the TJ strand. Claudins have four transmembrane fragments, two extracellular loops (the first extracellular segment [ECS1] and the second extracellular segment [ECS2]), two cytoplasmic termini (the COOH-terminus and the NH2-terminus). Among the claudins subtypes, ECS1 and ECS2 contain different amino acid sequences, respectively. However, ECS1 contains the claudins consensus motif, also called the “claudins signature sequence”, which performs its functions on stabilizing the structure of claudins and providing an interface for TJ[14]. About two cytoplasmic terminal fragments, the COOH-terminal fragment is almost 5 to 15 times longer than the NH2-terminal fragment. The number of residues of the COOH-terminal fragment is different among the claudins subtypes with residues between 21 and 63, except claudin-23 (106 residues). In claudins, the PSD-95/Dlg/ZO-1 (PDZ)-domainbinding motif at the COOH-terminal fragment can directly connects with TJ-related cytoplasmic molecules, such as ZOs and cingulin [14, 15]. The PDZ domain is a motif involved in protein binding and mediates protein-protein interactions, it maintains the connection between transmembrane and cytoplasmic parts to form a complete structure and ensures TJ's function. Furthermore, it participates in various processes, such as paracellular transport and signal transduction [13].
Occludin
Occludin is another important TJ strand component with a molecular weight around 65 kDa [16]. Also, occludin has four transmembrane fragments, two extracellular loops, and two cytoplasmic termini. But they share no similarity in structure and sequence [9]. In occludin, the COOH-terminal fragment has 254 residues and is about 3.88 times longer than the NH2-terminal fragment [17]. The PDZ domain at the COOH-terminal fragment binds to ZOs to perform its functions [18].
JAMs
JAM proteins have three main subtypes (JAM-A, JAM-B, and JAM-C), they show similar sequences and structures. JAM consists of a short NH2-terminal fragment, two extracellular immunoglobulin-like loops, a single transmembrane module, and a short COOH-terminal fragment in the cytoplasm with phosphorylation sites, and a COOH-terminal PDZ domain [19, 20]. They molecular weight is 36-41 kDa [21]. The extracellular domain functions as an adhesion structural component between cells. The phosphorylation sites are critical for JAMs to target TJs. Besides, the PDZ domain recruits TJ scaffold proteins, such as ZO-1, thus, it involves in the function of TJ through protein-protein interactions [13, 20, 22].
ZOs
The ZOs family, including ZO-1, ZO-2, and ZO-3, are TJ scaffold proteins. They localize to the cytoplasm and the molecular weight is 220 kDa, 160 kDa, and 130 kDa, respectively [9]. ZO-1, ZO-2, and ZO-3 have similar sequences. They have three PDZ domains, a Src homology 3 (SH3) domain and a guanylate kinase-like (GUK) domain. These domains are arranged in turn from NH2-terminus to COOH-terminus: PDZ1, PDZ2, PDZ3, SH3, and GUK [23]. Among them, PDZ1 and GUK directly connect with other TJ proteins. PDZ1 binds directly to claudins COOH-terminus [9], while GUK domain binds directly to occludin NH2-terminus [23]. Furthermore, COOH-terminal domain interacts with actin directly. But other domains do not directly connect with TJ proteins. For example, JAMs connect with the PDZ2 and PDZ3 of ZO-1 via ALL-1 fusion partner from chromosome 6 (AF-6) protein [23, 24]. The ZOs are very important for maintaining the TJ structural stability. As reported, a simultaneous absence of ZO-1 and ZO-2 can cause TJ transmembrane proteins to disperse along the cytoplasm and affect TJ functions [9].
Cingulin
Cingulin localizes in the cytoplasm. Cingulin is a parallel dimer of two subunits with a molecular weight of 140-160 kDa. Each subunit has a globular head, a coiled-coil rod, and a globular tail. Both myosin and ZO-3 can bind to the NH2-terminus and COOH-terminus of cingulin, while ZO-1 and ZO-2 can bind to the NH2-terminus [21]. In general, cingulin acts as a scaffold protein that connects and stabilizes the TJ structure through ZOs family and cytoskeletal proteins. Cingulin also takes part in signal transduction, such as regulating RhoA signaling to control epithelial cell proliferation [13, 25].
Actin
The cytoskeleton is composed of microtubules, microfilaments, and intermediate filaments. The cytoskeleton maintains cell morphology and bears external forces, it also helps maintain cell's internal structure. Microfilaments are cytoskeletal components that enable the opening and closing of TJs and are mainly composed of actin [26-28]. Actin binds to the cytoplasmic scaffold proteins to form a power system that controls the opening and closing of TJ strands composed channels [29].
TJ proteins can also participate in other physiological and pathological functions independently of TJ structure. Such as, the claudins family and ZO-2 act as markers of epithelial-mesenchymal transformation (EMT) and are related to organogenesis and differentiation (EMT type 1); claudin-1, occludin, and JAM-A are related to inflammation and fibrosis (EMT type 2); the claudins family are related to cancer metastasis/invasion (EMT type 3). Claudin-1 and occludin can serve as receptors for hepatitis C virus (HCV) entry into cells [30], and ZO-1 is a biomarker of multiple myeloma to identify patients who are most likely to benefit from proteasome inhibitors [31].
TJ functions
TJs are widely found in the skin and mucosal epithelial barriers [32], intestinal epithelial barrier [33], glomerular filtration barrier[34], bladder epithelial barrier [35], blood-brain barrier (BBB) [36, 37], brain-blood tumor barrier (BBTB) [38], and blood-testis barrier (BTB) [39, 40] (Figure 2). TJs can coordinate the entry and exit of molecules with different sizes and charges in an orderly manner, which plays an important role in paracellular barrier functions [41]. TJs also help prevent the invasion of viruses and other microorganisms [42], maintain the polarity of epithelial cells [43], and regulate cell proliferation [44]. Additionally, TJ proteins can also be expressed independently of TJ structure and play an essential role in signal transduction and regulating gene expression [31]. The TJ strands of adjacent cells interact to form two types of pathways that regulate the entry and exit of molecules. The first is a pore pathway consisting of five or more TJ strands that allow ions and uncharged small molecules with a radius of approximately 4 Å or less to pass through the epithelial barrier in large numbers simultaneously. The second is the leak pathway displayed only a single strand of adjacent cells, which allows larger ions and molecules to pass through regardless of their charges, but only a small number of large ions and macromolecules can pass through at one time [41, 45]. ZOs act as scaffold proteins to regulate the number of molecules passing through the two pathways [33]. The claudins family regulates the charge selectivity of the two pathways, with specific residues in the ECS1 region to determine the charge selection of the pathways. The claudins family can be divided into two types: sealing claudins (claudins-1, -3, -4, -5, -8, -11, and ect), and pore-forming claudins (claudins-2, -10, -14, -16, and ect). Sealing claudins inhibit molecules passing through the TJ barrier. Furthermore, JAM-A participates in coordinating regulating TJ barrier permeability with claudins. If JAM-A alters in distribution and/or expression alone, it does not influence TJ barrier permeability. However, when JAM-A and claudins alter in distribution and/or expression together, it strengthens the influences of claudins on TJ barrier permeability [46]. In contrast with sealing claudins, pore-forming claudins enable molecules passing through the TJ barrier easier [47-51]. Occludin is a critical structural and functional component of TJ [18]. Studies have shown that occludin constitutes a barrier function, and knockdown of occludin increases the paracellular pathway permeability [16, 52]. However, some findings are opposite to this. They suggest that occludin plays a key role in maintaining the normal function of the leak pathway. The overexpression of occludin enhances leak pathway function, while the molecular flux of the leak pathway without occludin is significantly lower [53, 54]. Cell polarity refers to the asymmetry of cell structure and function, and the subcellular structures are believed to be asymmetrically distributed along an axis, causing different regions inside the cell to perform different functions [55]. Epithelial cells polarize along the apical-basal axis, which is crucial for the epithelial cells to function as barriers [56]. In this manner, different cell-cell junctions are formed in different cell locations. TJs are located at the top of the epithelial cells, which is critical for maintaining cell polarity. For example, the absence of ZO-1 and ZO-2 induces JAM-A and claudins to be distributed widely along the apical and basolateral cell membranes, and ZO-3 and occludin are also displaced, destroying epithelial cell polarity [43]. Occludin regulates the polarity of epithelial cells by controlling the Par-3/Par-6/aPKC complex [57]. TJs can also regulate epithelial/endothelial cell proliferation through the Yes-associated protein 1 (YAP1) /HIPPO signaling pathway, ZO-1/zonula occludens 1‑associated nucleic acid binding protein (ZONAB) signaling pathway, and interact with integrin β1 signaling [58-62]. TJ proteins can also regulate the levels of immune proteasome subunits Low-molecular mass protein 7 (LMP7) and Low-molecular mass protein 2 (LMP2) via EGFR/JAK1/signal transducer and activator of transcription 3 (STAT3) signaling independent of TJ structure [31].
TJs are present in the skin and mucosal epithelial barriers, intestinal epithelial barrier, glomerular filtration barrier, bladder epithelial barrier, BBB, BTB, and BBTB. TJ dysfunction in these organs leads to a variety of diseases and morbid conditions.
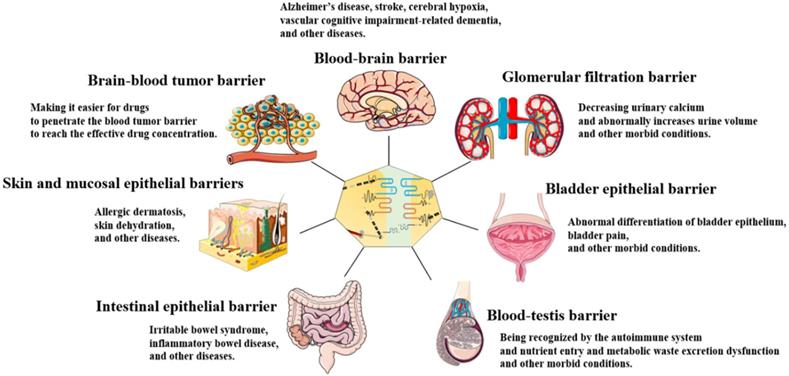
TJ functions and characteristics vary between organs. TJ barrier permeability is more highly selective in the skin and bladder epithelial barriers than in the intestinal epithelial barrier and glomerular filtration barrier. This is because a strong epidermal barrier is essential for homeostasis, and the bladder must stabilize urine concentration in the normal range and prevent the loss of ion gradients [35, 63]. In contrast to the skin and bladder epithelial barriers, intestinal epithelial barrier and glomerular filtration barrier drive paracellular absorption and secretion through selective TJ barrier penetration [33].
Role of TJ in the skin and mucosal epithelial barriers
The TJ is an essential barrier structure in skin and is the main barrier in skin appendages lacking stratum corneum, such as hair follicles and sweat glands [32]. The typical TJ structure is located in the granular layer of the skin [64]. TJ strands form channels between adjacent keratinocytes that regulate the entry and exit of water and solutes [32]. TJ deficiency leads to water and solutes losses in the skin and allows allergens entry, which in turn leads to dry skin and an overly active response to allergens. For example, atopic dermatitis is characterized by dry skin and highly active response to allergens, the two features are reported as the results of decreased claudin-1 and TJ deficiency [44, 65]. Down-regulation of occludin and ZO-1 in patients with allergic rhinitis leads to TJ deficiency in nasal mucosa, and the chance of allergens crossing the TJ barrier is increased [66, 67]. Defects on TJs make the skin appendages especially vulnerable to entry of viruses, such as herpes-simplex virus and infectious-molluscum virus; therefore, patients with atopic dermatitis are more likely to suffer from viral skin diseases [32]. Abnormality of TJ proteins is closely related to psoriasis progression. Linkage analysis identifies nine psoriatic susceptibility loci, some of which are located in the region encoding claudins. Patients with psoriasis have been shown to lack claudin-1 expression based on the detection of skin lesions [68]. In vitro experiments also show that the decreased expression of claudin-1 disturbs TJ functions and promotes keratinocyte proliferation [44].
Role of TJ in the intestinal epithelial barrier
TJs perform a more complicated task in the intestinal epithelial barrier than in the skin and mucosal epithelial barriers. They support the paracellular transport of water, ions (e.g., Na+, Ca2+, Mg2+, and Cl-) and nutrients (e.g., glucose and protein), and prevent the translocation of microorganisms [33]. TJs also inhibit apoptosis of intestinal epithelial cells and protect the intestinal epithelial barrier [69]. Transport of Na+ is essential for nutrients absorption. For instance, Na+-glucose cotransport through TJs are vital in maintaining Na+ concentration inside and outside the intestinal epithelial barrier, thereby enabling nutrients recycling. In the case of TJ deficiency, the intracellular and extracellular Na+ concentration gradient of the intestinal epithelial barriers is insufficient to drive the absorption of transcellular Na+-nutrient cotransport, which then affects nutrients absorption [33]. Thus, TJ deficiency can lead to intestinal epithelial absorption disorders and abnormal secretory function. TJ deficiency can cause oxalate secretion disorders, leading to hyperoxaluric acidemia, hyperoxaluria, and calcium oxalate stones. A study showed that modulation of the charge selectivity of the claudin-based pore pathway did not affect oxalate permeability, but knockdown of ZO-1 enhanced permeability to oxalate and mannitol in parallel [54]. TJ deficiency is associated with inflammatory bowel disease (IBD). For example, the expressions of ZO-1 and occludin are significantly down-regulated in the intestinal epithelial barrier of patients with irritable bowel syndrome (IBS) [70, 71]. Up-regulation of claudin-2, as well as down-regulation and redistribution of occludin, claudin-5, and claudin-8 lead to TJ deficiency in Crohn's disease [47]. IBD increases the risk of colorectal cancer, so protecting TJ integrity is significant for preventing colorectal cancer [72]. TJ defects also lead to the translocation of bacteria and bacterial products. When claudin-1 and occludin expression is down-regulated in the intestinal epithelial barrier, the barrier's permeability increases. Intestinal bacteria and their metabolites are then translocated to the liver, which can result in liver disease [42]. Down-regulation of occludin expression can also cause intestinal bacteria to enter the systemic circulation, increasing cardiovascular events after myocardial infarction [73]. It is worth noting that upregulation of TJ proteins can also damage the intestinal epithelial barrier, so TJ homeostasis is critical for maintaining intestinal epithelial functions.
Role of TJ in the glomerular filtration barrier
The reabsorption by epithelial cells in the glomerular filtration barrier depends on the selection of molecular size and charge by the pathways formed by the TJ strands [34]. The expression of claudins is variable in different nephron segments, which determines what will be reabsorbed in each nephron segment: claudin-2 forms cationic reabsorption pathways of proximal renal tubules; claudins-14, -16, and -19 constitute calcium transport pathways regulating the thick ascending branch of the Helen's loop; claudins-4, -7, and -8 form chloride pathways in the collecting duct; claudin-16 mediates Mg2+ transport protein [74], and claudins-8 and -15 mediate Na+-nutrient cotransport [30, 33]. TJ defects also lead to reabsorption disorders. The expressions of claudins-3, -4, and -8 are down-regulated in the renal collecting duct epithelial barrier, leading to urine dilution through water reabsorption [48]. Mutations in claudin-16 and claudin-19 lead to familial hypercalcemia and hypomagnesemia with renal calcinosis [74].
Role of TJ in the bladder epithelial barrier
The selection of molecular size and charge by the pathways formed by TJ strands between bladder epithelial cells stabilizes the composition and concentration of urine in the normal range [35]. Claudin-4 is a marker of bladder epithelial differentiation, and abnormal claudin-4 expression in bladder cancer is related to local invasion, lymph node metastasis, and distant metastasis [75]. A study howed that claudin-4 is abnormally expressed in the bladder epithelial barrier in many children with bladder valgus [76].
Role of TJ in the BBB
The TJ prevents harmful substances from entering the brain from the bloodstream. Down-regulation of claudin-5, ZO-1, ZO-2, and occludin causes viruses to cross the BBB [36, 37]. Meanwhile, many therapeutic drugs cannot reach an effective concentration in the central nervous system due to the relative impermeability of TJs. TJ proteins disappear in the BBB from 24 hours to seven days after a stroke; only claudin-5 reappear in newly formed brain endothelial cells, whereas other components do not so. Furthermore, only claudin-5 absence does not affect TJ structure, but leads to selective opening of the BBB for molecules that smaller than 800 Da [49, 77, 78].
Role of TJ in the BTB
TJs can protect specific antigens of germ cells from being recognized by the autoimmune system and regulate nutrient entry and metabolic waste excretion [39, 40]. Unlike the BBB, TJs in the BTB can be disassembled and reassembled periodically. The BTB must be opened regularly to allow germ cells to pass. Polar proteins, for example cell division control protein 42 homolog (Cdc42), Dishevelled-1/2/3 (Dvl1/2/3), and focal adhesion kinase (FAK), alter the connection between TJ strands and cytoskeletal proteins, leading to BTB remodeling to support germ cell passage during the epithelial cycle of spermatogenesis [79-82].
Regulatory mechanism of TJs
Studies currently have confirmed that a variety of signaling pathways can affect TJ by regulating TJ protein expression: Ras-mitogen-activated protein kinase (MAPK), epidermal growth factor receptor (EGFR)-Src family kinases (SFK), Toll-like receptor 2 (TLR2)-protein kinase C (PKC), Wnt- β-catenin, Nrf2, NF-κB, STAT5b, and other signaling pathways can regulate the expression of ZO-1[83-93]; mitogen-activated protein kinase (MAPK) kinase (MEK)-extracellular signal-regulated kinase (ERK), Nrf2, NF-κB, mediterranean fever gene (MEFV), and other signaling pathways can regulate occludin expression [72, 89-94]; the Nrf2 pathway regulates claudin-4 expression [90, 91]; the Wnt-β-catenin and c-myc pathways regulate claudin-5 expression [86-88, 95]; the STAT5b pathway regulates the expressions of ZO-2 and ZO-3 [96]; the Siah-Pard3A pathway regulates JAM-A expression [97]; and the liver kinase B1 (LKB1)/adenosine monophosphate-activated protein kinase (AMPK) pathway regulates cingulin expression [98]. Epigenetic regulation, such as DNA methylation and histone modifications, also plays an important role in maintaining TJ function. DNA methylation in the promoter regions of TJ-related genes can inhibit their expressions. For instance, DNA methylation inhibits claudin-11 expression by interfering with transcription activator GATA1 binding to its DNA promoter [99]. Histone modification in the promoter region of the TJ gene can also affect TJ gene transcription: both histone 3 lysine 9 trimethylation (H3K9me3) and histone 3 lysine 27 trimethylation (H3K27me3) in the ZO-1 DNA promoter inhibit ZO-1 transcriptional expression [100-102], while histone 3 lysine 4 trimethylation (H3K4me3) of the ZO-1 DNA promoter activates ZO-1 DNA transcription [101]; besides, H3K27me3 and histone 4 lysine 20 trimethylation (H4K20me3) of claudin-3 and claudin-4 DNA promoters inhibit their transcription [103]; and H3K27me3 of the occludin DNA promoter inhibits occludin transcriptional expression [101, 102]. Non-coding RNAs, including micro-RNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs) are also vital aspects of epigenetic regulation, they play an important role in the physiological and pathological regulation of various genes. In recent years, it has been reported that non-coding RNAs are also involved in TJ function regulation (Figure 3).
Non-coding RNAs
MiRNAs are small single-stranded RNAs with lengths of 21-23 nucleotides that can bind to the 3' UTR of mRNAs and result in mRNAs silencing or degradation [104, 105]. For example, miR-29 can participate in the pathogenesis of recessive dystrophic epidermolysis bullae by specifically silencing COL7A1 mRNA [106]. LncRNAs are long-stranded non-coding RNAs with lengths of 200-100,000 nucleotides that participate in the regulation of a variety of physiological and pathological processes [107, 108]. The regulation mechanisms of lncRNAs are highly complex, but they mainly work in the following ways [109-123]: (1) regulating gene transcription by affecting DNA promoter regions; (2) inhibiting RNA polymerase II or regulating chromatin remodeling and histone modification to affect downstream gene expression; (3) forming a complementary double strand with gene transcripts to interfere with mRNA splicing; (4) forming complementary double strands with the transcript of genes to produce endogenous siRNAs dependent on Dicer enzyme activity; (5) binding to a specific protein to regulate its activity; (6) binding to a specific protein to change its cellular location; (7) acting as a structural component, forming a nucleic acid-protein complex with protein; (8) a precursor of small RNAs such as miRNAs and piRNAs; (9) sponge-like absorbing of miRNAs to indirectly regulate gene expression that targeted by miRNA. CircRNA is a single-stranded RNA with a closed loop structure that is more stable than linear RNA. CircRNA exerts its biological functions mainly through sponging miRNAs, binding proteins, regulating gene transcription, or being translated into short polypeptide as templates [124-129]. For example, circ_0015756 can sponge miR-1250-3p to regulate the proliferation and invasiveness of hepatoblastoma [130]. Circ-human antigen R (HuR) has been shown to be down-regulated in gastric cancer and bound to the CCHC zinc-finger nucleic acid-binding protein (CNBP), which promotes HuR mRNA expression by interacting with the HuR DNA promoter. However, once bound to CNBP, circ-HuR competitively inhibits CNBP binding with the HuR DNA promoter. Thereby, it down-regulates HuR to inhibit the progression of gastric cancer [127]. Previous studies have shown that DNA methyltransferase 1 (DNMT1) contributes to the DNA methylation in CD4+ T cells, which leads to the production of autoantibodies. Down-regulation of circ_0012919 expression increases methyltransferase DNMT1 expression, which contributes to DNA methylation of CD11a and CD70 in CD4+T cells. Therefore, circ_0012919 can be regarded as a biomarker of systemic lupus erythematosus [131]. There are also small nuclear RNAs (snRNAs), transfer RNAs (tRNAs), small interfering RNAs (siRNAs), and other undiscovered non-coding RNAs in cells that regulate a variety of cellular processes.
TJ proteins are regulated by miRNAs, lncRNAs, and circRNAs.
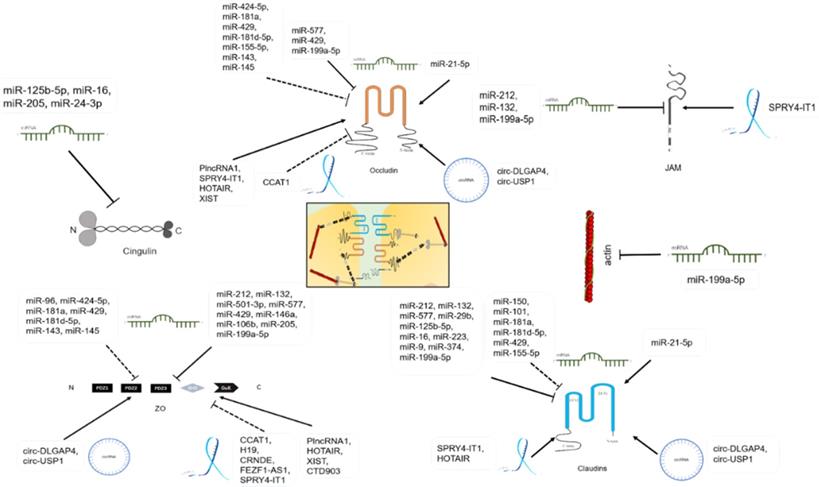
MiRNAs and TJs
The gene locations and sequences of miRNAs are highly conserved and are found throughout animals, plants, bacteria, fungi, and viruses [105, 132]. It has been reported that miRNAs participate in different kinds of physiological functions such as post-transcriptional gene-expression [133], cardiovascular biomarkers [134], angiogenesis, and inflammatory response [135]. Disordered miRNAs expressions also participate in the regulation of pathological processes such as tumor pathogenesis [136], drug resistance [137]. It has been found that miRNAs can regulate physiological and pathological processes by regulating TJs in the BBB, BBTB, intestinal epithelial barrier, skin epithelial barrier, glomerular filtration barrier, bladder epithelial barrier, and EMT via regulating TJ proteins (Table 1). The BBB is the barrier between plasma and brain cells formed by the cerebral capillary wall and glial cells, as well as the barrier between plasma and cerebrospinal fluid formed by the choroid plexus. The BBB protects brain tissue from harmful substances circulating in the blood and maintains the stability of the environment in the brain tissue [138]. The BBTB is the barrier between brain tumor tissue and plasma formed by the capillary wall in brain tumor tissue [138]. The intestinal epithelial barrier prevents bacteria and toxins from passing through the intestinal mucosa into other tissues, organs, and blood circulation in the human body. It can also promote the absorption of nutrients, such as proteins, into the bloodstream [139]. The skin and mucosal epithelial barriers are composed of the physical barrier and the chemical barrier. They both protect the internal organs/tissues from external damage, irritation, and sunlight entry, and they function in skin moisturization and immunomodulation. TJs are mainly important in the physical barrier of the skin and mucosal epithelial barriers [140-142]. Glomerular filtration barrier is the structure in which blood in the glomerular capillaries is filtered into Bowman space [143, 144]. The bladder epithelial barrier is the mucous membrane that lines the inner surface of the bladder [145]. EMT is the transformation from epithelial cells to mesenchymal cells, which gives cells the ability to migrate and invade other tissues. The EMT has a key role in tissue healing, organ fibrosis, carcinogenesis, and other processes [146].
Function and mechanism of miRNAs in TJs
| Name | Level | Function | Mechanism | Position | Reference |
|---|---|---|---|---|---|
| miR-150 | - | Increasing BBB permeability | Down-regulating claudin-5 by target inhibiting Tie-2 | BBB | [149] |
| miR-212 and miR-132 | Increased in hypoxic brain tissue | Destroying the integrity of BBB | Target inhibiting claudin-1, ZO-1, and JAM-C | BBB | [150] |
| miR-501-3p | - | Destructing the integrity of BBB | Target inhibiting ZO-1 | BBB | [151] |
| miR-96 | Increased in HBMEC of Alzheimer's disease | Increasing BBB permeability | Down-regulating ZO-1 by target inhibiting ERG | BBB | [148] |
| miR-424-5p | - | Increasing BBB permeability | Down-regulating ZO-1 and occludin by target promoting Endophilin-1 | BBB | [152] |
| miR-101 | - | Increasing BBB permeability | Down-regulating claudin-5 by target inhibiting VE-cadherin | BBB | [154] |
| miR-181a | Increased in Glioma | Increasing BBTB permeability | Down-regulating ZO-1, occludin, and claudin-5 by target inhibiting KLF6 | BBTB | [38] |
| miR-577 | - | Increasing BBTB permeability | Target inhibiting claudin-1, ZO-1, and occludin | BBTB | [155] |
| miR-429 | Decreased in Glioma | Increasing BBTB permeability | Target inhibiting ZO-1 and occludin. Also, down-regulating ZO-1, occludin, and claudin-5 by target inhibiting p70S6K-S6 | BBTB | [156] |
| miR-181d-5p | - | Increasing BBTB permeability | Down-regulating ZO-1, occludin, and claudin-5 by target inhibiting SOX5 | BBTB | [157] |
| miR-29b | - | Destroying the integrity of intestinal epithelial barrier | Target inhibiting claudin-1 | Intestinal epithelial barrier | [158] |
| miR-21-5p | Decreased in dysfunctional intestinal epithelial barrier | Protecting the integrity of intestinal epithelial barrier | Up-regulating occludin and claudin-4 by target inhibiting PTEN, PDCD4, SPRY1, and SPRY2 | Intestinal epithelial barrier | [159] |
| miR-125b-5p and miR-16 | Decreased in irritable bowel syndrome | Protecting the integrity of intestinal epithelial barrier | Target inhibiting cingulin and claudin-2 | Intestinal epithelial barrier | [160] |
| miR-223 | - | Destroying the integrity of intestinal epithelial barrier | Target inhibiting claudin-8 | Intestinal epithelial barrier | [161] |
| miR-155-5p | - | Increasing skin barrier permeability | Down-regulating occludin and claudin-1 by target inhibiting PKIα | Skin epithelial barrier | [162] |
| miR-146a and miR-106b | - | Increasing skin barrier permeability and TEWL | Target inhibiting ZO-1 and ZO-2 | Skin epithelial barrier | [163] |
| miR-9 and miR-374 | - | Increasing glomerular filtration barrier permeability leading to an increase in calcium reabsorption and a decrease in urinary calcium | Target inhibiting claudin-14 | Glomerular filtration barrier | [50, 164] |
| miR-205 | - | Regulating urinary tract epithelial cell differentiation | Target inhibiting cingulin and ZO-1 | Urinary tract epithelium | [166] |
| miR-199a-5p | Increased in the bladder pain syndrome | Increasing bladder epithelial permeability | Target inhibiting ZO-1, JAM-A, occludin, and actin | Bladder epithelium | [165] |
| miR-199a-5p | Increased in the bladder pain syndrome | Increasing bladder epithelial permeability | Target inhibiting claudin-1, JAM-A, and occludin | Bladder epithelium | [167] |
| miR-143 and miR-145 | - | Promoting EMT | Down-regulating occluding, ZO-1, and ZO-3 by target inhibiting CREB1 | Breast cancer cells | [168] |
| miR-24-3p | - | Promoting EMT | Target inhibiting cingulin | Malignant mesothelioma | [169] |
Abbreviation: BBB: blood-brain barrier; BBTB: brain blood-tumor barrier; CREB1: cAMP-response element binding protein 1; EMT: epithelial to mesenchymal transition; ERG: ETS-Related Gene; JAM-A: junctional adhesion molecule A; JAM-C: junctional adhesion molecule C; KLF6: Kruppel-like factor 6; p70S6K-S6: p70 S6 kinase-S6; PDCD4: programmed cell death factor 4; PKIα: protein kinase inhibitor α; PTEN: phosphatase and tensin homolog; SOX5: sex-determining region Y-box protein 5; SPRY1: sprouty 1; SPRY2: sprouty 2; TEWL: trans-epidermal water loss; VE-cadherin: vascular endothelial cell cadherin; ZO-1: zonula occludens-1; ZO-2: zonula occludens-2; ZO-3: zonula occludens-3.
MiRNAs and TJs in the BBB
The BBB damage will lead to stroke, vascular cognitive impairment-related dementia, Alzheimer's disease, cerebral hypoxia, and other conditions [147-152]. The most important TJ proteins in the BBB are claudin-5, ZO-1, and occludin. MiR-150 directly binds to angiopoietin receptor Tie-2 mRNA, resulting in the down-regulation of Tie-2 protein, which leads to the inhibition of claudin-5 protein, and an increase in the BBB permeability after ischemic stroke [149]. Hypoxia induces high miR-212 and miR-132 in the central nervous system. Both miR-212 and miR-132 can bind to the 3' UTR of claudin-1, ZO-1, and JAM-C mRNAs and inhibit their expressions, thereby damage the integrity of the BBB [150]. MiR-501-3p binds to the 3' UTR of ZO-1 mRNA to inhibit ZO-1 protein expression; this leads to damage of the BBB and promotes development of vascular cognitive impairment-related dementia [151]. Current studies have shown that Alzheimer's disease is closely related to an increase in monocyte migration across the BBB. High level of granulocyte-macrophage colony-stimulating factor (GM-CSF) in brain parenchyma promotes monocyte migration across the BBB. GM-CSF up-regulates the miR-96 level of human brain microvascular endothelial cells (HBMEC) via the phosphatidylinositol 3-kinase (PI3K)/serine-threonine protein kinase (AKT) signal pathway. MiR-96 targets the mRNA of the erythrocyte transformation specificity (ETS) transcription factor, ETS-related gene (ERG), to inhibit ERG protein expression. The interaction of ERG protein with the ZO-1 DNA promoter region enables ZO-1 mRNA expression. Therefore, miR-96 indirectly suppresses ZO-1 mRNA expression, which leads to an increase in monocyte migration across the BBB [148]. MiR-424-5p binds to the 3' UTR of endophilin-1 mRNA and up-regulates endophilin-1 mRNA. Endophilin-1 overexpression leads to inhibition of the EGFR-ERK and EGFR-Jun-N-terminal kinase (JNK) signaling pathways, then down-regulates ZO-1 and occludin protein in vascular endothelial cells. This increases the BBB permeability and induces development of Alzheimer's disease [152]. It is worth noted that the combination of miR-424-5p with the endophilin-1 mRNA 3' UTR does not decrease endophilin-1 mRNA. This may be due to a competitive interaction between miR-424-5p and multiple miRNAs, which reduce the inhibition of other miRNAs on endophilin-1 expression and indirectly increase endophilin-1 [153]. MiR-101 binds to the 3' UTR of vascular endothelial cadherin (VE-cadherin) mRNA to down-regulate it. VE-cadherin has been reported to promote claudin-5 expression. Therefore, miR-101 indirectly down-regulates claudin-5, resulting in the increase of BBB permeability [154].
MiRNAs and TJs in the BBTB
Research has focused on drugs or treatments that increase the permeability of the BBTB to enable an effective drug concentration to penetrate the BBTB and treat brain tumors. Studies have found that miRNAs can increase the BBTB permeability by regulating TJs to achieve increased drug efficacy, which is significant for clinical advancement. The expression of miR-181a is increased in glioma, and miR-181a can inhibit Kruppel-like factor 6 (KLF6) mRNA expression by directly binding to the 3' UTR of KLF6 mRNA; KLF6 can promote the transcriptional expressions of ZO-1, occludin, and claudin-5 mRNAs. Thus, miR-181a indirectly down-regulates ZO-1, occludin, and claudin-5, which further increases the BBTB permeability. MiR-181a may be a key miRNA for opening the BBTB and is a potential target to increase the therapeutic success rate for glioma [38]. MiR-577 directly binds to occludin, ZO-1, and claudin-1 mRNA 3' UTRs, down-regulating they and increasing the BBTB permeability to improve the chemosensitivity of malignant glioma [155]. MiR-429 expression in human glioma microvascular endothelial cells (GECs) is much lower than that in normal microvascular endothelial cells in human brain. MiR-429 regulates TJ proteins in GECs in two ways. First, miR-429 directly binds to ZO-1 and occludin mRNA 3' UTRs to inhibit the post-transcriptional processes. Second, miR-429 inhibits p70 S6 kinase-S6 (p70S6K-S6) expression by binding to the mRNA 3' UTR. The activation of the p70S6K-S6 signaling pathway can up-regulate ZO-1, occludin, and claudin-5. Therefore, miR-429 can also indirectly down-regulate ZO-1, occludin, and claudin-5 by inhibiting the p70S6K-S6 signaling pathway [156]. The expressions of ZO-1, occludin, and claudin-5 are also promoted by binding of Sex-determining region Y-box protein 5 (SOX5), whose expressions are inhibited by miR-181d-5p. Thus, miR-181d-5p could suppress SOX5 expression to reduce ZO-1, occludin, and claudin-5 expressions, thereby enhancing the BBTB permeability in glioma [157].
MiRNAs and TJs in the intestinal epithelial barrier
The destruction of the intestinal epithelial barrier leads to IBS, IBD, and other conditions. The function of the intestinal epithelial barrier is complex, and the TJ proteins, which predominate in the barrier structure, have intricate functions [158-161]. It has been found that miR-29b combines with the 3' UTR of claudin-1 mRNA to inhibit claudin-1 expression, resulting in intestinal epithelial barrier dysfunction [158]. Intestinal symbiotic floras induce the expression of miR-21-5p in intestinal epithelial cells. MiR-21-5p binds to phosphatase and tensin homolog (PTEN), programmed cell death factor 4 (PDCD4), sprouty 1 (SPRY1), and sprouty 2 (SPRY2) mRNAs to inhibit their expressions. These target genes are negative regulators of the phosphatidylinositol 3-kinase (PI3K)-Akt, JNK-activator protein 1 (AP-1), and ERK pathways, respectively. Therefore, miR-21-5p activates these pathways to up-regulate ADP ribosylation factor 4 (ARF4), promoting the expressions of claudin-4 and occludin and protecting the intestinal epithelial integrity [159]. MiR-125b-5p and miR-16 bind to cingulin and claudin-2 mRNAs and inhibit their expressions, respectively. In IBS, however, miR-125b-5p and miR-16 are down-regulated, which increase cingulin and claudin-2 in the intestinal epithelial barrier and leads to intestinal epithelial barrier dysfunction [160]. MiR-223 directly binds to the 3' UTR of claudin-8 mRNA and down-regulate it, increasing the permeability of intestinal epithelial barrier and inducing IBD occurrence [161].
MiRNAs and TJs in the skin and mucosal epithelial barriers
TJ destruction caused by abnormal miRNA expression induces allergic dermatosis, skin dehydration, and other diseases [162, 163]. For example, miR-155-5p binds to the protein kinase inhibitor α (PKI α) mRNA 3'UTR to inhibit PKI α expression. PKI α can promote claudin-1 and occludin expressions, while miR-155-5p can increase skin barrier permeability and induce atopic dermatitis by indirectly down-regulating claudin-1 and occluding [162]. Both miR-146a and miR-106b bind to the ZO-1 and ZO-2 mRNA 3'UTRs to inhibit their expressions, increasing skin barrier permeability, and increasing trans-epidermal water loss (TEWL) [163].
MiRNAs and TJs in the glomerular filtration barrier
The epithelium of the glomerular filtration barrier reabsorbs water, electrolytes, and nutrients in urine mainly through TJs. TJ destruction decreases urinary calcium abnormally increases urine volume and causes other morbid conditions [50, 164]. Both miR-9 and miR-374 recognize complementary binding sites in the claudin-14 mRNA 3' UTR. The miR-9/miR-374-claudin-14 pathway is directly regulated by Ca2+ sensor receptor (CaSR) and the pathway also constitutes an important part of the renal CaSR signal cascade system. CaSR activation decreases miR-9 and miR-374 expressions, then increases claudin-14, which increases calcium reabsorption and decreases urinary calcium by increasing TJ barrier permeability [50, 164].
MiRNAs and TJs in the bladder epithelial barrier
Damage to the TJs between adjacent bladder epithelial cells leads to abnormal differentiation of the bladder epithelial barrier, bladder pain, and other morbid conditions [165-167]. It was reported that miR-199a-5p increases bladder epithelial permeability in bladder pain syndrome by targeting the 3' UTRs of ZO-1, JAM-A, occludin, and actin mRNA [165]. Similarly, miR-199a-5p up-regulation in bladder pain syndrome leads to occludin, claudin-1, and JAM-A down-regulation, which is not conducive to the establishment of a tight bladder epithelial barrier and results in bladder chronic pain [167]. MiR-205 directly binds to ZO-1 and cingulin mRNAs to inhibit their expressions and participates in urothelial cell differentiation regulation [166].
MiRNAs regulate EMT via TJs
EMT leads to TJ destruction and the loss of cell adhesion and polarity. Damage to TJ structural integrity can also promote EMT [168, 169]. MiR-143 and miR-145 inhibit cAMP-response element binding protein 1 (CREB1) expression, which is the transcriptional activator of ZO-1, ZO-3, and occludin. So miR-143 and miR-145 down-regulate these TJ proteins, promoting breast cancer invasion and metastasis induced by EMT [168]. MiR-24-3p inhibits cingulin mRNA translation, which leads to invasion and metastasis of malignant mesothelioma induced by EMT [169].
LncRNAs and TJs
It has been reported that lncRNAs regulate the physiological functions of cell directional differentiation, biological growth and evolution [170]. Disordered lncRNAs expressions also participate in the regulation of pathological processes such as tumor progression, cardiovascular and cerebrovascular diseases, neurological diseases, autoimmune diseases, and congenital malformations [104, 105, 107, 108]. In recent years, it has been found that lncRNAs can regulate physiological and pathological processes by regulating TJs in the intestinal epithelial barrier, BBTB, and EMT (Table 2).
Function and mechanism of lncRNAs in TJs
| Name | Level | Function | Mechanism | Position | Reference |
|---|---|---|---|---|---|
| CCAT1 | - | Destroying intestinal epithelial barrier function and promoting the pathogenesis of inflammatory bowel disease. | Disturbing ZO-1 and occludin expressions by sponging miR-185-3p | Intestinal epithelial barrier | [171] |
| H19 | Increased in ulcerative colitis | Destroying the intestinal epithelial barrier | Down-regulating ZO-1 by releasing miR-675-5p | Intestinal epithelial barrier | [172,173] |
| PlncRNA1 | - | Protecting intestinal epithelial barrier function | Up-regulating ZO-1 and occludin by sponging miR-34c | Intestinal epithelial barrier | [174] |
| SPRY4-IT1 | - | Protecting intestinal epithelial barrier function | Target promoting occludin, claudin-1, claudin-3, and JAM-A translation | Intestinal epithelial barrier | [175] |
| HOTAIR | Increased in glioma | Decreasing BBTB permeability | Target up-regulating occludin, ZO-1, and claudin-5 | BBTB | [176] |
| XIST | - | Decreasing BBTB permeability | Up-regulating ZO-1, ZO-2, and occludin by sponging miR-137 | BBTB | [177] |
| CRNDE | Increased in osteosarcoma and hepatoma cells | Promoting EMT | Down-regulating ZO-1 by target activating Wnt/β-catenin signal pathway | Osteosarcoma and hepatoma | [178,179] |
| FEZF1-AS1 | Increased in NSCLC | Promoting EMT | Down-regulating ZO-1 by target activating Wnt/β-catenin signal pathway | NSCLC | [180] |
| CTD903 | Increased in colorectal cancer | Inhibiting EMT | Up-regulating ZO-1 by target inhibiting Wnt/β-catenin signal pathway | Colorectal cancer | [181] |
| SPRY4-IT1 | - | Promoting EMT | Down-regulating ZO-1 by target activating Snail transcription, expression, and nuclear localization | Esophageal squamous cell carcinoma | [182] |
Abbreviation: BBTB: brain blood-tumor barrier; CCAT1: colon cancer-associated transcript-1; CRNDE: colorectal neoplasia differentially expressed; EMT: epithelial to mesenchymal transition; FEZF1-AS1: FEZF1 antisense RNA 1; HOTAIR: HOX transcript antisense intergenic RNA; JAM-A: junctional adhesion molecule A; NSCLC: non-small cell lung cancer; SPRY4-IT1: SPRY4 intronic transcript 1; XIST: X inactivate-specific transcript; ZO-1: zonula occludens-1; ZO-2: zonula occludens-2.
LncRNAs and TJs in the intestinal epithelial barrier
Colon cancer-associated transcript-1 (CCAT1) performs an important role in tumorigenesis and progression. CCAT1 can also induce IBD by regulating the intestinal epithelial barrier. For example, inflammation-related genes are significantly enriched in colorectal cancer patients with high CCAT1 expression. Myosin light chain kinase (MLCK) can induce the disordered distribution of occludin and ZO-1 and can transform the intestine from a smooth arc shape into many irregular undulations. CCAT1 can adsorb miR-185-3p to promote MLCK expression, leading to disordered distribution of occludin and ZO-1 and destruction of the barrier function, which promotes IBD pathogenesis [171]. H19 expression increases in patients with ulcerative colitis. H19, as a precursor of miR-675-5p, inhibits ZO-1 mRNA expression and induces intestinal epithelial barrier function destruction in patients with ulcerative colitis [172, 173]. PlncRNA1 sponges miR-34c to remove the targeted inhibition of miR-34c on myc-associated zinc-finger protein (MAZ). MAZ promotes ZO-1 and occludin expressions by binding to ZO-1 and occludin promoter regions. Therefore, PlncRNA1 can up-regulate occludin and ZO-1 through the “miR-34c-MAZ” regulatory axis and enhance the intestinal epithelial barrier function [174]. SPRY4 intronic transcript 1 (SPRY4-IT1) directly interacts with occludin, claudin-1, claudin-3, and JAM-A mRNAs to promote their translation by binding to the RNA-binding protein HuR, and then enhances intestinal epithelial barrier function [175].
LncRNAs and TJs in the BBTB
HOX transcript antisense intergenic RNA (HOTAIR) is up-regulated in gliomas, and HOTAIR combines with the promoter regions of occludin, ZO-1, and claudin-5 to promote their expressions, which decreases BBTB permeability [176]. The adsorption of miR-137 by X inactivate-specific transcript (XIST) can antagonize the targeted inhibition of miR-137 on ZO-2. So XIST promotes ZO-2 expression. In addition, XIST can also promote forkhead box C1 (FOXC1) expression by adsorbing miR-137. FOXC1 is a gene transcription factor of ZO-1 and occludin. Therefore, XIST can up-regulate ZO-1, ZO-2, and occludin from multiple angles, which decreases BBTB permeability [177].
LncRNAs regulate tumor EMT through TJs
LncRNAs can participate in tumor EMT regulation by regulating TJ proteins. For example, colorectal neoplasia differentially expressed (CRNDE) is highly expressed in osteosarcoma and hepatocellular carcinoma cells. CRNDE activates the Wnt/β-catenin signaling pathway and down-regulates ZO-1, inducing EMT to promote osteosarcoma and hepatocellular carcinoma cell metastasis [178, 179]. FEZF1 antisense RNA 1 (FEZF1- AS1) expression is significantly up-regulated in non-small cell lung cancer (NSCLC) tissues. FEZF1-AS1 also down-regulates ZO-1, inducing EMT by activating the Wnt/β-catenin signal pathway [180]. CTD903 expression is significantly up-regulated in colorectal cancer tissue. CTD903 inhibits the Wnt/β-catenin signaling pathway to up-regulate ZO-1, inhibiting EMT in colorectal cancer [181]. SPRY4-IT1 down-regulates ZO-1 by promoting Snail transcription, expression, and nuclear localization, inducing EMT and metastasis of esophageal squamous cell carcinoma [182].
CircRNAs and TJs
There have been found a few studies of TJ regulation by circRNAs (Table 3). However, studies have introduced the role of circRNAs in the BBB and BBTB. For example, circ-DLGAP4 is highly expressed in the normal central nervous system, but significantly decreased after stroke, resulting in down-regulation of TJ protein and BBB destruction. The combination of miR-143 and HECT domain E3 ubiquitin protein ligase 1 (HECTD1) mRNA 3' UTR inhibits HECTD1 expression. HECTD1 can promote the expressions of occludin, ZO-1, and claudin-5. Circ-DLGAP4 can promote HECTD1 expression by sponging miR-143 and enhance BBB function [183]. As a miR-194-5p sponge, circ-ubiquitin specific peptidase 1 (USP1) relieves the inhibitory effect of miR-194-5p on occludin, ZO-1, and claudin-5 mRNAs and decreases the BBTB permeability [184]. CircRNAs are stable and have potential as molecular markers and drug targets, and their relationships with TJs deserve more extensive and in-depth researches.
Function and mechanism of circRNAs in TJs
| Name | Level | Function | Mechanism | Position | Reference |
|---|---|---|---|---|---|
| circ-DLGAP4 | Decreased in ischemic brain tissue | Protecting BBB | Up-regulating ZO-1, claudin-5 and occludin by sponging miR-143 | BBB | [183] |
| circ-USP1 | - | Decreasing BBTB permeability | Up-regulating ZO-1, claudin-5, and occludin by sponging miR-194-5p | BBTB | [184] |
Abbreviation: BBB: blood-brain barrier; BBTB: brain blood-tumor barrier; USP1: ubiquitin specific peptidase 1; ZO-1: zonula occludens-1.
Conclusion
A TJ is a “zippering up” junction structure. TJs maintain the relative stability of intracellular substances and functions by closing or opening paracellular pathways, coordinating the entry and exit of molecules of different sizes and charges, and regulating the permeability of paracellular barrier. TJs also prevent microbial invasion, maintain epithelial/endothelial cell polarity, and regulate cell proliferation. TJs are widely present in the skin and mucosal epithelial barriers, intestinal epithelial barrier, glomerular filtration barrier, bladder epithelial barrier, BBB, BBTB, and BTB. Because of TJ's critical role in organs and tissues, it is not surprising that diseases, such as IBD, gliomas, allergic dermatosis, and skin dehydration, are associated with disruption of TJs. A variety of non-coding RNAs, such as miRNAs, lncRNAs, and circRNAs, can directly or indirectly regulate TJ proteins, affecting the function of various organs or leading to diseases.
Prospect
Non-coding RNAs are important parts of epigenetic regulation. Current studies have confirmed that many miRNAs can affect the TJs by directly regulating TJ proteins or indirectly regulating upstream signal pathways or transcription factors of TJ proteins, thereby affecting the skin and mucosal epithelial barriers, intestinal epithelial barrier, BBB, EMT and others. However, the research on the roles of lncRNAs, circRNAs, and other non-coding RNAs in TJs is still in its infancy. Because of the diversity of mechanisms and functions of lncRNAs and circRNAs, more in-depth studies are needed to reveal their roles and specific mechanisms. The role of snRNAs, tRNAs and other non-coding RNAs in TJs also needs to be confirmed by follow-up research. Because the disorder of TJs can lead to a variety of diseases, such as skin barrier dysfunction, which will lead to the progression of atopic dermatitis, psoriasis, ichthyosis, melanoma and so on. TJ proteins and non-coding RNAs that regulate TJs are expected to become new therapeutic targets. Some drugs targeted at TJs may also promote the drugs to enter the lesion and increase the sensitivity of the drugs.
Abbreviations
AF-6: ALL-1 fusion partner from chromosome 6; AKT: serine-threonine protein kinase; AMPK: adenosine monophosphate-activated protein kinase; AP-1: activator protein 1; aPKC: atypical protein kinase C; ARF4: ADP ribosylation factor 4; BBB: blood-brain barrier; BBTB: brain blood-tumor barrier; CaSR: Ca-sensing receptor; CCAT1: colon cancer-associated transcript-1; Cdc42: cell division control protein 42 homolog; CNBP: CCHC Zinc finger nucleic acid-binding protein; COL7A1: collagen, type VII, alpha 1; CREB1: cAMP-response element binding protein 1; CRNDE: colorectal neoplasia differentially expressed; DNMT1: DNA methyltransferase 1; Dvl1/2/3: Dishevelled-1/2/3; ECS1: the first extracellular segment; ECS2: the second extracellular segment; EGFR: epidermal growth factor receptor; EMT: epithelial to mesenchymal transition; ERG: ETS-Related Gene; ERK: extracellular signal-regulated kinase; ETS: erythrocyte transformation specificity; FAK: focal adhesion kinase; FEZF1-AS1: FEZF1 antisense RNA 1; FOXC1: forkhead box C1; GECs: glioma microvascular endothelial cells; GM-CSF: granulocyte-macrophage colony-stimulating factor; GUK: guanylate kinase-like; HCV: hepatitis C virus; HBMEC: human brain microvascular endothelial cells; HECTD1: HECT domain E3 ubiquitin protein ligase 1; HOTAIR: HOX transcript antisense intergenic RNA; H3K4me3: histone 3 lysine 4 trimethylation; H3K9me3: histone 3 lysine 9 trimethylation; H3K20me3: histone 3 lysine 20 trimethylation; H3K27me3: histone 3 lysine 27 trimethylation; HuR: human antigen R; IBD: inflammatory bowel disease; IBS: irritable bowel syndrome; JAM-A: junctional adhesion molecule A; JAM-C: junctional adhesion molecule C; JNK: Jun-N-terminal kinase; KLF6: Kruppel-like factor 6; LKB1: liver kinase B1; LMP2: Low-molecular mass protein 2; LMP7: Low-molecular mass protein 7; MAPK: mitogen-activated protein kinase; MAZ: myc-associated zinc-finger protein; MEK: mitogen-activated protein kinase (MAPK) kinase; MLCK: myosin light chain kinase; NF-κB: nuclear factor-kappa B; NSCLC: non-small cell lung cancer; p70S6K-S6: p70 S6 kinase-S6; PDCD4: programmed cell death factor 4; PDZ: PSD-95/Dlg/ZO-1; PI3K: phosphatidylinositol 3-kinase; PKIα : protein kinase inhibitor α; PKC: protein kinase C; PTEN: phosphatase and tensin homolog; SH3: Src homology 3; snRNAs: small nuclear RNAs; siRNAs: small interfering RNA; SOX5: sex-determining region Y-box protein 5; SFK: Src family kinases; SPRY1: sprouty 1; SPRY2: sprouty 2; SPRY4-IT1: SPRY4 intronic transcript 1; STAT5b: signal transducer and activator of transcription 5b; TEWL: trans-epidermal water loss; TJ: tight junction; TLR2: Toll-like receptor 2; tRNAs: transfer RNAs; USP1: ubiquitin specific peptidase 1; VE-cadherin: vascular endothelial cell cadherin; XIST: X inactivate-specific transcript; YAP1: Yes-associated protein 1; ZO-1: zonula occludens-1; ZO-2: zonula occludens-2; ZO-3: zonula occludens-3; ZONAB: zonula occludens 1‑associated nucleic acid binding protein.
Acknowledgements
This work is supported by the National Natural Science Foundation of China (No. 81703101), the New Xiangya Talent Projects of the Third Xiangya Hospital of Central South University (No. JY201623 and No. 20170301), the Natural Science Foundation of Hunan Province (No. 2018JJ3788 and No. 2018JJ3793), the Health and Family Planning Commission of Hunan Province (No. C2019173).
Competing Interests
The authors have declared that no competing interest exists.
References
1. G B, E D. Endothelial cell-to-cell junctions: molecular organization and role in vascular homeostasis. Physiological reviews. 2004;84:869-901
2. DD M, B S, CY C. Anchoring junctions as drug targets: role in contraceptive development. Pharmacological reviews. 2008;60:146-80
3. M F, S T. Claudins in occluding junctions of humans and flies. Trends in cell biology. 2006;16:181-8
4. D B, J D, J H. Immunocytochemical localization of the lens main intrinsic polypeptide (MIP26) in communicating junctions. The Journal of cell biology. 1982;92:213-20
5. Tatari MN, De Craene B, Soen B. et al. ZEB2-transgene expression in the epidermis compromises the integrity of the epidermal barrier through the repression of different tight junction proteins. Cell Mol Life Sci. 2014;71:3599-609
6. Y S, H S, K T. et al. Tight junctions. Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin. Science (New York, NY). 2015;347:775-8
7. Zihni C, Mills C, Matter K. et al. Tight junctions: from simple barriers to multifunctional molecular gates. Nature reviews Molecular cell biology. 2016;17:564-80
8. Schwarz BT, Wang F, Shen L. et al. LIGHT signals directly to intestinal epithelia to cause barrier dysfunction via cytoskeletal and endocytic mechanisms. Gastroenterology. 2007;132:2383-94
9. K U, J I, S K-T. et al. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. 2006;126:741-54
10. Helfrich I, Schmitz A, Zigrino P. et al. Role of aPKC isoforms and their binding partners Par3 and Par6 in epidermal barrier formation. J Invest Dermatol. 2007;127:782-91
11. Zheng B, Cantley LC. Regulation of epithelial tight junction assembly and disassembly by AMP-activated protein kinase. Proc Natl Acad Sci U S A. 2007;104:819-22
12. Gunzel D, Yu AS. Claudins and the modulation of tight junction permeability. Physiological reviews. 2013;93:525-69
13. CM VI, JM A. Architecture of tight junctions and principles of molecular composition. Seminars in cell & developmental biology. 2014;36:157-65
14. S T, H T, A T. The Claudins: From Tight Junctions to Biological Systems. Trends in biochemical sciences. 2019;44:141-52
15. M L-N, PJ M. The claudins. Genome biology. 2009;10:235
16. CM M, D M, CY C. et al. Claudin and occludin expression and function in the seminiferous epithelium. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2010;365:1679-96
17. IE B, C B, J C. et al. Occludin protein family: oxidative stress and reducing conditions. Antioxidants & redox signaling. 2011;15:1195-219
18. GJ F, JM M, MP R. Occludin: structure, function and regulation. Advanced drug delivery reviews. 2005;57:883-917
19. D G, M F. Claudins and other tight junction proteins. Comprehensive Physiology. 2012;2:1819-52
20. KJ M, CA P. The JAM family of proteins. Advanced drug delivery reviews. 2005;57:857-67
21. Cordenonsi M, D'Atri F, Hammar E. et al. Cingulin contains globular and coiled-coil domains and interacts with ZO-1, ZO-2, ZO-3, and myosin. J Cell Biol. 1999;147:1569-82
22. Ebnet K. Junctional Adhesion Molecules (JAMs): Cell Adhesion Receptors With Pleiotropic Functions in Cell Physiology and Development. Physiological reviews. 2017;97:1529-54
23. L G-M, M Q, M D-C. ZO proteins and redox-dependent processes. Antioxidants & redox signaling. 2011;15:1235-53
24. K E, CU S, MK MZB. et al. Junctional adhesion molecule interacts with the PDZ domain-containing proteins AF-6 and ZO-1. The Journal of biological chemistry. 2000;275:27979-88
25. S C, S P, P P. et al. The tight junction protein cingulin regulates gene expression and RhoA signaling. Annals of the New York Academy of Sciences. 2009;1165:88-98
26. C S, DR K. Internetwork competition for monomers governs actin cytoskeleton organization. Nature reviews Molecular cell biology. 2016;17:799-810
27. A R, AS Y. A bigger picture: classical cadherins and the dynamic actin cytoskeleton. Nature reviews Molecular cell biology. 2012;13:673-9
28. S K, PA C. Emerging role for the cytoskeleton as an organizer and regulator of translation. Nature reviews Molecular cell biology. 2010;11:75-81
29. C S, S S, K P-F. et al. Mechanosensation of Tight Junctions Depends on ZO-1 Phase Separation and Flow. Cell. 2019;179:937-52.e18
30. Zeisel MB, Dhawan P, Baumert TF. Tight junction proteins in gastrointestinal and liver disease. Gut. 2018
31. XD Z, V B, H L. et al. Tight Junction Protein 1 Modulates Proteasome Capacity and Proteasome Inhibitor Sensitivity in Multiple Myeloma via EGFR/JAK1/STAT3 Signaling. Cancer cell. 2016;29:639-52
32. G E, K K. Multifactorial skin barrier deficiency and atopic dermatitis: Essential topics to prevent the atopic march. The Journal of allergy and clinical immunology. 2016;138:350-8.e1
33. JR T, MM B, I R-C. et al. The role of molecular remodeling in differential regulation of tight junction permeability. Seminars in cell & developmental biology. 2014;36:204-12
34. L P, G S, MT N. et al. Paracellular epithelial sodium transport maximizes energy efficiency in the kidney. The Journal of clinical investigation. 2016;126:2509-18
35. CO Z, JY W, KR K. et al. Regulation of tight junction proteins and bladder epithelial paracellular permeability by an antiproliferative factor from patients with interstitial cystitis. The Journal of urology. 2005;174:2382-7
36. JT H, APS R, G S. et al. Japanese encephalitis virus neuropenetrance is driven by mast cell chymase. Nature communications. 2019;10:706
37. HM L, BP D, AK P. et al. Interferon-λ restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Science translational medicine. 2015;7:284ra59
38. J M, Y Y, P W. et al. MiR-181a regulates blood-tumor barrier permeability by targeting Krüppel-like factor 6. Journal of cerebral blood flow and metabolism: official journal of the International Society of Cerebral Blood Flow and Metabolism. 2014;34:1826-36
39. H C, DD M, WM L. et al. Regulation of spermatogenesis by a local functional axis in the testis: role of the basement membrane-derived noncollagenous 1 domain peptide. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2017;31:3587-607
40. K H, Y S. Retinoic acid signaling in Sertoli cells regulates organization of the blood-testis barrier through cyclical changes in gene expression. Development (Cambridge, England). 2012;139:4347-55
41. CR W JR T. Dynamic modeling of the tight junction pore pathway. Annals of the New York Academy of Sciences. 2017;1397:209-18
42. T S, C Z, F L. et al. Intestinal HIF-1α deletion exacerbates alcoholic liver disease by inducing intestinal dysbiosis and barrier dysfunction. Journal of hepatology. 2018;69:886-95
43. Otani T, Nguyen TP, Tokuda S. et al. Claudins and JAM-A coordinately regulate tight junction formation and epithelial polarity. J Cell Biol. 2019;218:3372-96
44. A DB, NM R, LY M. et al. Tight junction defects in patients with atopic dermatitis. The Journal of allergy and clinical immunology. 2011;127:773-86.e1 -7
45. Shen L, Weber C, Raleigh D. et al. Tight junction pore and leak pathways: a dynamic duo. Annual review of physiology. 2011;73:283-309
46. T O, TP N, S T. et al. Claudins and JAM-A coordinately regulate tight junction formation and epithelial polarity. The Journal of cell biology. 2019;218:3372-96
47. S Z, N B, D G. et al. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn's disease. Gut. 2007;56:61-72
48. C H, J R, K W. et al. GRHL2 Is Required for Collecting Duct Epithelial Barrier Function and Renal Osmoregulation. Journal of the American Society of Nephrology: JASN. 2018;29:857-68
49. J L, W H, Z Y. et al. Focusing on claudin-5: A promising candidate in the regulation of BBB to treat ischemic stroke. Progress in neurobiology. 2018;161:79-96
50. J H. Claudins and mineral metabolism. Current opinion in nephrology and hypertension. 2016;25:308-13
51. G A, A B, SS W. et al. Claudins 6, 9, and 13 are developmentally expressed renal tight junction proteins. American journal of physiology Renal physiology. 2006;291:F1132-41
52. SM K, JD S, M F. Tight junction, selective permeability, and related diseases. Seminars in cell & developmental biology. 2014;36:166-76
53. L S, CR W, DR R. et al. Tight junction pore and leak pathways: a dynamic duo. Annual review of physiology. 2011;73:283-309
54. F K, N K, Z J. et al. Net intestinal transport of oxalate reflects passive absorption and SLC26A6-mediated secretion. Journal of the American Society of Nephrology: JASN. 2011;22:2247-55
55. F M-B, M P-M. Epithelial cell polarity, stem cells and cancer. Nature reviews Cancer. 2011;12:23-38
56. Wen W, Zhang M. Protein Complex Assemblies in Epithelial Cell Polarity and Asymmetric Cell Division. Journal of molecular biology. 2018;430:3504-20
57. Du D, Xu F, Yu L. et al. The tight junction protein, occludin, regulates the directional migration of epithelial cells. Dev Cell. 2010;18:52-63
58. SJ H, LH A, Y Z. et al. Loss of Tight Junction Protein Claudin 18 Promotes Progressive Neoplasia Development in Mouse Stomach. Gastroenterology. 2018;155:1852-67
59. B Z, P F, J L. et al. Claudin-18-mediated YAP activity regulates lung stem and progenitor cell homeostasis and tumorigenesis. The Journal of clinical investigation. 2018;128:970-84
60. DN K. Claudin-18: unexpected regulator of lung alveolar epithelial cell proliferation. The Journal of clinical investigation. 2018;128:903-5
61. BP F, Z C, M B. et al. Impaired autophagy bridges lysosomal storage disease and epithelial dysfunction in the kidney. Nature communications. 2018;9:161
62. Z L, DH K, J F. et al. A non-tight junction function of claudin-7-Interaction with integrin signaling in suppressing lung cancer cell proliferation and detachment. Molecular cancer. 2015;14:120
63. A H, AR K, T R. et al. Permeation of hypericin in spheroids composed of different grade transitional cell carcinoma cell lines and normal human urothelial cells. The Journal of urology. 2005;174:69-72
64. Brandner JM, Kief S, Grund C. et al. Organization and formation of the tight junction system in human epidermis and cultured keratinocytes. European journal of cell biology. 2002;81:253-63
65. Hadj-Rabia S, Brideau G, Al-Sarraj Y. et al. Multiplex epithelium dysfunction due to CLDN10 mutation: the HELIX syndrome. Genet Med. 2018;20:190-201
66. I KK, SF S, G L. et al. Nasal epithelial barrier dysfunction increases sensitization and mast cell degranulation in the absence of allergic inflammation. Allergy. 2019
67. Steelant B, Farre R, Wawrzyniak P. et al. Impaired barrier function in patients with house dust mite-induced allergic rhinitis is accompanied by decreased occludin and zonula occludens-1 expression. J Allergy Clin Immunol. 2016;137:1043-53.e5
68. Watson RE, Poddar R, Walker JM. et al. Altered claudin expression is a feature of chronic plaque psoriasis. The Journal of pathology. 2007;212:450-8
69. WT K, L S, L Z. et al. Inflammation-induced Occludin Downregulation Limits Epithelial Apoptosis by Suppressing Caspase-3 Expression. Gastroenterology. 2019;157:1323-37
70. Vivinus-Nebot M, Frin-Mathy G, Bzioueche H. et al. Functional bowel symptoms in quiescent inflammatory bowel diseases: role of epithelial barrier disruption and low-grade inflammation. Gut. 2014;63:744-52
71. Coeffier M, Gloro R, Boukhettala N. et al. Increased proteasome-mediated degradation of occludin in irritable bowel syndrome. Am J Gastroenterol. 2010;105:1181-8
72. D S, A M, CS G. et al. Pyrin Inflammasome Regulates Tight Junction Integrity to Restrict Colitis and Tumorigenesis. Gastroenterology. 2018;154:948-64.e8
73. X Z, J L, J G. et al. Gut-dependent microbial translocation induces inflammation and cardiovascular events after ST-elevation myocardial infarction. Microbiome. 2018;6:66
74. J H, M R, AS Y. Claudins and the kidney. Annual review of physiology. 2013;75:479-501
75. M K, Y C, Y L. et al. Pro-chemotherapeutic effects of antibody against extracellular domain of claudin-4 in bladder cancer. Cancer letters. 2015;369:212-21
76. PC R, F E, AK E. et al. Expression and potential clinical significance of urothelial cytodifferentiation markers in the exstrophic bladder. The Journal of urology. 2012;187:1806-11
77. N P, E D, S D. et al. Opioid antagonists as potential therapeutics for ischemic stroke. Progress in neurobiology. 2019;182:101679
78. J N, HH T, KK H. et al. Aberrant oligodendroglial-vascular interactions disrupt the blood-brain barrier, triggering CNS inflammation. Nature neuroscience. 2019;22:709-18
79. Y G, X X, WY L. et al. Cell polarity proteins and spermatogenesis. Seminars in cell & developmental biology. 2016;59:62-70
80. W S, CY C. Cdc42 is involved in NC1 peptide-regulated BTB dynamics through actin and microtubule cytoskeletal reorganization. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2019;33:14461-78
81. L L, B M, M Y. et al. Planar cell polarity protein Dishevelled 3 (Dvl3) regulates ectoplasmic specialization (ES) dynamics in the testis through changes in cytoskeletal organization. Cell death & disease. 2019;10:194
82. PP L, DD M, KW M. et al. Focal adhesion kinase-Tyr407 and -Tyr397 exhibit antagonistic effects on blood-testis barrier dynamics in the rat. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12562-7
83. C Y, S T, D F. et al. A tight junction-associated Merlin-angiomotin complex mediates Merlin's regulation of mitogenic signaling and tumor suppressive functions. Cancer cell. 2011;19:527-40
84. Y W, D D, L F. et al. Tyrosine phosphorylated Par3 regulates epithelial tight junction assembly promoted by EGFR signaling. The EMBO journal. 2006;25:5058-70
85. E C, G G, DK P. Toll-like receptor 2 enhances ZO-1-associated intestinal epithelial barrier integrity via protein kinase C. Gastroenterology. 2004;127:224-38
86. J C, MR M, C M. et al. Gpr124 is essential for blood-brain barrier integrity in central nervous system disease. Nature medicine. 2017;23:450-60
87. J C, A S, NM K. et al. Wnt signaling mediates pathological vascular growth in proliferative retinopathy. Circulation. 2011;124:1871-81
88. Zhu L, Yang N, Du G. et al. LncRNA CRNDE promotes the epithelial-mesenchymal transition of hepatocellular carcinoma cells via enhancing the Wnt/beta-catenin signaling pathway. Journal of cellular biochemistry. 2018
89. T G, Z W, Y D. et al. Role of melatonin in sleep deprivation-induced intestinal barrier dysfunction in mice. Journal of pineal research. 2019;67:e12574
90. R S, S C SR B. et al. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nature communications. 2019;10:89
91. H C, Y H, Y F. et al. Nrf2 deficiency impairs the barrier function of mouse oesophageal epithelium. Gut. 2014;63:711-9
92. DA R, J W, K M. et al. Brain endothelial TAK1 and NEMO safeguard the neurovascular unit. The Journal of experimental medicine. 2015;212:1529-49
93. W Z, Q C, Y P. et al. FoxO4 inhibits NF-kappaB and protects mice against colonic injury and inflammation. Gastroenterology. 2009;137:1403-14
94. T B, TT V, J L. et al. Inhibition of RAF1 kinase activity restores apicobasal polarity and impairs tumour growth in human colorectal cancer. Gut. 2017;66:1106-15
95. PL M, Y G, JM S. et al. Brain endothelial dysfunction in cerebral adrenoleukodystrophy. Brain: a journal of neurology. 2015;138:3206-20
96. X H, X R, I J. et al. Regulation of intestinal barrier function by signal transducer and activator of transcription 5b. Gut. 2009;58:49-58
97. JK F, N T, D H. et al. Siah regulation of Pard3A controls neuronal cell adhesion during germinal zone exit. Science (New York, NY). 2010;330:1834-8
98. N P-S, AJ T, CM VI. et al. Liver kinase B1 regulates hepatocellular tight junction distribution and function in vivo. Hepatology (Baltimore, Md). 2016;64:1317-29
99. HP L, CC P, CC W. et al. Inactivation of the tight junction gene CLDN11 by aberrant hypermethylation modulates tubulins polymerization and promotes cell migration in nasopharyngeal carcinoma. Journal of experimental & clinical cancer research: CR. 2018;37:102
100. N I, M K, N O-S. et al. Abrogated Caveolin-1 expression via histone modification enzyme Setdb2 regulates brain edema in a mouse model of influenza-associated encephalopathy. Scientific reports. 2019;9:284
101. X S, Q Y, CJ R. et al. AMPK improves gut epithelial differentiation and barrier function via regulating Cdx2 expression. Cell death and differentiation. 2017;24:819-31
102. N K, DV P, L B. et al. Immunosuppressant MPA Modulates Tight Junction through Epigenetic Activation of MLCK/MLC-2 Pathway via p38MAPK. Frontiers in physiology. 2015;6:381
103. MJ K, SS K, YL C. et al. Derepression of CLDN3 and CLDN4 during ovarian tumorigenesis is associated with loss of repressive histone modifications. Carcinogenesis. 2010;31:974-83
104. DP B. Metazoan MicroRNAs. Cell. 2018;173:20-51
105. D S, M A. MicroRNAs in development and disease. Physiological reviews. 2011;91:827-87
106. Vanden Oever M, Muldoon D, Mathews W. et al. miR-29 Regulates Type VII Collagen in Recessive Dystrophic Epidermolysis Bullosa. J Invest Dermatol. 2016;136:2013-21
107. P W, J C, Q C. et al. Lnc-TALC promotes O-methylguanine-DNA methyltransferase expression via regulating the c-Met pathway by competitively binding with miR-20b-3p. Nature communications. 2019;10:2045
108. Q S, V T, JH Y. et al. MIR100 host gene-encoded lncRNAs regulate cell cycle by modulating the interaction between HuR and its target mRNAs. Nucleic acids research. 2018;46:10405-16
109. J A, F C. Long non-coding RNAs in corticogenesis: deciphering the non-coding code of the brain. The EMBO journal. 2015;34:2865-84
110. N G, D K, M D. Lnc-ing non-coding RNAs with metabolism and diabetes: roles of lncRNAs. Cellular and molecular life sciences: CMLS. 2018;75:1827-37
111. JH B, DL S. Long non-coding RNAs: modulators of nuclear structure and function. Current opinion in cell biology. 2014;26:10-8
112. T N, M T, J F. et al. Deregulated Expression of Mammalian lncRNA through Loss of SPT6 Induces R-Loop Formation, Replication Stress, and Cellular Senescence. Molecular cell. 2018;72:970-84.e7
113. Q M, K W, T B. et al. DGCR5The long noncoding RNA may regulate expression of several schizophrenia-related genes. Science translational medicine. 2018 10
114. J P, H L, N H. et al. Long non-coding RNA ChRO1 facilitates ATRX/DAXX-dependent H3.3 deposition for transcription-associated heterochromatin reorganization. Nucleic acids research. 2018;46:11759-75
115. S D, E Z, P S. et al. A Novel Angiotensin II-Induced Long Noncoding RNA Giver Regulates Oxidative Stress, Inflammation, and Proliferation in Vascular Smooth Muscle Cells. Circulation research. 2018;123:1298-312
116. Xiao G, Yao J, Kong D. et al. The Long Noncoding RNA TTTY15, Which Is Located on the Y Chromosome, Promotes Prostate Cancer Progression by Sponging let-7. European urology. 2019;76:315-26
117. CJ L, Y X, M Y. et al. Long noncoding RNA Bmncr regulates mesenchymal stem cell fate during skeletal aging. The Journal of clinical investigation. 2018;128:5251-66
118. N L, Q L, X Y. et al. Hepatitis B Virus-Upregulated LNC-HUR1 Promotes Cell Proliferation and Tumorigenesis by Blocking p53 Activity. Hepatology (Baltimore, Md). 2018;68:2130-44
119. MS SHA, X C, M M. et al. LncRNA Meg3 protects endothelial function by regulating the DNA damage response. Nucleic acids research. 2019;47:1505-22
120. AA A, KR M-G, MM K. et al. Finely tuned conformational dynamics regulate the protective function of the lncRNA MALAT1 triple helix. Nucleic acids research. 2019;47:1468-81
121. Y Y, P N-M, L F. et al. A novel mechanism of lncRNA and miRNA interaction: CCAT2 regulates miR-145 expression by suppressing its maturation process in colon cancer cells. Molecular cancer. 2017;16:155
122. B L, L S, Q L. et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer cell. 2015;27:370-81
123. J L, A P, M S. et al. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Molecular cell. 2014;55:138-47
124. WW D, W Y, Y C. et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. European heart journal. 2017;38:1402-12
125. X J, H H, J Z. et al. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Molecular cancer. 2020;19:20
126. H Y, X L, Q M. et al. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Molecular cancer. 2020;19:13
127. F Y, A H, D L. et al. Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Molecular cancer. 2019;18:158
128. H L, F Y, A H. et al. Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO molecular medicine. 2019;11:e10835
129. X X, X L, F L. et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Molecular cancer. 2019;18:131
130. Liu BH, Zhang BB, Liu XQ. et al. Expression Profiling Identifies Circular RNA Signature in Hepatoblastoma. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2018;45:706-19
131. Zhang C, Wang X, Chen Y. et al. The down-regulation of hsa_circ_0012919, the sponge for miR-125a-3p, contributes to DNA methylation of CD11a and CD70 in CD4(+) T cells of systemic lupus erythematous. Clinical science (London, England: 1979). 2018;132:2285-98
132. BR C. MicroRNAs as mediators of viral evasion of the immune system. Nature immunology. 2013;14:205-10
133. Kaucsár T, Rácz Z, Hamar P. Post-transcriptional gene-expression regulation by micro RNA (miRNA) network in renal disease. Advanced drug delivery reviews. 2010;62:1390-401
134. Halushka P, Goodwin A, Halushka M. Opportunities for microRNAs in the Crowded Field of Cardiovascular Biomarkers. Annual review of pathology. 2019;14:211-38
135. Mastropasqua R, Toto L, Cipollone F. et al. Role of microRNAs in the modulation of diabetic retinopathy. Progress in retinal and eye research. 2014;43:92-107
136. Abdi J, Rastgoo N, Li L. et al. Role of tumor suppressor p53 and micro-RNA interplay in multiple myeloma pathogenesis. Journal of hematology & oncology. 2017;10:169
137. Leonetti A, Assaraf Y, Veltsista P. et al. MicroRNAs as a drug resistance mechanism to targeted therapies in EGFR-mutated NSCLC: Current implications and future directions. Drug resistance updates: reviews and commentaries in antimicrobial and anticancer chemotherapy. 2019;42:1-11
138. CD A, GB F, RK J. The blood-brain barrier and blood-tumour barrier in brain tumours and metastases. Nature reviews Cancer. 2020;20:26-41
139. MA O JR T. The intestinal epithelial barrier: a therapeutic target? Nature reviews Gastroenterology & hepatology. 2017;14:9-21
140. T K, S N, K N. Choreographing Immunity in the Skin Epithelial Barrier. Immunity. 2019;50:552-65
141. S E, K E, C T-H. et al. Cutaneous Barriers and Skin Immunity: Differentiating A Connected Network. Trends in immunology. 2018;39:315-27
142. T D, S H, K K. Classification of inflammatory skin diseases: a proposal based on the disorders of the three-layered defense systems, barrier, innate immunity and acquired immunity. Journal of dermatological science. 2014;76:81-9
143. C F, K A, E Y-A. et al. Actin up: regulation of podocyte structure and function by components of the actin cytoskeleton. Trends in cell biology. 2007;17:428-37
144. V V, SC T. Renal function in diabetic disease models: the tubular system in the pathophysiology of the diabetic kidney. Annual review of physiology. 2012;74:351-75
145. K S, J L, N G. et al. Hedgehog/Wnt feedback supports regenerative proliferation of epithelial stem cells in bladder. Nature. 2011;472:110-4
146. I P, A B, A S. et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463-8
147. H W. Blood-brain barrier pathology linked to epilepsy in Alzheimer disease. Nature reviews Neurology. 2020;16:66
148. H Z, S Z, J Z. et al. ZO-1 expression is suppressed by GM-CSF via miR-96/ERG in brain microvascular endothelial cells. Journal of cerebral blood flow and metabolism: official journal of the International Society of Cerebral Blood Flow and Metabolism. 2018;38:809-22
149. Z F, QW H, Q L. et al. MicroRNA-150 regulates blood-brain barrier permeability via Tie-2 after permanent middle cerebral artery occlusion in rats. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2016;30:2097-107
150. M B, A K, M L. et al. Hypoxia-Induced MicroRNA-212/132 Alter Blood-Brain Barrier Integrity Through Inhibition of Tight Junction-Associated Proteins in Human and Mouse Brain Microvascular Endothelial Cells. Translational stroke research. 2019;10:672-83
151. K T, JM S, AC D. et al. MicroRNA-Mediated Therapy Modulating Blood-Brain Barrier Disruption Improves Vascular Cognitive Impairment. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:1392-406
152. Lin M, Zhu L, Wang J. et al. miR-424-5p maybe regulate blood-brain barrier permeability in a model in vitro with Abeta incubated endothelial cells. Biochemical and biophysical research communications. 2019;517:525-31
153. D N, CJ G. Mathematical modeling of combinatorial regulation suggests that apparent positive regulation of targets by miRNA could be an artifact resulting from competition for mRNA. RNA (New York, NY). 2015;21:307-19
154. R M, SK S. HIV-1 Tat C modulates expression of miRNA-101 to suppress VE-cadherin in human brain microvascular endothelial cells. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2013;33:5992-6000
155. Wu P, Gao Y, Shen S. et al. KHDRBS3 regulates the permeability of blood-tumor barrier via cDENND4C/miR-577 axis. Cell death & disease. 2019;10:536
156. L C, Y X, J Z. et al. MiR-429 Regulated by Endothelial Monocyte Activating Polypeptide-II (EMAP-II) Influences Blood-Tumor Barrier Permeability by Inhibiting the Expressions of ZO-1, Occludin and Claudin-5. Frontiers in molecular neuroscience. 2018;11:35
157. J G, H C, J Z. et al. Long non-coding RNA NEAT1 regulates permeability of the blood-tumor barrier via miR-181d-5p-mediated expression changes in ZO-1, occludin, and claudin-5. Biochimica et biophysica acta Molecular basis of disease. 2017;1863:2240-54
158. JY W, YH C, L X, et al. uc.173Regulation of Intestinal Epithelial Barrier Function by Long Noncoding RNA through Interaction with MicroRNA 29b. Molecular and cellular biology. 2018; 38
159. K N, Y S, H N. et al. Commensal microbiota-induced microRNA modulates intestinal epithelial permeability through the small GTPase ARF4. The Journal of biological chemistry. 2017;292:15426-33
160. Martinez C, Rodino-Janeiro BK, Lobo B. et al. miR-16 and miR-125b are involved in barrier function dysregulation through the modulation of claudin-2 and cingulin expression in the jejunum in IBS with diarrhoea. Gut. 2017;66:1537-8
161. Wang H, Chao K, Ng SC. et al. Pro-inflammatory miR-223 mediates the cross-talk between the IL23 pathway and the intestinal barrier in inflammatory bowel disease. Genome biology. 2016;17:58
162. X W, Y C, W Y. et al. MicroRNA-155-5p is a key regulator of allergic inflammation, modulating the epithelial barrier by targeting PKIα. Cell death & disease. 2019;10:884
163. S R, H E, M S. et al. Mixed-species biofilm compromises wound healing by disrupting epidermal barrier function. The Journal of pathology. 2014;233:331-43
164. Y G, J H. Claudin-14 underlies Ca⁺⁺-sensing receptor-mediated Ca⁺⁺ metabolism via NFAT-microRNA-based mechanisms. Journal of the American Society of Nephrology: JASN. 2014;25:745-60
165. A HG, FC B, H R. et al. MicroRNA MiR-199a-5p regulates smooth muscle cell proliferation and morphology by targeting WNT2 signaling pathway. The Journal of biological chemistry. 2015;290:7067-86
166. PJ C, LM C, CL C. et al. MicroRNA-205 targets tight junction-related proteins during urothelial cellular differentiation. Molecular & cellular proteomics: MCP. 2014;13:2321-36
167. K M, V S-F, A HG. et al. miR-199a-5p regulates urothelial permeability and may play a role in bladder pain syndrome. The American journal of pathology. 2013;182:431-48
168. L A, D I, A S. et al. MicroRNAs-143 and -145 induce epithelial to mesenchymal transition and modulate the expression of junction proteins. Cell death and differentiation. 2017;24:1750-60
169. S O, R A, A M. et al. A Polysome-Based microRNA Screen Identifies miR-24-3p as a Novel Promigratory miRNA in Mesothelioma. Cancer research. 2018;78:5741-53
170. JJ Q, HY C. Unique features of long non-coding RNA biogenesis and function. Nature reviews Genetics. 2016;17:47-62
171. Ma D, Cao Y, Wang Z. et al. CCAT1 lncRNA Promotes Inflammatory Bowel Disease Malignancy by Destroying Intestinal Barrier via Downregulating miR-185-3p. Inflammatory bowel diseases. 2019;25:862-74
172. T Z, SK J, L L. et al. H19 Long Noncoding RNA Regulates Intestinal Epithelial Barrier Function via MicroRNA 675 by Interacting with RNA-Binding Protein HuR. Molecular and cellular biology. 2016;36:1332-41
173. SW C, PY W, YC L. et al. Effect of Long Noncoding RNA H19 Overexpression on Intestinal Barrier Function and Its Potential Role in the Pathogenesis of Ulcerative Colitis. Inflammatory bowel diseases. 2016;22:2582-92
174. Chen T, Xue H, Lin R. et al. MiR-34c and PlncRNA1 mediated the function of intestinal epithelial barrier by regulating tight junction proteins in inflammatory bowel disease. Biochemical and biophysical research communications. 2017;486:6-13
175. Xiao L, Rao JN, Cao S. et al. Long noncoding RNA SPRY4-IT1 regulates intestinal epithelial barrier function by modulating the expression levels of tight junction proteins. Molecular biology of the cell. 2016;27:617-26
176. Sa L, Li Y, Zhao L. et al. The Role of HOTAIR/miR-148b-3p/USF1 on Regulating the Permeability of BTB. Frontiers in molecular neuroscience. 2017;10:194
177. Yu H, Xue Y, Wang P. et al. Knockdown of long non-coding RNA XIST increases blood-tumor barrier permeability and inhibits glioma angiogenesis by targeting miR-137. Oncogenesis. 2017;6:e303
178. Q D, F M, X C. et al. LncRNA CRNDE is activated by SP1 and promotes osteosarcoma proliferation, invasion, and epithelial-mesenchymal transition via Wnt/β-catenin signaling pathway. Journal of cellular biochemistry. 2020
179. L Z, N Y, G D. et al. LncRNA CRNDE promotes the epithelial-mesenchymal transition of hepatocellular carcinoma cells via enhancing the Wnt/β-catenin signaling pathway. Journal of cellular biochemistry. 2018
180. R H, FH Z, N S. LncRNA FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through suppressing E-cadherin and regulating WNT pathway in non-small cell lung cancer (NSCLC). Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2017;95:331-8
181. Z Y, X Y, B N. et al. Overexpression of long non-coding RNA-CTD903 inhibits colorectal cancer invasion and migration by repressing Wnt/β-catenin signaling and predicts favorable prognosis. International journal of oncology. 2016;48:2675-85
182. Zhang CY, Li RK, Qi Y. et al. Upregulation of long noncoding RNA SPRY4-IT1 promotes metastasis of esophageal squamous cell carcinoma via induction of epithelial-mesenchymal transition. Cell biology and toxicology. 2016;32:391-401
183. Bai Y, Zhang Y, Han B. et al. Circular RNA DLGAP4 Ameliorates Ischemic Stroke Outcomes by Targeting miR-143 to Regulate Endothelial-Mesenchymal Transition Associated with Blood-Brain Barrier Integrity. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2018;38:32-50
184. Gao Y, Wu P, Ma Y. et al. Circular RNA USP1 regulates the permeability of blood-tumour barrier via miR-194-5p/FLI1 axis. J Cell Mol Med. 2020;24:342-55
Author contact
![]() Corresponding authors: Jing Chen, Department of Dermatology, Third Xiangya Hospital, Central South University, 138 Tongzipo Road, Changsha, Hunan 410013, P.R. China. E-mail: 43700351com; Qinghai Zeng, Department of Dermatology, Third Xiangya Hospital, Central South University, 138 Tongzipo Road, Changsha, Hunan 410013, P.R. China. E-mail: zengqinghaiedu.cn.
Corresponding authors: Jing Chen, Department of Dermatology, Third Xiangya Hospital, Central South University, 138 Tongzipo Road, Changsha, Hunan 410013, P.R. China. E-mail: 43700351com; Qinghai Zeng, Department of Dermatology, Third Xiangya Hospital, Central South University, 138 Tongzipo Road, Changsha, Hunan 410013, P.R. China. E-mail: zengqinghaiedu.cn.

 Global reach, higher impact
Global reach, higher impact