Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(8):2069-2079. doi:10.7150/ijbs.60631 This issue Cite
Review
Progress of Breast Cancer basic research in China
1. Department of Pharmacology, Nanjing Medical University, Nanjing, Jiangsu 210029, China.
2. Department of Thoracic Surgery, Jiangsu Cancer Hospital, Nanjing Medical University Affiliated Cancer Hospital, Nanjing, Jiangsu 210009, China.
3. Department of Hematology and Oncology, Zhongda Hospital, School of Medicine, Southeast University, Nanjing, Jiangsu 210009, China.
4. Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
#These authors contribute equally to this work.
Received 2021-3-19; Accepted 2021-4-28; Published 2021-5-11
Abstract
Breast cancer is the most commonly diagnosed and the most lethal cancer in females both in China and worldwide. Currently, the origin of cancer stem cells, the heterogeneity of cancer cells, the mechanism of cancer metastasis and drug resistance are the most important issues that need to be addressed. Chinese investigators have recently made new discoveries in basic breast cancer researches, especially regarding cancer stem cells, cancer metabolism, and microenvironments. These efforts have led to a deeper understanding of drug resistance and metastasis and have also indicated new biomarkers and therapeutic targets. These findings emphasized the importance of the cancer stem cells for targeted therapy. In this review, we summarized the latest important findings in this field in China.
Keywords: breast cancer, cancer stem cells, tumor microenvironment, metastasis, drug resistance
Introduction
Breast cancer is the most common malignant disease in women worldwide. According to the data released in 2019, breast cancer accounted for 30% of newly diagnosed malignant tumors in females, and led to 15% of female deaths from cancer [1]. The incidence and mortality of breast cancer are still rising, both in developing and developed countries [2]. In China, breast cancer morbidity and mortality rank first among malignant diseases in females. Death rate reached about 6.9% among all female malignant tumors [3]. The lack of early-stage screening and detection methods and cost-effective therapies makes breast cancer as one of the most severe disease burden globally [4]. According to the international consensus guideline, the current treatment for breast cancer mainly included chemotherapy, radiotherapy, targeted therapy, immunity therapy, and endocrine therapy before and after surgery [5]. Nowadays, therapies targeted to cancer stem cells have been a hot spot in breast cancer treatment, which is a supplement to the traditional chemotherapeutic drugs that are unable to eradicate tumor dissemination and metastasis [6, 7]. Great advances in cancer cell-of-origin, somatic mutation, epigenetic alteration, and tumor microenvironment were revealed in recent studies, both basic and clinical, leading to a better understanding of the mechanism, the pathogenesis, the diagnosis, and the treatment of breast cancer. Particularly, Chinese investigators have made great achievements in cancer stem cells, microenvironment, metastasis, drug resistance, tumor biomarkers, and new targets, as well as drugs for breast cancer therapy. In this review, we summarized the important findings on breast cancer basic research in China from 2019 to 2020.
Cancer stem cells (CSCs)
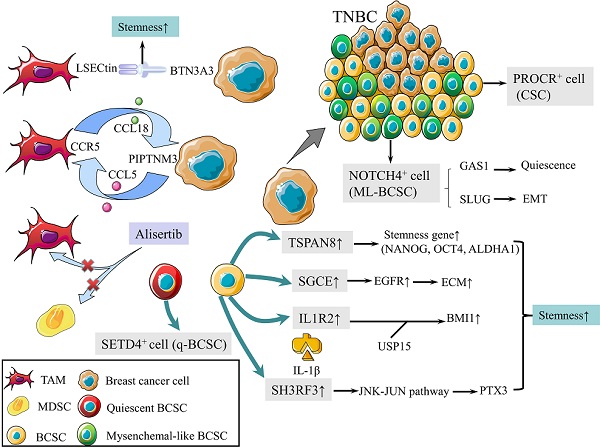
CSCs are defined as a group of undifferentiated cells that possess properties of self-renewal and pluripotent differentiation. Although breast cancer stem cells (BCSCs) make up only a small subpopulation within tumors, they are responsible for tumor initiation, recurrence, metastasis, and therapy resistance [8]. Previous studies revealed a panel of biomarkers for BCSCs identification, among which CD44high, CD24low and aldehyde dehydrogenase 1 (ALDH1)+ were the most commonly used biomarkers [9]. Triple-negative breast cancer (TNBC) contains more BCSCs than other subtypes, which is associated with worse outcomes [10, 11]. In 2019, protein C receptor (PROCR) was identified as a novel marker for BCSCs, which could stratify TNBC into clinically relevant subgroups and be a therapeutic target (Fig. 1) [12]. NOTCH4 receptor was not only implicated in the regulation of BCSCs in TNBC, but also regarded as a better marker for BCSCs than CD24-CD44+ in 2020 (Fig. 1). Mechanistically, NOTCH4 drives mesenchymal-like BCSCs into a quiescent state and induces epithelial-mesenchymal transition (EMT) via upregulating GAS1 and SLUG, respectively. Zhou et al. also demonstrated the importance of NOTCH4-SLUG-GAS1 circuit in maintaining mesenchymal-like BCSCs [13].
Schematic diagram of BCSCs and TME. (1) PROCR is a marker of BCSCs for stratifying TNBC into subgroups. (2) NOTCH4 is also a marker for BCSCs, driving ML-BCBCs into a quiescent state via GAS1 and inducing EMT via SLUG. (3) TAMs with elevated LSECtin expression interacts with BTN3A3, promoting the stemness of breast cancer cells. (4) The following pathways increase cell stemness. IL1R2 interacts with USP15 to induce deubiquitination and stabilization of BMI1. SGCE stabilizes EGFR. TSPAN-8 enhances stemness genes, including NANOG, OCT4, and ALDHA1. SH3RF3 activates PTX3 via the JNK-JUN pathway. (5) BCSCs are divided into e-BCSCs and q-BCSCs. SETD4 is important in the maintenance of qBCSCs. (6) A positive feedback loop between CCL5-CCR5 and CCL18-PIPTNM3 is shown. TAMs secret CCL18, binding to PIPTNM3 on breast cancer cells. Then breast cancer cells secret CCL5, inducing macrophages to a TAM-like phenotype CCR5. (7) Alisertib eliminates tumor-promoting MDSCs and TAMs to reshape the immune microenvironment.
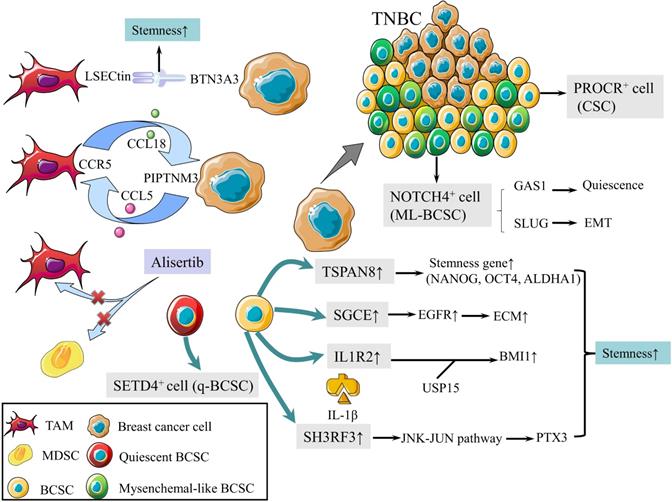
Tumor microenvironment (TME) plays an essential role in regulating the activity of BCSCs [14]. Tumor-associated macrophages (TAMs) with elevated LSECtin expression interacts with its receptor BTN3A3, expressed in cancer cells, to promote the stemness of breast cancer cells in a cell-cell contact dependent manner (Fig. 1) [15]. In addition, the Jag1 expression in endothelial cells activates Notch 1 to upregulate Zeb1 expression and to increase vascular endothelial growth factor A (VEGFA) production. In turn, VEGFA induces Jag1 expression in endothelial cells. Thus, the positive feedback loop in the tumor perivascular niche promotes the stemness of breast cancer cells [16].
There are other pathways to enhance the stemness of BCSC. Activated interleukin-1 receptor type 2 (IL1R2) interacts with deubiquitinase USP15 (ubiquitin-specific protease 15) to induce deubiquitination and stabilization of BMI1, which facilitates the self-renewal of BCSCs (Fig. 1) [17]. BCSCs were also promoted by SGCE via an interaction with c-Cbl and the inhibition of c-Cbl-mediated epidermal growth factor receptor (EGFR) lysosomal degradation (Fig. 1) [18]. The elevated stress sensor in response to hypoxia, Tribble 3 (TRIB3) promoted BCSCs through AKT1-FOXO1 (forkhead box O1)-SOX2 (sry-related high mobility box 2) axis [19]. Phospholipid scramblase 1 (PLSCR1) enhanced stem cell-like properties through upregulating signal transducer as well as activator of transcription 1 (STAT1) expression in basal-like breast cancer (BLBC) [20]. Additionally, loss of SIRT4 promoted self-renewal and expansion of BCSCs through suppressing glutamine metabolism in mitochondrial and SIRT1-mediated BRCA1 transcription in nucleus, which provided a novel cross-talk between mitochondrial and nuclear sirtuins [21].
Signaling pathways activated in BCSCs mainly include Wnt, Notch, and Hedgedog (Hh) [22]. The Hh signaling pathway is activated by the binding of Hh ligands, such as Sonic Hedgehog (SHH), and their cognate receptors, such as Ptch1. Tetraspanin-8 (TSPAN-8) recruits the ATXN3 deubiquitinating enzyme to reduce the ubiquitination of PTCH1 and to inhibit the degradation of the SHH/PTCH1 complex (Fig. 1). Therefore, TSPAN8 enhances the Hh pathway and breast cancer cell stemness [23]. In addition, SH3 domain containing ring finger 3 (SH3RF3) promotes BCSCs by activating the JNK (c-Jun N-terminal kinase)-JUN pathway, as well as the expression of pentraxin 3 (PTX3) (Fig. 1) [24].
BCSCs are divided into two categories according to the cell cycle rate: energetic BCSCs (e-BCSCs) [25] and quiescent BCSCs (q-BCSCs) [26]. Q-BCSCs play critical roles in resistance to chemoradiotherapy and disease relapse. SET domain-containing protein 4 (SETD4) is important in the maintenance of qBCSCs (Fig. 1). SETD4 facilitates heterochromatin formation via H4K20me3 (trimethylation of lysine 20 of histone 4) catalysis on certain promoter regions which leads to the silencing of qBCSCs-suppressing genes. Notably, SETD4-defined qBCSCs maintain quiescent state by asymmetric division, producing a small qBCSC and a big active daughter cell; the latter generates normal cancer cells [27]. These findings on stem cell status expanded our knowledge on the epigenetic determinants of q-BCSCs and provided new therapeutic targets for drug-resistant q-BCSCs.
Metabolism
Altered metabolism is an emerging hallmark of cancer. Unlike normal cells, cancer cells prefer aerobic glycolysis, which is accompanied by increased lactate production, also known as the Warburg effect [28]. Based on the law of conservation of electrons in chemical reactions, Li et al. built up an electron balance model to outline metabolic plasticity under hypoxia. According to the model, both proline biosynthesis and lipogenesis can act as alternative electron acceptor. Blocking them synergistically suppresses tumor growth [29]. Moreover, tumor associated macrophages (TAMs) enhance aerobic glycolysis and induce apoptosis resistance of breast cancer cells via extracellular vesicle (EV) transmission of HIF-1α-stabilizing long noncoding RNA (HISLA). Reciprocally, lactate released from glycolytic cancer cells upregulates HISLA in TAMs, constituting a feed-forward loop between TAMs and cancer cells [30].
Psychological factors induced metabolic alterations was revealed to cancer progression. Lactate dehydrogenase A (LDHA) executes the final step of the Warburg effect by converting pyruvate to lactate. Breast cancer stem-like properties can be promoted by chronic stress-induced epinephrine via LDHA-dependent metabolic rewiring. Interestingly, vitamin C may reverse the chronic stress-induced cancer stem-like phenotype [31].
Tumor microenvironment (TME)
The TME of breast cancer is composed of multiple stromal cells, soluble factors, and physical properties, including intratumor and metastatic microenvironment representing local and distant lesions, respectively [32].
In malignant phyllodes tumors (PT), TAMs promote malignant progression by secreting large amount of CCL18, which then binds to its receptor PIPTNM3 on myofibroblasts [33]. Reciprocally, malignant PT recruits and induces macrophages to a TAM-like phenotype by secreting CCL5. Thus, a positive feedback loop between CCL5-CCR5 and CCL18-PIPTNM3 is constituted, which represents the communication of myofibroblasts with TAMs and plays a central role in tumorigenesis of PT (Fig. 1) [34].
Tumor development is accompanied by the occurrence and persistence of immunosuppressive microenvironment. Myeloid-derived suppressor cells (MDSCs) and TAMs interact with CD4+ and CD8+ T lymphocytes and subsequently attenuate the anti-tumor immunity response, which is one of the factors that most responsible for immune evasion. Alisertib, a selective Aurora A kinase inhibitor, could reshape the immune microenvironment through selectively eliminating tumor-promoting myeloid cells, including MDSCs and TAMs, as well as restoring the anti-tumor immunity of T lymphocytes (Fig. 1). Intriguingly, combining alisertib with anti-programmed cell death-ligand 1 (PD-L1) therapy showed a synergistic efficacy in the treatment of advanced breast cancer [35].
According to the “seed and soil” theory, premetastatic niches in destination organs are key driving force for tumor dissemination and colonization [36]. Additionally, primary tumor is capable of inducing B cell accumulation in draining lymph nodes. These tumor-educated B cells produce pathogenic IgG that targets the tumor membrane antigen HSPA4 (heat shock protein family A member 4) to activate the HSPA4-binding protein ITGB5 (integrin β5) and the downstream Src/NF-κB pathway. These findings illustrated the role of B cells in promoting the establishment of premetastatic niches in draining lymph nodes and accelerating breast cancer lymph node metastasis [37].
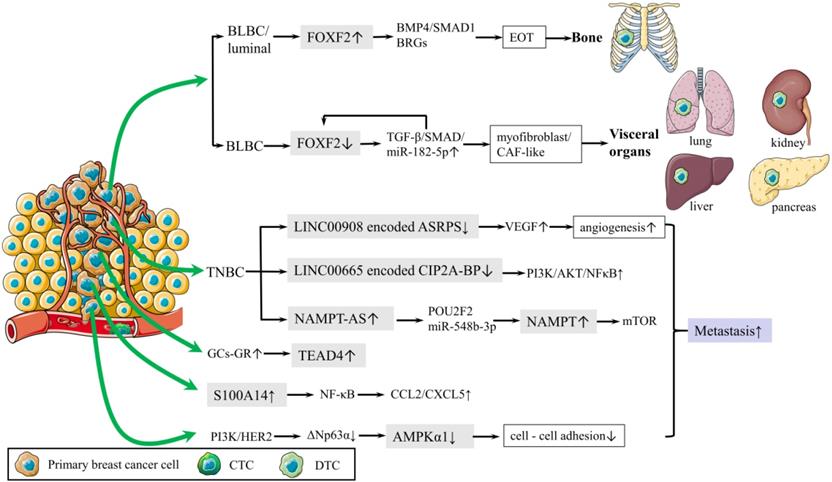
Schematic diagram of metastasis. (1) Mechanism of bone metastasis is shown.FOXF2 overexpressed by BLBC/luminal regulates EOT via BMP4/SMAD1 pathway and BRGs, leading to bone metastasis. (2) Mechanism of visceral metastasis is shown. Deficiency of FOXF2 in BLBC transdifferentiates to myofibroblast/cancer associated fibroblast (CAF)-like phenotype and metastasizes to visceral organs via TGF-β/SMAD/miR-182-5p, which in turn repress FOXF2. (3) PI3K and HER2 activation downregulates AMPKα1 expression via ΔNp63α, reducing cell-cell adhesion. (4) S100A14 promotes cancer metastasis by upregulating the secretion of CCL2 and CXCL5 via NF-κB pathway. (5) Mechanisms of lncRNA in regulating TNBC metastasis are shown below. Oncogenic lncRNA, NAMPT-AS activates NAMPT via POU2F2 miR-548b-3p.The reduction of LINC00665 encoded CIP2A-BP increases PI3K/AKT/NFκB to reduce lung metastasis. LINC00908 encodes ASRPS inhibits angiogenesis via VEGF.
Tumor metastasis
Distant metastasis is the main cause of breast cancer related death. A recent study deepened our understanding of EMT in metastasis by revealing that breast cancer cells were capable of recapitulating various epithelial and mesenchymal phenotypes. Epithelial-type circulating tumor cells (CTCs) and disseminated tumor cells (DTCs) with a restricted mesenchymal transition show the most metastatic traits, whereas mesenchymal-type CTCs and DTCs display limited metastatic ability [34]. Bone is the most common site of distant metastasis. To further explain the mechanisms underlying the predilection of bone metastasis has always been a focus of breast cancer research. It was proved that Forkhead box F2 (FOXF2) regulated the epithelium-to-osteomimicry transition (EOT) via pleiotropic transactivation of the BMP4/SMAD1 signaling pathway and bone-related genes (BRGs), thus giving the metastatic tendency to bone (Fig. 2) [38]. The same group also revealed that FOXF2 deficiency BLBC tended to transdifferentiate to a myofibroblast/cancer associated fibroblast (CAF)-like phenotype and metastasized to visceral organs. The underlying mechanism involves the reciprocal repression loop between FOXF2 and transforming growth factor-β (TGF-β) (Fig. 2) [39]. These groundbreaking studies provided theoretical basis for preventing breast cancer metastasis.
Metastasis-initiating cells (MIC) are a tiny population of cells, estimated less than 0.02% of disseminated tumor cells, and are responsible for forming secondary tumors. Yang et al found platelet-derived growth factor receptor (PDGFR) inhibition blocked AKT activation but not serum and glucocorticoid induced kinase 1 (SGK1) signaling in MIC, which resulted in suppressing lung metastasis of breast cancer; however, primary tumor burden was not affected. Co-targeting PDGFR and SGK1 showed synergistic anti-cancer effects and led to further inhibition of pulmonary metastases and primary breast tumors [40]. Reduction of cell-cell adhesion is crucial for cancer cells departing from the primary tumor. AMP-activated protein kinase (AMPK) is well known for maintaining energy homeostasis. A recent study revealed that PI3K and HER2 activation transcriptionally downregulated AMPKα1 expression via ΔNp63α and highlighted that transcriptional control was another layer of AMPK regulation, which suggested the pivotal role of AMPKα1 in cell-cell adhesion and cancer metastasis (Fig. 2) [41]. S100 calcium binding protein A14 (S100A14) promoted cancer metastasis by upregulating the expression and secretion of CCL2 and CXCL5 via RAGE/NF-κB pathway (Fig. 2) [42]. These findings provided new therapeutic strategies for inhibiting breast cancer metastasis.
Long noncoding RNA (lncRNA) plays an important role in regulating TNBC metastasis. Antisense strand of nicotinamide phosphoribosyltransferase (NAMPT-AS) was an oncogenic lncRNA that epigenetically activated NAMPT to promote breast cancer metastasis (Fig. 2) [43]. LncRNA LINC00665, encoding a micropeptide CIP2A-BP (CIP2A binding peptide), significantly reduced lung metastasis of breast cancer (Fig. 2) [44]. LncRNA LINC00908, encoding a polypeptide ASRPS (a small regulatory peptide of STAT3), inhibits angiogenesis and metastasis of TNBC (Fig. 2) [45].
In addition, Nie et al. raised new precautions in dexamethasone therapy for breast cancer patients. Glucocorticoids (GCs) could activate TEA domain transcription factor 4 (TEAD4) in the Hippo signaling pathway for cancer metastasis and chemo-resistance by enhancing the interaction of the glucocorticoid receptor (GR) with TEAD4 to form a transcriptional complex (Fig. 2) [46].
Drug resistance
Drug resistance, both intrinsic and acquired, remains the main obstacle for effective anticancer therapies.
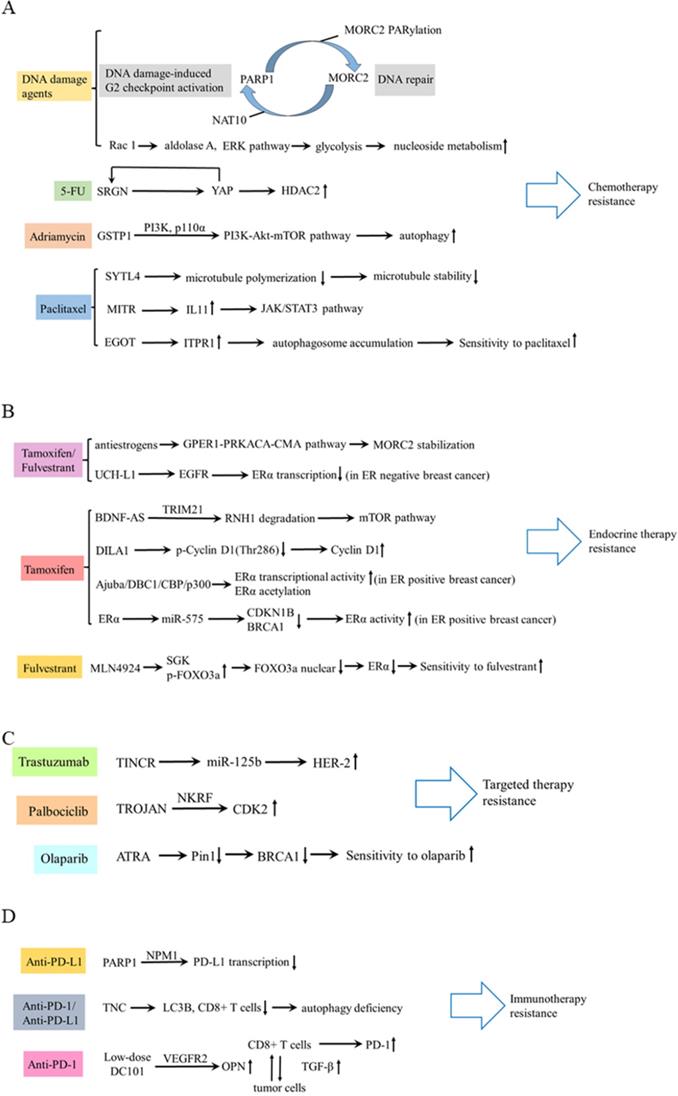
In terms of chemotherapy, cancer cells can develop drug resistance by activating DNA damage repair signaling pathways [47]. Following DNA damage, poly ADP-ribose polymerase 1 (PARP1) recruits MORC2 (MORC family CW-type zinc finger 2) to DNA damage sites and catalyzes MORC2 PARylation, which enhances its chromatin remodeling activities, thereby facilitating efficient DNA repair. In turn, MORC2 stabilizes PARP1 through a crosstalk between N-acetyltransferase (NAT10)-mediated acetylation and CHFR-mediated ubiquitination [48]. DNA-damaging chemotherapeutic agents stimulate MORC2 acetylation in a NAT10 dependent manner, then acetylated MORC2 activates G2 DNA damage checkpoint, enhancing cell survival following DNA damage [49]. Therefore, depleting MORC2 or inhibiting NAT10 sensitizes cancer cells to chemotherapy by abrogating DNA damage repair. Moreover, Rac 1 enhances the activity of non-oxidative pentose phosphate pathway via activating aldolase A and ERK signaling pathway, thereby inhibiting the nucleotide metabolism and DNA damage caused by chemotherapy in breast cancer [50]. Additionally, Zhang et al revealed that a feed-forward circuit between serglycin (SRGN) and YES-associated protein (YAP) induced HDAC2 expression, promoting chemoresistance to 5-FU [51]. Dong et al. found glutathione S-transferases P1 (GSTP1) contributed to adriamycin resistance [52]. In TNBC, synaptotagmin-like 4 (SYTL4) contributes to taxane resistance via attenuating the stability of microtubule network and increasing the growth rate of microtubule [53] The antisense intronic lncRNA (ai-lncRNA) EGOT (eosinophil granule ontogeny transcript) sensitizes breast cancer cells to paclitaxel by enhancing autophagy[54]. MEF2-interacting transcriptional repressor (MITR), as a truncated isoform of HDAC9, induces resistance of paclitaxel via increasing interleukin-11 (IL11) expression and subsequent activating JAK/STAT3 signaling pathway [55].
In terms of endocrine therapy, antiestrogens stabilize MORC2 in a GPER1 (G protein coupled estrogen receptor 1) dependent manner and decrease MORC2 enhanced cellular sensitivity to tamoxifen and fulvestrant [56]. In estrogen receptor (ER) positive breast cancer, lncRNA BDNF-AS, driven by a MEF2A regulated enhancer, induces endocrine resistance of breast cancer by activating the RNH1 (ribonuclease inhibitor 1)/TRIM21/mTOR (mechanistic target of rapamycin) cascade [57]. This is a novel mechanism other than the canonical PI3K (phosphatidylinositol-3-kinase)/AKT/mTOR axis. LncRNA DILA1 contributes to tamoxifen resistance in breast cancer via interacting with Cyclin D1 and blocking its phosphorylation and degradation [58]. Ajuba recruits DBC1 and CBP/p300, forming a ternary complex, to increase transcriptional activity of ERα and to potentiate the ERα target gene expression, thereby contributing to tamoxifen resistance [59]. MiR-575 enhances ERα activity and tamoxifen resistance via targeting cyclin dependent kinase inhibitor 1B (CDKN1B) and BRCA1 [60]. Moreover, inactivation of neddylation with MLN4924 transcriptionally inhibits ERα and significantly improves the fulvestrant sensitivity [61]. However, in ER negative breast cancer, targeting ubiquitin carboxyl terminal hydrolase-L1 (UCH-L1) upregulates ERα expression via enhancing ubiquitination and degradation of EGFR, and subsequently increases the sensitivity of tamoxifen and fulvestrant [62].
In terms of targeted therapy, lncRNA TINCR induces trastuzumab resistance by regulating miR-125b/HER-2 (human epidermal growth factor receptor 2) pathways [63]. LncRNA TROJAN promotes ER+ breast cancer resistance to palbociclib, a cyclin-dependent kinase 4 and 6 (CDK4/6) inhibitor, through TROJAN-NKRF-CDK2 axis [64]. PARP inhibitor olaparib is effective in treating breast cancer patients with BRCA1 mutations. All-trans retinoic acid (ATRA) sensitizes BRCA1-proficient breast cancer to PARP inhibition by inhibiting Pin1 and destabilizing BRCA1, which could extend the use of PARP inhibitors [65].
In regard to immunotherapy, breast cancers have poor response to immunotherapy because of poor T-cell infiltration and heightened immunosuppression within TME [66]. A low-dose vascular endothelial growth factor receptor 2 (VEGFR2) blockade can increase immune cell infiltration and upregulate programmed cell death protein-1 (PD-1) expression on immune cells, thus sensitizing breast cancer to a PD-1 blockade [67]. PARP1 suppresses PD-L1 expression via interaction with nucleophosmin (NPM1), which abolishes the binding of NPM1 at the PD-L1 gene promoter in TNBC. Olaparib elevates PD-L1 expression and leads to better anti-cancer efficacy in combination with the anti-PD-L1 antibody [68]. In addition, as a strong candidate for autophagy deficiency mediated immunosuppression, Tenascin-C (TNC) is overexpressed and is inversely correlated with LC3B expression and CD8+ T cells in TNBC patients. Inhibition of TNC in autophagy impaired TNBC cells sensitizes T cell mediated tumor killing and boosts anti-tumor effects of anti-PD-1/PD-L1 therapy [69]. These findings provided new approaches to potentially combine immunotherapy with molecular-targeting agents. These drug resistance mechanisms aforementioned are summarized in Table 1 and Figure 3.
Tumor biomarkers
The prognosis of breast cancer mainly depends on early detection and intervention. Recently, accumulated evidence indicates that circulating tumor DNA (ctDNA) could be a sensitive and specific biomarker for monitoring breast cancer progression and predicting relapse in early-stage [70]. A study showed the positive detection rate of ctDNA in early-stage breast cancer could reach 74.2%. Moreover, positive ctDNA after surgery indicated potential recurrence and remote metastasis [71]. Serum LRP6 ectodomain (LRP6N) could be another promising metastatic diagnosis marker for its binding to CXCR4 and competitively preventing SDF-1/CXCR4-induced lung metastasis [72].
Drug resistance mechanisms of breast cancer
| Drug resistance | Drugs | Mechanism | Reference |
|---|---|---|---|
| Chemotherapy | DNA damage agents | A feedback loop between MORC2 and PARP1 facilitates efficient DNA repair | 48 |
| NAT10-mediated MORC2 acetylation contributes to DNA damage-induced G2 checkpoint activation | 49 | ||
| Rac1 promotes the glycolysis, especially non-oxidative pentose phosphate pathway and nucleoside metabolism | 50 | ||
| 5-FU | A feed-forward circuit between SRGN and YAP induces HDAC2 expression to maintain stemness and chemoresistance | 51 | |
| adriamycin | GSTP1 promotes autophagy by interacting with PI3K, p110α, and then preventing PI3K-Akt-mTOR pathway signaling | 52 | |
| paclitaxel | SYTL4 decreases microtubule stability via inhibiting microtubule polymerization | 53 | |
| EGOT enhances autophagosome accumulation via the upregulation of ITPR1 expression in cis and in trans | 54 | ||
| MITR increases IL11 expression and activation of downstream JAK/STAT3 signaling pathway | 55 | ||
| Endocrine therapy | tamoxifen/fulvestrant | Estrogen receptor antagonists stabilize MORC2 via the GPER1-PRKACA-CMA pathway | 56 |
| UCH-L1 contributes to loss or reduction of ERα by the deubiquitinase-mediated stability of EGFR | 62 | ||
| tamoxifen | BDNF-AS promotes RNH1 degradation via TRIM21-mediated ubiquitination and sustains the activation of mTOR signaling | 57 | |
| DILA1 blocks phosphorylation and degradation of Cyclin D1 | 58 | ||
| Ajuba/DBC1/CBP/p300 ternary complex co-activates ERα transcriptional activity and enhances ERα acetylation | 59 | ||
| miR-575 enhances ERα activity by targeting CDKN1B and BRCA1 | 60 | ||
| fulvestrant | Inactivation of neddylation with MLN4924 inhibits ERα via delaying SGK degradation and inducing FOXO3a nuclear export | 61 | |
| Targeted therapy | trastuzumab | TINCR promotes HER-2 expression by sponging miR-125b and promotes EMT by targeting Snail-1 | 63 |
| palbociclib | TROJAN binds to NKRF and inhibits its interaction with RELA, upregulating the expression of CDK2 | 64 | |
| olaparib | ATRA sensitizes BRCA1-proficient breast cancer to PARP inhibition by inhibiting Pin1 and destabilizing BRCA1 | 65 | |
| Immunotherapy | anti-PD-1 | Low-dose VEGFR2 blockade sensitizes tumors to anti-PD-1 therapy via upregulation of PD-1 on immune cells through stimulating the secretion of OPN and TGF-β | 67 |
| anti-PD-L1 | PARP1 suppresses PD-L1 transcription through interacting with NPM1 and abolishing the binding of NPM1 at the PD-L1 promoter | 68 | |
| anti-PD-1/anti-PD-L1 | TNC contributes to autophagy deficiencymediated immunosuppression via suppressing LC3B and CD8+ T cells | 69 |
PRKACA protein kinase cAMP-activated catalytic subunit alpha, CMA chaperone-mediated autophagy, SGK serum and glucocorticoid-induced protein kinase, FOXO3a forkhead box O3a, OPN osteopontin.
Schematic diagram of drug resistance. (A) The mechanism of drug resistance in chemotherapy, including DNA damage agents, 5-FU, Adriamycin, and paclitaxel. (B) Drug resistance mechanism of tamoxifen/fulvestrant, tamoxifen and fulvestrant. (C) Drug resistance mechanism of trastuzumab, palbociclib and olaparib. (D) Drug resistance mechanism of immunotherapy.
Because of the highly heterogeneity of TNBC, appropriate classification will provide effective prediction for outcomes. TNBC were classified into three heterogeneous clusters based on the microenvironment phenotypes: cluster 1, the “immune-desert” cluster, with low microenvironment cells infiltration; cluster 2, the “innate immune-inactivated” cluster, with resting innate immune cells and nonimmune stromal cells infiltration; and cluster 3, the “immune-inflamed” cluster, with abundant adaptive and innate immune cells infiltration. Cluster 1 and 2 were both called “cold tumors” while cluster 3 was a “hot tumor”. Immune checkpoint inhibitors might be effective for “immune-inflamed” clusters, indicating that these distinct phenotypes were potential biomarkers for predicting therapeutic efficacy. The transformation of “cold tumors” into “hot tumors” should also be considered when dealing with “immune-desert” and “innate immune-inactivated” clusters [73].
The molecular typing of breast cancer has been revealed deeply by novel detecting techniques. A study divided Chinese TNBC into four subtypes based on multi-omics data: luminal androgen receptor, immunomodulatory, basal-like immune-suppressed, and mesenchymal-like subtypes, which could serve as a reference in order to further advance the understanding and precision treatment of TNBC [74]. Further, the same group conducted a phase Ib/II trial, demonstrating the clinical benefit of molecular subtyping-based and targeted sequencing-based therapy for refractory metastatic TNBC. Specifically, nab-paclitaxel in combination with anti-PD-1 therapy or with anti-VEGFR therapy showed favorable outcomes on IM (immunomodulatory) subtype and BRCA1/2 wild type-BLIS (basal-like immune-suppressed) subtypes, respectively [75]. Moreover, based on distinct metabolic dysregulation, TNBC was classified into three subtypes via multi-omics database analysis. MPS1 (heterogeneous metabolic-pathway-based subtype 1) is the lipogenic subtype with upregulated lipid metabolism. MPS2 is the glycolytic subtype with upregulated carbohydrate and nucleotide metabolism. MPS3 is the mixed subtype with partial pathway dysregulation. Different subtypes respond to metabolic inhibitors with distinct sensitivity which enables the development of personalized cancer therapy by targeting unique metabolic profiles [76].
Novel therapeutic targets and new drugs
With the increasing popularity of precision therapy for breast cancer, it is urgent to develop new therapeutic targets and drugs.
Recent studies showed the effects of FTO[77], protein arginine methyltransferase 1 (PRMT1) [78], coactivator associated arginine methyltransferase 1 (CARM1)/PRMT4 [79], Otubain-2 (OTUB2) [80], RING finger protein 144A (RNF144A) [81], plant homeodomain finger protein 20-like protein 1 (PHF20L1) [82], heat shock transcription factor 1 (HSF1) [83], epithelial cell transforming sequence 2 (ECT2) [84], pescadillo homolog 1 (PES1) [85], PHACTR2-AS1 (PAS1) [86], breast cancer-related transcript 1 (BCRT1) [87], and circCDYL [88] on progression of breast cancer and they provided novel therapeutic targets. TNBC is especially hard to treat due to the lack of targets. A recent study demonstrated NOTCH1-ATR-CHK1 cascade and cisplatin displayed good synergy in inhibiting TNBC by targeting cell cycle checkpoint, DNA damage, and EMT [89]. In addition, TNBC progression was also promoted by TROJAN [90], circSEPT9 [91], and moesin (MSN) [92], which could be potent options for this fatal disease. These potential novel therapeutic targets are summarized in Table 2.
Potential novel therapeutic targets of breast cancer
| Target | Function | Mechanism | Reference |
|---|---|---|---|
| PES1 | Promotes breast cancer growth | Forms a complex with TERT and the TR, regulating telomerase activity, telomere length maintenance, and senescence | 85 |
| MSN | Stimulates TNBC cells proliferation and invasion | Phosphorylated MSN interacts with the nucleoprotein NONO and promotes the nuclear localization of PKC interacting with MSN, which leads to the phosphorylation of CREB and the up-regulation of downstream gene expression | 92 |
| PHF20L1 | Maintains the proliferative state of breast cancer cells | recognizes H3K27me2 and collaborates with PRC2 and the NuRD complex in regulating H3K27 modifications to suppress a series of tumor suppressors | 82 |
| TROJAN | Promotes TNBC cells proliferation and invasion | Increases ZMYND8 degradation and epigenetically upregulates metastasis-related genes | 90 |
| PAS1 | Inhibits breast cancer cells proliferation and metastasis | Binds to rDNA genes and recruits histone methyltransferase SUV39H1, triggering H3K9 methylation of these genes, resulting in the suppression of ribosome synthesis | 86 |
| BCRT1 | Promotes breast cancer cells proliferation and mobility | competitively binding with miR-1303 to protect PTBP3 from degradation; promotes M2 polarization; facilitates hypoxia-induced EMT | 87 |
| circCDYL | Promotes breast cancer cells proliferation | promotes breast cancer malignant progression via the miR-1275-ATG7/ULK1-autophagic axis | 88 |
| circSEPT9 | Promotes TNBC cells proliferation, migration, invasion and inhibits TNBC cells apoptosis and autophagy | E2F1 and EIF4A3 mediated circSEPT9 regulates the expression of LIF via sponging miR-637 and activates LIF/Stat3 signaling pathway | 91 |
| OTUB2 | Promotes breast cancer stemness and metastasis | EGF and KRAS mutation induce OTUB2 poly-SUMOylation, thereby deubiquitinates and activates YAP/TAZ | 80 |
| NOTCH1 | Induces the TNBC formation | Promotes the EMT and regulates the cell cycle through activation of ATR-CHK1 signalling pathway | 89 |
| PRMT1 | Promotes breast cancer cells proliferation | PRMT1-dependent methylation of C/EBPα promotes the expression of cyclin D1 by blocking the interaction between C/EBPα and HDAC3 | 78 |
| CARM1 | Promotes ERα-positive breast cancer cells proliferation | Transcriptional activates cognate estrogen-induced genes and methylates a large cohort of proteins | 79 |
| FTO | Promotes breast cancer cells proliferation, colony formation and metastasis | Demethylates N6-methyladenosine in the 3'UTR of BNIP3 and causes its degradation | 77 |
| RNF114A | Suppresses breast cancer cells proliferation, colony formation, migration, and invasion | Interacts with and targets HSPA2 for ubiquitination and degradation | 81 |
| HSF1 | Promotes breast cancer cells proliferation, migration, and invasion | PIM2-mediated HSF1 phosphorylation at Thr120 promotes proteostasis and carboplatin-induced autophagy, and enhances PD-L1 expression | 83 |
| ECT2 | promotes breast cancer cells survival | Forms a positive feedback loop with USP7 to promote stabilization of each other, ultimately sustains the expression of MDM2 | 84 |
TERT telomerase reverse transcriptase, TR telomerase RNA, PKC protein kinase C, CREB cAMP response element-binding protein, PRC2 polycomb repressive complex 2, NuRD Mi-2/nucleosome remodeling and deacetylase, ZMYND8 zinc finger MYND-type containing 8, rDNA ribosome DNA, LIF leukemia inhibitory factor, TAZ WW domain-containing transcription factor, C/EBPα CCAAT/enhancer binding protein α, HSPA2 heat-shock protein family A member 2,PIM proviral integration site for moloney murine leukemia virus.
As we know, tumor metabolism is important in cancer progression. Nevertheless, the lack of effective and selective anti-metabolism drugs hinders clinical application. Chen et al reported a small molecule ZY-444 bound to pyruvate carboxylase, which was a key anaplerotic enzyme of the tricarboxylic acid cycle in promoting cancer growth and metastasis through Wnt/β-catenin/Snail signaling pathways. ZY-444 suppressed breast cancer cell proliferation with low cytotoxicity in normal cells by inactivating catalytic activity of pyruvate carboxylase [93]. Flubendazole, a broad-spectrum anthelmintic drug belonging to benzimidazole group, has been repurposed as a promising anti-cancer agent [94]. Zhen et al revealed flubendazole induced autophagic cell death by targeting Eva-1 homolog A (EVA1A) and suppressed TNBC proliferation and migration [95]. Moreover, resistomycin, QN-1 and emodin attenuates TNBC progression via binding to Pellino-1 to promote FBXO11-mediated SNAIL/SLUG degradation [96], down-regulating c-MYC transcription [97] and reducing VEGFA transcription [98], respectively. Accordingly, these studies provided alternative strategies for treating TNBC.
Conclusion
In summary, Chinese investigators have made significant progress in all areas of breast cancer research over the last two years, particularly in the field of BCSCs. A number of new BCSC membrane biomarkers, including PROCR, NOTCH4, SGCE, TSPAN8, and IL1R2, were identified. They may be used for diagnosis and potential therapeutic targets. In addition, stemness of BCSCs could be regulated by tumor microenvironment, such as TAM, CAF, and signaling molecules. Due to the increased patient numbers, limited treatment options, and unsatisfactory treatment efficacy in China, investigators focused on the exploration of drug resistance and metastasis mechanisms and made great efforts to identify new tumor biomarkers and therapeutic targets in order to improve the diagnosis and treatment of breast cancer. However, there is still a lack of milestone discovery in terms of breast cancer cell origin, pathological typing, and targeted metastatic therapy. Thus, multi-party cooperation is strongly recommended to make a breakthrough in basic research and clinical translation.
Acknowledgements
This work was supported by the National Key Research and Development Program of China [2020YFA0112300 and 2018YFC2000400 to CC], National Natural Science Foundation of China [81830087 and 31771516 for CC, 81972763 and 81473241 for XW, 81872378 for LX] and Jiangsu Social Development Project [BE2018711 for BC].
Author contribution Statement
CW and XW wrote the manuscript. CC, LX, JG and BC conceived the study and edited the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A. et al. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69:438-51
2. Fitzmaurice C, Akinyemiju TF, Al Lami FH, Alam T, Alizadeh-Navaei R, Allen C. et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2018;4:1553-68
3. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32
4. Ginsburg O, Bray F, Coleman MP, Vanderpuye V, Eniu A, Kotha SR. et al. The global burden of women's cancers: a grand challenge in global health. Lancet. 2017;389:847-60
5. Cardoso F, Paluch-Shimon S, Senkus E, Curigliano G, Aapro MS, André F. et al. 5th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 5). Ann Oncol. 2020;31:1623-49
6. Dittmer J. Breast cancer stem cells: Features, key drivers and treatment options. Semin Cancer Biol. 2018;53:59-74
7. Yang F, Xu J, Tang L, Guan X. Breast cancer stem cell: the roles and therapeutic implications. Cell Mol Life Sci. 2017;74:951-66
8. Garcia-Mayea Y, Mir C, Masson F, Paciucci R, ME LL. Insights into new mechanisms and models of cancer stem cell multidrug resistance. Seminars in cancer biology. 2020;60:166-80
9. Zhou J, Chen Q, Zou Y, Chen H, Qi L, Chen Y. Stem Cells and Cellular Origins of Breast Cancer: Updates in the Rationale, Controversies, and Therapeutic Implications. Frontiers in oncology. 2019;9:820
10. Ma F, Li H, Wang H, Shi X, Fan Y, Ding X. et al. Enriched CD44(+)/CD24(-) population drives the aggressive phenotypes presented in triple-negative breast cancer (TNBC). Cancer letters. 2014;353:153-9
11. Li H, Ma F, Wang H, Lin C, Fan Y, Zhang X. et al. Stem cell marker aldehyde dehydrogenase 1 (ALDH1)-expressing cells are enriched in triple-negative breast cancer. The International journal of biological markers. 2013;28:e357-64
12. Wang D, Hu X, Liu C, Jia Y, Bai Y, Cai C. et al. Protein C receptor is a therapeutic stem cell target in a distinct group of breast cancers. Cell Res. 2019;29:832-45
13. Zhou L, Wang D, Sheng D, Xu J, Chen W, Qin Y. et al. NOTCH4 maintains quiescent mesenchymal-like breast cancer stem cells via transcriptionally activating SLUG and GAS1 in triple-negative breast cancer. Theranostics. 2020;10:2405-21
14. Bhat V, Allan AL, Raouf A. Role of the Microenvironment in Regulating Normal and Cancer Stem Cell Activity: Implications for Breast Cancer Progression and Therapy Response. Cancers. 2019 11
15. Bu W, Liu Z, Jiang W, Nagi C, Huang S, Edwards DP. et al. Mammary Precancerous Stem and Non-Stem Cells Evolve into Cancers of Distinct Subtypes. Cancer Res. 2019;79:61-71
16. Jiang H, Zhou C, Zhang Z, Wang Q, Wei H, Shi W. et al. Jagged1-Notch1-deployed tumor perivascular niche promotes breast cancer stem cell phenotype through Zeb1. Nature communications. 2020;11:5129
17. Zhang L, Qiang J, Yang X, Wang D, Rehman AU, He X. et al. IL1R2 Blockade Suppresses Breast Tumorigenesis and Progression by Impairing USP15-Dependent BMI1 Stability. Adv Sci (Weinh). 2020;7:1901728
18. Zhao L, Qiu T, Jiang D, Xu H, Zou L, Yang Q. et al. SGCE Promotes Breast Cancer Stem Cells by Stabilizing EGFR. Advanced science (Weinheim, Baden-Wurttemberg, Germany). 2020;7:1903700
19. Yu JM, Sun W, Wang ZH, Liang X, Hua F, Li K. et al. TRIB3 supports breast cancer stemness by suppressing FOXO1 degradation and enhancing SOX2 transcription. Nat Commun. 2019;10:5720
20. Huang P, Liao R, Chen X, Wu X, Li X, Wang Y. et al. Nuclear translocation of PLSCR1 activates STAT1 signaling in basal-like breast cancer. Theranostics. 2020;10:4644-58
21. Du L, Liu X, Ren Y, Li J, Li P, Jiao Q. et al. Loss of SIRT4 promotes the self-renewal of Breast Cancer Stem Cells. Theranostics. 2020;10:9458-76
22. Scioli MG, Storti G, D'Amico F, Gentile P, Fabbri G, Cervelli V. et al. The Role of Breast Cancer Stem Cells as a Prognostic Marker and a Target to Improve the Efficacy of Breast Cancer Therapy. Cancers. 2019 11
23. Nie L, Wei Y, Zhang F, Hsu YH, Chan LC, Xia W. et al. CDK2-mediated site-specific phosphorylation of EZH2 drives and maintains triple-negative breast cancer. Nat Commun. 2019;10:5114
24. Zhang P, Liu Y, Lian C, Cao X, Wang Y, Li X. et al. SH3RF3 promotes breast cancer stem-like properties via JNK activation and PTX3 upregulation. Nature communications. 2020;11:2487
25. Fiorillo M, Sotgia F, Lisanti MP. "Energetic" Cancer Stem Cells (e-CSCs): A New Hyper-Metabolic and Proliferative Tumor Cell Phenotype, Driven by Mitochondrial Energy. Frontiers in oncology. 2018;8:677
26. Talukdar S, Bhoopathi P, Emdad L, Das S, Sarkar D, Fisher PB. Dormancy and cancer stem cells: An enigma for cancer therapeutic targeting. Advances in cancer research. 2019;141:43-84
27. Ye S, Ding YF, Jia WH, Liu XL, Feng JY, Zhu Q. et al. SET Domain-Containing Protein 4 Epigenetically Controls Breast Cancer Stem Cell Quiescence. Cancer Res. 2019;79:4729-43
28. Liberti MV, Locasale JW. The Warburg Effect: How Does it Benefit Cancer Cells? Trends in biochemical sciences. 2016;41:211-8
29. Liu M, Wang Y, Yang C, Ruan Y, Bai C, Chu Q. et al. Inhibiting both proline biosynthesis and lipogenesis synergistically suppresses tumor growth. The Journal of experimental medicine. 2020 217
30. Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y. et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nature cell biology. 2019;21:498-510
31. Cui B, Luo Y, Tian P, Peng F, Lu J, Yang Y. et al. Stress-induced epinephrine enhances lactate dehydrogenase A and promotes breast cancer stem-like cells. J Clin Invest. 2019;129:1030-46
32. Mittal S, Brown NJ, Holen I. The breast tumor microenvironment: role in cancer development, progression and response to therapy. Expert review of molecular diagnostics. 2018;18:227-43
33. Nie Y, Chen J, Huang D, Yao Y, Chen J, Ding L. et al. Tumor-Associated Macrophages Promote Malignant Progression of Breast Phyllodes Tumors by Inducing Myofibroblast Differentiation. Cancer Res. 2017;77:3605-18
34. Liu X, Li J, Cadilha BL, Markota A, Voigt C, Huang Z. et al. Epithelial-type systemic breast carcinoma cells with a restricted mesenchymal transition are a major source of metastasis. Sci Adv. 2019;5:eaav4275
35. Yin T, Zhao ZB, Guo J, Wang T, Yang JB, Wang C. et al. Aurora A Inhibition Eliminates Myeloid Cell-Mediated Immunosuppression and Enhances the Efficacy of Anti-PD-L1 Therapy in Breast Cancer. Cancer Res. 2019;79:3431-44
36. Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G. et al. Pre-metastatic niches: organ-specific homes for metastases. Nature reviews Cancer. 2017;17:302-17
37. Gu Y, Liu Y, Fu L, Zhai L, Zhu J, Han Y. et al. Tumor-educated B cells selectively promote breast cancer lymph node metastasis by HSPA4-targeting IgG. Nat Med. 2019;25:312-22
38. Ishay-Ronen D, Diepenbruck M, Kalathur RKR, Sugiyama N, Tiede S, Ivanek R. et al. Gain Fat-Lose Metastasis: Converting Invasive Breast Cancer Cells into Adipocytes Inhibits Cancer Metastasis. Cancer Cell. 2019;35:17-32 e6
39. Lu JT, Tan CC, Wu XR, He R, Zhang X, Wang QS. et al. FOXF2 deficiency accelerates the visceral metastasis of basal-like breast cancer by unrestrictedly increasing TGF-β and miR-182-5p. Cell death and differentiation. 2020;27:2973-87
40. Yang L, Li N, Xue Z, Liu LR, Li J, Huang X. et al. Synergistic therapeutic effect of combined PDGFR and SGK1 inhibition in metastasis-initiating cells of breast cancer. Cell death and differentiation. 2020;27:2066-80
41. Yi Y, Chen D, Ao J, Zhang W, Yi J, Ren X. et al. Transcriptional suppression of AMPKα1 promotes breast cancer metastasis upon oncogene activation. Proceedings of the National Academy of Sciences of the United States of America. 2020;117:8013-21
42. Li X, Wang M, Gong T, Lei X, Hu T, Tian M. et al. A S100A14-CCL2/CXCL5 signaling axis drives breast cancer metastasis. Theranostics. 2020;10:5687-703
43. Zhang H, Zhang N, Liu Y, Su P, Liang Y, Li Y. et al. Epigenetic Regulation of NAMPT by NAMPT-AS Drives Metastatic Progression in Triple-Negative Breast Cancer. Cancer research. 2019;79:3347-59
44. Guo B, Wu S, Zhu X, Zhang L, Deng J, Li F. et al. Micropeptide CIP2A-BP encoded by LINC00665 inhibits triple-negative breast cancer progression. The EMBO journal. 2020;39:e102190
45. Wang Y, Wu S, Zhu X, Zhang L, Deng J, Li F. et al. LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. The Journal of experimental medicine. 2020 217
46. Nie Y, Huang H, Guo M, Chen J, Wu W, Li W. et al. Breast Phyllodes Tumors Recruit and Repolarize Tumor-Associated Macrophages via Secreting CCL5 to Promote Malignant Progression, Which Can Be Inhibited by CCR5 Inhibition Therapy. Clin Cancer Res. 2019;25:3873-86
47. Aktas BY, Guner G, Guven DC, Arslan C, Dizdar O. Exploiting DNA repair defects in breast cancer: from chemotherapy to immunotherapy. Expert review of anticancer therapy. 2019;19:589-601
48. Zhang L, Li DQ. MORC2 regulates DNA damage response through a PARP1-dependent pathway. Nucleic Acids Res. 2019;47:8502-20
49. Liu HY, Liu YY, Yang F, Zhang L, Zhang FL, Hu X. et al. Acetylation of MORC2 by NAT10 regulates cell-cycle checkpoint control and resistance to DNA-damaging chemotherapy and radiotherapy in breast cancer. Nucleic Acids Res. 2020;48:3638-56
50. Li Q, Qin T, Bi Z, Hong H, Ding L, Chen J. et al. Rac1 activates non-oxidative pentose phosphate pathway to induce chemoresistance of breast cancer. Nature communications. 2020;11:1456
51. Zhang Z, Qiu N, Yin J, Zhang J, Liu H, Guo W. et al. SRGN crosstalks with YAP to maintain chemoresistance and stemness in breast cancer cells by modulating HDAC2 expression. Theranostics. 2020;10:4290-307
52. Dong X, Yang Y, Zhou Y, Bi X, Zhao N, Zhang Z. et al. Glutathione S-transferases P1 protects breast cancer cell from adriamycin-induced cell death through promoting autophagy. Cell death and differentiation. 2019;26:2086-99
53. Liu XY, Jiang W, Ma D, Ge LP, Yang YS, Gou ZC. et al. SYTL4 downregulates microtubule stability and confers paclitaxel resistance in triple-negative breast cancer. Theranostics. 2020;10:10940-56
54. Xu S, Wang P, Zhang J, Wu H, Sui S, Zhang J. et al. Ai-lncRNA EGOT enhancing autophagy sensitizes paclitaxel cytotoxicity via upregulation of ITPR1 expression by RNA-RNA and RNA-protein interactions in human cancer. Mol Cancer. 2019;18:89
55. Lian B, Pei YC, Jiang YZ, Xue MZ, Li DQ, Li XG. et al. Truncated HDAC9 identified by integrated genome-wide screen as the key modulator for paclitaxel resistance in triple-negative breast cancer. Theranostics. 2020;10:11092-109
56. Yang F, Xie HY, Yang LF, Zhang L, Zhang FL, Liu HY. et al. Stabilization of MORC2 by estrogen and antiestrogens through GPER1- PRKACA-CMA pathway contributes to estrogen-induced proliferation and endocrine resistance of breast cancer cells. Autophagy. 2020;16:1061-76
57. Lin X, Dinglin X, Cao S, Zheng S, Wu C, Chen W. et al. Enhancer-Driven lncRNA BDNF-AS Induces Endocrine Resistance and Malignant Progression of Breast Cancer through the RNH1/TRIM21/mTOR Cascade. Cell reports. 2020;31:107753
58. Shi Q, Li Y, Li S, Jin L, Lai H, Wu Y. et al. LncRNA DILA1 inhibits Cyclin D1 degradation and contributes to tamoxifen resistance in breast cancer. Nature communications. 2020;11:5513
59. Xu B, Li Q, Chen N, Zhu C, Meng Q, Ayyanathan K. et al. The LIM protein Ajuba recruits DBC1 and CBP/p300 to acetylate ERα and enhances ERα target gene expression in breast cancer cells. Nucleic acids research. 2019;47:2322-35
60. Liu SS, Li Y, Zhang H, Zhang D, Zhang XB, Wang X. et al. The ERα-miR-575-p27 feedback loop regulates tamoxifen sensitivity in ER-positive Breast Cancer. Theranostics. 2020;10:10729-42
61. Jia X, Li C, Li L, Liu X, Zhou L, Zhang W. et al. Neddylation Inactivation Facilitates FOXO3a Nuclear Export to Suppress Estrogen Receptor Transcription and Improve Fulvestrant Sensitivity. Clin Cancer Res. 2019;25:3658-72
62. Chen XS, Wang KS, Guo W, Li LY, Yu P, Sun XY. et al. UCH-L1-mediated Down-regulation of Estrogen Receptor α Contributes to Insensitivity to Endocrine Therapy for Breast Cancer. Theranostics. 2020;10:1833-48
63. Dong H, Hu J, Zou K, Ye M, Chen Y, Wu C. et al. Activation of LncRNA TINCR by H3K27 acetylation promotes Trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast Cancer. Molecular cancer. 2019;18:3
64. Jin X, Ge LP, Li DQ, Shao ZM, Di GH, Xu XE. et al. LncRNA TROJAN promotes proliferation and resistance to CDK4/6 inhibitor via CDK2 transcriptional activation in ER+ breast cancer. Molecular cancer. 2020;19:87
65. Luo ML, Zheng F, Chen W, Liang ZM, Chandramouly G, Tan J. et al. Inactivation of the Prolyl Isomerase Pin1 Sensitizes BRCA1-Proficient Breast Cancer to PARP Inhibition. Cancer research. 2020;80:3033-45
66. Basu A, Ramamoorthi G, Jia Y, Faughn J, Wiener D, Awshah S. et al. Immunotherapy in breast cancer: Current status and future directions. Advances in cancer research. 2019;143:295-349
67. Li Q, Wang Y, Jia W, Deng H, Li G, Deng W. et al. Low-Dose Anti-Angiogenic Therapy Sensitizes Breast Cancer to PD-1 Blockade. Clinical cancer research: an official journal of the American Association for Cancer Research. 2020;26:1712-24
68. Qin G, Wang X, Ye S, Li Y, Chen M, Wang S. et al. NPM1 upregulates the transcription of PD-L1 and suppresses T cell activity in triple-negative breast cancer. Nature communications. 2020;11:1669
69. Li ZL, Zhang HL, Huang Y, Huang JH, Sun P, Zhou NN. et al. Autophagy deficiency promotes triple-negative breast cancer resistance to T cell-mediated cytotoxicity by blocking tenascin-C degradation. Nature communications. 2020;11:3806
70. Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ. et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Science translational medicine. 2015;7:302ra133
71. Zhang X, Zhao W, Wei W, You Z, Ou X, Sun M. et al. Parallel Analyses of Somatic Mutations in Plasma Circulating Tumor DNA (ctDNA) and Matched Tumor Tissues in Early-Stage Breast Cancer. Clin Cancer Res. 2019;25:6546-53
72. Zhang J, Chen J, Wo D, Yan H, Liu P, Ma E. et al. LRP6 Ectodomain Prevents SDF-1/CXCR4-Induced Breast Cancer Metastasis to Lung. Clin Cancer Res. 2019;25:4832-45
73. Gong Y, Ji P, Yang YS, Xie S, Yu TJ, Xiao Y. et al. Metabolic-Pathway-Based Subtyping of Triple-Negative Breast Cancer Reveals Potential Therapeutic Targets. Cell Metab. 2021;33:51-64 e9
74. Jiang YZ, Ma D, Suo C, Shi J, Xue M, Hu X. et al. Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell. 2019;35:428-40 e5
75. Jiang YZ, Liu Y, Xiao Y, Hu X, Jiang L, Zuo WJ. et al. Molecular subtyping and genomic profiling expand precision medicine in refractory metastatic triple-negative breast cancer: the FUTURE trial. Cell Res. 2021;31:178-86
76. Xiao Y, Ma D, Zhao S, Suo C, Shi J, Xue MZ. et al. Multi-Omics Profiling Reveals Distinct Microenvironment Characterization and Suggests Immune Escape Mechanisms of Triple-Negative Breast Cancer. Clin Cancer Res. 2019;25:5002-14
77. Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L. et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Molecular cancer. 2019;18:46
78. Liu LM, Sun WZ, Fan XZ, Xu YL, Cheng MB, Zhang Y. Methylation of C/EBPα by PRMT1 Inhibits Its Tumor-Suppressive Function in Breast Cancer. Cancer research. 2019;79:2865-77
79. Peng BL, Li WJ, Ding JC, He YH, Ran T, Xie BL. et al. A hypermethylation strategy utilized by enhancer-bound CARM1 to promote estrogen receptor α-dependent transcriptional activation and breast carcinogenesis. Theranostics. 2020;10:3451-73
80. Zhang Z, Du J, Wang S, Shao L, Jin K, Li F. et al. OTUB2 Promotes Cancer Metastasis via Hippo-Independent Activation of YAP and TAZ. Mol Cell. 2019;73:7-21 e7
81. Yang YL, Zhang Y, Li DD, Zhang FL, Liu HY, Liao XH. et al. RNF144A functions as a tumor suppressor in breast cancer through ubiquitin ligase activity-dependent regulation of stability and oncogenic functions of HSPA2. Cell Death Differ. 2020;27:1105-18
82. Hou Y, Liu W, Yi X, Yang Y, Su D, Huang W. et al. PHF20L1 as a H3K27me2 reader coordinates with transcriptional repressors to promote breast tumorigenesis. Science advances. 2020;6:eaaz0356
83. Yang T, Ren C, Lu C, Qiao P, Han X, Wang L. et al. Phosphorylation of HSF1 by PIM2 Induces PD-L1 Expression and Promotes Tumor Growth in Breast Cancer. Cancer research. 2019;79:5233-44
84. Zhang Q, Cao C, Gong W, Bao K, Wang Q, Wang Y. et al. A feedforward circuit shaped by ECT2 and USP7 contributes to breast carcinogenesis. Theranostics. 2020;10:10769-90
85. Cheng L, Yuan B, Ying S, Niu C, Mai H, Guan X. et al. PES1 is a critical component of telomerase assembly and regulates cellular senescence. Sci Adv. 2019;5:eaav1090
86. Chu W, Zhang X, Qi L, Fu Y, Wang P, Zhao W. et al. The EZH2-PHACTR2-AS1-Ribosome Axis induces Genomic Instability and Promotes Growth and Metastasis in Breast Cancer. Cancer research. 2020;80:2737-50
87. Liang Y, Song X, Li Y, Chen B, Zhao W, Wang L. et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Molecular cancer. 2020;19:85
88. Liang G, Ling Y, Mehrpour M, Saw PE, Liu Z, Tan W. et al. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Molecular cancer. 2020;19:65
89. Miao K, Lei JH, Valecha MV, Zhang A, Xu J, Wang L. et al. NOTCH1 activation compensates BRCA1 deficiency and promotes triple-negative breast cancer formation. Nature communications. 2020;11:3256
90. Jin X, Xu XE, Jiang YZ, Liu YR, Sun W, Guo YJ. et al. The endogenous retrovirus-derived long noncoding RNA TROJAN promotes triple-negative breast cancer progression via ZMYND8 degradation. Sci Adv. 2019;5:eaat9820
91. Zheng X, Huang M, Xing L, Yang R, Wang X, Jiang R. et al. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Molecular cancer. 2020;19:73
92. Qin Y, Chen W, Jiang G, Zhou L, Yang X, Li H. et al. Interfering MSN-NONO complex-activated CREB signaling serves as a therapeutic strategy for triple-negative breast cancer. Sci Adv. 2020;6:eaaw9960
93. Lin Q, He Y, Wang X, Zhang Y, Hu M, Guo W. et al. Targeting Pyruvate Carboxylase by a Small Molecule Suppresses Breast Cancer Progression. Advanced science (Weinheim, Baden-Wurttemberg, Germany). 2020;7:1903483
94. Hou ZJ, Luo X, Zhang W, Peng F, Cui B, Wu SJ. et al. Flubendazole, FDA-approved anthelmintic, targets breast cancer stem-like cells. Oncotarget. 2015;6:6326-40
95. Zhen Y, Zhao R, Wang M, Jiang X, Gao F, Fu L. et al. Flubendazole elicits anti-cancer effects via targeting EVA1A-modulated autophagy and apoptosis in Triple-negative Breast Cancer. Theranostics. 2020;10:8080-97
96. Liu SS, Qi J, Teng ZD, Tian FT, Lv XX, Li K. et al. Resistomycin attenuates triple-negative breast cancer progression by inhibiting E3 ligase Pellino-1 and inducing SNAIL/SLUG degradation. Signal transduction and targeted therapy. 2020;5:133
97. Hu MH, Wu TY, Huang Q, Jin G. New substituted quinoxalines inhibit triple-negative breast cancer by specifically downregulating the c-MYC transcription. Nucleic Acids Res. 2019;47:10529-42
98. Zou G, Zhang X, Wang L, Li X, Xie T, Zhao J. et al. Herb-sourced emodin inhibits angiogenesis of breast cancer by targeting VEGFA transcription. Theranostics. 2020;10:6839-53
Author contact
![]() Corresponding authors: Ceshi Chen, Ph.D, E-mail: chenckiz.ac.cn; Tel: +86-871-65181944; Fax: +86-871-65181945; Lin Xu, M.D, E-mail: xulin_83com; Tel: +86-25-83283571; Fax: +86-25-83641062.
Corresponding authors: Ceshi Chen, Ph.D, E-mail: chenckiz.ac.cn; Tel: +86-871-65181944; Fax: +86-871-65181945; Lin Xu, M.D, E-mail: xulin_83com; Tel: +86-25-83283571; Fax: +86-25-83641062.

 Global reach, higher impact
Global reach, higher impact