10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(13):3554-3572. doi:10.7150/ijbs.62168 This issue Cite
Research Paper
Identification and Characterization of Alcohol-related Hepatocellular Carcinoma Prognostic Subtypes based on an Integrative N6-methyladenosine methylation Model
1. Department of Hepatobiliary Surgery II, Guangdong Provincial Research Center for Artificial Organ and Tissue Engineering, Guangzhou Clinical Research and Transformation Center for Artificial Liver, Institute of Regenerative Medicine, ZhuJiang Hospital, Southern Medical University, Guangzhou, Guangdong Province, China.
2. State Key Laboratory of Organ Failure Research, Southern Medical University, Guangzhou, China.
#These authors contributed equally to this work.
Abstract
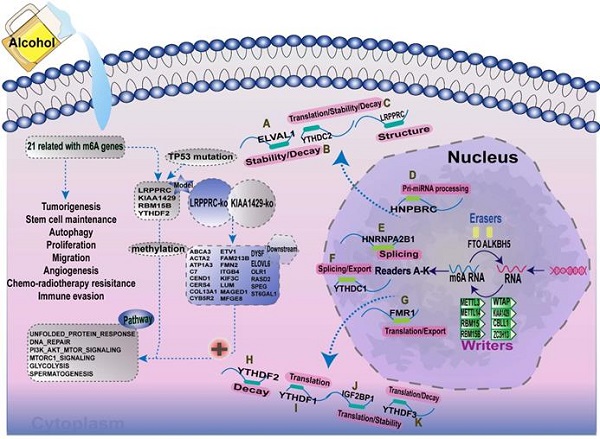
Background: Alcohol consumption increases the risk of hepatocellular carcinoma (HCC), and associated with a high mortality rate and poor prognosis. N6-methyladenosine (m6A) methylations play key roles in tumorigenesis and progression. However, our current knowledge about m6A in alcohol-related HCC (A-HCC) remains elucidated. Herein, the authors construct an integrative m6A model based on A-HCC subtyping and mechanism exploration workflow.
Methods: Based on the m6A expressions of A-HCC and in vivo experiment, different prognosis risk A-HCC subtypes are identified. Meanwhile, multiple interdependent indicators of prognosis including patient survival rate, clinical pathological prognosis and immunotherapy sensitivity.
Results: The m6A model includes LRPPRC, YTHDF2, KIAA14219, and RBM15B, classified A-HCC patients into high/low-risk subtypes. The high-risk subtype compared to the low-risk subtype showed phenotypic malignancy, poor prognosis, immunosuppression, and activation of tumorigenesis and proliferation-related pathways, including the E2F target, DNA repair, and mTORC1 signalling pathways. The expression of Immunosuppressive cytokines DNMT1/EZH2 was up-regulated in A-HCC patients, and teniposide may be a potential therapeutic drug for A-HCC.
Conclusion: Our model redefined A-HCC prognosis risk, identified potential m6As linking tumour progress and immune regulations and selected possible therapy target, thus promoting understanding and clinical applications about A-HCC.
Keywords: Hepatocellular carcinoma, N6-methyladenosine, tumour immune microenvironment, treatment sensitivity, teniposide

 Global reach, higher impact
Global reach, higher impact