10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(15):5713-5723. doi:10.7150/ijbs.77141 This issue Cite
Review
Functional and Structural Diversity of Insect Glutathione S-transferases in Xenobiotic Adaptation
1. Department of Entomology, Pennsylvania State University, University Park, PA 16802, USA.
2. Huck Institutes of the Life Sciences, Pennsylvania State University, University Park, PA 16802, USA.
Received 2022-7-15; Accepted 2022-8-29; Published 2022-9-11
Abstract
As a superfamily of multifunctional enzymes that is mainly associated with xenobiotic adaptation, glutathione S-transferases (GSTs) facilitate insects' survival under chemical stresses in their environment. GSTs confer xenobiotic adaptation through direct metabolism or sequestration of xenobiotics, and/or indirectly by providing protection against oxidative stress induced by xenobiotic exposure. In this article, a comprehensive overview of current understanding on the versatile functions of insect GSTs in detoxifying chemical compounds is presented. The diverse structures of different classes of insect GSTs, specifically the spatial localization and composition of their amino acid residues constituted in their active sites are also summarized. Recent availability of whole genome sequences of numerous insect species, accompanied by RNA interference, X-ray crystallography, enzyme kinetics and site-directed mutagenesis techniques have significantly enhanced our understanding of functional and structural diversity of insect GSTs.
Keywords: enzyme, metabolic detoxification, host adaptation, oxidative stress, pesticide resistance
Introduction
Insects constitute the largest class of animals encompassing about 53% of all living species on our planet [1]. Many of these species (about 45%) are herbivores by partly or completely feeding on plants and represent a significant proportion of pests or pollinators for economically important crops. Annually, the economic association of these herbivores with food production in the U.S. exceeds $50 billion [1, 2]. The arms race between plants and insect herbivores have driven their coevolution for hundreds of millions of years. To defend against insect herbivores, plants produce a wide range of chemical compounds, such as terpenoids, alkaloids, anthocyanins, glucosinolates, phenols, quinones, plant protease inhibitors (PIs), and herbivore-induced plant volatiles (HIPVs). These chemicals either directly reduce herbivores fitness or indirectly attract herbivores' natural enemies and enhance the effectiveness of their natural enemies [3, 4]. In response, herbivores have simultaneously developed countermeasures against plant defense compounds [5]. Such adaptive capability has been proposed to be co-opted by herbivore arthropod pests for pesticide resistance when they are exposed to the pressure of recently introduced synthetic pesticides [6-8]. The similarities in modes of action between various naturally occurring chemical substances released by plants and synthetic pesticides further supports the possible linkage between host plant adaptation and currently prevailed pesticide resistance [9]. In fact, more than 50% of all agrochemicals are natural products or derived from natural products [10-12].
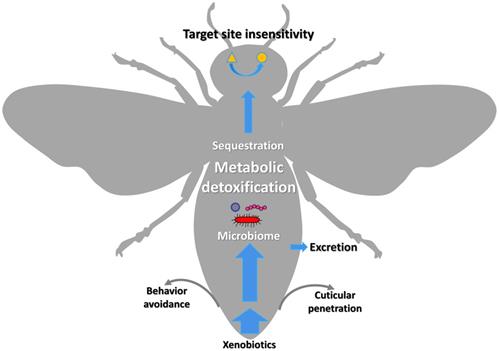
The xenobiotic adaptation in arthropods evolves through multiple mechanisms (Figure 1) [13, 14], including reduced penetration through the cuticle, behavioral avoidance [15, 16], microbiome-mediated detoxification [17-20], enhanced metabolic detoxification [21-25], enhanced sequestration or excretion [13, 19, 26, 27], and target site insensitivity [28-32]. Among them, enhanced metabolic detoxification and target site insensitivity are the most common mechanisms [5, 33-35].
Graphic representation of the xenobiotic adaptations in arthropods that have evolved through different mechanisms. The thickness of the blue arrows represents the concentration of xenobiotics.
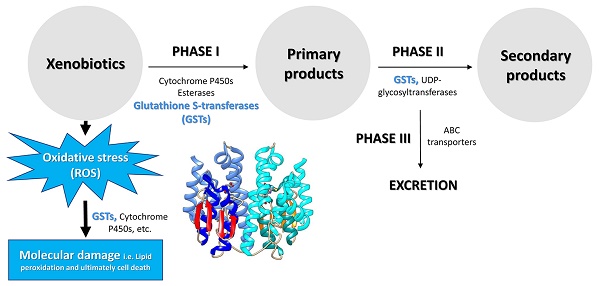
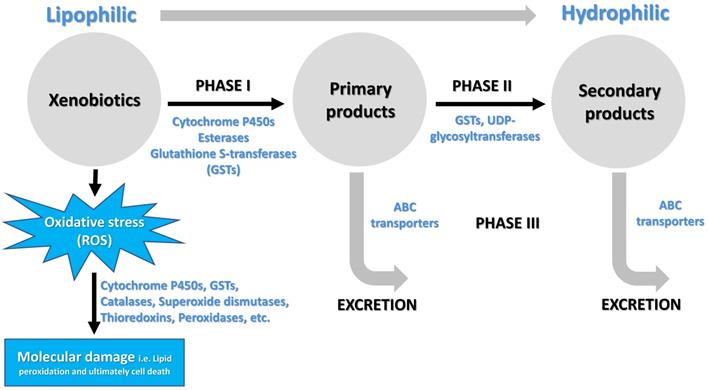
There are several categories of enzymes involved in the metabolism of lipophilic xenobiotics and their conversions into less toxic compounds exhibiting increased hydrophilicity (Figure 2). The major enzyme superfamilies comprise cytochrome P450 monooxygenases (P450s), glutathione S-transferases (GSTs), carboxylesterases (COEs), ATP-binding cassette (ABC) transporters, and UDP-glycosyltransferases (UGTs) [5, 26, 36-39] (Figure 2). In general, three phases of metabolic detoxification of xenobiotics have been often described in the literature. Phase I detoxification includes oxidation, reduction, and hydrolysis of lipophilic substances carried out by a variety of enzymes. Phase II reactions involve conjugation of hydrophilic compounds (i.e. glutathione) to xenobiotics and/or phase I products to produce more hydrophilic products. In Phase III, products of phases I and/or II are excreted from cells by multidrug resistance proteins and other ABC transporters [37]. Among metabolic detoxification enzymes, GST is a family of multifunctional enzymes that are ubiquitously present in eukaryotes and prokaryotes, playing an important role in the detoxification of numerous endogenous and exogenous compounds. As phase II enzymes, GSTs detoxify chemical compounds through catalyzing nucleophilic attack by the thiol group in reduced glutathione (GSH) on a wide range of electrophilic substrates [37, 40, 41]. These substrates can be plant allelochemicals, pesticides, environmental pollutants, or byproducts of oxidative stress [40, 42]. GSTs are also involved in the phase I detoxification process such as dehydrochlorination of 1,1,1-Trichloro-2,2-bis(p-chlorophenyl) ethane (DDT) to less toxic 1,l-Dichloro-2,2-bis(p-chlorophenyl) ethylene (DDE) [22, 43, 44]. In addition, GSTs may participate in the passive non-catalytic binding of substrates and sequestration, which prevents the binding of xenobiotics to their target proteins [45-48].
Besides triggering a sequence of events that cause toxic outcomes, exposure to xenobiotics leads to induced oxidative stress, generating an over production of reactive oxygen species (ROS) [49] and consequently triggering oxidative damage to macromolecules such as proteins, lipids, and nucleic acids [50, 51]. To cope with oxidative stress, arthropods evolve antioxidant enzymes for removing excess ROS to maintain intracellular redox homeostasis and avoid oxidative damage. These antioxidant enzymes include GSTs, catalases, superoxide dismutases, thioredoxins, glutathione peroxidases, glutaredoxins and thioredoxin peroxidases [52, 53] (Figure 2). Insect GSTs not only are involved in xenobiotic conjugation but also play roles in protection against oxidative stress caused by exposure to pesticides [46], plant allelochemicals [54], as well as various other abiotic factors [55, 56]. Recent reviews had summarized functions of insect GSTs in insecticide resistance [22, 57]. Therefore, the current review focuses on the structural and functional divergence of GST enzymes in arthropods and their potential roles in xenobiotic adaptation.
Classification of GSTs
In eukaryotes and aerobic prokaryotes, GSTs are grouped into at least four major protein families: cytosolic GSTs, mitochondrial GSTs, microsomal GSTs, and bacterial Fosfomycin-resistance proteins [40, 42, 58-59]. Mitochondrial GSTs are known as the kappa class in mammals and are mostly found in the mitochondrial matrix [60] and peroxisomes [61]. Research has indicated that mitochondrial GSTs in humans play important roles in the detoxification of lipid peroxide and lipid metabolism [61]. Microsomal GSTs belong to the MAPEG family (membrane‐associated proteins in eicosanoid and glutathione metabolism), which play a significant role in the reduction of lipid peroxidation and xenobiotic detoxification [62, 63]. In contrast to mitochondrial and microsomal GSTs, cytosolic GSTs are present in the cytoplasm and are soluble [44]. Both microsomal and cytosolic GSTs are found in arthropod species; however, the gene numbers in microsomal GSTs are fewer than the cytosolic GSTs (Table 1) [62, 64, 65]. Moreover, cytosolic GSTs, which are typically 200-250 amino acids in length, form homo- or hetero-dimers, whereas microsomal GSTs are smaller (nearly 150 amino acids) and form trimers [62, 66]. Arthropod cytosolic GSTs are classified into several classes according to the sequence similarities and structural properties: Delta, Epsilon, Omega, Sigma, Theta, Zeta, and unclassified classes (Table 1). Among these classes, Omega, Sigma, Theta, and Zeta classes are identified in most metazoans [67] and some aerobic prokaryotes [58, 68]. Epsilon and Delta classes are insect-specific [62, 69]. These two classes of cytosolic GSTs have undergone species-specific gene expansion to a great extent [41, 64, 65]. It was hypothesized that such expansion might have occurred during adaptation to environmental selection pressure. This expansion or duplication of genes resulted in sequence variations that expanded substrate functionality and/or responses to environmental stresses [62, 70, 71].
Schematic illustrating the process of xenobiotic metabolism, which encompasses three phases I, II, III (Adopted from [137]) as well as xenobiotic induced oxidative stress and molecular damage.
Arthropod cytosolic GSTs are mainly involved in xenobiotic adaptation. With genomes of arthropod species available, gene number variation in each class of cytosolic GSTs has been observed in different species (Table 1). It has been hypothesized that a smaller number of cytosolic GST genes in the European honey bee (Aphis mellifera) than in other insect species might be associated with pesticide sensitivity and reduction in vitality [72]. Besides, predator Orius laevigatus, monophagous or oligophagous agricultural pests Nilaparvata lugens and Diaphorina citri possess a low number of Delta, Epsilon, and total GSTs in their genomes (Table 1). The deficit in the number of GST genes is likely due to the low degree of exposure to xenobiotics in their natural environment.
General structure of cytosolic GSTs
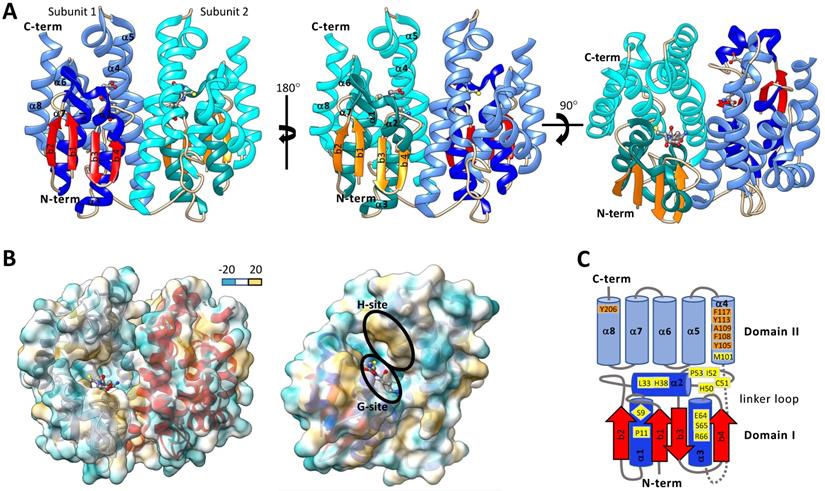
Typically, cytosolic GSTs are hetero- or homo-dimeric proteins and are about 23-30 kDa per monomer. It has been proposed that heterodimer formation is restricted to both subunits being from the same class due to dimer interface compatibility interactions. Crystallographic evidence shows that homodimer subunits are related by a two-fold symmetry axis (Figure 3A&B) [73]. Each monomer of a cytosolic GST is composed of an N-terminal domain (domain I) and C-terminal domain (domain II). N-terminal domain has β strands and α helices, and the C-terminal domain consists of helices [42, 62, 74]. Domain I exhibits the structurally conserved thioredoxin-like fold motif βαβαββα (Figure 3A&C) [44, 68, 75]. The N-terminal domain I is connected to the C-terminal domain II by a linker loop region consisting of around 10 amino acids [42, 62, 76]. The C-term domain II consists of 4-8 helices depending on the GST class [42, 62, 73, 76]. One of the striking features of GST is that each subunit has two ligand-binding sites - “G” site and “H” site (Figure 3), which together constitute the catalytic active site [62, 77]. The G-site is more hydrophilic and exhibits a higher degree of sequence conservation within GST families than the H-site [42]. The G-site is predominantly contained in the N-terminal domain and binds GSH and primes the thiol sulfur for nucleophilic attack on an electrophilic substrate [77-79]. In contrast, the hydrophobic H-site is predominantly contained in the C-terminal domain adjacent to the G-site and binds electrophilic substrates [62, 80]. The amino acid residues that make up the H-site are involved in recognizing and binding various exogenous and endogenous compounds and positioning their electrophilic centers for attack by the nucleophilic GSH.
G-site
The type and position of amino acids in the active site of GSTs (G-site and H-site) play important roles in substrate binding affinity and catalytic function [74]. It is thus important to make a comparison among different GSTs to understand their evolution and functions in the detoxification of diverse chemical substrates. With the aid of X-ray crystallography and site-directed mutagenesis techniques, the roles of GST active site amino acid residues were identified and evaluated [62, 81]. In Anopheles dirus, a delta GST GSTD3-3 (PDB: 1JLV), G-site residues Ser-9, Pro-11, Leu-33, His-38, His-50, Cys-51, Ile-52, Pro-53, Glu-64, Ser-65, Arg-66, and Met-101 are within a 4.0 Å distance cutoff of GSH (Figure 3C) [82, 83]. Among them, the Ser-65 residue was generally conserved across all GST classes. Ser-65 forms a hydrogen bond with the GSH γ-glutamyl carboxylate [80, 83]. Additionally, Ile-52 and Glu-64 were generally maintained as either hydrophobic or polar residues across GST classes [82]. The Ile-52 backbone amide forms a hydrogen bond with the backbone carbonyl of the GSH cysteinyl group and Glu-64 forms a salt-bridge with the amino group of γ-glutamyl moiety of GSH. In delta and epsilon GSTs, His-38 is maintained in most cases as a polar or charged residue and His-50 is conserved as part of an NPQHTVPTL motif. His-38 and His-50 are located within polar interaction distance of the glycyl carboxylate moiety of GSH [80, 84, 85]. Ser-9 is conserved in epsilon, delta, theta, and unclassified GSTs and works to stabilize the GSH thiolate through a hydrogen bonding interaction [42, 73, 80, 83, 84, 86]. In a zeta class GST of Homo sapiens, the GSH thiolate is stabilized by interaction with Cys-16, Ser-15, Gln-111, and Ser-14 [73]. In omega GSTs, BmGST-O Cys-38 is located adjacent to the GSH thiolate and dmGST-S1 Tyr-54 plays a major role in stabilizing the GSH thiolate [76, 87]. The remaining amino acids that make up the core of the G-site are more variable across GSTs but are thought to aid in the positioning of GSH in the G-Site [84].
Structures of representative insect cytosolic GSTs. A. Ribbon diagram of Drosophila melanogaster dmGSTD1 (PDB: 3MAK). In subunit 1, the N-terminal domain I helices are shown in dark blue, and β-strands are shown in red, and the C-terminal domain II helices are shown in light blue. In subunit 2 the domain I helices are dark cyan β-strands are orange, and the domain II helices are light cyan. Glutathione is colored by the element and is shown in ball and stick format. B. Dimer (left) and monomer (right) ribbon diagrams of dmGSTD1 (PDB: 3MAK) overlayed with lipophilic surface representation. C. Secondary structure map of Anopheles dirus GSTD3-3 (PDB: 1JLV). Domain I helices are shown in dark blue and beta strands are shown in red. Domain II helices are shown in light blue. Loop regions for both domains I and II are shown in grey. The link region loop is dashed. Ribbon and surface diagrams were generated with UCSF ChimeraX.
GST gene number in diverse species across six insect orders
| Order | Name | Type | Delta | Epsilon | Omega | Sigma | Theta | Zeta | Unclassified | Microsomal | Total | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coleoptera | Leptinotarsa decemlineata | Pest (Oligophagous) | 3 | 10 | 5 | 4 | 4 | 1 | 2 | 1 | 30 | [66] |
| Tribolium castenaum | Pest (Polyphagous) | 3 | 19 | 3 | 7 | 1 | 1 | 2 | 5 | 41 | [64] | |
| Diptera | Aedes aegypti | Pest (Oligophagous) | 8 | 8 | 1 | 1 | 4 | 1 | 3 | - | 26 | [138] |
| Anopheles gambiae | Pest (Sanguivorous, Oligophagous) | 12 | 8 | 1 | 1 | 2 | 1 | 3 | 3 | 31 | [72] | |
| Bactrocera dorsalis | Pest (Polyphagous) | 4 | 8 | 2 | 0 | 1 | 1 | 1 | - | 17 | [139] | |
| Chironomus riparius | Pest (Sanguivorous, Oligophagous) | 3 | 1 | 1 | 4 | 1 | 1 | 2 | - | 13 | [140] | |
| Culex quinquefasciatus | Pest (Sanguivorous, Oligophagous) | 14 | 9 | 1 | 1 | 6 | 0 | 4 | 5 | 40 | [141] | |
| Drosophila melanogaster | Pest (Polyphagous) | 11 | 14 | 5 | 1 | 4 | 2 | 0 | 1 | 38 | [72, 149] | |
| Hemiptera | Bemisia tabaci | Pest (Polyphagous) | 14 | 0 | 1 | 6 | 0 | 2 | - | 2 | 25 | [142] |
| Diaphorina citri | Pest (Oligophagous) | 2 | 2 | 0 | 3 | 0 | 0 | 1 | 2 | 11 | [142] | |
| Myzus persicae | Pest (Polyphagous) | 8 | 0 | 0 | 8 | 2 | 0 | 0 | 2 | 21 | [143] | |
| Nilaparvata lugens | Pest (Monophagous) | 2 | 1 | 1 | 3 | 1 | 1 | 0 | 2 | 11 | [144] | |
| Orius laevigatus | Predator (Polyphagous) | 1 | 0 | 2 | 16 | 1 | 1 | 0 | 3 | 24 | [145] | |
| Homoptera | Acyrthosiphon pisum | Pest (Oligophagous) | 10 | 0 | 2 | 6 | 2 | 0 | 0 | 2 | 22 | [144] |
| Hymenoptera | Apis mellifera | Pollinator (polyphagous) | 1 | 0 | 1 | 4 | 1 | 1 | 0 | 2 | 10 | [72] |
| Nasonia vitripennis | Parasitoid (Monophagous) | 5 | 0 | 2 | 8 | 3 | 1 | 0 | - | 19 | [146] | |
| Lepidoptera | Bombyx mori | Economic (Monophagous) | 4 | 8 | 4 | 2 | 1 | 2 | 2 | - | 23 | [147] |
| Plutella xylostella | Pest (Oligophagous) | 5 | 5 | 5 | 2 | 1 | 2 | 2 | - | 22 | [65] | |
| Spodoptera litura | Pest (Polyphagous) | 5 | 21 | 3 | 7 | 1 | 5 | 3 | 2 | 47 | [148] |
-: There is no known gene in these classes.
H-site
In the GST H-site, the amino acids that contribute to the binding of multiple substrates ultimately facilitate the tolerance that an organism exhibits in a specific stress environment. Amino acid mutations in the H-site can significantly alter the catalytical activity of GST enzymes towards their substrates [88, 89]. However, the sequence variability in GST active sites across species and enzyme families result in differing enzyme activities for various substrates [58]. In contrast to the G-site that binds GSH across GST classes, the H-sites that bind various substrates have distinct variations in amino acid sequence and structural conformation [90]. While the G-site is more hydrophilic in nature compared to the H-site, the extent of hydrophobicity of the H-site varies across GST classes and amongst individual GSTs [77, 78, 91].
In general, hydrophilic amino acids contribute to the formation of a hydrophobic pocket in the H-site adjacent to the GSH-binding site (Figure 3B&C) [80, 85]. In Anopheles gambiae, residues in the H-site of AgGSTe2 were presumptively responsible for DDT binding and they were mostly hydrophobic residues [84]. In Plutella xylostella, the amino acids Phe-9, Pro-10, Ile-11, Leu-14, Gly-49, Pro-52, Ala-100, and Tyr-107 are the putative H-site residues in a sigma class GST, PxGSTσ [77]. Site-directed mutagenesis and inhibition assays revealed that Phe-9 is potentially an important residue for the binding of the inhibitor S-hexyl glutathione (GTX) [77]. In Blattella germanica, Tyr-107, Tyr-115, Phe-119, and Phe-206 constitute the H-site of BgGSTD1. Purified BgGSTD1 had the highest cumene peroxidase activity among insect GSTs reported at that time that played a vital role in defending against oxidative stress [92]. Studies have shown that the H-sites of different classes of GSTs are dissimilar. Diverse H-sites allow for binding and catalytic activity towards a wider range of xenobiotic substrates [93]. Despite lifetime exposure to a wide variety of toxic chemicals, the presence of multiple GST classes with diverse substrate specificities facilitates an organism adaptation to adverse environments.
Functions of insect GSTs in host plant adaptation and pesticide resistance
Many studies have found that plant allelochemicals are inducers of phase II detoxification enzymes in herbivorous arthropods [5-7]. In Choristoneura fumiferana, the expression of CfGST was induced by balsam fir foliage and other multiple stresses suggesting its potential role in xenobiotic detoxification [94]. The isothiocyanates produced from the breakdown of glucosinolates by the action of the enzyme myrosinase [95] are highly electrophilic, a property of a compound that makes it readily available for the nucleophilic GSH when in the presence of GST [96]. Gonzalez et al. reported that the expression of GSTD2 in Drosophila melanogaster was significantly higher in the taste organs (labellum and forelegs) when exposed to an isothiocyanate, insecticidal compounds naturally present in cruciferous plants [91]. In addition, the mechanism of detoxification by GSTD2 was revealed via its strong affinity towards isothiocyanate and catalysis of the conjugation between GSH and isothiocyanate. Zou and others showed that glucosinolate and xanthotoxin present in Brassica juncea stimulated the expression of GSTE1 in the midgut of Spodoptera littoralis larvae after feeding. The conjugation activity towards these allelochemicals was reduced when suppressing GSTE1 gene expression via RNA interference (RNAi), suggesting a role for GSTE1 in host plant adaptation [97]. In the Hessian fly, Mayetiola destructor, feeding on wheat varieties led to increased production of deterrent allelochemicals and the consequent upregulation of delta class GST genes [98]. The enhanced expression of MdesGST-1 (Delta group) in the midgut and fat body of Hessian fly larvae might explain its involvement in the detoxification of plant defense compounds such as flavonoids and scavenging endogenous ROS. Indeed, based on evidence from GST activity and RNAi studies, three GSTs are thought to have contributed to the adaptation of N. lugens to the host rice plant allelochemical (gramine) [99]. Recently, Ma et al. identified two Lymantria dispar GST genes, LdGSTe4 and LdGSTo1 induced by host poplar allelochemicals. After silencing these two GST genes individually, the adaptation of L. dispar to host poplar allelochemicals was depleted [100].
Plant volatile compounds play roles in host selection by insects. For example, herbivore-induced plant volatile compounds could serve as repellents of some insects and reduce their activities, which is termed allelochemical nonpreference [4]. Even for the adapted herbivore species, these volatile compounds can cause direct physiological damage to herbivores due to their neurotoxic properties at high concentrations [101, 102]. As odorant degrading enzymes (ODEs), GSTs play an important role in chemoreception for the adaptation to host plant volatiles and termination of stimulation from signals (i.e., sex pheromones and plant volatiles). Antenna expressed GSTs present in the sensillar lymph of insect antennae, function in signal termination and odorant clearance, enhancing olfactory and neuron sensitivity [103-106]. In Manduca sexta, an antenna specific GST, GST-msolf1 is expressed in the sex-pheromone-sensitive sensilla and can modify trans-2-hexenal, a plant derived green leaf aldehyde, suggesting its dual role in protecting sphinx moth olfactory system from harmful xenobiotics and pheromone inactivation [107]. Likewise, in male silk moth (Bombyx mori), the antennae specific BmGSTD4 had high GSH-conjugating activity towards 1-chloro-2, 4-dinitrobenzene (CDNB), indicating its potential role in the metabolism of xenobiotics [108]. Recently, the antenna expressed GmolGSTD1 was found to exhibit high degradation activity to both the sex pheromone ((Z)-8-dodecenyl alcohol) and the host plant volatile butyl hexanoate in Grapholita molesta [109]. Most recently, the high abundance of a delta GST, SzeaGSTd1 in Sitophilus zeamais antennae, inhibition of SzeaGSTd1 catalytic activity by capryl alcohol, along with the degradation of capryl alcohol by recombinant SzeaGSTd1 were observed [110]. Since capryl alcohol is a volatile component generated during grain storage, the inhibitory effects and degradation of capryl alcohol by the antenna specific SzeaGSTd1 suggest its functions in locating food and favorable oviposition site locations [110].
As phase II detoxification enzymes, arthropod GSTs confer pesticide resistance through direct metabolism or sequestration of pesticides and indirectly by providing protection against oxidative stress induced by synthetic pesticides [22]. In Rynchophorus phoenicis, the enhanced glutathione transferase activity was associated with degradation of dichlorvos, an organophosphate insecticide [111]. Yu and Killiny reported upregulation of DcGSTe2 and DcGSTd1 in the Asian citrus psyllid (Di. citri) when exposed to thiamethoxam and fenpropathrin treatment. Silencing of these GST genes enhanced mortality of Asian citrus psyllid [112]. In Tetranychus cinnabarinus, GST TcGSTm02 was overexpressed in a cyflumetofen resistant strain compared to a susceptible one. The activity of recombinant TcGSTm02 could be inhibited by cyflumetofen and the enzyme catalyzed the conjugation of GSH to cyflumetofen [113]. Recently, RNAi-mediated knockdown of four overexpressed GST genes in the imidacloprid resistant N. lugens resulted in increased sensitivities to the insecticide, suggesting the roles of these GSTs in imidacloprid resistance of N. lugens [114]. One P. xylostella GST, GSTu1 upregulated in several chlorantraniliprole-resistant P. xylostella strains was confirmed to contribute to chlorantraniliprole resistance [115]. In that study, GSTu1 was suggested to be regulated by a novel noncoding RNA-mediated pathway [115]. In Locusta migratoria, LmGSTE4 was found to metabolize malathion and DDT. However, insecticide bioassay showed that after suppression by RNAi, L. migratoria insect mortality was increased in malathion treated insects but not in deltamethrin- or DDT-treated insects [116]. Most recently, 25 GST genes including 22 cytosolic and 3 microsomal genes were identified in insecticide resistance to lambda-cyhalothrin in Cydia pomonella. Among these GSTs, recombinant CpGSTd1, CpGSTd3, CpGSTe3, and CpGSTs2 could bind and metabolize lambda-cyhalothrin, however, no metabolites were detected. Therefore, the authors suggested that the involvement of these GSTs in lambda-cyhalothrin resistance might be through sequestration [117].
Functions of insect GSTs in defense of xenobiotics induced oxidative stress
Eukaryotic cells have evolved to respond against a range of environmental stresses. Oxidative stress is a compromised state for the lipidic cell membrane due to its peroxidation by different free radicals. Pesticides produce oxidative stress in the cell, which in turn generates several ROS free radicals [50]. Free radicals are atoms or molecules with unpaired electrons [118]. In the quest for electronic stability, free radicals attack other molecules to stabilize their electronic state and thereby alter chemical structures and disrupt biomolecular functions [50, 118]. A buildup of ROS such as H2O2 (hydrogen peroxide) and O2- (superoxide anion) can lead to changes in metal homeostasis or oxidation states of protein metal complexes, such as the release of Fe from ferratin or the reduction of iron in cytochrome C [119]. Additionally, exposure to ROS can lead to modifications that cause genomic DNA mutations, negatively affect protein activity, damage cellular membranes, and eventually leading to cell death. Evolutionarily, GSH has been one of the key nucleophilic chemicals in living organisms that convert a range of electrophilic compounds into a less toxic form [120, 121]. In the case of redox stress, two molecules of GSH reduce one molecule of hydrogen peroxide in the presence of glutathione peroxidases, generating one molecule of glutathione disulfide (GSSG), an oxidized form of GSH, and two molecules of water [122, 123]. The glutathione peroxidase, which is responsible for protecting lipids and proteins from oxidation, is regulated by the essential trace metal element Selenium (Se) [124]. The Se-dependent glutathione peroxidase metabolizes hydrogen peroxides and hydroperoxides [40, 125]. In the absence of Se, GST performs glutathione peroxidase activity mostly towards organic hydroperoxides [121, 126, 127]. Once GSSG is formed, flavin adenine dinucleotide (FAD)-dependent enzyme glutathione reductase transfers electrons from NADPH, regenerating two molecules of GSH [121].
Many Se-independent peroxidase reactions performed by GSTs in insects have been reported. In Dr. melanogaster, DmGSTS1-1 exhibited glutathione peroxidase activity towards cumene hydroperoxide (CHP, oxidative stress inducer). Since DmGSTS1-1 was highly expressed in the flight muscle, the localization of the corresponding GST enzyme might provide a protective role against oxidative stress generated from mitochondrial respiration [128]. Similarly, Sawicki and others found six delta class GST genes (GSTD1, GSTD2, GSTD3, GSTD7, GSTD9, and GSTD10), one epsilon class GST (GSTE1), and one sigma class GST gene (GSTS1) in Dr. melanogaster that could conjugate 4-hydroxynonenal (4-HNE), an electrophilic end-product of lipid peroxidation [129]. The role of GSTs in attenuating pyrethroid-induced oxidative stress, which conferred insecticide resistance in the rice brown planthopper (N. lugens) was highlighted by Vontas et al. [46]. It was reported that the increase in GST-based peroxidase activity and the increased amount of GSH indicated the role of GST in reducing the damage from pesticide-induced oxidative stress. Zhang and others showed GSTO2 in Apis cerana cerana had peroxidase activity toward CHP and t-butylhydroperoxide [130]. Similarly, a defensive role against oxidative stress by RpGSTO1 towards different concentrations of CHP was observed in the bird cherry-oat aphid, Rhopalosiphum padi [131]. The GST antioxidant role has also been highlighted in an urban pest, the German cockroach B. germanica. Cockroaches exhibited high GSTD1 peroxidase activity against CHP, indicating a role in insecticide metabolism and reduction of redox stress [92]. Similarly, GSTE1-1 in both DDT resistance and susceptible An. gambia, showed peroxidase activity with CHP but was unable to perform dehydrochlorination activity. The opposite result was obtained for GSTE2-2, indicating these two GSTs play an important role in gaining resistance to DDT via conjugation and peroxidase activity, respectively [49]. In two-week-old adults of Ap. cerana cerana, the expression of AccGSTS1 was high when exposed to various environmental stressors such as temperature (cold and heat shock), heavy metal (HgCl2), pesticides (phoxim, cyhalothrin, and acaricide), H2O2, and ultraviolet [45] radiation which are known for their property to generate oxidative stress [55]. The researchers observed dose-dependent removal of H2O2, indicating AccGSTS1 functions in the elimination of oxidative stress [55]. A similar result was obtained for AccGSTZ1 in Ap. cerana cerana when exposed to varying temperatures and H2O2, suggesting a protective function against oxidative stress [132].
During evolution, insects have adapted to stresses posed by plant-derived toxic chemicals. When feeding on plant species in the Apiaceae or Rutaceae families, which contain furanocoumarin- a toxic photoactive pro-oxidant, Papilio polyxenes exhibited significantly higher GST-mediated peroxidase activity. This is indicative of an insect adaptive mechanism against oxidative stress generated by the plant-derived toxic chemical substances [54, 133], suggesting many GSTs are responsible for protecting tissues and reducing the mortality rate of insects caused by oxidative stress. There are also some cases where specific GSTs are not able to conduct peroxidase activity, such as theta class GSTs [134, 135]. Interestingly, some insects do not have Se-dependent glutathione peroxidases or have enzymes with limited expression and/or activity [49, 54, 127, 136]. Finding evidence on how insects eliminate oxidative stress in the absence of Se-dependent glutathione peroxidase for survival or adaptations to environmental stressors is the direction of future research.
Conclusions
GSTs play a vital role in detoxifying or metabolizing a diverse range of chemical compounds, of xenobiotic or endobiotic origin. GST mediated detoxification is critical for adaptation against xenobiotics including plant allelochemicals and synthesized pesticides. GSTs confer adaptation to a diverse range of xenobiotics through metabolism or sequestration of chemicals and protection against chemical induced oxidative stress. The key to the diverse roles of different classes of GSTs is due to their structure, specifically the composition and spatial localization of amino acid residues composed in the enzymatic active sites. Through a combination of arthropod structural biology, enzyme kinetics and site-directed mutagenesis techniques, our understanding of such diversity in GST structural and functional complexity can be improved.
Abbreviations
GSTs: glutathione S-transferases; PIs: protease inhibitors; HIPVs: herbivore-induced plant volatiles; P450s: cytochrome P450 monooxygenases; COEs: carboxylesterases; ABC transporters: ATP-binding cassette transporters; UGTs: UDP-glycosyltransferases; GSH: glutathione; ROS: reactive oxygen species; RNAi: RNA interference; ODEs: odorant degrading enzymes; GSSG: glutathione disulfide; CHP: cumene hydroperoxide; 4-HNE: 4-hydroxynonenal; CDNB: 1-chloro-2, 4-dinitrobenzene.
Acknowledgements
This research was supported by a faculty start-up fund from Pennsylvania State University, NSF CAREER IOS-2144082, the USDA National Institute of Food and Federal Appropriations under Hatch Project #PEN04770 and Accession #1010058. T.M. was supported by USDA NIFA postdoctoral fellowship, grant #2020-67034-31780/project accession#1022959.
Author Contributions
FZ conceived and designed the study. SKBK wrote the original draft. SKBK, TM, FZ summarized the table and made the figures. TM and FZ revised the manuscript. All authors approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Robinson GE, Hackett KJ, Purcell-Miramontes M, Brown SJ, Evans JD, Goldsmith MR. et al. Creating a buzz about insect genomes. Science (New York, NY). 2011;331:1386
2. Gallaia N, Sallesc J, Setteled J, Vaissièrea BE. Economic valuation of the vulnerability of world agricultureconfronted with pollinator decline. Ecol Econ. 2009;68:810-21
3. War AR, Sharma HC. Induced resistance in plants and counter-adaptation by insect pests. In: Chandrasekar R, Tyagi BK, Gui Z, Reeck GR, editors. Short Views on Insect Biochemistry and Molecular Biology. Manhattan, USA: Academic Publisher. 2014 p. 1-16
4. War AR, Taggar GK, Hussain B, Taggar MS, Nair RM, Sharma HC. Plant defence against herbivory and insect adaptations. AoB PLANTS. 2018 10
5. Despres L, David JP, Gallet C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol Evol. 2007;22:298-307
6. Alyokhin A, Chen YH. Adaptation to toxic hosts as a factor in the evolution of insecticide resistance. Curr Opin Insect Sci. 2017;21:33-8
7. Dermauw W, Wybouw N, Rombauts S, Menten B, Vontas J, Grbic M. et al. A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae. Proc Natl Acad Sci U.S.A. 2013;110:E113-E22
8. Zhu F, Moural TW, Nelson DR, Palli SR. A specialist herbivore pest adaptation to xenobiotics through up-regulation of multiple Cytochrome P450s. Sci Rep. 2016;6:20421
9. Schuler MA. The role of cytochrome P450 monooxygenases in plant-insect interactions. Plant Physiol. 1996;112:1411-9
10. Sparks TC, Nauen R. IRAC: Mode of action classification and insecticide resistance management. Pestic Biochem Physiol. 2015;121:122-8
11. Loso MR, Garizi N, Hegde VB, Hunter JE, Sparks TC. Lead generation in crop protection research: a portfolio approach to agrochemical discovery. Pest Manag Sci. 2017;73:678-85
12. Lorsbach BA, Sparks TC, Cicchillo RM, Garizi NV, Hahn DR, Meyer KG. Natural products: a strategic lead generation approach in crop protection discovery. Pest Manag Sci. 2019;75:2301-9
13. Zhu F, Cui Y, Walsh DB, Lavine LC. Application of RNAi towards insecticide resistance management. In: Chandrasekar R, Tyagi BK, Gui Z, Reeck GR, editors. Short Views on Insect Biochemistry and Molecular Biology. Manhattan, USA: Academic Publisher. 2014 p. 595-619
14. Brattsten LB. Potential role of plant allelochemicals in the development of insecticide resistance. In: Barbosa P, editor. Novel Aspects of Insect-Plant Interactions. New York: John Wiley & Sons, Inc. 1988 p. 313-48
15. Adesanya AW, Beauchamp MJ, Lavine MD, Lavine LC, Zhu F, Walsh DB. Physiological resistance alters behavioral response of Tetranychus urticae to acaricides. Sci Rep. 2019;9:19308
16. Zalucki MP, Furlong MJ. Behavior as a mechanism of insecticide resistance: evaluation of the evidence. Curr Opin Insect Sci. 2017;21:19-25
17. Kikuchi Y, Hayatsu M, Hosokawa T, Nagayama A, Tago K, Fukatsu T. Symbiont-mediated insecticide resistance. Proc Natl Acad Sci U.S.A. 2012;109:8618-22
18. Cheng D, Guo Z, Riegler M, Xi Z, Liang G, Xu Y. Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (Hendel). Microbiome. 2017;5:13
19. Irwin RE, Cook D, Richardson LL, Manson JS, Gardner DR. Secondary compounds in floral rewards of toxic rangeland plants: impacts on pollinators. J Agric Food Chem. 2014;62:7335-44
20. Koch H, Cisarovsky G, Schmid-Hempel P. Ecological effects on gut bacterial communities in wild bumblebee colonies. J Anim Ecol. 2012;81:1202-10
21. Zhu F, Gujar H, Gordon JR, Haynes KF, Potter MF, Palli SR. Bed bugs evolved unique adaptive strategy to resist pyrethroid insecticides. Sci Rep. 2013;3:1456
22. Pavlidi N, Vontas J, Van Leeuwen T. The role of glutathione S-transferases (GSTs) in insecticide resistance in crop pests and disease vectors. Curr Opin Insect Sci. 2018;27:97-102
23. Nauen R, Bass C, Feyereisen R, Vontas J. The Role of Cytochrome P450s in Insect Toxicology and Resistance. Annu Rev Entomol. 2022;67:105-24
24. Xia J, Guo Z, Yang Z, Han H, Wang S, Xu H. et al. Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell. 2021;184:1693-705.e17
25. Van Leeuwen T, Dermauw W. The molecular evolution of xenobiotic metabolism and resistance in Chelicerate mites. Annu Rev Entomol. 2016;61:475-98
26. Strauss AS, Peters S, Boland W, Burse A. ABC transporter functions as a pacemaker for sequestration of plant glucosides in leaf beetles. eLife. 2013;2:e01096
27. Hopkins DH, Fraser NJ, Mabbitt PD, Carr PD, Oakeshott JG, Jackson CJ. Structure of an insecticide sequestering carboxylesterase from the disease vector Culex quinquefasciatus: What makes an enzyme a good insecticide sponge? Biochemistry. 2017;56:5512-25
28. Bass C, Puinean AM, Andrews M, Cutler P, Daniels M, Elias J. et al. Mutation of a nicotinic acetylcholine receptor β subunit is associated with resistance to neonicotinoid insecticides in the aphid Myzus persicae. BMC Neurosci. 2011;12:51
29. Dobler S, Dalla S, Wagschal V, Agrawal AA. Community-wide convergent evolution in insect adaptation to toxic cardenolides by substitutions in the Na,K-ATPase. Proc Natl Acad Sci U.S.A. 2012;109:13040-5
30. Ffrench-Constant RH. The molecular genetics of insecticide resistance. Genetics. 2013;194:807-15
31. Adesanya AW, Lavine MD, Moural TW, Lavine LC, Zhu F, Walsh DB. Mechanisms and management of acaricide resistance for Tetranychus urticae in agroecosystems. J Pest Sci. 2021;94:639-63
32. Wu MX, Adesanya AW, Morales MA, Walsh DB, Lavine LC, Lavine MD. et al. Multiple acaricide resistance and underlying mechanisms in Tetranychus urticae on hops. J Pest Sci. 2019;92:543-55
33. Scott JG. Cytochrome P450 monooxygenases and insecticide resistance: lessons from CYP6D1. In: Ishaaya I, editor. Biochemical Sites of Insecticide Action and Resistance Berlin, Heidelberg: Springer. 2001 p. 255-67
34. Adesanya AW, Morales MA, Walsh DB, Lavine LC, Lavine MD, Zhu F. Mechanisms of resistance to three mite growth inhibitors of Tetranychus urticae in hops. Bull Entomol Res. 2018;108:23-34
35. Bass C, Puinean AM, Zimmer CT, Denholm I, Field LM, Foster SP. et al. The evolution of insecticide resistance in the peach potato aphid, Myzus persicae. Insect Biochem Mol Biol. 2014;51:41-51
36. Rand EED, Smit S, Beukes M, Apostolides Z, Pirk CWW, Nicolson SW. Detoxification mechanisms of honey bees (Apis mellifera) resulting in tolerance of dietary nicotine. Sci Rep. 2015;5:11779
37. Berenbaum MR, Johnson RM. Xenobiotic detoxification pathways in honey bees. Curr Opin Insect Sci. 2015;10:51-8
38. Li X, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231-53
39. Amezian D, Nauen R, Le Goff G. Transcriptional regulation of xenobiotic detoxification genes in insects - An overview. Pestic Biochem Physiol. 2021;174:104822
40. Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51-88
41. Ranson H, Hemingway J. Mosquito glutathione transferases. Methods Enzymol. 2005;401:226-41
42. Liu Y, Moural T, Koirala B K S, Hernandez J, Shen Z, Alyokhin A. et al. Structural and functional characterization of one unclassified glutathione S-transferase in xenobiotic adaptation of Leptinotarsa decemlineata. Int J Mol Sci. 2021;22:11921
43. Clark AG, Shamaan NA. Evidence that DDT-dehydrochlorinase from the house fly is a glutathione S-transferase. Pestic Biochem Physiol. 1984;22:249-61
44. Che-Mendoza A, Penilla RP, Rodriguez DA. Insecticide resistance and glutathione S-transferases in mosquitoes: A review. Afr J Biotechnol. 2009;8:1386-97
45. Kostaropoulos I, Papadopoulos AI, Metaxakis A, Boukouvala E, Papadopoulou-Mourkidou E. Glutathione S-transferase in the defence against pyrethroids in insects. Insect Biochem Mol Biol. 2001;31:313-9
46. Vontas JG, Small GJ, Hemingway J. Glutathione S-transferases as antioxidant defence agents confer pyrethroid resistance in Nilaparvata lugens. Biochem J. 2001;357:65-72
47. Wilding CS, Weetman D, Rippon EJ, Steen K, Mawejje HD, Barsukov I. et al. Parallel evolution or purifying selection, not introgression, explains similarity in the pyrethroid detoxification linked GSTE4 of Anopheles gambiae and An. arabiensis. Mol Genet Genomics. 2015;290:201-15
48. Townsend DM, Tew KD. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene. 2003;22:7369-75
49. Ortelli F, Rossiter LC, Vontas J, Ranson H, Hemingway J. Heterologous expression of four glutathione transferase genes genetically linked to a major insecticide-resistance locus from the malaria vector Anopheles gambiae. Biochem J. 2003;373:957-63
50. Abdollahi M, Ranjbar A, Shadnia S, Nikfar S, Rezaie A. Pesticides and oxidative stress: a review. Med Sci Monit. 2004;10:Ra141-7
51. Burcham PC. Toxicodynamics: how chemicals harm cells. In: Burcham PC, editor. An Introduction to Toxicology. Verlag London: Springer. 2014 p. 91-125
52. Liu Y, Zhu F, Shen Z, Moural TW, Liu L, Li Z. et al. Glutaredoxins and thioredoxin peroxidase involved in defense of emamectin benzoate induced oxidative stress in Grapholita molesta. Pestic Biochem Physiol. 2021 176
53. Corona M, Robinson GE. Genes of the antioxidant system of the honey bee: annotation and phylogeny. Insect Mol Biol. 2006;15:687-701
54. Weinhold LC, Ahmad S, Pardini RS. Insect glutathione-S-transferase: A predictor of allelochemical and oxidative stress. Comp Biochem Physiol B: Comp Biochem. 1990;95:355-63
55. Yan H, Jia H, Gao H, Guo X, Xu B. Identification, genomic organization, and oxidative stress response of a sigma class glutathione S-transferase gene (AccGSTS1) in the honey bee, Apis cerana cerana. Cell Stress Chaperones. 2013;18:415-26
56. Yan H, Jia H, Wang X, Gao H, Guo X, Xu B. Identification and characterization of an Apis cerana cerana Delta class glutathione S-transferase gene (AccGSTD) in response to thermal stress. Naturwissenschaften. 2013;100:153-63
57. Hilliou F, Chertemps T, Maïbèche M, Le Goff G. Resistance in the genus Spodoptera: key insect detoxification genes. Insects. 2021;12:544
58. Allocati N, Federici L, Masulli M, Di Ilio C. Glutathione transferases in bacteria. FEBS J. 2009;276:58-75
59. Enayati AA, Ranson H, Hemingway J. Insect glutathione transferases and insecticide resistance. Insect Mol Biol. 2005;14:3-8
60. Ladner JE, Parsons JF, Rife CL, Gilliland GL, Armstrong RN. Parallel evolutionary pathways for glutathione transferases: structure and mechanism of the mitochondrial class kappa enzyme rGSTK1-1. Biochemistry. 2004;43:352-61
61. Morel F, Rauch C, Petit E, Piton A, Theret N, Coles B. et al. Gene and protein characterization of the human glutathione S-transferase kappa and evidence for a peroxisomal localization. J Biol Chem. 2004;279:16246-53
62. Frova C. Glutathione transferases in the genomics era: new insights and perspectives. Biomol Eng. 2006;23:149-69
63. Kim JH, Raisuddin S, Rhee JS, Lee YM, Han KN, Lee JS. Molecular cloning, phylogenetic analysis and expression of a MAPEG superfamily gene from the pufferfish Takifugu obscurus. Comp Biochem Physiol C: Toxicol Pharmacol. 2009;149:358-62
64. Shi H, Pei L, Gu S, Zhu S, Wang Y, Zhang Y. et al. Glutathione S-transferase (GST) genes in the red flour beetle, Tribolium castaneum, and comparative analysis with five additional insects. Genomics. 2012;100:327-35
65. You Y, Xie M, Ren N, Cheng X, Li J, Ma X. et al. Characterization and expression profiling of glutathione S-transferases in the diamondback moth, Plutella xylostella (L.). BMC Genomics. 2015;16:152
66. Han JB, Li GQ, Wan PJ, Zhu TT, Meng QW. Identification of glutathione S-transferase genes in Leptinotarsa decemlineata and their expression patterns under stress of three insecticides. Pestic Biochem Physiol. 2016;133:26-34
67. Wongsantichon J, Robinson RC, Ketterman AJ. Epsilon glutathione transferases possess a unique class-conserved subunit interface motif that directly interacts with glutathione in the active site. Biosci Rep. 2015 35
68. Sheehan D, Meade G, Foley VM, Dowd CA. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem J. 2001;360:1-16
69. Ranson H, Rossiter L, Ortelli F, Jensen B, Wang X, Roth CW. et al. Identification of a novel class of insect glutathione S-transferases involved in resistance to DDT in the malaria vector Anopheles gambiae. Biochem J. 2001;359:295-304
70. da Fonseca RR, Johnson WE, O'Brien SJ, Vasconcelos V, Antunes A. Molecular evolution and the role of oxidative stress in the expansion and functional diversification of cytosolic glutathione transferases. BMC Evol Biol. 2010;10:281
71. Ranson H, Claudianos C, Ortelli F, Abgrall C, Hemingway J, Sharakhova MV. et al. Evolution of supergene families associated with insecticide resistance. Science (New York, NY). 2002;298:179-81
72. Claudianos C, Ranson H, Johnson RM, Biswas S, Schuler MA, Berenbaum MR. et al. A deficit of detoxification enzymes: pesticide sensitivity and environmental response in the honeybee. Insect Mol Biol. 2006;15:615-36
73. Polekhina G, Board PG, Blackburn AC, Parker MW. Crystal structure of maleylacetoacetate isomerase/glutathione transferase Zeta reveals the molecular basis for its remarkable catalytic promiscuity. Biochemistry. 2001;40:1567-76
74. Wiktelius E, Stenberg G. Novel class of glutathione transferases from cyanobacteria exhibit high catalytic activities towards naturally occurring isothiocyanates. Biochem J. 2007;406:115-23
75. Allocati N, Masulli M, Di Ilio C, Federici L. Glutathione transferases: substrates, inihibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis. 2018;7:8
76. Yamamoto K, Suzuki M, Higashiura A, Nakagawa A. Three-dimensional structure of a Bombyx mori Omega-class glutathione transferase. Biochem Biophys Res Commun. 2013;438:588-93
77. Liu J, Li Y, Tian Z, Sun H, Chen X, Zheng S. et al. Identification of key residues associated with the interaction between Plutella xylostella Sigma-class glutathione S-transferase and the inhibitor S-hexyl glutathione. J Agric Food Chem. 2018;66:10169-78
78. Armstrong RN. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem Res Toxicol. 1997;10:2-18
79. Tars K, Larsson A-K, Shokeer A, Olin B, Mannervik B, Kleywegt GJ. Structural basis of the suppressed catalytic activity of wild-type human glutathione transferase T1-1 compared to its W234R mutant. J Mol Biol. 2006;355:96-105
80. Low WY, Feil SC, Ng HL, Gorman MA, Morton CJ, Pyke J. et al. Recognition and detoxification of the insecticide DDT by Drosophila melanogaster glutathione S-transferase D1. J Mol Biol. 2010;399:358-66
81. Udomsinprasert R, Pongjaroenkit S, Wongsantichon J, Oakley AJ, Prapanthadara LA, Wilce MC. et al. Identification, characterization and structure of a new Delta class glutathione transferase isoenzyme. Biochem J. 2005;388:763-71
82. Winayanuwattikun P, Ketterman AJ. Catalytic and structural contributions for glutathione-binding residues in a Delta class glutathione S-transferase. Biochem J. 2004;382:751-7
83. Oakley AJ, Harnnoi T, Udomsinprasert R, Jirajaroenrat K, Ketterman AJ, Wilce MCJ. The crystal structures of glutathione S-transferases isozymes 1-3 and 1-4 from Anopheles dirus species B. Protein Sci. 2001;10:2176-85
84. Wang Y, Qiu L, Ranson H, Lumjuan N, Hemingway J, Setzer WN. et al. Structure of an insect epsilon class glutathione S-transferase from the malaria vector Anopheles gambiae provides an explanation for the high DDT-detoxifying activity. J Struct Biol. 2008;164:228-35
85. Scian M, Le Trong I, Mazari AMA, Mannervik B, Atkins WM, Stenkamp RE. Comparison of epsilon- and delta-class glutathione S-transferases: the crystal structures of the glutathione S-transferases DmGSTE6 and DmGSTE7 from Drosophila melanogaster. Acta Crystallogr D Biol Crystallogr. 2015;71:2089-98
86. Rossjohn J, McKinstry WJ, Oakley AJ, Verger D, Flanagan J, Chelvanayagam G. et al. Human theta class glutathione transferase: the crystal structure reveals a sulfate-binding pocket within a buried active site. Structure. 1998;6:309-22
87. Agianian B, Tucker PA, Schouten A, Leonard K, Bullard B, Gros P. Structure of a Drosophila Sigma class glutathione S-transferase reveals a novel active site topography suited for lipid peroxidation products. J Mol Biol. 2003;326:151-65
88. Vuilleumier S. Vuilleumier S. Bacterial glutathione S-transferases: what are they good for? J Bacteriol. 1997;179:1431-41
89. Caccuri AM, Antonini G, Nicotra M, Battistoni A, Lo Bello M, Board PG. et al. Catalytic mechanism and role of hydroxyl residues in the active site of theta class glutathione S-transferases. Investigation of Ser-9 and Tyr-113 in a glutathione S-transferase from the Australian sheep blowfly, Lucilia cuprina. J Biol Chem. 1997;272:29681-6
90. Sue M, Yajima S. Crystal structure of the delta-class glutathione transferase in Musca domestica. Biochem Biophys Res Commun. 2018;502:345-50
91. Gonzalez D, Fraichard S, Grassein P, Delarue P, Senet P, Nicolai A. et al. Characterization of a Drosophila glutathione transferase involved in isothiocyanate detoxification. Insect Biochem Mol Biol. 2018;95:33-43
92. Ma B, Chang FN. Purification and cloning of a Delta class glutathione S-transferase displaying high peroxidase activity isolated from the German cockroach Blattella germanica. FEBS J. 2007;274:1793-803
93. Ralat LA, Colman RF. Glutathione S-transferase Pi has at least three distinguishable xenobiotic substrate sites close to its glutathione-binding site. J Biol Chem. 2004;279:50204-13
94. Feng Q, Davey KG, A SDP, Ladd TR, Retnakaran A, Tomkins BL. et al. Developmental expression and stress induction of glutathione S-transferase in the spruce budworm, Choristoneura fumiferana. J Insect Physiol. 2001;47:1-10
95. Munday R, Munday CM. Induction of phase II detoxification enzymes in rats by plant-derived isothiocyanates: comparison of allyl isothiocyanate with sulforaphane and related compounds. J Agric Food Chem. 2004;52:1867-71
96. Gloss AD, Vassão DG, Hailey AL, Nelson Dittrich AC, Schramm K, Reichelt M. et al. Evolution in an ancient detoxification pathway is coupled with a transition to herbivory in the drosophilidae. Mol Biol Evol. 2014;31:2441-56
97. Zou X, Xu Z, Zou H, Liu J, Chen S, Feng Q. et al. Glutathione S-transferase SlGSTE1 in Spodoptera litura may be associated with feeding adaptation of host plants. Insect Biochem Mol Biol. 2016;70:32-43
98. Mittapalli O, Neal JJ, Shukle RH. Tissue and life stage specificity of glutathione S-transferase expression in the Hessian fly, Mayetiola destructor: implications for resistance to host allelochemicals. J Insect Sci. 2007;7:1-13
99. Yang J, Kong X-D, Zhu-Salzman K, Qin Q-M, Cai Q-N. The key glutathione S-transferase family genes involved in the detoxification of rice gramine in Brown planthopper Nilaparvata lugens. Insects. 2021;12:1055
100. Ma J, Sun L, Zhao H, Wang Z, Zou L, Cao C. Functional identification and characterization of GST genes in the Asian gypsy moth in response to poplar secondary metabolites. Pestic Biochem Physiol. 2021;176:104860
101. von Merey GE, Veyrat N, D'Alessandro M, Turlings TC. Herbivore-induced maize leaf volatiles affect attraction and feeding behavior of Spodoptera littoralis caterpillars. Front Plant Sci. 2013;4:209
102. Chiu CC, Keeling CI, Bohlmann J. Toxicity of pine monoterpenes to mountain pine beetle. Sci Rep. 2017;7:8858
103. Durand N, Pottier M-A, Siaussat D, Bozzolan F, Maïbèche M, Chertemps T. Glutathione-S-transferases in the olfactory organ of the Noctuid moth Spodoptera littoralis, diversity and conservation of chemosensory clades. Front Physiol. 2018;9:1283 -
104. Younus F, Chertemps T, Pearce SL, Pandey G, Bozzolan F, Coppin CW. et al. Identification of candidate odorant degrading gene/enzyme systems in the antennal transcriptome of Drosophila melanogaster. Insect Biochem Mol Biol. 2014;53:30-43
105. Kang Z, Liu F, Xu Y, Cheng J, Lin X, Jing X. et al. Identification of candidate odorant-degrading enzyme genes in the antennal transcriptome of Aphidius gifuensis. Entomol Res. 2020;51:36-54
106. Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373-91
107. Rogers ME, Jani MK, Vogt RG. An olfactory-specific glutathione-S-transferase in the sphinx moth Manduca sexta. J Exp Biol. 1999;202:1625-37
108. Tan X, Hu XM, Zhong XW, Chen QM, Xia QY, Zhao P. Antenna-specific glutathione S-transferase in male silkmoth Bombyx mori. Int J Mol Sci. 2014;15:7429-43
109. Li GW, Chen XL, Xu XL, Wu JX. Degradation of sex pheromone and plant volatile components by an antennal glutathione S-transferase in the oriental fruit moth, Grapholita molesta Busck (Lepidoptera: Tortricidae). Arch Insect Biochem Physiol. 2018;99:e21512
110. Xia D, Zheng R, Huang J, Lu S, Tang Q. Identification and functional analysis of glutathione S-transferases from Sitophilus zeamais in olfactory organ. Insects. 2022;13:259
111. Bamidele OS, Ajele JO, Olajuyigbe FM. An evaluation of glutathione transferase associated with Dichlorvos degradation in African palm weevil (Rynchophorus phoenicis) larva. Cogent Biol. 2017;3:1286764
112. Yu X, Killiny N. RNA interference of two glutathione S-transferase genes, Diaphorina citri DcGSTe2 and DcGSTd1, increases the susceptibility of Asian citrus psyllid (Hemiptera: Liviidae) to the pesticides fenpropathrin and thiamethoxam. Pest Manag Sci. 2018;74:638-47
113. Feng K, Yang Y, Wen X, Ou S, Zhang P, Yu Q. et al. Stability of cyflumetofen resistance in Tetranychus cinnabarinus and its correlation with glutathione-S-transferase gene expression. Pest Manag Sci. 2019;75:2802-9
114. Yang B, Lin X, Yu N, Gao H, Zhang Y, Liu W. et al. Contribution of glutathione S-transferases to imidacloprid resistance in Nilaparvata lugens. J Agri Food Chem. 2020;68:15403-8
115. Zhu B, Li L, Wei R, Liang P, Gao X. Regulation of GSTu1-mediated insecticide resistance in Plutella xylostella by miRNA and lncRNA. PLoS Genet. 2021;17:e1009888
116. Zhang J, Ma W, Yin F, Park Y, Zhu KY, Zhang X. et al. Evaluations of two glutathione S-transferase epsilon genes for their contributions to metabolism of three selected insecticides in Locusta migratoria. Pestic Biochem Physiol. 2022;183:105084
117. Hu C, Liu JY, Wang W, Mota-Sanchez D, He S, Shi Y. et al. Glutathione S-transferase genes are involved in lambda-cyhalothrin resistance in Cydia pomonella via sequestration. J Agric Food Chem. 2022;70:2265-79
118. Cortés-Iza SC, Rodríguez AI. Oxidative stress and pesticide disease: a challenge for toxicology. Rev Fac Med. 2018;66:261-7
119. Phaniendra A, Jestadi DB, Periyasamy L. Free radicals: properties, sources, targets, and their implication in various diseases. Indian J Clin Biochem. 2015;30:11-26
120. Kwon DH, Cha HJ, Lee H, Hong SH, Park C, Park SH. et al. Protective effect of glutathione against oxidative stress-induced cytotoxicity in RAW 264.7 macrophages through activating the nuclear factor erythroid 2-related factor-2/heme oxygenase-1 pathway. Antioxidants. 2019;8:82
121. Deponte M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim Biophys Acta. 2013;1830:3217-66
122. Kültz D. Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol. 2005;67:225-57
123. Gaikwad Y, Gaikwad S, Bhawane G. Effect of induced oxidative stress and herbal extracts on acid phosphatase activity in lysosomal and microsomal fractions of midgut tissue of the silkworm, Bombyx mori. J Insect Sci. 2010;10:113
124. Nogales F, Ojeda ML, Fenutría M, Murillo ML, Carreras O. Role of selenium and glutathione peroxidase on development, growth, and oxidative balance in rat offspring. Reproduction. 2013;146:659-67
125. Wendel A. Glutathione peroxidase. Methods Enzymol. 1981;77:325-33
126. Ketterer B, Meyer D. Gluthathione transferases: A possible role in the detoxication and repair of DNA and lipid hydroperoxides. Mutat Res- Fundam Mol Mech Mutagen. 1989;214:33-40
127. Parkes TL, Hilliker AJ, Phillips JP. Genetic and biochemical analysis of glutathione-S-transferase in the oxygen defense system of Drosophila melanogaster. Genome. 1993;36:1007-14
128. Singh SP, Coronella JA, Beneš H, Cochrane BJ, Zimniak P. Catalytic function of Drosophila melanogaster glutathione S-transferase DmGSTS1-1 (GST-2) in conjugation of lipid peroxidation end products. Eur J Biochem. 2001;268:2912-23
129. Sawicki R, Singh SP, Mondal AK, Benes H, Zimniak P. Cloning, expression and biochemical characterization of one Epsilon-class (GST-3) and ten Delta-class (GST-1) glutathione S-transferases from Drosophila melanogaster, and identification of additional nine members of the Epsilon class. Biochem J. 2003;370:661-9
130. Zhang Y, Yan H, Lu W, Li Y, Guo X, Xu B. A novel Omega-class glutathione S-transferase gene in Apis cerana cerana: molecular characterisation of GSTO2 and its protective effects in oxidative stress. Cell Stress Chaperones. 2013;18:503-16
131. Balakrishnan B, Su S, Wang K, Tian R, Chen M. Identification, expression, and regulation of an Omega class glutathione S-transferase in Rhopalosiphum padi (L.) (Hemiptera: Aphididae) under insecticide stress. Front Physiol. 2018;9:427
132. Yan H, Meng F, Jia H, Guo X, Xu B. The identification and oxidative stress response of a zeta class glutathione S-transferase (GSTZ1) gene from Apis cerana cerana. J Insect Physiol. 2012;58:782-91
133. Ahmad S. Oxidative stress from environmental pollutants. Arch Insect Biochem Physiol. 1995;29:135-57
134. Liu S, Liu F, Jia H, Yan Y, Wang H, Guo X. et al. A glutathione S-transferase gene associated with antioxidant properties isolated from Apis cerana cerana. Sci Nat. 2016;103:1-12
135. Hossain MT, Yamada N, Yamamoto K. Glutathione-binding site of a Bombyx mori theta-class glutathione transferase. PLoS One. 2014;9:e97740
136. Simmons TW, Jamall IS, Lockshin RA. Selenium modulates peroxidation in the absence of glutathione peroxidase in Musca domestica. Biochem Biophys Res Commun. 1989;165:158-63
137. Brattsten LB. Biochemical defense mechansims in herbivores against plant allelochemicals. In: Rosenthal GA, Janzen DH, editors. Herbivores-Their Interaction with Secondary Plant Metabolites. New York: Academic Press. 1979 p. 199-270
138. Strode C, Wondji CS, David J-P, Hawkes NJ, Lumjuan N, Nelson DR. et al. Genomic analysis of detoxification genes in the mosquito Aedes aegypti. Insect Biochem Mol Biol. 2008;38:113-23
139. Hu F, Dou W, Wang JJ, Jia FX, Wang JJ. Multiple glutathione S-transferase genes: identification and expression in oriental fruit fly, Bactrocera dorsalis. Pest Manag Sci. 2014;70:295-303
140. Nair PMG, Choi J. Identification, characterization and expression profiles of Chironomus riparius glutathione S-transferase (GST) genes in response to cadmium and silver nanoparticles exposure. Aquat Toxicol. 2011;101:550-60
141. Niranjan Reddy B, Prasad G, Raghavendra K. In silico characterization and comparative genomic analysis of the Culex quinquefasciatus glutathione S-transferase (GST) supergene family. Parasitol Res. 2011;109:1165-77
142. Aidlin Harari O, Santos-Garcia D, Musseri M, Moshitzky P, Patel M, Visendi P. et al. Molecular evolution of the glutathione S-transferase family in the Bemisia tabaci species complex. Genome Biol Evol. 2020;12:3857-72
143. Ramsey JS, Rider DS, Walsh TK, De Vos M, Gordon K, Ponnala L. et al. Comparative analysis of detoxification enzymes in Acyrthosiphon pisum and Myzus persicae. Insect Mol Biol. 2010;19:155-64
144. Xue J, Zhou X, Zhang C-X, Yu L-L, Fan H-W, Wang Z. et al. Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 2014;15:1-20
145. Bailey E, Field L, Rawlings C, King R, Mohareb F, Pak K-H. et al. A scaffold-level genome assembly of the pirate bug, Orius laevigatus, and a comparative analysis of insecticide resistance-related gene families with hemipteran crop pests. BMC Genomics. 2022;23:45
146. Oakeshott JG, Johnson RM, Berenbaum MR, Ranson H, Cristino AS, Claudianos C. Metabolic enzymes associated with xenobiotic and chemosensory responses in Nasonia vitripennis. Insect Mol Biol. 2010;19:147-63
147. Yu Q, Lu C, Li B, Fang S, Zuo W, Dai F. et al. Identification, genomic organization and expression pattern of glutathione S-transferase in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2008;38:1158-64
148. Cheng T, Wu J, Wu Y, Chilukuri RV, Huang L, Yamamoto K. et al. Genomic adaptation to polyphagy and insecticides in a major East Asian noctuid pest. Nat Ecol Evol. 2017;1:1747-56
149. Gonis E, Fraichard S, Chertemps T, Hecker A, Schwartz M, Canon F, Neiers F. Expression patterns of Drosophila Melanogaster glutathione transferases. Insects. 2022;13:612
Author contact
![]() Corresponding author: Dr. Fang Zhu, Department of Entomology and Huck Institutes of the Life Sciences, Pennsylvania State University, University Park, PA 16802, USA. Phone: +1-814-863-4432; Fax: +1- 814-865-3048; E-mail: fuz59edu.
Corresponding author: Dr. Fang Zhu, Department of Entomology and Huck Institutes of the Life Sciences, Pennsylvania State University, University Park, PA 16802, USA. Phone: +1-814-863-4432; Fax: +1- 814-865-3048; E-mail: fuz59edu.

 Global reach, higher impact
Global reach, higher impact