10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2025; 21(9):3934-3948. doi:10.7150/ijbs.102309 This issue Cite
Research Paper
WNT5B regulates myogenesis and fiber type conversion by affecting mRNA stability
1. Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518124, China.
2. Kunpeng Institute of Modern Agriculture at Foshan, Agricultural Genomics Institute, Chinese Academy of Agricultural Sciences, Foshan 528226, China.
3. Key Laboratory of Livestock and Poultry Multi-Omics of MARA, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518124, China.
4. Guangxi Key Laboratory of Livestock Genetic Improvement, Guangxi Institute of Animal Sciences, Nanning 530001, China.
#These authors equally contribute to this work.
Received 2024-8-13; Accepted 2025-4-26; Published 2025-6-9
Abstract
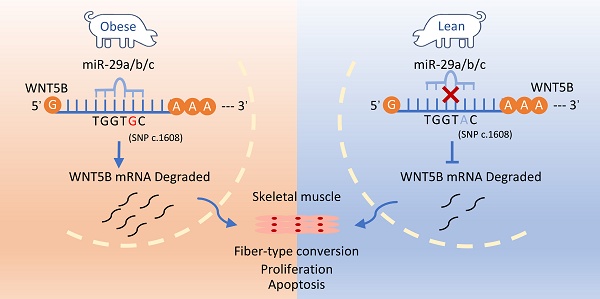
The wingless-integrated (WNT) signaling pathway is known to play a critical role in myogenesis. WNT5B, a member of the WNT family, is essential for determining cell fate and development. However, the molecular mechanisms by which WNT5B regulates myogenesis remain unclear. This study observed that WNT5B, which is conserved between mice and pigs, is highly expressed in the skeletal muscle. The expression of WNT5B was varied in the skeletal muscle between the Tongcheng (obese-type) and Landrace (lean-type) pigs. In vitro, WNT5B promoted skeletal muscle cell proliferation and cell cycle progression, while inhibited cell apoptosis. In vivo, WNT5B promoted myofiber thickness and increased slow-type muscle fibers in porcine skeletal muscles. Mechanically, a SNP site (c.1608 A > G) located in the 3' untranslated region (3'UTR) of WNT5B regulated transcript attenuation and acts through AU-rich element mediation (ARE) to influence myogenesis. Also, the SNP was located in the miRNA response element of miR-29a/b/c to influence WNT5B expression. However, ARE sites did not affect the binding relationship of miR-29a/b/c. In conclusion, this study demonstrated that the SNP (c.1608 A > G) affects WNT5B mRNA stability by protecting the 3'UTR of the WNT5B from the degrading effects of miR-29a/b/c. This study reveals the critical role of WNT5B in myogenesis and indicates that it is a novel candidate gene for pig breeding.
Keywords: WNT5B, single nucleotide polymorphisms, mRNA stability, myogenesis, fiber type conversion
Introduction
Skeletal muscle, which constitute the largest metabolic and endocytic organ system in the body, plays a crucial role in maintaining physiological characteristics and homeostasis [1, 2]. The growth and development of skeletal muscle are highly precise, tightly coordinated, and well-organized multistep biological processes [3], involving myoblast proliferation [4, 5], differentiation [6, 7], and apoptosis [8, 9]. Skeletal muscle growth and development are regulated by multiple signaling pathways. Among the key signaling pathways that orchestrate skeletal muscle development, the WNT signaling pathway has emerged as a critical regulator due to its involvement in cell fate determination, tissue patterning, and myogenic progression [10-12]. However, the role of the WNT pathway, a profound signaling pathway [13], in skeletal muscle growth and development is not fully understood.
WNT5B (WNT Family Member 5B) is classified as a non-canonical WNT ligand and mainly functions through the non-canonical WNT pathway [14-16]. It plays a key role in the development and physiological processes of various tissues, including bone [17, 18], adipose [19], and heart [20]. Previous studies have demonstrated that WNT5B affects cell proliferation and migration and is involved in cancer and vascular smooth muscle cell differentiation [21, 22]. Also, WNT5B regulate tissue polarity and cell migration [23, 24], and WNT5B-Ror2 complexes form in the producing cell and are handed over from these cytonemes to the receiving cell [25]. Most studies on WNT5B have centered on its expression patterns and links to disease. However, the role of WNT5B in myogenesis is not well understood.
WNT5B is also a pivotal regulatory factor in mammalian health and disease and is regulated by genetic and epigenetic factor [26, 27]. The SNP (rs2887571) at the WNT5B enhancer alters ERα binding, affecting WNT5B expression in osteoporosis [28]. DNA methylation [29, 30], histone modification factors [31, 32], and non-coding RNAs (ncRNAs) [33, 34] have been shown to regulate WNT5B expression. The binding of ncRNA tsRMST to the PRC2 component SUZ12 inhibits WNT5B expression during human embryonic stem cell differentiation [35]. Moreover, miR-5587-3P, miR-587, and miR-149-5P have been found to modulate WNT5B expression by binding to its 3' untranslated region (3'UTR) in, respectively, cancer, fat and chondrocytes cells [36-38]. However, the specific regulatory mechanisms of WNT5B during myogenesis require further investigation.
In this study, the expression profiles of WNT5B were comparatively analyzed in different pig breeds at different developmental stages to investigate the regulatory role of WNT5B in myogenesis. A functional SNP site (c.1608 A > G) was identified and located in the 3'UTR of WNT5B, which regulates myogenesis through genetic and epigenetic mechanisms. Our findings suggest that WNT5B is a promising candidate gene for the regulation of myogenesis and has potential benefits in improving skeletal muscle health and animal breeding.
Materials and Methods
Samples collection
A total of 200 sows were included in this study. DNA samples were obtained from obese-type breeds (Tongcheng sows, n = 40; Bama sows, n = 35; Luchuan sows, n = 25) and lean-type breeds (Landrace sows, n = 100), the age of sows selected are all in 180 days.
WNT5B-related Gene Set Enrichment Analysis (GSEA)
To explore the expression and enrichment of WNT5B, we employed an RNA-seq dataset spanning 27 developmental time points ranging from embryonic day 33 to postnatal day 180 in Tongcheng and Landrace pigs (NCBI: GSE157045). The WNT family genes were shown by heatmap via R “heatmap” package. In addition, GSEA-Gene Ontology (Gene Ontology) and Kyoto Encyclopedia of Genes and Genomes (GSEA-KEGG) analyses were performed by ranking the correlations between WNT5B and all genes and calculating the enrichment of the set of genes [39]. GSEA was performed using the “ClusterProfiler” package in R (v4.2.3).
Isolation and culture of porcine primary myoblasts
Porcine primary myoblasts were isolated from the biceps femoris of piglets that were less than one week of age. They have a good proliferation rate and myogenic stem cell properties, and are widely used in molecular and cellular experimental studies in vitro [7, 40]. The skeletal muscle tissue was minced and digested using type II collagenase (300 U/mL; Gibco, USA) in an oscillating water bath at 37°C for 30 min. The digestion process was terminated by adding a high-glucose medium (DMEM; Gibco, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, USA) and 2% double antibody. The mixture was then filtered through 40, 70, and 100 μm filters to remove tissue debris. The resulting cell mass was resuspended and cultured in RPMI-1640 medium. Purified satellite cells were transferred to plates coated with Matrigel (BD Biosciences, USA) for proliferation. When the porcine primary myoblasts reached 90% confluence, differentiation was induced by adding 5% horse serum (HS, Gibco, USA) to the DMEM.
Cell culture and transfection
For the in vitro experiments, C2C12 myoblasts and HEK293T cells were obtained from the American Type Culture Collection. Porcine skeletal muscle cells were obtained from Guangzhou Xinyuan Technology Co. Ltd., China. C2C12 myoblasts and HEK293T cells were cultured in Dulbecco's Modified Eagle's medium (DMEM; Sigma, USA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, China), 1% penicillin, 100 mg/mL streptomycin, and 1% glutamine (Mediatech, USA). The cells were incubated at 37℃ with 5% CO2. Plasmids and oligonucleotides were transfected into cells using Lipofectamine 3000 according to the manufacturer's instructions (Thermo Fisher, USA). Typically, transfection was performed when the cells reached 50-60% confluence.
SNP sequencing
The 3'UTR regions of Tongcheng, Bama, Luchuan and Landrace porcine WNT5B were amplified by PCR using genomic DNA as a template. The amplified regions were sequenced using Sanger sequencing (Sangon Biotech, China).
Vectors construction and oligonucleotides
The plasmids of pcDNA3.1 and pcDNA3.1-WNT5B were synthesized by GeneCreate, China. These plasmids were used for the overexpression studies. siRNA-NC, siRNA-WNT5B, all-miRNA mimics, mimic NC, miRNA inhibitors, and inhibitor-NC were obtained from RiboBio, China. These reagents were used for the knockdown and modulation studies. The small molecule RNA sequences are detailed in Table S1.
To construct the WNT5B 3'UTR dual luciferase reporter vector, the full-length 3'UTR of WNT5B was amplified and inserted into the PGL3 reporter vector (Promega, USA) downstream of firefly gene. The resulting vector contained two ARE sites at WNT5B 3'UTR position 1608 with A or G. To mutate the ARE sites, ATTTA < ATGTA mutations were introduced using overlap extension PCR and homologous recombination. The sequences used in this process are listed in Table S2.
Luciferase activity assay
In the experiment, cells were transfected with a total of 2 μg each of the luciferase reporter constructs using LipofectaminTM 3000 (Thermo Fisher, USA). Transfection was performed according to the manufacturer's instructions. After 36 h of transfection, the cells were harvested, and luciferase activity was analyzed using the Dual Luciferase Reporter Assay System (Promega, USA). Firefly luciferase was used as the primary reporter to monitor miRNA regulation and Renilla luciferase was used as the control reporter for normalization and screening.
Lentivirus packaging and transduction
To package lentivirus, 1.5×106 HEK293T cells were seeded in complete medium and incubated overnight at 37°C until reaching 70% confluence at 24 h. The next day, the target plasmids psPAX2 and PMD2.G (Addgene, USA) was mixed with Opti-MEM according to the corresponding ratios for transfection. The transfection complex was then added dropwise to the wells containing the HEK293T and incubated for 72 h at 37°C with CO2. After 72 h, the viral supernatants were collected. The harvested supernatant was centrifuged at 500✕g for 4 min to remove cell debris. The resulting supernatant was filtered using a 0.45 μm low-protein retention syringe filter (Sartorius, France), and 0.5 mL aliquots were stored at -80°C.
For lentivirus transduction, lentiviral particles containing the target gene (500 uL) were mixed with complete medium (500 uL). This mixture was added to each well of a twelve-well plate containing transduced cells. Polybrene (1 uL), a co-staining reagent, was also add to enhance transduction efficiency. The plates were incubated at 37°C with CO2 for 60 h.
Real-time quantitative PCR (qRT-PCR)
Cytoplasmic, nuclear, and tissue RNA were extracted using TRIzol (Invitrogen, China) and a Cytoplasmic, Nuclear RNA Extraction Kit (Norgen Biotek, USA). cDNA reverse transcription of mRNA was performed using the HiScript III 1st Strand cDNA Synthesis kit (+gDNA wiper) (Vazyme, China) following the manufacturer's instructions. qRT-PCR was performed using Fast ChamQ Universal SYBR qPCR Master Mix (Vazyme, China) according to the manufacturer's instructions. The relative expression of each example was calculated using the 2-ΔΔCT method. NEAT1 is a known nuclear lncRNA and GAPDH is a cytoplasmic-enriched gene. The primer sequences are listed in Table S3.
Western blot
Protein samples were extracted from treated cells or tissues using a protein lysis buffer consisting of RIPA buffer (Beyotime Biotechnology, China) and PMSF (Solarbio, China). Concentrations of the extracted proteins were determined using the BCA kit (Beyotime Biotechnology, China). To denature the protein samples, sodium dodecyl sulfate (SDS; CWBIO, China) was added and the samples were heated at 100°C for 20 min. Precast 10% and 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (EpiZyme, China) were used with a sample volume of 10 µL. After electrophoresis, the proteins were transferred onto a 0.45 μm hybridized nitrocellulose filter membrane (NC, Merck & Co, USA) and sealed with 5% skimmed milk powder. The membrane was then incubated with the primary and secondary antibodies. Primary antibodies, including WNT5B (1:1000, Abcam, ab164311, Cambridge, UK), Cyclin A2 (1:2000, Abcam ab181591, Cambridge, UK), MYH4 (1:1000, Proteintech 20140-1-AP, China) and MYH7 (1:1000, Proteintech 22280-1-AP, China), BAX (1:1000, Abcam ab32503, Cambridge, UK), BCL2 (1:2000, Abcam ab182858, Cambridge, UK), and GAPDH (1:1000, Abcam ab9484, Cambridge, UK). Secondary antibodies were used HRP-labeled Goat Anti-Mouse IgG(H+L) (1:1000, Beyotime, A0216, China) and HRP-labeled Goat Anti-Rabbit IgG(H+L) (1:1000, Beyotime, A0208, China). Primary and secondary antibodies were diluted in 1× Tween (TBST) buffer (EpiZyme, China) as per the manufacturer's instructions. The grayscale values of the protein bands were analyzed using ImageJ software (NIH, USA).
Cell Counting Kit-8 assay
The Cell Counting Kit-8 assay (CCK-8) was performed to measure cell proliferation in primary porcine skeletal muscle cells and C2C12 myoblasts, which were seeded in a 96-well plate for transfection. The Cell Counting Kit-8 (Beyotime Biotechnology, China) was used to detect cell growth at 0 h, 24 h, 36 h, 48 h, and 72 h post-transfection. For the assay, a mixture of CCK-8 reagent and complete culture medium was prepared in a 1:9 ratio and added to the wells. The plate was incubated at 37°C for 40 minutes. Subsequently, the absorbance at 450 nm was measured using a microplate reader, and the optical density (OD) values obtained were plotted to create a growth curve.
5-Ethynyl-2'-deoxyuridine (EdU) staining
Cells were cultured in 12-well plates and when they reached 50% confluence, they were transfected with plasmids, siRNA, miRNA mimics, miRNA inhibitor or corresponding controls. After 24 h of transfection, the cells were treated with 50 μM EdU (Beyotime Biotechnology, China) and incubated at 37°C for 1 or 2 h. Following this, the cells were fixed with 4% paraformaldehyde for 30 min, neutralized with a 2 mg/mL glycine solution and washed with 0.5% Triton X-100. The cells were then incubated with EdU working solution as per the manufacturer's instructions for 30 min at room temperature. Hoechst 33342 (Beyotime Biotechnology, China) was added to visualize the cell nuclei. The number of EdU-stained cells was determined using a confocal fluorescence microscope (A1HD25; Nikon, Japan). Three areas were randomly selected for the analysis.
Cell cycle assay
After 48 h of transfection, cells were collected and fixed with 70% ethanol at 4°C for 2 h. Cells were then stained with a solution containing propidium iodide (0.05 mg/mL), RNase A (1 mg/mL), and 0.3% Triton X-100 for 30 min in the dark. To determine the percentage of cells in different cell cycle phases, DNA content (propidium iodide staining) was measured using a flow cytometer (CytoFLEX, USA). The G1, S, and G2/M phase cell populations were analyzed using ModFitLT5 software (Verity Software House, USA). A total of 10,000 cells were analyzed per sample. Each experiment was repeated three times.
Apoptosis assay
Apoptosis was assessed using the FITC Annexin V Apoptosis Detection Kit (Yeasen Biotech, China) and analyzed by flow cytometry. Briefly, cells were harvested by digestion with EDTA-free trypsin 48 h after transfection. Double staining was performed using FITC Annexin V and propidium iodide according to the manufacturer's instructions, followed by flow cytometry (CytoFLEX, USA). Fluorescence data were analyzed using FlowJo software (Becton, Dickinson & Company, USA) for a total of 10,000 cells per sample. Each experiment was repeated three times.
Immunofluorescence staining
Porcine primary myoblasts were induced to differentiate for four days using 5% horse serum. The cells were incubated overnight at 4°C with MYH4 (1:1000, Proteintech, 20140-1-AP, China) and MYH7 (1:1000, Proteintech, 22280-1-AP, China) primary antibodies. Fluorescent secondary antibodies (FITC-conjugated Donkey Anti-Rabbit IgG (H+L) (1:200, GB22403, Servicebio, China) and Cy3 conjugated Donkey Anti-Mouse IgG (H+L) (1:200, GB21401, Servicebio, China) were then used in combination with the primary antibodies. After DAPI staining of cell nuclei, images were acquired using a fluorescent confocal microscope (A1HD25; Nikon, Japan). For immunofluorescence of tissue sections, 10 µm sections were incubated overnight at 4°C with laminin (1:1000, Abcam, ab11575, Cambridge, UK), MYH4 (1:1000, Proteintech, 20140-1-AP, China) and MYH7 (1:1000, Proteintech, 22280-1-AP, China) primary antibodies. Fluorescent secondary antibodies (FITC-conjugated Donkey Anti-Rabbit IgG (H+L) (1:200; Servicebio, GB22403, China) and Cy3 conjugated Donkey Anti-Mouse IgG (H+L) (1:200; Servicebio, GB21401, China) were used in combination with the primary antibody. After DAPI staining of cell nuclei, images were acquired using a fluorescence confocal microscope (A1HD25; Nikon, Japan).
Succinate Dehydrogenase (SDH) staining
Frozen sections (10 µm) were immersed in a 0.2 M sodium phosphate buffer solution (pH = 7.6) containing 0.6 mM nitro blue tetrazolium and 50 mM sodium succinate (Sigma-Aldrich, USA). The sections were then incubated at 37°C for 30 min. Images were captured using a confocal fluorescence microscope (A1HD25; Nikon, Japan).
Statistical analysis
GraphPad Prism 9.0 software (GraphPad Software, USA) was used for statistical analysis. All experiments were repeated at least three times. All data are expressed as means ± S.E.M and analyzed with an unpaired Student's t test for statistical significance. p <0.05 was considered statistically significant. *p < 0.05, **p < 0.01, ***p < 0.001, p ≥ 0.05: ns (Not significant).
Results
WNT5B is a potential regulator for myogenesis
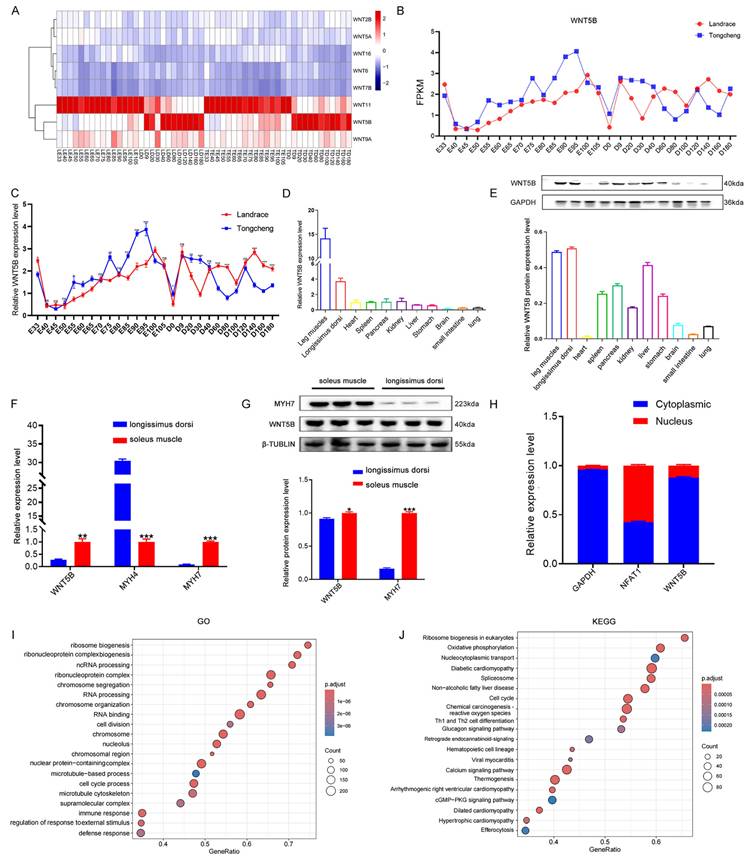
To investigate the regulatory role of WNT family genes in porcine myogenesis, we conducted RNA-seq analysis and observed the differential expression of WNT5B and WNT11 at different developmental stages (Figure 1A). Additionally, WNT5B was differentially expressed at 27 time points during skeletal muscle development in Tongcheng and Landrace pigs, whereas WNT11 was not (Figure 1B and S1A). The qRT-PCR results further confirmed that WNT5B was differentially expressed at different time points in Tongcheng and Landrace pigs (Figure 1C). Tissue expression analysis revealed that WNT5B displayed specific highly expressed in the leg muscle and longissimus dorsi (Figure 1D-E). Moreover, WNT5B was highly expressed in the soleus muscle (Figure 1F-G), and mainly in the myoblast cytoplasm of Tongcheng and Landrace pigs (Figure 1H and S1B). To further explore the WNT5B function, GO and KEGG analyses were performed, revealing that WNT5B and its co-expressed genes were associated with various biological processes and signaling pathways, including chromosome organization, cell division, the cGMP-PKG signaling pathway, the glucagon signaling pathway, and the cell cycle (Figure 1I-J). Furthermore, the GO terms and KEGG pathways were significantly enriched in the cell cycle, which is directly related to skeletal muscle cell proliferation, regeneration, myogenesis, and development [41-43]. These results suggest that WNT5B is a candidate factor involved in the regulation of skeletal muscle development.
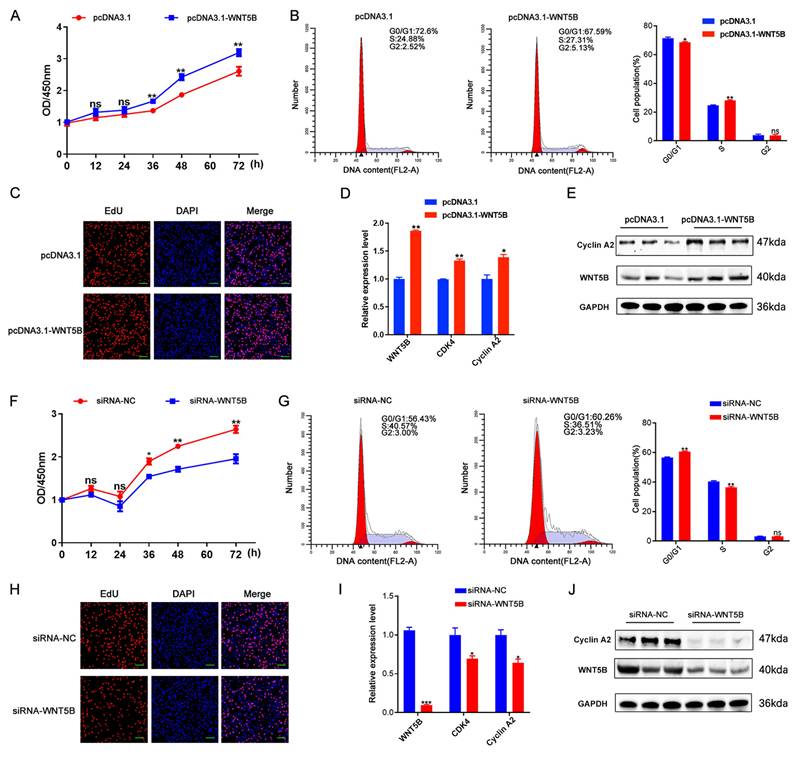
WNT5B regulates cell proliferation and cell cycle
The effects of WNT5B on the proliferation and cell cycle of porcine myoblasts were subsequently examined. To elucidate its functional role, siRNA and overexpression vectors targeting WNT5B were constructed and transfected into porcine myoblasts. The results showed that WNT5B overexpression significantly decreased the percentage of cells in the G0/G1 phase and increased the percentage of cells in the S phase, as well as the number of proliferating and EdU-positive cells (Figure 2A-C). Moreover, WNT5B overexpression upregulated cell proliferation and the expression of cell cycle markers (CDK4 and Cyclin A2) (Figure 2D-E). Conversely, knockdown of WNT5B resulted in the opposite effects compared to its overexpression (Figure 2F-J). Sequence conservation analysis revealed that WNT5B is highly conserved in pigs and mice (Figure S2A). Consequently, the effects on cell proliferation and cell cycle were examined by transfecting porcine WNT5B overexpression vectors and siRNA into C2C12 myoblasts. The results showed that the effects of knockdown and overexpression of WNT5B in C2C12 myoblasts on cell proliferation and the cell cycle were consistent with those observed in porcine myoblasts. Overexpression of WNT5B promoted cell proliferation and cell cycle progression, while knockdown of WNT5B inhibited the proliferation and cell cycle progression of C2C12 myoblasts (Figure S3). These results suggest that WNT5B regulates myogenesis and demonstrates functional conservation across species.
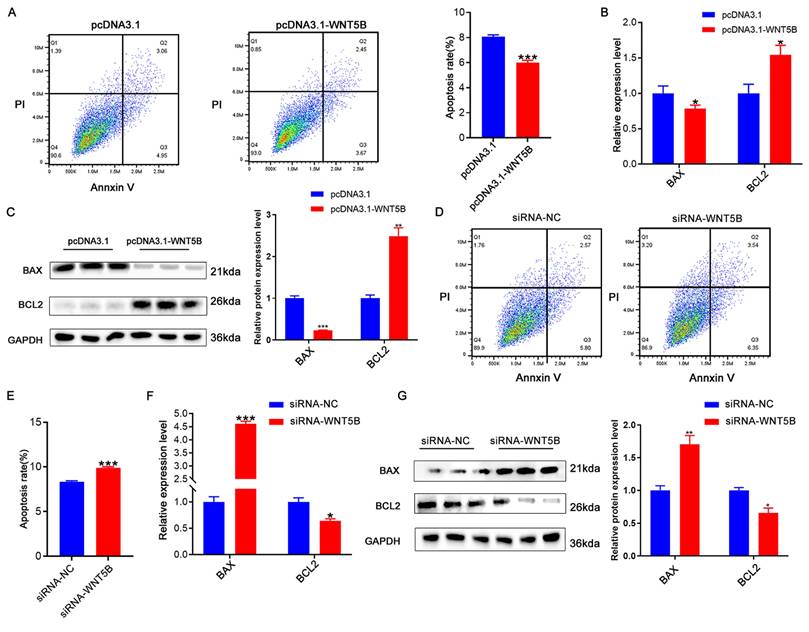
WNT5B regulates cell apoptosis in myoblasts
To investigate the role of WNT5B in myogenesis, we examined its effect on apoptosis. Overexpression of WNT5B led to a reduction in apoptosis rates in porcine and C2C12 myoblasts (Figure 3A and S4A). Moreover, at both the mRNA and protein levels, WNT5B overexpression resulted in decreased BAX expression and increased BCL2 expression (Figure 3B-C and S4B-C). Conversely, WNT5B knockdown promoted apoptosis in porcine and C2C12 myoblasts (Figure 3D-E and S4D-E). BAX expression was upregulated, while BCL2 expression was downregulated (Figure 3F-G and S4F-G). Collectively, these results suggest that WNT5B promotes cell proliferation and cell cycle progression and suppresses myoblast apoptosis.
WNT5B regulates fiber type conversion of porcine skeletal muscle
The results presented in Figure 1F-G suggest that WNT5B may play a role in regulating the conversion of myofiber types. The effects of WNT5B on pig myofiber myogenesis were examined. In vitro, knockdown of WNT5B down-regulated the expression of slow muscle-specific genes (MYH7, TNNI1, and TNNT1) and upregulated the expression of fast muscle-specific genes (MYH4, TNNI2, and TNNT3) (Figure 4A). Immunofluorescence experiments confirmed that WNT5B knockdown inhibited the generation of slow muscle fiber (Figure 4B). Furthermore, the WNT5B overexpression vector was packaged using a lentivirus. Its impact on the myofiber area and myofiber type conversion in porcine skeletal muscle was examined 28 days post-injection into quadriceps. In vivo, slow muscle-specific genes (MYH7, TNNI1, and TNNT1) were upregulated and fast muscle-specific genes (MYH4, TNNI2, and TNNT3) downregulated after WNT5B overexpression (Figure 4C). Western blot and immunofluorescence assays further revealed that WNT5B overexpression resulted in increased MYH7 expression, decreased MYH4 expression, and an increase in the number of slow muscle fibers in vivo (Figure 4D-F). Additionally, SDH and Laminin staining demonstrated that WNT5B overexpression promoted the formation of slow fibers and increased the muscle fiber area in vivo (Figure 4G-H). These results indicate that WNT5B plays a significant role in fiber type conversion.
Expression patterns of WNT5B gene. (A) The heatmap of expression pattern of WNT family-related genes in developmental skeletal muscle across 27 time points in Landrace and Tongcheng pigs (LE: the embryonic stage of Landrace pig; LD: the postnatal day of Landrace pig; TE: the embryonic stage of Tongcheng pig; TD: the postnatal day of Tongcheng pig) based on the RNA-seq data. (B) RNA-seq analysis the expression level of WNT5B changes at 27 different developmental time points. (C) qRT-PCR result for the expression pattern of WNT5B in skeletal muscle from Landrace and Tongcheng pigs across 27 developmental time points. (D-E) The expression of WNT5B at mRNA (D) and protein (E) level in eleven different tissues of Tongcheng pigs. (F-G) The expression of WNT5B at mRNA (F) and protein (G) level in fast muscle fibers and slow muscle fibers. (H) qRT-PCR analysis WNT5B expression in the cytoplasm and nucleus of Landrace pig myoblasts. (I-J) GO (I) and KEGG (J) analysis of WNT5B co-expressed genes. Data are presented as mean ± SEM and analyzed for statistical differences between groups using unpaired two-tailed t-tests. *p < 0.05, **p < 0.01, ***p < 0.001, ns means no significant differences.
WNT5B promotes skeletal muscle cell proliferation and cycle in porcine. (A-C) The results of CCK-8 (A), cell cycle (B) and cell proliferation status (C) of porcine skeletal muscle cells after transfection with pcDNA3.1 and pcDNA3.1-WNT5B vectors. Scale bar, 50 μm. (D-E) mRNA (D) and protein (E) expression levels of proliferation marker genes in porcine skeletal muscle cells after WNT5B overexpression. (F-H) The results of CCK-8 (F), cell cycle (G) and cell proliferation status (H) of porcine skeletal muscle cells after transfection with siRNA-NC and siRNA-WNT5B. Scale bar, 50 μm. (I-J) mRNA (I) and protein (J) expression levels of proliferation marker genes in porcine skeletal muscle cells after WNT5B knockdown. Data are presented as mean ± SEM and analyzed for statistical differences between groups using unpaired two-tailed t-tests. *p < 0.05, **p < 0.01, ***p < 0.001, ns means no significant differences.
The SNP c.1608 A > G in the 3'UTR affects WNT5B mRNA stability
Given WNT5B gene varied in the transcriptomic data from 27 different developmental time points in the skeletal muscle of obese and lean pig breeds, we aimed to investigate the reasons behind the differences. Gene expression is influenced by various regulatory factors, and the 3'UTR sequence contains key elements that regulate the spatial and temporal expression of gene mRNAs, thereby determining cellular fates [44, 45]. Previous studies have demonstrated the association between genetic variations in the 3'UTR and livestock and poultry production performance [46, 47]. This study investigated whether there was SNP site in the 3'UTR of the WNT5B gene that could regulate its expression. The 3'UTR of the WNT5B gene was amplified in different breeds of obese-type (Tongcheng, Bama, Luchuan) and lean-type (Landrace) porcine, and a SNP site, named c.1608 A > G, was identified at position 1608 of the WNT5B 3'UTR. WNT5B was most highly expressed in the AA genotype, followed by the AG genotype, and least highly expressed in the GG genotype (Figure 5A-B). To further understand the effects of the c.1608 A > G SNP on the post-transcriptional regulation and mRNA stability of WNT5B, a dual-luciferase vector was constructed containing the full-length sequence of WNT5B 3'UTR with either A (WNT5B-A) or G (WNT5B-G) at the SNP site (Figure 5C). The dual-luciferase activity assay results revealed that the luciferase activity of the WNT5B-G 3'UTR was 30-40% lower than that of the WNT5B-A 3'UTR (Figure 5D). Furthermore, analysis of luciferase mRNA levels showed that luciferase mRNA containing the WNT5B-G 3'UTR decayed twice as fast as mRNA containing the WNT5B-A 3'UTR, indicating that a single-nucleotide change at the 3'UTR of WNT5B affects the overall stability of mRNA transcripts (Figure 5E-F).
The 3'UTR of WNT5B contains two adenosine-uridine repeats (AREs) in the cis-regulatory region, which are known to be involved in mRNA stability by binding to degradative RNA-binding proteins (Figure 5G). To investigate the role of AREs in the differential regulation of WNT5B-A and WNT5B-G, dual-luciferase expression vectors were constructed: one with the wild-type sequence and the other with a mutation that altered the original AUUUA sequence to AUGUA. The dual-luciferase activity assays showed that the WNT5B-A ΔARE and WNT5B-G ΔARE 3'UTRs exhibited similar rescue efficiency compared to the WNT5B-A and WNT5B-G 3'UTRs (Figure 5H). Furthermore, the differential regulation of WNT5B-A versus WNT5B-G was maintained in the presence of ARE2 alone, independent of ARE1 (Figure 5I-J and S5).
The effects of WNT5B on cell apoptosis. (A) The results of cell apoptosis of porcine skeletal muscle cells after WNT5B overexpression. (B-C) mRNA (B) and protein (C) expression levels of cell apoptosis markers genes in porcine skeletal muscle cells after WNT5B overexpression. (D-E) The results of cell apoptosis of porcine skeletal muscle cells after WNT5B knockdown. (F-G) mRNA (F) and protein (G) expression levels of cell apoptosis marker genes in porcine skeletal muscle cells after WNT5B knockdown. Data are presented as mean ± SEM and analyzed for statistical differences between groups using unpaired two-tailed t-tests. *p < 0.05, **p < 0.01, ***p < 0.001, ns means no significant differences.
The effects of WNT5B on the muscle fiber type conversion. (A) The mRNA expression level of muscle fiber type transformation markers in porcine primary myoblast after WNT5B knockdown. (B) Immunofluorescence staining results of slow and fast muscle fiber transformation in porcine primary myoblasts after WNT5B knockdown. Scale bar, 50 μm. (C-D) The results of WNT5B overexpression of mRNA (C) and protein (D) expression levels of muscle fiber type transformation markers in vivo. (E-F) Immunofluorescence (E) results of the regeneration of slow and fast fibers after overexpression of WNT5B in vivo. Scale bar, 20 μm. The quantity (F) results of slow- and fast-fibers regeneration. (G) Succinate dehydrogenase (SDH) staining results of the slow muscle fiber formation after WNT5B overexpression in vivo. (H) Laminin staining results of muscle fiber thickness after WNT5B overexpression in vivo. Data are presented as mean ± SEM and analyzed for statistical differences between groups using unpaired two-tailed t-tests. *p < 0.05, **p < 0.01, ***p < 0.001, ns means no significant differences.
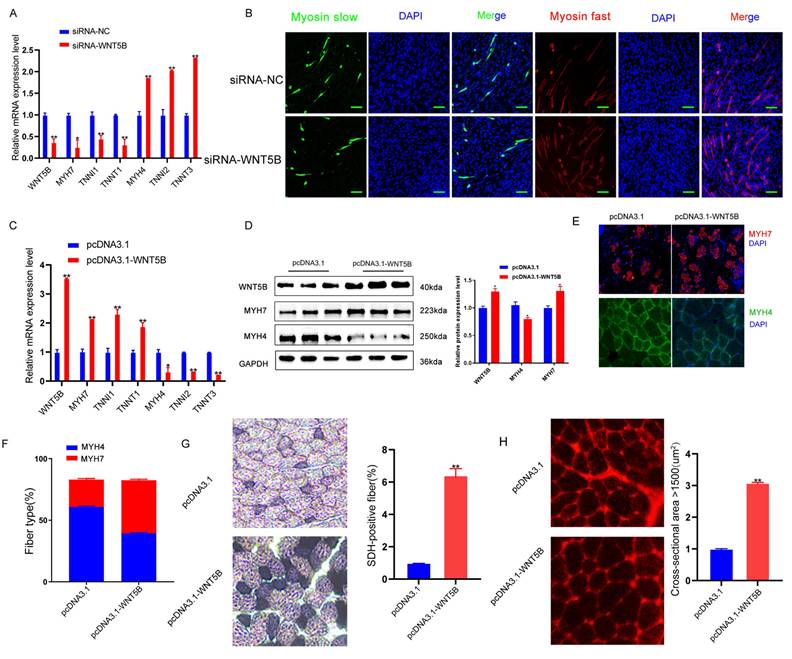
miR-29a/b/c regulates myogenesis
Recent studies have shown that miRNAs collaborate with ARE-binding proteins to destabilize cytokines [48-50], miRNA play a role in silencing target genes by recognizing the 3'UTR region of target mRNAs and forming a silencing complex [51]. This study further analyzed whether the WNT5B 3'UTR SNP affects miRNA binding. Interestingly, this SNP was located in the MRE (microRNA Response Elements) region of miR-29a/b/c (Figure S6A). Additionally, miR-29a/b/c sequences were conserved among various species (Figure S2B). Dual-luciferase activity assays confirmed that miR-29a/b/c bound to the G site but not to the A site (Figure 5K-L). Following overexpression of miR-29a/b/c into WNT5B-G porcine myoblasts, both mRNA and protein levels of WNT5B were downregulated, indicating that miR-29a/b/c can indeed bind to the WNT5B 3'UTR (Figure 5M-N).
We subsequently investigated the effects of miR-29a/b/c on cell proliferation, cell cycle, and apoptosis. EdU staining, cell cycle analysis, and apoptosis assays demonstrated that knockdown of miR-29a/b/c enhanced cell proliferation and cell cycle progression, while suppressing apoptosis in porcine skeletal muscle cells (Figure 6A-F and S7A-B). Additionally, inhibition of miR-29a/b/c led to increased mRNA expression of CDK4, Cyclin A2, and BCL2, and decreased expression of BAX and CASP3 (Figure 6G-H). Western blot analysis further revealed that knockdown of miR-29a/b/c resulted in elevated expression of Cyclin A2 and BCL2 proteins, as well as decreased expression of BAX protein (Figure 6I). Overexpression of miR-29a/b/c had effects on cell proliferation, cell cycle, and apoptosis that were opposite to those observed with knockdown (Figure S8A-I). Furthermore, the binding site for miR-29a/b/c was conserved between pig and mouse in WNT5B 3'UTR and the function of miR-29a/b/c in C2C12 myoblasts proliferation, cell cycle and apoptosis were consistent with those observed in porcine myoblast (Figure S6B, S7C-D and S9A-L).
miR-29a/b/c regulates myogenesis by targeting WNT5B-G 3'UTR
Next, we tested whether miR-29a/b/c could regulate cell proliferation, cell cycle, and apoptosis through WNT5B by co-transfection experiments. Results from EdU staining, cell cycle analysis, and apoptosis assays demonstrated that miR-29a/b/c knockdown significantly impacted these cellular processes, but these effects were rescued by WNT5B siRNA (Figure 7A-F). Additionally, the effect of miR-29a/b/c inhibitor on the mRNA expression of CDK4, Cyclin A2, BAX, CASP3, and BCL2 was reversed by WNT5B knockdown (Figure 7G-H). This rescue was also observed in C2C12 myoblasts (Figure S10A-H).
Effects of SNPs on WNT5B mRNA stability. (A) The sequencing results indicate the 3'UTR of the WNT5B gene in obese-type and lean-type pigs. A SNP site, named c.1608 A > G. (B) The mRNA expression of WNT5B in AA, AG, GG genotype pigs. (C) Schematic of a dual-luciferase reporter vector construction containing WNT5B-A-3'UTR or WNT5B-G-3'UTR vectors. (D) Dual-luciferase activity was analyzed after transfection with WNT5B-A-3'UTR or WNT5B-G-3'UTR vector in HEK-293T cells. (E) The luciferase mRNA expression level at 2 hours after transfection of WNT5B-A and WNT5B-G. (F) The expression levels of WNT5B-A and WNT5B-G at various time points following treatment with actinomycin D. (G) Positional pattern of the ARE sites on the WNT5B 3'UTR. (H-I) The results of the dual-luciferase activity assay after mutation ARE sites in WNT5B-A-3'UTR or WNT5B-G-3'UTR vectors. (J) The effects of WT and ARE2 (right) site mutations on WNT5B-G mRNA secondary structure. (K-L) Dual-luciferase were used to analyze the effect of miR-29a/b/c on the SNP (c.1608 A > G) in WNT5B. (M-N) Expression levels of WNT5B mRNA (M) and protein (N) after miR-29a/b/c overexpression in porcine myoblasts. Data are presented as mean ± SEM and analyzed for statistical differences between groups using unpaired two-tailed t-tests. *p < 0.05, **p < 0.01, ***p < 0.001, ns means no significant differences.
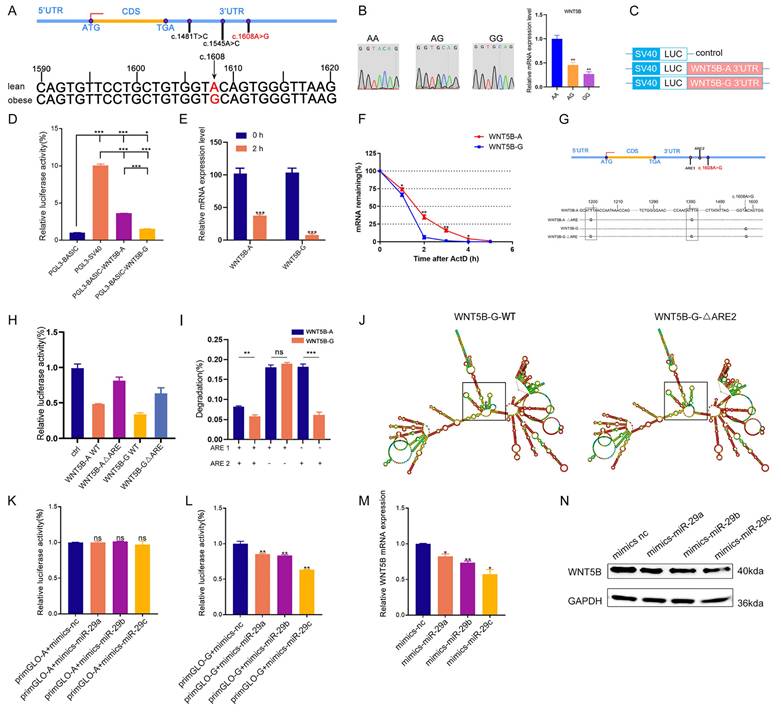
The effects of miR-29a/b/c knockdown on proliferation and apoptosis in porcine skeletal muscle cells. (A-C) The results of cell proliferation (A), cell cycle (B), and cell apoptosis (C) after transfection with miR-29a/b/c inhibitor in porcine skeletal muscle cells. Scale bars, 50 μm. (D-F) Quantitative results of cell proliferation (D), cell cycle (E), and cell apoptosis (F). (G-H) The mRNA expression of cell cycle (G) and cell apoptosis (H) markers expression after miR-29a/b/c knockdown in porcine skeletal muscle cells. (I) The protein expression of cell cycle and cell apoptosis markers after miR-29a/b/c knockdown in porcine skeletal muscle cells. Data are presented as mean ± SEM and analyzed for statistical differences between groups using unpaired two-tailed t-tests. *p < 0.05, **p < 0.01, ***p < 0.001, ns means no significant differences.
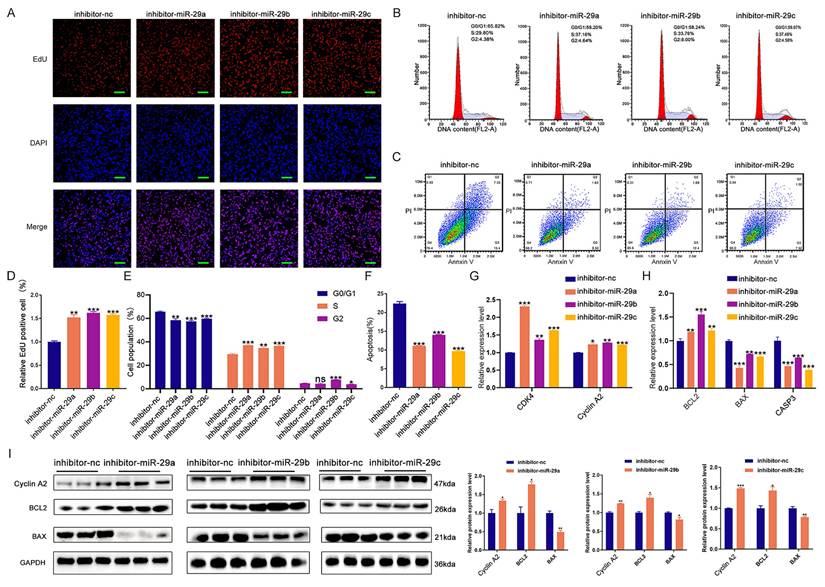
We further investigated whether ARE-binding sites are involved in miRNA-induced silencing complex (miRISC) recruitment. miR-29a/b/c mimics were co-transfected with WNT5B-A 3'UTR and WNT5B-G 3'UTR along with the WNT5B ΔARE luciferase reporter vector. Similar to the effects observed with the WNT5B-A and WNT5B-G 3'UTR, miR-29a/b/c inhibited the WNT5B-G ΔARE 3'UTR but had no significant effect on the WNT5B-A ΔARE 3'UTR (Figure 7I). Thus, miRISC recruitment to the WNT5B-G 3'UTR does not require an ARE-binding protein. Overall, these results indicate that miR-29a/b/c can directly target and mediate the degradation of the WNT5B-G 3'UTR. The A-to-G position SNP have a protective effect against inhibition by miR-29a/b/c.
Discussion
WNT5B plays crucial roles in various biological processes, including osteogenic differentiation, cartilage formation, adipose differentiation, etc. [16, 52]. However, its role in myogenesis has not yet been fully elucidated. A previous study indicated differential expression of WNT5B during myogenesis induction in the P19 cell line [53]. In this study, differential expression of WNT5B was observed in obese and lean pigs during skeletal muscle development. Its expression was higher in the soleus muscle. These findings suggest that WNT5B plays an important role in skeletal muscle development.
The study demonstrates that WNT5B promotes the proliferation and cell cycle of porcine skeletal muscle cells. This is consistent with previous reports showing that WNT5B promotes cell proliferation and cell cycle progression in cancer and LAD cells [21, 36]. Additionally, overexpression of WNT5B inhibited CASP3/7 activity and apoptosis in HEK293T cells [54]. Our study further demonstrated that WNT5B inhibits apoptosis in porcine skeletal muscle cells. Moreover, the effects of WNT5B on cell proliferation, cell cycle, and apoptosis are functionally conserved in pigs and mice. Importantly, WNT5B expression was higher in slow muscle fibers than in fast muscle fibers, and promoted the transformation of fast to slow fibers. Different muscle fiber types are associated with muscle thickness and meat quality [55-58]. Therefore, the results suggest that WNT5B regulates myogenesis and plays an important role in meat quality traits.
miR-29a/b/c regulates cell proliferation and apoptosis through binding to WNT5B. (A-C) The results of cell proliferation (A), cell cycle (B), and cell apoptosis (C) after miR-29a/b/c and WNT5B knockdown in porcine skeletal muscle cells. Scale bar, 50 μm. (D-F) Quantitative results of cell proliferation (D), cell cycle (E), and cell apoptosis (F). (G-H) The expression of cell cycle (G) and cell apoptosis markers (H) in mRNA level after co-transfection with miR-29a/b/c inhibitor and WNT5B siRNA in porcine skeletal muscle cells. (I) The effects of co-transfection with miR-29a/b/c mimics and mutation of ARE1 and ARE2 sites on WNT5B-A-3'UTR or WNT5B-G-3'UTR vectors. Data are presented as mean ± SEM and analyzed for statistical differences between groups using unpaired two-tailed t-tests. *p < 0.05, **p < 0.01, ***p < 0.001, ns means no significant differences.
c.1608 A > G SNP protects the WNT5B gene 3'UTR from degradation by the miR-29 family of genes, affecting proliferation, cell cycle and apoptosis in skeletal muscle cells.
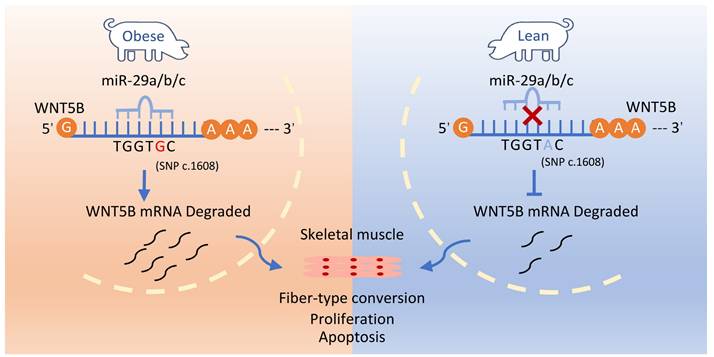
To investigate the regulatory mechanism of WNT5B on myogenesis, we analyzed the SNP c.1608 A > G located at the 3'UTR of WNT5B in porcine. We found that the G allele at the c.1608 A > G SNP site led to a decrease in WNT5B mRNA stability. Additionally, the SNP reduced the miR-29a/b/c miRNA response element motif. It was discovered that miR-29a/b/c inhibited the activity of the WNT5B-G 3'UTR luciferase reporter vector, but had no effect on the activity of the WNT5B-A 3'UTR luciferase reporter vector. Moreover, the SNP attenuated the inhibitory effect of miR-29a/b/c on WNT5B 3'UTR activity. These findings are consistent with previous reports indicating that SNPs can alter miRNA interactions with the target 3'UTRs, resulting in reduced the degradation of the target gene [59]. Previous studies have shown that SNPs located within the miRNA-binding motifs disrupt their interactions, thereby altering the expression of target genes and influencing cellular processes [60, 61]. In our study, miR-29a/b/c regulated the proliferation and apoptosis of porcine and mouse skeletal muscle cells by targeting WNT5B.
Mutations in RNA regulatory elements have been shown to impact gene expression and function by affecting interactions between RNA-binding proteins, miRNAs, and gene 3'UTRs [61-63]. For example, in the BMP2 3'UTR, the SNP rs15705 has been identified as a variant associated with osteoporosis [64]. Furthermore, the SNP located in the ARE binding domain of BMP2 has been shown to affect its 3'UTR activity and post-transcriptional regulation [65-67]. This study examined the 3'UTR of WNT5B gene and classified it as an ARE-containing mRNA due to its possession of two copies of the pentameric AUUUA motif. Further experiments revealed that the inhibitory effect of miR-29a/b/c on the WNT5B 3'UTR was abolished upon mutation of the two ARE sites. Notably, a mutation in the second ARE site near the c.1608 A > G SNP was observed to directly rescue the luciferase activity of WNT5B. This outcome may be due to the alteration of ARE sites, which can influence the secondary structure of WNT5B mRNA. However, further studies are required to confirm these regulatory mechanisms. miRNAs cooperate with ARE-binding proteins to destabilize mRNAs that encode cytokines. AREs serve as a signal for miRNA-activated translation and under miRNA guidance, miRISC complex members such as AGO and FMRP are recruited to ARE to activate translation and regulate gene expression [68].
Conclusions
This study identified and confirmed a functional SNP site (c.1608 A > G) in the WNT5B 3'UTR, which influences WNT5B mRNA stability through the ARE binding site. This functional SNP is located within the miR-29a/b/c miRNA response element motif. The c.1608 A > G SNP protects the WNT5B gene 3'UTR from degradation by miR-29 family genes, leading to an increase in WNT5B expression. This upregulation subsequently regulates the proliferation, cell cycle, and apoptosis of skeletal muscle cells. Our results suggest that WNT5B has significant and promising effects on porcine skeletal muscle production traits (Figure 8).
Abbreviations
ARE: Adenosine-uridine Repeats Element
MRE: MicroRNA Response Elements
ATCC: American Type Culture Collection
BSA: Bovine Serum Albumin
circRNA: Circular no-coding RNA
DAPI: 4',6-Diamidino-2-phenylindole dihydrochloride
DEPC: Diethyl Pyrocarbonate
DMEM: Dulbecco's Modified Eagle Medium
DPBS: Dulbecco's Phosphate-Buffered Saline
EDTA: Ethylene Diamine Tetraacetic Acid
EdU: 5-Ethynyl-2'-deoxyuridine
FBS: Fetal Bovine Serum
GO: Gene Ontology
GSEA: Gene Set Enrichment Analysis
KEGG: Kyoto Encyclopedia of Genes and Genomes
lncRNA: Long no-coding RNA
miRISC: miRNA-induced silencing complex
miRNA: MicroRNA
MYH: Myosin Heavy Chains
PBS: Phosphate Buffered Saline
PI: Propidium Iodide
PS: Penicillin-Streptomycin
PMSF: Phenylmethanesulfonylfluoride
qRT-PCR: quantitative real-time PCR
SDS: Sodium dodecyl sulfate
siRNA: Small interfering RNA
SNP: Single nucleotide polymorphism
TBST: TBS with Tween-20
UTR: Untranslated region
WNT: Wingless-integrated
WNT5B: WNT Family Member 5B
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by Science and Technology Major Project from Shenzhen (KJZD20230923115003006), the National Natural Science Foundation of China (U23A20229 and 32302718), the Guangxi Science and Technology Plan project (GK-AB21196060) and Agricultural Science and Technology Innovation Program (CAAS-ZDRW202406).
Author contributions
Z.T. designed and supervised the study. D.F., Yil. Y., C.Y., F.L. and B. X. performed the experiment. D.F. and Yil. Y. statistically analyzed and summarized results. D.F., Yil. Y., and C.Y. wrote the manuscript. Z.T., Yil. Y., C.Y. and Z.T. revised the manuscript. C.Y., and Yal. Y. performed the bioinformatic analysis. All authors read and approved the final manuscript.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
All experimental animal procedures were reviewed and approved by the guidelines of Institutional Animal Care and Use Committee of Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, the animal Ethics Committee approval number is AGIS2020.04.17.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chen M, Wang Y, Deng S, Lian Z, Yu K. Skeletal muscle oxidative stress and inflammation in aging: Focus on antioxidant and anti-inflammatory therapy. Front Cell Dev Biol. 2022;10:964130
2. Mukund K, Subramaniam S. Skeletal muscle: A review of molecular structure and function, in health and disease. Wiley Interdiscip Rev Syst Biol Med. 2020;12:e1462
3. Iberite F, Gruppioni E, Ricotti L. Skeletal muscle differentiation of human iPSCs meets bioengineering strategies: perspectives and challenges. npj Regener Med. 2022;7:23
4. Vishal K, Lovato TL, Bragg C, Chechenova MB, Cripps RM. FGF signaling promotes myoblast proliferation through activation of wingless signaling. Dev Biol. 2020;464:1-10
5. Luo W, Chen J, Li L, Ren X, Cheng T, Lu S. et al. c-Myc inhibits myoblast differentiation and promotes myoblast proliferation and muscle fibre hypertrophy by regulating the expression of its target genes, miRNAs and lincRNAs. Cell Death Differ. 2019;26:426-42
6. Liu Y, Yao Y, Zhang Y, Yan C, Yang M, Wang Z. et al. MicroRNA-200c-5p Regulates Migration and Differentiation of Myoblasts via Targeting Adamts5 in Skeletal Muscle Regeneration and Myogenesis. Int J Mol Sci. 2023;24:4995
7. Yang Y, Yan J, Fan X, Chen J, Wang Z, Liu X. et al. The genome variation and developmental transcriptome maps reveal genetic differentiation of skeletal muscle in pigs. PLos Genet. 2021;17:e1009910
8. Chen B, Wang Y, Hou D, Zhang Y, Zhang B, Niu Y. et al. Transcriptome-Based Identification of the Muscle Tissue-Specific Expression Gene CKM and Its Regulation of Proliferation, Apoptosis and Differentiation in Chicken Primary Myoblasts. Animals. 2023;13:2316
9. Sun J, Ruan Y, Xu J, Shi P, Xu H. Effect of Bovine MEF2A Gene Expression on Proliferation and Apoptosis of Myoblast Cells. Genes. 2023;14:1498
10. Xue C, Chu Q, Shi Q, Zeng Y, Lu J, Li L. Wnt signaling pathways in biology and disease: mechanisms and therapeutic advances. Signal Transduction Targeted Ther. 2025;10:106
11. De A. Wnt/Ca 2+ signaling pathway: a brief overview. Acta Biochim Biophys Sin. 2011;43:745-56
12. Katoh M. WNT/PCP signaling pathway and human cancer. Oncol Rep. 2005;14:1583-8
13. Hayat R, Manzoor M, Hussain A. Wnt signaling pathway: A comprehensive review. Cell Biol Int. 2022;46:863-77
14. Xiao Q, Chen Z, Jin X, Mao R, Chen Z. The many postures of noncanonical Wnt signaling in development and diseases. Biomed Pharmacother. 2017;93:359-69
15. Chen Y, Chen Z, Tang Y, Xiao Q. The involvement of noncanonical Wnt signaling in cancers. Biomed Pharmacother. 2021;133:110946
16. Suthon S, Perkins RS, Bryja V, Miranda-Carboni GA, Krum SA. WNT5B in Physiology and Disease. Front Cell Dev Biol. 2021;9:667581
17. He J, Wang X, Liang C, Yao X, Zhang Z, Yang R. et al. Wnt5b/Ryk-mediated membrane trafficking of P2X3 receptors contributes to bone cancer pain. Exp Neurol. 2020;334:113482
18. Hendrickx G, Boudin E, Steenackers E, Nielsen TL, Andersen M, Brixen K. et al. Genetic Screening of WNT4 and WNT5B in Two Populations with Deviating Bone Mineral Densities. Calcif Tissue Int. 2017;100:244-9
19. Maegawa H, Maeda S. Role of TFAP2B and Wnt5B in adipose tissue. Nihon Rinsho. 2006;64(Suppl 9):244-8
20. Ren J, Han P, Ma X, Farah EN, Bloomekatz J, Zeng XI. et al. Canonical Wnt5b Signaling Directs Outlying Nkx2.5+ Mesoderm into Pacemaker Cardiomyocytes. Dev Cell. 2019;50:729-43.e5
21. Harada T, Yamamoto H, Kishida S, Kishida M, Awada C, Takao T. et al. Wnt5b-associated exosomes promote cancer cell migration and proliferation. Cancer Sci. 2017;108:42-52
22. Wang F, Zhen Y, Si C, Wang C, Pan L, Chen Y. et al. WNT5B promotes vascular smooth muscle cell dedifferentiation via mitochondrial dynamics regulation in chronic thromboembolic pulmonary hypertension. J Cell Physiol. 2022;237:789-803
23. Tada M, Heisenberg C-P. Convergent extension: using collective cell migration and cell intercalation to shape embryos. Development. 2012;139:3897-904
24. Yang Y, Mlodzik M. Wnt-Frizzled/planar cell polarity signaling: cellular orientation by facing the wind (Wnt). Annu Rev Cell Dev Biol. 2015;31:623-46
25. Zhang C, Brunt L, Ono Y, Rogers S, Scholpp S. Cytoneme-mediated transport of active Wnt5b-Ror2 complexes in zebrafish. Nature. 2024;625:126-33
26. Ge X, Ma H, Zheng X, Ruan H, Liao X, Xue W. et al. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. 2013;104:1675-82
27. Roy B, Dunbar M, Agrawal J, Allen L, Dwivedi Y. Amygdala-Based Altered miRNome and Epigenetic Contribution of miR-128-3p in Conferring Susceptibility to Depression-Like Behavior via Wnt Signaling. Int J Neuropsychopharmacol. 2020;23:165-77
28. Suthon S, Lin J, Perkins RS, Crockarell JR Jr, Miranda-Carboni GA, Krum SA. Estrogen receptor alpha and NFATc1 bind to a bone mineral density-associated SNP to repress WNT5B in osteoblasts. Am J Hum Genet. 2022;109:97-115
29. Smith AR, Smith RG, Pishva E, Hannon E, Roubroeks JAY, Burrage J. et al. Parallel profiling of DNA methylation and hydroxymethylation highlights neuropathology-associated epigenetic variation in Alzheimer's disease. Clin Epigenet. 2019;11:52
30. Küpers LK, Fernández-Barrés S, Nounu A, Friedman C, Fore R, Mancano G. et al. Maternal Mediterranean diet in pregnancy and newborn DNA methylation: a meta-analysis in the PACE Consortium. Epigenetics. 2022;17:1419-31
31. Sasidharan Nair V, Saleh R, Toor SM, Taha RZ, Ahmed AA, Kurer MA. et al. Transcriptomic profiling disclosed the role of DNA methylation and histone modifications in tumor-infiltrating myeloid-derived suppressor cell subsets in colorectal cancer. Clin Epigenet. 2020;12:13
32. Hatta M, Naganuma K, Kato K, Yamazaki J. 3-Deazaneplanocin A suppresses aggressive phenotype-related gene expression in an oral squamous cell carcinoma cell line. Biochem Biophys Res Commun. 2015;468:269-73
33. Chen G, Wang Q, Li Z, Yang Q, Song Y. Circular RNA CDR1as promotes adipogenic and suppresses osteogenic differentiation of BMSCs in steroid-induced osteonecrosis of the femoral head. Bone. 2020;133:115258
34. Feng H, Yousuf S, Liu T, Zhang X, Huang W, Li A. et al. The comprehensive detection of miRNA and circRNA in the regulation of intramuscular and subcutaneous adipose tissue of Laiwu pig. Sci Rep. 2022;12:16542
35. Yu C, Kuo H. The Trans-Spliced Long Noncoding RNA tsRMST Impedes Human Embryonic Stem Cell Differentiation Through WNT5A-Mediated Inhibition of the Epithelial-to-Mesenchymal Transition. Stem Cells. 2016;34:2052-62
36. Zhang Q, Fan H, Liu H, Jin J, Zhu S, Zhou L. et al. WNT5B exerts oncogenic effects and is negatively regulated by miR-5587-3p in lung adenocarcinoma progression. Oncogene. 2020;39:1484-97
37. Zhang T, Liu H, Mao R, Yang H, Zhang Y, Zhang Y. et al. The lncRNA RP11-142A22.4 promotes adipogenesis by sponging miR-587 to modulate Wnt5β expression. Cell Death Dis. 2020;11:475
38. Zhu C, Shen K, Zhou W, Wu H, Lu Y. Exosome-mediated circ_0001846 participates in IL-1β-induced chondrocyte cell damage by miR-149-5p-dependent regulation of WNT5B. Clin Immunol. 2021;232:108856
39. Cui X, Zhang X, Liu M, Zhao C, Zhang N, Ren Y. et al. A pan-cancer analysis of the oncogenic role of staphylococcal nuclease domain-containing protein 1 (SND1) in human tumors. Genomics. 2020;112:3958-67
40. Fan D, Yao Y, Liu Y, Yan C, Li F, Wang S. et al. Regulation of myo-miR-24-3p on the Myogenesis and Fiber Type Transformation of Skeletal Muscle. Genes. 2024;15:269
41. Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ. et al. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018-21
42. Nader GA, McLoughlin TJ, Esser KA. mTOR function in skeletal muscle hypertrophy: increased ribosomal RNA via cell cycle regulators. Am J Physiol Cell Physiol. 2005;289:C1457-65
43. Wang J, Song C, Cao X, Li H, Cai H, Ma Y. et al. MiR-208b regulates cell cycle and promotes skeletal muscle cell proliferation by targeting CDKN1A. J Cell Physiol. 2019;234:3720-9
44. Li Q, Zhang Z, Xia P, Wang Y, Wu Z, Jia Y. et al. A SNP in the 3′-UTR of HSF1 in dairy cattle affects binding of target bta-miR-484. Genet Mol Res. 2015;14:12746-55
45. An X, Song Y, Bu S, Ma H, Gao K, Hou J. et al. Association of polymorphisms at the microRNA binding site of the caprine KITLG 3′-UTR with litter size. Sci Rep. 2016;6:25691
46. Stachowiak M, Szydlowski M, Flisikowski K, Flisikowska T, Bartz M, Schnieke A. et al. Polymorphism in 3' untranslated region of the pig PPARA gene influences its transcript level and is associated with adipose tissue accumulation. J Anim Sci. 2014;92:2363-71
47. Hou J, An X, Song Y, Gao T, Lei Y, Cao B. Two Mutations in the Caprine MTHFR 3'UTR Regulated by MicroRNAs Are Associated with Milk Production Traits. PLoS One. 2015;10:e0133015
48. Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J. et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623-34
49. Ma F, Liu X, Li D, Wang P, Li N, Lu L. et al. MicroRNA-466I Upregulates IL-10 Expression in TLR-Triggered Macrophages by Antagonizing RNA-Binding Protein Tristetraprolin-Mediated IL-10 mRNA Degradation. J Immunol. 2010;184:6053-9
50. McFarland AP, Horner SM, Jarret A, Joslyn RC, Bindewald E, Shapiro BA. et al. IFNL3 (IL28B) favorable genotype escapes hepatitis C virus-induced microRNAs and mRNA decay. Nat Immunol. 2014;15:72-9
51. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
52. Perkins RS, Suthon S, Miranda-Carboni GA, Krum SA. WNT5B in cellular signaling pathways. Semin Cell Dev Biol. 2022;125:11-6
53. Ridgeway AG, Petropoulos H, Wilton S, Skerjanc IS. Wnt signaling regulates the function of MyoD and myogenin. J Biol Chem. 2000;275:32398-405
54. Wang C, Ruan L, Shi H, Xu X. Wnt5b regulates apoptosis in Litopenaeus vannamei against white spot syndrome virus. Fish Shellfish Immunol. 2018;74:318-24
55. Yao Y, Wang Z, Chen Y, Liu L, Wang L, Yi G. et al. Single-cell analysis reveals the lncRNA-MEG3/miRNA-133a-3p/PRRT2 axis regulates skeletal muscle regeneration and myogenesis. Genes Dis. 2023;10:359-62
56. Yan J, Yang Y, Fan X, Liang G, Wang Z, Li J. et al. circRNAome profiling reveals circFgfr2 regulates myogenesis and muscle regeneration via a feedback loop. J Cachexia Sarcopenia Muscle. 2022;13:696-712
57. Du Y, Ding Q, Li Y, Fang W. Identification of Differentially Expressed Genes and Pathways for Myofiber Characteristics in Soleus Muscles between Chicken Breeds Differing in Meat Quality. Anim Biotechnol. 2017;28:83-93
58. Ismail I, Joo ST. Poultry Meat Quality in Relation to Muscle Growth and Muscle Fiber Characteristics. Korean J Food Sci Anim Resour. 2017;37:873-83
59. Deng Q, Hu H, Yu X, Liu S, Wang L, Chen W. et al. Tissue-specific microRNA expression alters cancer susceptibility conferred by a TP53 noncoding variant. Nat Commun. 2019;10:5061
60. Nicoloso MS, Sun H, Spizzo R, Kim H, Wickramasinghe P, Shimizu M. et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70:2789-98
61. Kedde M, Van Kouwenhove M, Zwart W, Oude Vrielink JAF, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014-20
62. Wei W, Gao W, Li Q, Liu Y, Chen H, Cui Y. et al. Comprehensive characterization of posttranscriptional impairment-related 3′-UTR mutations in 2413 whole genomes of cancer patients. npj Genomic Med. 2022;7:34
63. Kristjánsdóttir K, Fogarty EA, Grimson A. Systematic analysis of the Hmga2 3′ UTR identifies many independent regulatory sequences and a novel interaction between distal sites. RNA. 2015;21:1346-60
64. Styrkarsdottir U, Cazier JB, Kong A, Rolfsson O, Larsen H, Bjarnadottir E. et al. Linkage of osteoporosis to chromosome 20p12 and association to BMP2. PLoS Biol. 2003;1:E69
65. Su Y, Hou C, Yang W. Control of hair cell development by molecular pathways involving Atoh1, Hes1 and Hes5. Gene. 2015;558:6-24
66. Wang L, Du X, Li Q, Wu W, Pan Z, Li Q. miR-2337 induces TGF-β1 production in granulosa cells by acting as an endogenous small activating RNA. Cell Death Discov. 2021;7:253
67. Chao Y, Jiang Y, Zhong M, Wei K, Hu C, Qin Y. et al. Regulatory roles and mechanisms of alternative RNA splicing in adipogenesis and human metabolic health. Cell Biosci. 2021;11:1-16
68. Zhao S, Liu M-F. Mechanisms of microRNA-mediated gene regulation. Sci China C Life Sci. 2009;52:1111-6
Author contact
![]() Corresponding author: tangzhonglincn. Address: Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Pengfei Road 7, Da Peng New District, Shenzhen, Guangdong Province, China.
Corresponding author: tangzhonglincn. Address: Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Pengfei Road 7, Da Peng New District, Shenzhen, Guangdong Province, China.

 Global reach, higher impact
Global reach, higher impact